Abstract
Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) are the main cytosolic sensors of single-stranded RNA viruses, including paramyxoviruses, and are required to initiate a quick and robust innate antiviral response. Despite different ligand-binding properties, the consensus view is that RIG-I and MDA5 trigger common signal(s) to activate interferon regulatory factor 3 (IRF-3) and NF-κB, and downstream antiviral and proinflammatory cytokine expression. Here, we performed a thorough analysis of the temporal involvement of RIG-I and MDA5 in the regulation of IRF-3 during respiratory syncytial virus (RSV) infection. Based on specific RNA interference-mediated knockdown of RIG-I and MDA5 in A549 cells, we confirmed that RIG-I is critical for the initiation of IRF-3 phosphorylation, dimerization and downstream gene expression. On the other hand, our experiments yielded the first evidence that knockdown of MDA5 leads to early ubiquitination and proteasomal degradation of active IRF-3. Conversely, ectopic expression of MDA5 prolonged RIG-I-induced IRF-3 activation. Altogether, we provide novel mechanistic insight into the temporal involvement of RIG-I and MDA5 in the innate antiviral response. While RIG-I is essential for initial IRF-3 activation, engagement of induced MDA5 is essential to prevent early degradation of IRF-3, thereby sustaining IRF-3-dependent antiviral gene expression. MDA5 plays a similar role during Sendai virus infection suggesting that this model is not restricted to RSV amongst paramyxoviruses.
Key Words: Pathogen recognition receptors, Host defense, Respiratory syncytial virus, Innate immunity, Retinoic acid-inducible gene I, Melanoma differentiation-associated gene 5, Interferon regulatory factor 3
Introduction
The ability of the host to mount a quick and robust innate immune antiviral response relies upon the recognition of highly conserved pathogen-associated molecular patterns (PAMPs) by a heterogeneous group of pathogen recognition receptors (PRRs) that detect viral RNA species [1]. Amongst PRRs, the retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family plays a key role in the cytosolicsensing of single-stranded RNA viruses. The founding member, RIG-I, and its structural homologue melanoma differentiation-associated gene 5 (MDA5) positively contribute to the formation of an antiviral state [2]. RIG-I and MDA5 contain a DExD/H ATP-dependent helicase domain, two N-terminal caspase-recruiting domains (CARD) responsible for signal transduction, and a regulatory C-terminal domain [3]. Biochemical and structural studies have started to elucidate preferences of RIG-I and MDA5 for distinct nucleic acid ligands. A 5′-triphosphate (5′-ppp) together with a blunt-ended double-stranded (ds) motif defines a sufficient molecular pattern for recognition by RIG-I [4]. Deep-sequencing analysis revealed that short defective interfering RNA generated through mistakes in the replication of Sendai virus (SeV) and Influenza virus constitute the privileged ligand for RIG-I [5]. MDA5 ligands are less well defined, but MDA5 responds more specifically to long dsRNA that contain branched or higher order structures [6] and senses the absence of 2′-O-methylation at the 5′ cap structure of viral RNA [7]. Direct binding of viral RNA to MDA5 was not yet demonstrated, but colocalization of MDA5 and the replicative dsRNA of enteroviruses was observed using fluorescent imaging [8].
Despite differences in their ligand-binding properties, the current modelpostulates that RIG-I and MDA5 trigger common downstream signal(s). The binding of RNA moieties to RIG-I or MDA5 forces a conformational change allowing their interaction with the mitochondrial antiviral signaling (MAVS) adaptor, also known as IPS-1, CARDIF or VISA. Activated MAVS provides an interface for the binding of various signaling proteins to elicit the activation of the NF-κB and interferon (IFN) regulatory factor 3 (IRF-3) transcription factors, culminating in the production of proinflammatory and antiviral cytokines, mainly type-I IFNs [9, 10]. IFNβ autocrine and paracrine action induces the expression of numerous IFN-stimulated genes (ISGs) encoding antiviral proteins that modulate viral replication, protein synthesis, growth arrest and apoptosis [11]. IRF-3 also mediates an early IFN-independent antiviral state via the expression of a subset of key antiviral genes, including genes encoding IFN-induced protein with tetratricopeptide repeats (IFIT) 1/ISG56, IFIT2/ISG54 and IFIT3/ISG60 [12].
While it was initially thought that RIG-I and MDA5 recognize selective families of viruses, it is now documented that several viruses, including Dengue virus, West Nile virus, Reovirus and SeV can be sensed by both receptors to induce type-I IFN production [13, 14, 15]. We previously found that both RIG-I and MDA5 play a role in respiratory syncytial virus (RSV)-mediated activation of NF-κB. Interestingly, while both RIG-I and MDA5 downregulation significantly impaired RSV-induced NF-κB-dependent promoter activity, only RIG-I acts upstream of the classical NF-κB activation pathway involving phosphorylation of the IκBα inhibitor and of the p65 subunit [16]. This points to a distinct role of MDA5 in the regulation of the NF-κB-dependent innate immune response via an alternative pathway that remains to be characterized.
In the present study, we compared the role of RIG-I and MDA5 in the regulation of IRF-3 during RSV infection using specific siRNA-mediated silencing of each receptor in A549 cells. Consistent with previous reports [13, 17], we found that RIG-I is essential for RSV-induced IRF-3 activation measured through phosphorylation and dimerization, and downstream IFIT1 antiviral gene expression. In contrary, MDA5 was not essential for early IRF-3 activation. Termination of IRF-3 activation is essential to prevent a prolonged antiviral response. We and others have shown that hyperphosphorylated IRF-3 is subjected to degradation by the proteasome [18, 19, 20]. Interestingly, proteasome-mediated degradation of IRF-3 occurred at earlier time points when MDA5 was silenced compared to control cells. Moreover, ectopic expression of MDA5 resulted in a prolonged activation of IRF-3 induced by the RIG-I-specific ligand 5′ppp-dsRNA. Consistently, expression of the IRF-3 target gene IFIT1 was strongly impaired in the absence of RIG-I, while in the absence of MDA5 it was initially induced, but its expression was transient. Altogether, these results support a novel model in which activation of RIG-I is essential to activate IRF-3, while induction of MDA5 during infection prevents early degradation of IRF-3, thereby sustaining IRF-3-dependent antiviral gene expression. The observation that MDA5 plays a similar role in response to SeV suggests that this model is not restricted to RSV amongst paramyxoviruses. These results shed a new light on our understanding of the distinct functions of two closely related PRRs in the regulation of the antiviral response.
