Abstract
As synthetic biology approaches are extended to diverse applications throughout medicine, biotechnology and basic biological research, there is an increasing need to engineer yeast, plant and mammalian cells. Eukaryotic genomes are regulated by the diverse biochemical and biophysical states of chromatin, which brings distinct challenges, as well as opportunities, over applications in bacteria. Recent synthetic approaches, including `epigenome editing', have allowed the direct and functional dissection of many aspects of physiological chromatin regulation. These studies lay the foundation for biomedical and biotechnological engineering applications that could take advantage of the unique combinatorial and spatiotemporal layers of chromatin regulation to create synthetic systems of unprecedented sophistication.
Synthetic biology provides a powerful framework to understand and harness biology. Biological components are assembled into well-controlled systems, enabling the systematic study of emergent properties and complex behaviours. Synthetic components and systems and the regulatory behaviours they encode can then be adapted by engineers for numerous applications — for example, controlling the dynamic expression of biosynthetic genes in industrial organisms, engineering sensors of environmental state and creating therapeutically relevant cell types. The core components that synthetic biologists have come to rely on, particularly transcriptional repressors and inducible promoters, were assembled in bacteria by early pioneers into genetic networks with switching1 and oscillating behaviours2, and these components have since been applied to engineer bacterial cells that can sense and `remember' the presence of antibiotics in the mouse gut3. Importantly, by engineering connections between basic genetic units, these pioneering studies demonstrated the functional power of synthetic systems to directly test hypotheses about how complex regulatory behaviours arise and to create useful cellular devices.
As we confront new challenges in biology, medicine and biotechnology, there are great opportunities to apply synthetic biology approaches to higher-order organisms such as yeast, plants and mammals. Indeed, many of the synthetic components and gene networks developed in bacterial systems have been demonstrated to have utility in eukaryotic systems4. However, it is becoming increasingly clear that these types of regulatory systems alone are unlikely to drive and recapitulate the biological complexity of eukaryotic organisms. Eukaryotic genes are regulated in fundamentally different ways from bacterial genes5. A central distinguishing feature is the packaging of eukaryotic DNA into chromatin. Chromatin underlies the greater complexity of eukaryotic gene regulation and has been implicated in a broad range of industrially and biomedically relevant behaviours, including cellular responses to environmental stresses, cancer and stem cell differentiation6–12. More than a decade ago, synthetic biology provided a functional approach to test hypotheses surrounding genetic networks. It now has bright prospects for functionally testing and expanding our understanding of chromatin and harnessing its diverse roles in cellular regulation.
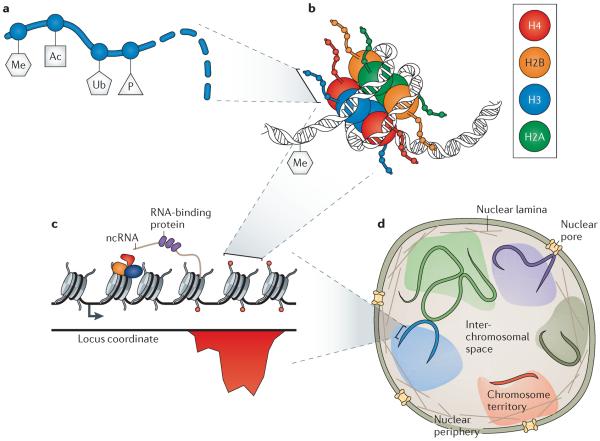
Chromatin is a constellation of DNA, proteins and RNA components that exists in diverse biochemical and conformational states (FIG. 1). Genomic DNA is wound around octamers of histone proteins (FIG. 1b), called nucleosomes, which are arrayed to form the `bead-on-a-string' backbone of chromatin (FIG. 1c). Nucleosomes can be biochemically modified (FIG. 1a) and spatially positioned on DNA, which itself can be methylated13, by the actions of hundreds of chromatin-modifying proteins. Nucleosomes also provide binding surfaces for these modifying proteins, as well as for transcription factors and nucleic acid polymerases14. Furthermore, nucleosomes alter the affinity of transcription factors for the underlying DNA through steric interactions15,16. In fact, there is intimate crosstalk between transcription factors and the chromatin state, and the genomic locations of one are a significant predictor of the other17. At larger scales (FIG. 1d), regions of chromatin on the same or different chromosomes can interact with each other and with subnuclear structures, thus sharing regulatory factors that can influence and coordinate gene expression18. Much of our understanding of chromatin comes from decades of work in molecular cell biology and biophysics, and it has recently accelerated through the generation of many genome-wide maps of chromatin components and structure19,20. This work has revealed a rich realm of spatiotemporal and combinatorial regulatory potential19,21. Synthetic approaches are providing functional complements to these high-resolution maps and revealing the roles of different chromatin features in generating complex regulatory behaviours. Moreover, by expanding our understanding of chromatin and ability to functionally manipulate it, synthetic systems could soon be engineered for a wide variety of human health and industrial applications.