Materials and Methods
Reagents and Plasmids
Dimethyl sulfoxide (DMSO) and bovine serum albumin were from Sigma-Aldrich. Lactacystin and MG132 were from Calbiochem. TransIT-LT1 transfection reagent was from Mirus BIO, and Oligofectamine and Lipofectamine 2000 transfection reagents were from Invitrogen. Previously described RNA interference (RNAi) oligonucleotides [16] were obtained from Dharmacon. The 5′ppp-dsRNA was from Invivogen. The pRL-null reporter plasmid was from Promega. The IFIT1prom-pGL3 luciferase reporter construct has previously been described [12]. The hMDA5-flag-pCMV6c plasmid was graciously supplied by Dr. S. Akira, Osaka University, Japan.
Cell Culture and Infections
All media and supplements were from GIBCO. A549 cells were obtained from the American Type Culture Collection (ATCC) and grown in F-12 nutrient mixture (Ham) medium supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS). HEC1B cells (ATCC) were grown in MEM supplemented with 10% HI-FBS, sodium pyruvate and non-essential amino acids. All cells were grown without antibiotics.
The initial stock of RSV A2 strain was from Advanced Biotechnologies Inc. Amplification and purification were performed as previously described [16]. Infection of subconfluent A549 cells with RSV A2 was performed in culture medium containing 2% HI-FBS at a multiplicity of infection (MOI) of 3, except in luciferase reporter assays where a MOI of 6 was used. Where indicated, lactacystin, MG132 or DMSO (vehicle) were added in serum-free medium for 1 h before the addition of RSV and 2% HI-FBS.
Transfection of Plasmids, RNAi and 5′ppp-dsRNA
Transfection of plasmids was performed using the TransIT-LT1 Transfection Reagent according to the manufacturer's instructions. Cells were plated to reach 60–70% confluency at the time of transfection and were transfected using a DNA/transfection reagent ratio of 1:2 or 1:3. RNAi oligonucleotide transfection was performed as previously described [21] using Oligofectamine reagent and pursued for 63-66 h before RSV infection, except for plaque-forming unit assay where infection was performed at 24 h post-RNAi transfection. Where required, plasmid transfection was performed 36 h after RNAi transfection. For transfection of 5′ppp-dsRNA, A549 cells at 80–90% confluency in 35-mm plates were transfected with 1.5-μg 5′ppp-dsRNA using Lipofectamine 2000 (Invitrogen) with a ligand/transfection reagent ratio of 1:1.
Multiplex ELISA
RSV infection at an MOI of 3 was conducted in Opti-MEM Reduced Serum media (GIBCO) 48 h post-RNAi transfection. According to the manufacturer's instructions, 50 µl of supernatant were analyzed using the VeriPlex™ Human Interferon Multiplex ELISA (PBL Interferon). Acquisition of chemiluminescence was performed with a LAS4000mini CCD camera apparatus (GE Healthcare) using an NP Tray. Data analysis was performed with the Q-View Software from Quansys Biosciences.
Immunoblot Analysis
Whole-cell extracts (WCE) were prepared on ice in Nonidet P-40 (Igepal; Sigma) lysis buffer [22], quantified using a Bio-Rad protein assay (Bio-Rad), resolved by SDS-PAGE electrophoresis and transferred onto nitrocellulose membrane followed by immunoblot. Proteins were immunodetected using anti-IRF-3Ser396 phosphospecific (anti-IRF-3-P-Ser396 [22]), anti-IRF-3 (Active Motif or IBL), anti-RIG-I (Alexis Biochemicals), anti-MDA5 (Alexis Biochemicals), anti-Flag M2 (Sigma-Aldrich), anti-actin (Chemicon International), and anti-RSV (Chemicon International) antibodies diluted in phosphate-buffered saline containing 0.5% Tween and either 5% non-fat dry milk or 5% bovine serum albumin. The membranes were further incubated with HRP-conjugated goat anti-rabbit or anti-mouse IgG, or rabbit anti-goat IgG (Kirkegaard & Perry or Jackson Laboratories). Immunoreactive bands were visualized by enhanced chemiluminescence using the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences) and detected using an LAS4000mini CCD camera apparatus (GE Healthcare).
Native Gel Electrophoresis
To separate IRF-3 monomer and dimer, native-PAGE was conducted as described previously [23] using 10 µg of WCE prepared as described above. After transfer onto nitrocellulose membrane, IRF-3 was detected by immunoblot as described above.
Luciferase Reporter Assays
A549 cells in 24-well plates were cotransfected with the pRL-null renilla (internal control, 50 ng) and the IFIT1prom-pGL3 luciferase reporter (100 ng). Where RNAi transfection preceded a luciferase reporter gene assay, transfection with reporter plasmids was performed 48 h post-RNAi transfection. Quantification of luciferase activities was performed 24 h post-transfection of reporter plasmids using the dual luciferase reporter assay kit (Promega) according to the manufacturer's instructions. Relative luciferase activities were calculated as the ratio of luciferase over renilla activities.
In vivo IRF-3 Polyubiquitination Assay
The state of endogenous IRF-3 polyubiquitination was evaluated in vivo as previously described [20]. Briefly, cells were lysed in RIPA buffer containing 2 mMN-ethylmaleimide and protease inhibitors mixture (Sigma-Aldrich), and passed through a 25-gauge needle five times. Clarified supernatant were subjected to immunoprecipitation using anti-IRF-3 antibodies as described previously [22] and immunocomplexes were resolved by SDS-PAGE electrophoresis, followed by immunoblot as described above using anti-IRF-3 and P4D1 monoclonal antibodies against ubiquitin (Santa Cruz Biotechnology).
Virus Titration
Quantification of RSV infectious virions was performed by methylcellulose plaque-forming unit assays. Briefly, supernatant from RNAi-transfected A549 cells infected with RecRSV-GFP (a kind gift from Dr. P. Collins, NIH) at an MOI of 0.01 was harvested at 48 hpi and used to infect confluent Vero cells (ATCC) for 2 h. The medium was then replaced with 1% methylcellulose in DMEM/2% HI-FBS. Infection was pursued for 7 days and GFP-positive lysis plaques were visualized using a Typhoon apparatus and quantified using Imagequant software (Molecular Dynamics).
Statistical Analyses
Data are presented as the mean ± standard error of the mean (SEM). Statistical significance was assessed by the indicated test using Prism 5 software (GraphPad). Statistical relevance was evaluated using the following p values: * p < 0.05, ** p < 0.01 or *** p < 0.001.