Figure 1. Regulatory features of chromatin at multiple length scales.
a | The amino termini of histone proteins have numerous amino acid residues that can be biochemically modified, such as by the addition of methyl (Me), acetyl (Ac), ubiquitin (Ub) and phosphate (P) groups. These modifications influence the binding of DNA and regulatory proteins26. b | Genomic DNA, which itself can be methylated on cytosine residues, is wound around 4 pairs of histone proteins, which collectively comprise a nucleosome15. c | The positioning of nucleosomes on DNA influences the accessibility of transcription factors to regions such as the promoter. Regulatory proteins (orange, blue, red and purple) bind to nucleosomes, DNA and transcribed non-coding RNA (ncRNA). Histone marks (red circles) often appear in large spatial domains; their occupancy as a function of genomic position (red histogram) can be quantified using chromatin immunoprecipitation followed by DNA sequencing (ChIP–seq)19,21. d | Chromosomes exist in spatial territories in the nucleus. There are interactions within and between chromosomes, as well as between chromosomes and nuclear structures such as the nuclear pore, inner nuclear membrane and nuclear lamina18.
This Review discusses recent progress at the intersection of chromatin biology and synthetic biology, highlighting synthetic approaches to study and control three major features of chromatin structure: biochemical chromatin modifications, including their contribution to heritable and stable gene silencing through heterochromatin; nucleosome positioning; and spatial and topological conformations. We also consider the regulatory potential of non-coding RNAs in BOX 1. We focus on how fundamental regulatory motifs are revealed by the synthetic manipulation of chromatin. Native chromatin biology has been reviewed in greater depth and detail elsewhere21,22. We conclude by discussing potential applications in medicine and biotechnology, as well as new technologies that may accelerate future work in synthetic chromatin biology.
Editing chromatin biochemistry
In 1964, Vincent Allfrey and colleagues radioactively labelled histones in vitro with 14C-acetyl and 14C-methyl groups, which provided evidence for the post-translational modification of histones23. They went on to hypothesize that lysine acetylation reduces the electrostatic attraction of positively charged histones to negatively charged DNA: “as a charge neutralization mechanism, acetylation of the histones would be expected to modify DNA–histone interactions, and this may offer a molecular basis for the pronounced changes in histone acetylation and RNA synthesis during the course of gene activation” (REF. 24). This was an exciting hypothesis not only because of the mechanism it posited but also because it advocated a functional relationship between chromatin structure and gene expression. We now know that there are more than a dozen distinct types of histone modifications25, more than 60 modified amino acid residues of histones25, 2 of each histone type per nucleosome, and several different forms of DNA methylation. This combinatorial diversity creates a challenging problem for studying the individual and combinatorial roles of histone modifications; yet, it could underlie a powerful approach to synthetically set and tune expression states of synthetic genes without altering their DNA sequence. Thus, understanding the roles of these chromatin modifications has been a central goal in the past 50 years and will continue to be so in the foreseeable future.
Is there a histone code?