Results
Both RIG-I and MDA5 Are Involved in Proinflammatory and Antiviral Cytokine Production during RSV Infection
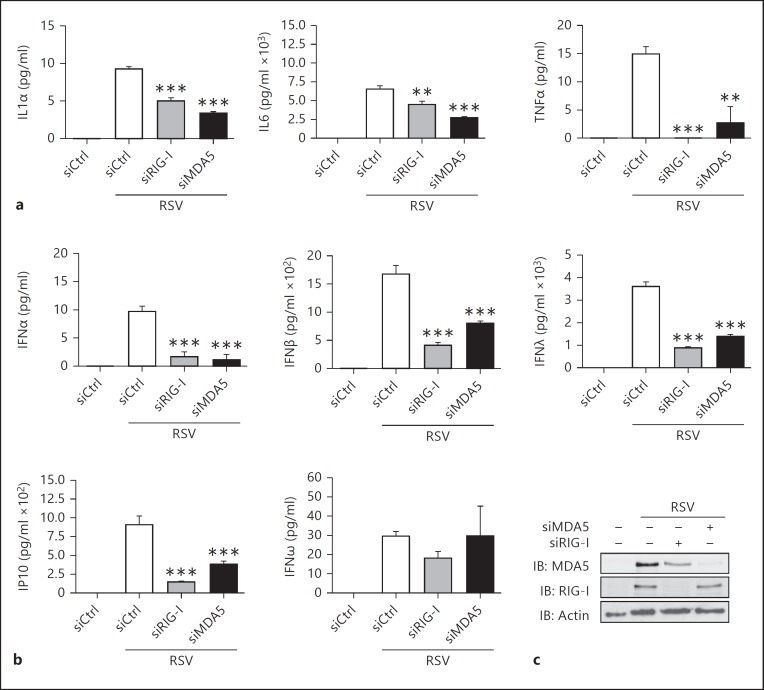
In a previous study, we observed that both RIG-I and MDA5 are involved in the control of NF-κB activation during RSV infection in A549 cells [16]. Consistently, levels of the NF-κB-dependent IL-1α, IL-6 and TNFα proinflammatory cytokines induced at 24 h post-RSV infection, quantified using a multiplex ELISA assay, were significantly decreased when either RIG-I or MDA5 were downregulated by RNAi in A549 cells (fig. 1a). Considering the partial overlap of NF-κB and IRF-3 regulation pathways in response to virus infection, concomitant analysis of IRF-3-dependent antiviral cytokines expression was performed. As shown in figure 1b, silencing of either RIG-I or MDA5 significantly inhibited RSV-induced production of type-I (α and β) IFNs, type-III (λ) IFNs and IP-10. Of note, IFNω expression was neither dependent on RIG-I nor MDA5. Altogether these results suggest that efficient NF-κB and IRF-3 proinflammatory and antiviral genes expression involves RSV sensing by both RIG-I and MDA5.
Fig. 1.
RIG-I and MDA5 are essential for efficient proinflammatory and antiviral cytokine production in RSV-infected A549 cells. Expression of the proinflammatory cytokines (a) and antiviral cytokines (b) was monitored in the supernatant of A549 cells transfected with Ctrl-, RIG-I- or MDA5-specific RNAi and infected with RSV (MOI = 3, 24 h) using the VeriPlex Human Interferon Multiplex ELISA. Results were analyzed as mean ± SEM of triplicate experiments and a one-way ANOVA followed by a Dunnett post hoc test using siCtrl-RSV-infected cells as controls. c Efficiency of RIG-I and MDA5 knockdown was monitored by immunoblot (IB) using anti-MDA5 and anti-RIG-I and anti-actin antibodies.
RIG-I, but Not MDA5, Is Essential for Early IRF-3 Phosphorylation and Dimerization
In uninfected cells, IRF-3 is present as non-phosphorylated and hypophosphorylated forms (form I and II detected by SDS-PAGE [24]). Following virus infection, IRF-3 is subjected to hyperphosphorylation (forms III and IV detected by SDS-PAGE [24]). Phosphorylated forms of IRF-3 constitute the active forms that dimerize, accumulate in the nucleus, bind the promoter of target genes, associate with coactivator and transactivate genes [25]. Thus, to further characterize the role of RIG-I and MDA5 in RSV-induced IRF-3 activation, A549 cells were transfected with control (Ctrl), RIG-I and MDA5 RNAi before infection with RSV for various times. Detailed analysis of IRF-3 phosphorylation was performed by immunoblot using anti-IRF-3Ser396 phosphospecific antibodies [22] (fig. 2a, b) and detection of forms III and IV using anti-IRF-3 antibodies (fig. 2a-c). Additionally, dimerization was analyzed by native-PAGE electrophoresis (fig. 2d, e). Consistent with the previously reported role of RIG-I in RSV-mediated IRF-3 activation [13, 17], RSV-induced phosphorylation of IRF-3 was markedly reduced in the absence of RIG-I, with IRF-3 remaining mostly present as unphosphorylated/hypophosphorylated forms I and II throughout the infection (fig. 2a, c). Accordingly, dimerization of IRF-3 was also significantly impaired and IRF-3 remained mostly as a monomer throughout the infection (fig. 2d). Interestingly, the profiles of phosphorylation and dimerization of IRF-3 in the absence of MDA5 were different. At early time points of RSV infection (4 h), phosphorylation and dimerization of IRF-3 were similar in the presence or absence of MDA5 (fig. 2b, e). However, during the course of the infection, the levels of phosphorylated forms (fig. 2b, c) and of dimers (fig. 2d) were significantly diminished in the absence of MDA5 compared to control cells. Importantly, these decreased levels of active forms neither correlated with a recovery of the unphosphorylated/hypophosphorylated forms I and II nor with the accumulation of IRF-3 monomer, as observed in the absence of RIG-I (fig. 2b, c, e). Consistent with previous observations that MDA5 is induced in an IFN-dependent manner, downregulation of RIG-I using RNAi impairs RSV-mediated induction of MDA5. Thus, to ensure that the observed effect of RIG-I on IRF-3 phosphorylation and dimerization was not due to the concomitant inhibition of MDA5 expression, Flag-MDA5 was ectopically expressed in cells previously transfected with RIG-I RNAi before infection with RSV. As shown in figure 2f, ectopic expression of MDA5 was not capable of rescuing IRF-3 phosphorylation in the absence of RIG-I, suggesting that MDA5 and RIG-I do not have redundant functions and that the phenotype observed through downregulation of RIG-I is not due to its effect on MDA5 expression.
Fig. 2.
RIG-I is required for initiation of IRF-3 activation, while MDA5 is required for sustained IRF-3 activation following RSV infection. A549 cells were transfected with Ctrl-, RIG-I- (a, c, d, f) or MDA5- (b, c, e) specific RNAi. f RNAi-transfected cells were further transfected with empty plasmid or plasmid expressing Flag-MDA5. RNAi-transfected cells were infected with RSV (MOI of 3) for the indicated times. a-c, f WCE were analyzed by immunoblot (IB) using anti-IRF-3-Ser396 phosphospecific (IRF-3-P-Ser396), anti-IRF-3, anti-RIG-I, anti-MDA5, anti-RSV (the nucleocapsid N is shown) and anti-actin antibodies. d, e IRF-3 dimerization was analyzed by native PAGE followed by immunoblot using anti-IRF-3 antibodies. D = Dimer; M = monomer; hpi = hours post-infection. The data are representative of at least three different experiments with similar results.
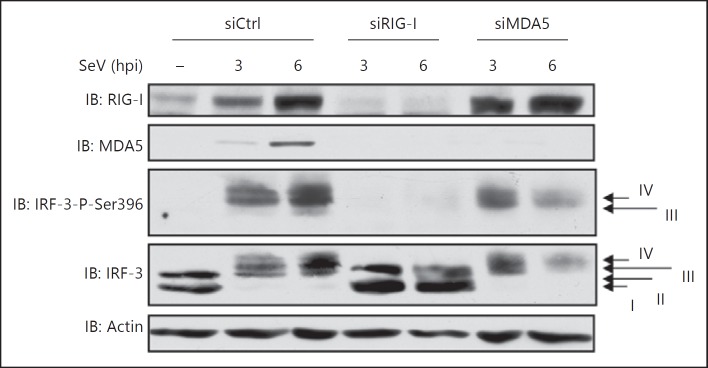
To determine whether differences in the pattern of IRF-3 activation observed in the absence of RIG-I or MDA5 were specific to RSV infection or if it was more general amongst paramyxoviruses, the impact of RIG-I and MDA5 silencing was also studied during SeV infection (fig. 3). Consistent with previous reports [26], SeV-induced IRF-3 Ser396 phosphorylation was dramatically impaired following RIG-I knockdown with IRF-3 remaining in form I and II over the infection. Similar to the observation in the context of RSV infection, SeV-induced Ser396 phosphorylation of IRF-3 was similar in Ctrl and MDA5 RNAi-transfected cells at an early time of infection (3 h), but at a later time (6 h) the levels of IRF-3 phosphorylated forms were significantly decreased in the absence of MDA5 without a corresponding recovery of unphosphorylated/hypophosphorylated forms. Taken together, these results suggest that RIG-I controls the formation of the phosphorylated dimeric forms of IRF-3 from early times of infection. MDA5 is not essential for the initial formation of phosphorylated dimeric forms of IRF-3, but is required to sustain their levels over the course of infection.
Fig. 3.
SeV-mediated IRF-3 activation requires RIG-I and is sustained in an MDA5-dependent manner. A549 cells were transfected with Ctrl-, RIG-I- or MDA5-specific RNAi before being infected with SeV (40 HAU/106 cells) for the indicated times. WCE were resolved by SDS-PAGE and analyzed by immunoblot (IB) using anti-IRF-3-Ser396 phosphospecific (IRF-3-P-Ser396), anti-IRF-3, anti-RIG-I and anti-MDA5 antibodies. Equal loading was analyzed using anti-actin antibodies. Hpi = Hours post-infection. The data are representative of at least three different experiments with similar results.
Reduced Levels of Active IRF-3 during the Course of Infection Observed in the Absence of MDA5 Is Proteasome Dependent
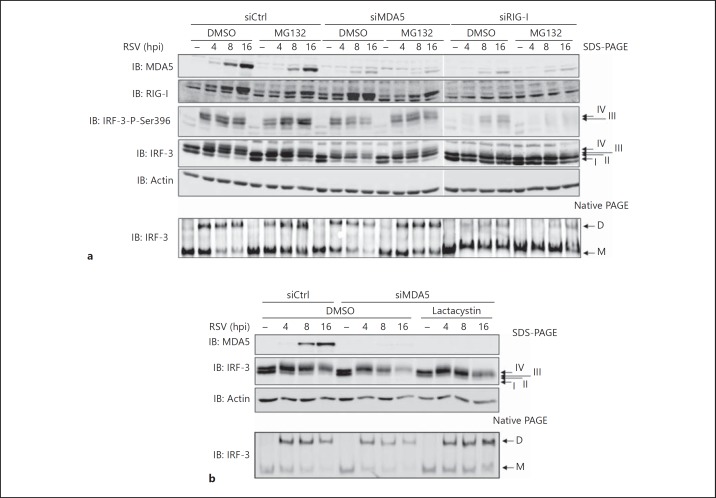
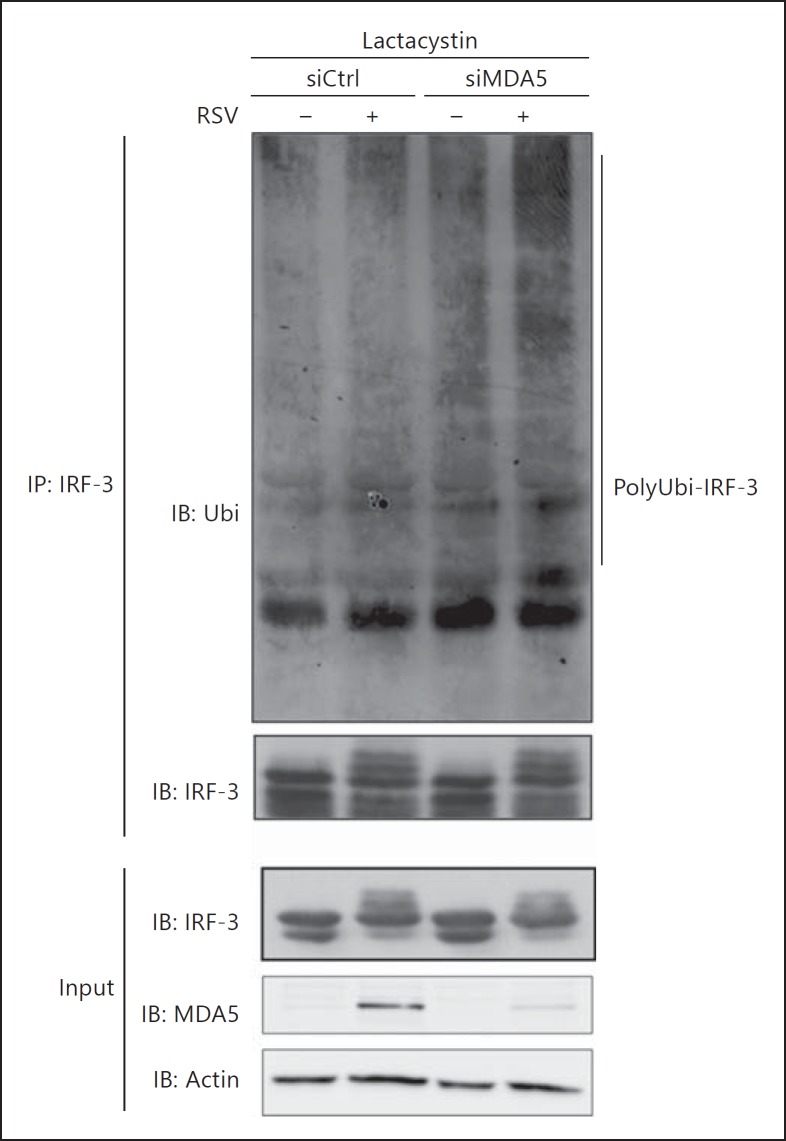
Inhibition of IRF-3 activation typically results in IRF-3 remaining in unphosphorylated/hypophosphorylated monomeric forms, similar to what is observed in the absence of RIG-I (fig. 2, 3). In contrast, the observation that in the absence of MDA5 the levels of active IRF-3 were decreased during the course of infection without a recovery of unphosphorylated/hypophosphorylated monomeric forms (fig. 2, 3) is similar to the profile observed at late time points of infection, when IRF-3 is subjected to polyubiquitination and proteasomal degradation [18, 20]. This suggests that in the absence of MDA5, reduced levels of active IRF-3 could result from proteasome-mediated degradation occurring at earlier time points compared to control cells. Thus, we sought to verify whether the decreased levels of active IRF-3 in the absence of MDA5 could be rescued