An attractive hypothesis called the `histone code' posits that particular combinations of histone modifications are `read' by proteins to influence downstream functions26. There have been impressive efforts that mapped the genome-wide locations of chromatin modifications and correlated them to transcriptional states21. In addition, common experimental perturbations knock out or pharmacologically inhibit histone-modifying proteins to alter chromatin state. However, correlation and pleiotropy resulting from genome-wide perturbations often confound these approaches. Thus, for many specific histone modifications, it remains unclear whether they cause, result from or are unrelated to transcriptional activity27,28. Two complementary synthetic approaches that tackle these challenges are the generation of synthetic histone proteins to study the effects of modifications on specific amino acid residues, and the biochemical modification of nucleosomes at specific genomic locations in cells. In addressing the histone code hypothesis, these minimal synthetic approaches may also provide a `blueprint' for assembling chromatin components to derive useful regulatory logic and behaviours from the vast combinatorial space of histone modifications.
Residue-specific modifications in synthetic histones
Synthetic histones have been extremely powerful tools to control the specific type and residue of histone modifications (FIG. 2A). The most comprehensive approaches were derived from chemical biology and unnatural amino acid techniques, and are able to mimic almost all modification types at any histone residue29. Chemical ligation of peptides with specific post-translational modifications to recombinant histones provides control over the modification type and the modified residue30 (FIG. 2Aa). These histones can be reconstituted into nucleosomes for in vitro studies31. A high-throughput method was recently developed to synthesize 54 chemically defined nucleosomes with distinct combinations of post-translational modifications32. These nucleosomes were incubated in vitro with histone-binding and histone-modifying proteins to assess the effects of histone modifications on the binding affinity of these proteins and on their catalytic activity towards other histone residues. Binding of the nucleosomes to barcoded DNA sequences followed by next-generation sequencing allowed all the different synthetic nucleosomes to be pooled for each assay. This increased throughput to thousands of experimental samples. Unnatural amino acids also provide a strategy to control the modification of specific histone residues33,34 (FIG. 2Ab). For example, Nguyen and colleagues replaced the histone H3 lysine 9 (H3K9) residue with unnatural amino acids that could be chemically deprotected in vitro to reveal monomethyl35 and dimethyl36 lysines, and they used these recombinant histones to study the binding of heterochromatin protein 1 (HP1).
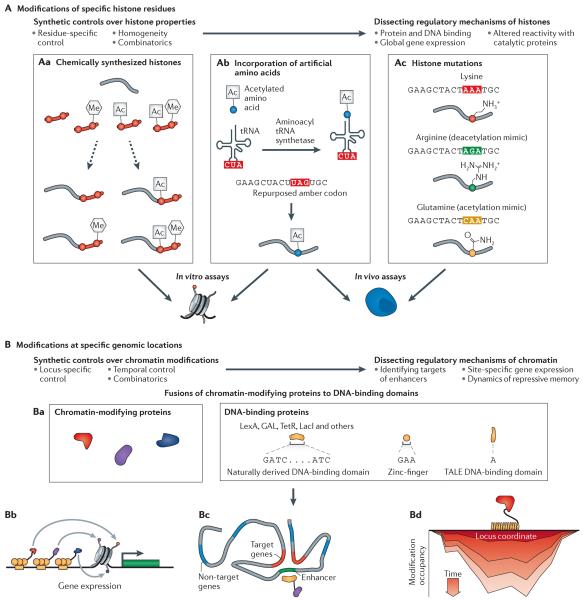
Figure 2. Synthetic control of biochemical chromatin modifications.
Synthetic approaches provide two types of specificity: specificity in the histone residues that are modified (part A) and specificity in the genomic locations of histone modifications (part B). Aa | Synthetic histones can be created by ligating chemically defined peptides (red) to the amino termini of partial histone proteins that are produced recombinantly (grey)30–32. Ab | Artificial amino acids that are already biochemically modified can be incorporated into specific residues of histones that are recombinantly produced in bacteria33–37. This approach requires expression of an aminoacyl tRNA synthetase and an acetyl-lysine transporter to import exogenously supplied acetyl-lysine. Ac | Genetic mutations can change specific residues into ones that partially mimic the charge and shape of a modified histone residue38–40. Histones that are chemically synthesized or that contain artificial amino acids can be used in in vitro assays to measure changes in binding affinities to regulatory proteins or altered reactivity of specific histone residues due to the presence of modifications on other residues. Histones that were genetically mutated can be used in cellular assays to measure the effects of specific residues on global gene expression profiles. Ba | Chromatin-modifying proteins with catalytic properties such as acetyltransferase or methyltransferase activities can be fused to DNA-binding proteins42–45,47,48,52–56. Bb–c | The fusion proteins bind to specific locations in the genome, altering gene expression of a downstream gene (part Bb) or genes that are targeted by a bound enhancer (part Bc). Bd | Chromatin modifiers can also be dynamically recruited to a genomic locus, allowing temporal measurements of changes in chromatin modifications53,66,70. TALE, transcription activator-like effector.