through inhibition of the proteasome. For this purpose, Ctrl, RIG-I and MDA5 RNAi-transfected A549 cells were pretreated with the proteasome inhibitor MG132 or DMSO (vehicle) before infection with RSV for various times. As shown in figure 4a, pretreatment of cells lacking MDA5 with MG132 resulted in the stabilization of Ser396 phosphorylated and total IRF-3 levels during the course of the infection. Consistently, IRF-3 dimer levels were stabilized over the course of RSV infection in the presence of MG132 (fig. 4a). In contrast, in the absence of RIG-I, IRF-3 activation, detected by anti-phosphoSer396 immunoblot, form III and IV and dimer formation, was highly impaired and MG132 treatment did not affect these different parameters (fig. 4a). Treatment with lactacystin, another structurally unrelated proteasome inhibitor, also resulted in the accumulation of activated forms of IRF-3 (forms III and IV and dimers) following virus infection in the absence of MDA5 (fig. 4b). Additionally, analysis of IRF-3 ubiquitination status revealed that in the absence of MDA5, RSV-induced IRF-3 polyubiquitination is significantly increased compared to control cells (fig. 5). Altogether, these results strongly support the role of MDA5 induction in the prevention of early polyubiquitination and subsequent proteasomal degradation of activated IRF-3.
Fig. 4.
Inhibition of the proteasome prevents early diminution of active IRF-3 levels observed in the absence of MDA5 during the course of RSV infection. A549 cells were transfected with Ctrl, RIG-I or MDA5 RNAi and pretreated either with DMSO (vehicle) or 5 μM MG132 (a) or 10 μM lactacystin (b) proteasome inhibitors prior to infection with RSV (MOI of 3) for the indicated times. WCE were resolved by SDS-PAGE and analyzed by immunoblot (IB) using anti-MDA5, anti-RIG-I, anti-IRF-3-Ser396 phosphospecific (IRF-3-P-Ser396), anti-IRF-3 and anti-actin antibodies or resolved by native gel electrophoresis and immunoblotted with anti-IRF-3 antibodies. D = Dimer; M = monomer; hpi = hours post-infection. The data are representative of three different experiments with similar results.
Fig. 5.
Absence of MDA5 results in enhanced IRF-3 polyubiquitination during RSV infection. A549 cells were transfected with Ctrl or MDA5 RNAi and pretreated with lactacystin (10 μM) prior to infection with RSV (MOI of 3) for 16 h. WCE were subjected to immunoprecipitation (IP) using anti-IRF-3 antibodies. Immunocomplexes and input WCE were analyzed by immunoblot (IB) using anti-ubiquitin (Ubi), anti-MDA5, anti-IRF-3 and anti-actin antibodies. The data are representative of two independent experiments with similar results.
RIG-I-Induced IRF-3 Activation Is Prolonged by Ectopic Expression of MDA5
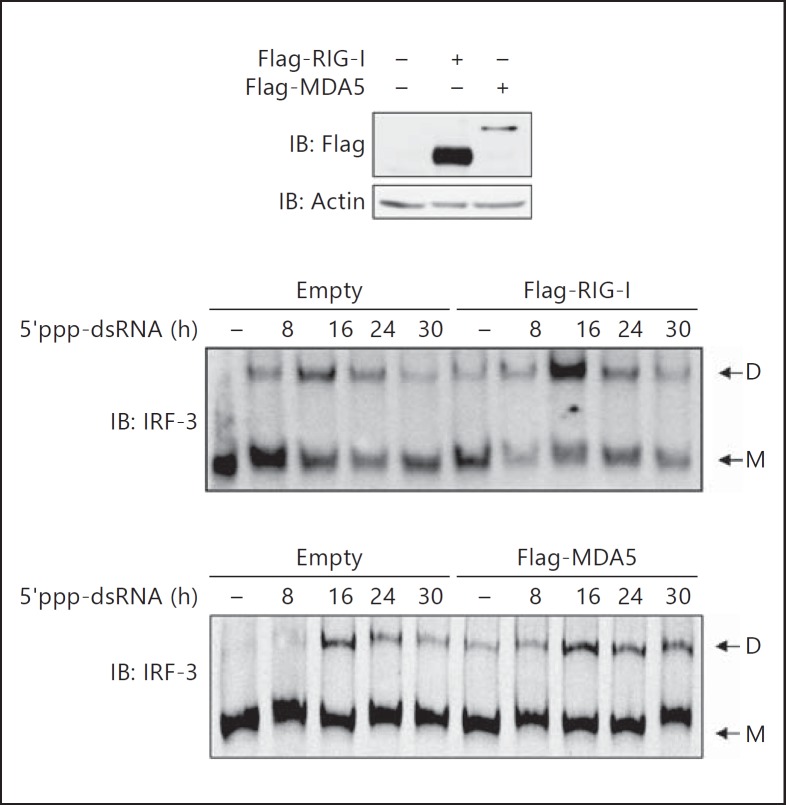
To further demonstrate that MDA5 expression is involved in the stabilization of active IRF-3 levels, the RIG-I-specific ligand 5′ppp-dsRNA was transfected in A549 cells ectopically expressing either control, Flag-RIG-I or Flag-MDA5 (fig. 6). As expected, 5′ppp-dsRNA induced IRF-3 dimerization in control cells with a peak of dimer levels observed at 16 h post-transfection. In cells ectopically expressing Flag-RIG-I, formation of IRF-3 dimer also peaked at 16 h, but the levels of dimer was increased compared to control cells. Interestingly, in Flag-MDA5-expressing cells, 5′ppp-dsRNA induced IRF-3 dimerization with maximum levels similar to those observed in control cells at 16 h post-stimulation. However, IRF-3 dimer levels were sustained for up to 30 h compared to control cells. Altogether, these results demonstrate that RIG-I-induced IRF-3 activation is prolonged by a mechanism that is dependent on MDA5 expression.
Fig. 6.
MDA5 ectopic expression sustains RIG-I-induced active IRF-3 levels. A549 cells were transfected with either empty, Flag-RIG-I- or Flag-MDA5-encoding plasmids before being transfected with 5′ppp-dsRNA for the indicated times. WCE were analyzed by immunoblot (IB) using anti-Flag and anti-actin antibodies to control the expression of Flag-tagged proteins. IRF-3 dimerization was analyzed by native PAGE followed by immunoblot using anti-IRF-3 antibodies. D = Dimer; M = monomer. The data are representative of three different experiments with similar results.
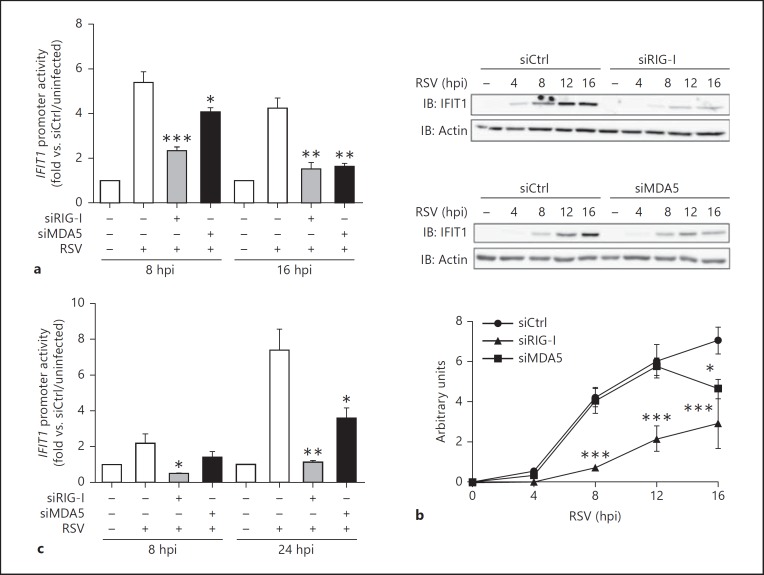
MDA5 Is Essential for Persistent IRF-3-Dependent IFIT1 Antiviral Gene Expression
The observation that active IRF-3 is subjected to proteasomal degradation at earlier times in the absence of MDA5 compared to control cells suggests that MDA5 is required for prolonged IRF-3-dependent antiviral gene expression. To further characterize the relative roles of RIG-I and MDA5 in IRF-3-dependent gene expression, IFIT1 gene (encoding for IFIT1/ISG56) promoter activity and endogenous IFIT1 protein expression were investigated at various time points in RSV-infected A549 cells transfected with Ctrl, RIG-I or MDA5 RNAi. As shown in figure 7a, RSV-mediated stimulation of the IFIT1-promoter activity, measured by luciferase reporter assay, was highly decreased in RIG-I-depleted cells compared to cells transfected with siCtrl at early (8 h) and late (16 h) times post-infection. Interestingly, downregulation of MDA5 only weakly interfered with IFIT1-promoter activity at an early time point (8 h) post-infection, while it dramatically decreased it at a late time point (16 h; fig. 7a). Similar profiles were observed when endogenous IFIT1 expression was analyzed by immunoblot. IFIT1 expression was strongly abrogated at every time point during RSV infection (0-16 h) in the absence of RIG-I compared to control cells. In the absence of MDA5, IFIT1 expression was similar to control cells up to 12 h post-infection, while it was significantly lower at 16 h post-infection (fig. 7b). To exclude the contribution of type-I IFN in RIG-I- or MDA5-mediated effects on IFIT1 expression, the same experiment was performed in HEC1B cells deficient in type-I IFN receptor [27]. Similar to results observed in A549 cells, RIG-I downregulation resulted in inhibition of RSV-induced IFIT1 promoter induction at all time points of the infection, while in the absence of MDA5, IFIT1 promoter activity was comparable to control cells at 8 h, but was significantly impaired at 24 h (fig. 7c). These data suggest that RIG-I is essential for IRF-3 target gene expression, but MDA5 induction is only required for persistent expression. In agreement with a role of both RIG-I and MDA5 in IRF-3 activation, silencing of RIG-I or MDA5 resulted in the increase of RSV replication as measured by plaque-forming unit assay. Interestingly, increase in RSV replication was higher following MAVS knockdown, which acts downstream of both RIG-I and MDA5, (fig. 8). Altogether, these results support a model in which MDA5-dependent signaling plays a major role in preventing early degradation of active IRF-3 by the proteasome, thereby sustaining the IRF-3-dependent antiviral response.
Fig. 7.
MDA5 is required for sustained IRF-3-dependent gene expression. A549 (a, b) or HEC1B (c) cells were transfected with Ctrl, RIG-I or MDA5 RNAi and infected with RSV (MOI of 6) for the indicated times. a, c At 48 h post-RNAi transfection, cells were further transfected with the IFIT1prom-pGL3 firefly luciferase and the pRL-null renilla luciferase (internal control) reporter constructs and either left uninfected or infected with RSV. Luciferase activities were expressed as fold activation over the corresponding uninfected condition and analyzed as mean ± SEM of triplicate experiments by a one-way ANOVA-Dunnett post hoc test using siCtrl-infected cells as controls. b WCE were analyzed by immunoblot (IB) using anti-IFIT1 and anti-actin antibodies. Representative immunoblots of three independent experiments are shown. Immunoblots were quantified by densitometric analysis using the ImageJ software. Quantification data (arbitrary units) are expressed as mean ± SEM. Statistical comparison was performed using a two-way ANOVA post hoc Bonferroni test using siCtrl-infected cells as controls. hpi = Hours post-infection.
Fig. 8.
Silencing of RIG-I, MDA5 or MAVS results in increased RSV replication. Released RSV infectious virions were quantified from A549 cells transfected with Ctrl-, RIG-I-, MDA5- or MAVS-specific RNAi by plaque-forming unit (pfu) assay. The virus titers were analyzed as mean ± SEM of independent triplicates followed by a one-way ANOVA-Dunnett post hoc test using siCtrl cells as controls.
Discussion
Accumulating evidence indicates that most or all RNA viruses produce several PAMPs that are detected by different PRRs of the RLR and Toll-like receptor families [28, 29]. The current consensus view of the cooperation of different families of PRRs in the recognition of a single pathogen is that it provides multiple redundant mechanisms in the immune responses, thereby offering effective protection of the host. It is currently thought that this concept of redundancy also applies to RLRs, as several viruses, including the paramyxoviruses SeV and RSV, are recognized by both RIG-I and MDA5 [13, 15, 16, 26]. Although their distinct role in ligand recognition starts to be better understood [30], the current model proposes that RIG-I and MDA5 activate a common downstream signaling cascade(s) initiated through their interaction with MAVS that leads to the activation of IRF-3 and NF-κB and the subsequent induction of antiviral and proinflammatory cytokines. In contrast to this model, our experiments yielded interesting results supporting the belief that temporal involvement of RIG-I and MDA5 contributes to the control of the initiation and the duration of IRF-3 activation during RSV and SeV infections. Based on specific RNAi-mediated knockdown of RIG-I and MDA5 coupled to a detailed kinetic analysis, we demonstrated that IRF-3 activation and downstream IFIT1 gene expression are severely impaired in the absence of RIG-I. On the other hand, we provided the first evidence that expression of MDA5 while viral replication proceeds is not essential for the initiation of IRF-3 activation, but is rather implicated in the prevention of active IRF-3 degradation, thereby sustaining IRF-3 activation and downstream gene expression. Importantly, ectopic expression of MDA5 was able to prolong IRF-3 activation triggered by the RIG-I ligand 5′ppp-dsRNA.
Spatiotemporal changes in the localization of RIG-I and MDA5 during RSV infection recently highlighted that both sensors colocalize with RSV in cytoplasmic inclusion bodies that contain RSV N and genomic RNA at an early time of infection. At a late time of infection, only MDA5 colocalized with viral RNA filaments [31]. These specific subcellular localizations have not yet been correlated with activation of the innate antiviral response activation, but rather with the capacity of RSV proteins to inhibit this response. Other reports that have analyzed the role of RIG-I and MDA5 in RSV- and SeV-mediated IRF-3 activation have studied their temporal involvement with far less experimental scrutiny. Our results are consistent with previous reports showing that RIG-I controls the early antiviral responses against RSV and SeV [13, 16, 26], a result that was reinforced by the successful detection of RSV and SeV RNA binding to RIG-I [5, 17]. Conflicting results have been reported concerning the role of MDA5 in RSV sensing. In HeLa cells, RNAi-mediated knockdown of either RIG-I or MDA5 slightly decreased RSV-induced IFNβ expression [32]. In murine embryonic fibroblasts deficient in MDA5 (MEF MDA5−/−), RSV-induced ISGs expression was relatively delayed compared to control wild-type MEF cells [13], hence supporting an accessory physiological function of MDA5 in amplifying the RSV-induced innate immune response. However, MEF MDA5−/− cells were found less permissive to RSV infection. Thus, it cannot be excluded that delayed expression of ISGs was a result of decreased RSV replication, as NF-κB and IRF-3 activation is dependent on virus replication [[16;] N.G., unpubl. data]. In primary nasal epithelial cells and A549 cells, RNAi-mediated knockdown of MDA5 did not decrease IRF-3-dependent IFN-λ1 gene induction following RSV infection [33]. However, this study was limited to a single time point and thereby was not appropriate for evaluating the potential role of MDA5 in the sustainability of the response. In mice infected by SeV, MDA5 was shown to be required for sustained expression of type-I and III IFN expression, suggesting a role of MDA5 at late time points of infection [15]. Similarly, in a mouse model of West Nile virus infection, MDA5 was shown to contribute to the innate immune response at late time points of infection likely due to the generation of RIG-I and MDA5-specific PAMPs with differential kinetics over the course of viral replication [34]. These observations are consistent with our results. However, we have provided the first mechanistic study highlighting the importance of MDA5 activation to sustain RIG-I-induced IRF-3 activation and the thereof mediated antiviral gene expression.
RIG-I and MDA5 exhibit distinct substrate specificity but other known functional divergences are restricted to their regulation at basal level, a key feature to prevent unnecessary innate immune responses that might promote the development of allergy, necrosis, chronic inflammatory and autoimmune diseases [35]. Specifically, in the unliganded state, the CARD domains of RIG-I are involved in a strong inhibitory intramolecular interaction with the Hel2i domain, which is relieved upon binding of the ligand [36, 37, 38]. In contrast, structural data have recently confirmed the previous biochemical evidence showing that MDA5 does not undergo inhibitory intramolecular interaction in the absence of RNA [36, 39]. Currently, the only known negative regulator specific to MDA5 is the DAK kinase that is thought to keep MDA5 inactive in the steady state through specific interaction with its CARD domain [40]. To our knowledge, there is currently no data reporting distinct signaling cascade events downstream of RIG-I and MDA5 that could provide a likely mechanism for their differential role in the regulation of IRF-3. RIG-I and MDA5 are thought to act in parallel to elicit identical signaling cascades via their CARD-mediated interaction with MAVS [10]. Ultimately, this leads to activation of the IκB-kinase (IKK)-related kinases, TANK-binding kinase-1 and IKKε, which play a key role in a complex set of phosphorylation events that regulate IRF-3 dimerization, nuclear accumulation, transactivation capacities and finally degradation via a proteasome-dependent mechanism [18, 20, 41, 42]. Here, consistent with previous reports, we have shown that downregulation of RIG-I impaired IRF-3 phosphorylation, dimerization and transactivation capacities. However, we demonstrated for the first time that in the absence of MDA5, initial IRF-3 phosphorylation, dimerization and transactivation capacities were not affected compared to control cells, but that IRF-3 polyubiquitination was increased and proteasome-mediated degradation occurred at earlier time points, ultimately resulting in the incapacity of the cell to sustain IRF-3-dependent antiviral gene expression. Several E3 ubiquitin ligases, including the suppressor of cytokine signaling (SOCS1), the RBCC protein interacting with PKC1 (RBCK1), Ro52, also known as TRIM21, RAUL and an uncharacterized Cullin1-based E3 ligase, have been implicated in the polyubiquitination and degradation of IRF-3 [20, 43, 44, 45, 46]. However, whereas the putative Cullin1-based E3 ligase recognizes the active phosphorylated forms of IRF-3 by a mechanism dependent on the binding of the prolyl isomerase Pin1 [19], the other E3 ligases target IRF-3 independently of its phosphorylation status, thereby regulating the basal turnover of the protein. Importantly, in this study, we did not observe a reduction in IRF-3 levels in the absence of MDA5 in uninfected cells. This points to a specific role of MDA5-dependent signaling in the regulation of the degradation of active IRF-3. The precise nature of the signal eliciting polyubiquitination of activated IRF-3 still remains elusive, but might be part of an intricate interrelation between highly complex phosphorylation, SUMOylation, and ISGylation modifications, which were all shown to regulate IRF-3 activation intensity and duration [20, 47, 48, 49]. It will be of great interest to evaluate whether MDA5-dependent signaling controls the activity of the Cullin1-based IRF-3 E3 ligase that specifically mediates polyubiquitination of the active forms of IRF-3 once it is identified. A complete picture of the biochemical processes regulating all the post-translational modifications of IRF-3 and their dynamics involved in the control of IRF-3 ubiquitination and stability is required to further unveil the specific function of RIG-I and MDA5 in the regulation of the antiviral response.
Altogether, our study reveals that the initial trigger of IRF-3-dependent innate immunity through recognition of paramyxoviruses, RSV and SeV, by RIG-I is sustained through the upregulation of MDA5 that prevents early degradation of IRF-3. Our work uncovers novel mechanistic insight into the temporal involvement of RIG-I and MDA5 in the innate antiviral response, and thereby provides a new interesting avenue for therapeutic modulation of the intensity and duration of the innate immune response during the infection.
Acknowledgements
The authors thank Dr. S. Akira, Osaka University, and Dr. P. Collins, NIH, for reagents used in this study. The authors are also grateful to E. Mukawera for technical assistance and RSV propagation and purification. The present work was funded by grants from the CIHR (Canada) to N.G.; N.Z. is the recipient of a fellowship from FRSQ/INSERM (Quebec, Canada/France); K.F. is the recipient of a studentship from CIHR; X.G. and F.Y. are recipients of a scholarship from the Faculty of Graduate Studies, Université de Montréal. M.S. and N.G. are recipients of Tier II Canada Research Chairs.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 4.Kolakofsky D, Kowalinski E, Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e., Sousa C Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. Visualisation of direct interaction of Mda5 and the dsrna replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 9.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, Kalinke U, Pasparakis M, Tschopp J. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Sen GC, Peters GA. Viral stress-inducible genes. Adv Virus Res. 2007;70:233–263. doi: 10.1016/S0065-3527(07)70006-4. [DOI] [PubMed] [Google Scholar]
- 12.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoboua F, Martel A, Duval A, Mukawera E, Grandvaux N. Respiratory syncytial virus-mediated NF-kappa B p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J Virol. 2010;84:7267–7277. doi: 10.1128/JVI.00142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic acid-inducible gene I mediates early antiviral response and toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 20.Bibeau-Poirier A, Gravel SP, Clement JF, Rolland S, Rodier G, Coulombe P, Hiscott J, Grandvaux N, Meloche S, Servant MJ. Involvement of the IκB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol. 2006;177:5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- 21.Fink K, Duval A, Martel A, Soucy-Faulkner A, Grandvaux N. Dual role of nox2 in respiratory syncytial virus- and sendai virus-induced activation of NF-κB in airway epithelial cells. J Immunol. 2008;180:6911–6922. doi: 10.4049/jimmunol.180.10.6911. [DOI] [PubMed] [Google Scholar]
- 22.Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem. 2003;278:9441–9447. doi: 10.1074/jbc.M209851200. [DOI] [PubMed] [Google Scholar]
- 23.Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, Okabe Y, Namiki H, Fujita T. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- 24.Servant MJ, Grandvaux N, Hiscott J. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem Pharmacol. 2002;64:985–992. doi: 10.1016/s0006-2952(02)01165-6. [DOI] [PubMed] [Google Scholar]
- 25.Clement JF, Bibeau-Poirier A, Gravel SP, Grandvaux N, Bonneil E, Thibault P, Meloche S, Servant MJ. Phosphorylation of IRF-3 on Ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J Virol. 2008;82:3984–3996. doi: 10.1128/JVI.02526-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 27.Fuse A, Ashino-Fuse H, Kuwata T. Binding of 125I-labeled human interferon to cell lines with low sensitivity to interferon. Gann. 1984;75:379–384. [PubMed] [Google Scholar]
- 28.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann NY Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 30.Baum A, Garcia-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2010;38:1283–1299. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE, Jr, Santangelo PJ. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol. 2012;86:8245–8258. doi: 10.1128/JVI.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasai M, Shingai M, Funami K, Yoneyama M, Fujita T, Matsumoto M, Seya T. NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J Immunol. 2006;177:8676–8683. doi: 10.4049/jimmunol.177.12.8676. [DOI] [PubMed] [Google Scholar]
- 33.Okabayashi T, Kojima T, Masaki T, Yokota S, Imaizumi T, Tsutsumi H, Himi T, Fujii N, Sawada N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011;160:360–366. doi: 10.1016/j.virusres.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 36.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 39.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, Wang RP, Chen D, Zhai Z, Zhong B, Tien P, Shu HB. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 42.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 43.Oliere S, Douville R, Sze A, Belgnaoui SM, Hiscott J. Modulation of innate immune responses during human T-cell leukemia virus (HTLV-1) pathogenesis. Cytokine Growth Factor Rev. 2011;22:197–210. doi: 10.1016/j.cytogfr.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 2008;18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 45.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 2010;33:863–877. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu G, Reinert JT, Pitha-Rowe I, Okumura A, Kellum M, Knobeloch KP, Hassel B, Pitha PM. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol Biol (Noisy-le-grand) 2006;52:29–41. [PubMed] [Google Scholar]
- 48.Shi HX, Yang K, Liu X, Liu X, Wei B, Shan YF, Zhu LH, Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]