Although chemical ligation and artificial amino acid strategies generate highly homogeneous histones, they are difficult to implement in cells, but this remains a tantalizing possibility37. Instead, most methods in cells have relied on the generation of residue-specific histone mutants or the partial deletion of histone tails38,39 (FIG. 2Ac). In one of the most comprehensive studies so far, Dai et al.40 used gene synthesis to create a library of 486 Saccharomyces cerevisiae mutants of histones H3 and H4. They substituted every amino acid in the complete histone proteins with alanine and every alanine with serine, and further substituted all lysine residues with arginine or glutamine to mimic the electrostatic charges of constitutively deacetylated or acetylated states, respectively. They also deleted segments of varying lengths from the amino-terminal tails of histones H3 and H4. By DNA barcoding each mutant, the yeast strains could be efficiently screened for a range of phenotypes, including fitness, sensitivity to heat shock, sensitivity to DNA damage, altered transcriptional elongation and altered transcriptional silencing. Fitness-related genetic interactions were also identified between specific histone mutants and Ubp8 and Set2, which regulate H2BK123 ubiquitylation and H3K36 methylation, respectively. In the future, this type of high-throughput approach could be extended to identify additional interdependencies between histone residues and chromatin-regulating proteins. It is also possible that particular histone modifications have different and potentially counterproductive effects on industrially relevant cellular properties, such as fitness, heat shock response and ethanol tolerance. Libraries of industrial yeast strains expressing mutant histones could be phenotypically screened to identify biosynthetic strains with desirable characteristics. Thus, residue-specific modifications in synthetic histones provide an important avenue both to study the combinatorial complexity of the histone code and to generate useful cellular properties and industrial cell strains.
Site-specific chromatin modifications: evolving technologies
Synthetic histones provide unparalleled control over the types and residues of post-translational modifications. However, they are limited to in vitro studies or to cellular studies of effects globally across the genome. As a complementary strategy, chromatin modifiers can be recruited to a particular genomic locus; such site-specific `epigenome editing' approaches help to avoid pleiotropic effects and provide evidence of whether chromatin modifications directly cause changes in gene expression (FIG. 2B). This approach also has broader functionality in terms of its ability to recruit activities besides histone modifications, including DNA methylation and nucleosome remodelling. Furthermore, site-specific control will be necessary to regulate specific genes or genomic regions for biomedical41 and biotechnological applications. The origin of synthetic site-specific regulators can be traced to early work in synthetic yeast transcription factors, in which the DNA-binding bacterial repressor LexA was fused to the transcriptional activator Gal4 (REF. 42). Since then, many chromatin modifiers have also been fused to DNA-binding domains (DBDs), as reviewed elsewhere43 (FIG. 2Ba).
More recently, DBDs that can be programmed to bind to almost any DNA sequence have been fused to chromatin modifiers (FIG. 2Ba). Programmable DBDs, such as zinc-finger (ZF) and transcription activator-like effector (TALE) proteins44, are important for several reasons. They enable modification of chromatin at essentially any endogenous locus without the need to insert synthetic DNA sequences, which is a key advancement over the use of artificial episomal or integrated DNA sequences (FIG. 2Bb). For example, endogenous genes were activated by ZF–Tet and TALE–Tet fusion proteins (which induce active demethylation of methylated cytosine)45,46 and were suppressed by ZF–DNA methyltransferases47–49 and ZF–H3K9 methyltransferases50,51. These studies provide strong evidence that DNA and H3K9 methylation are causative of gene repression, especially with the use of minimal H3K9 methyltransferase catalytic domains and catalytic mutants that failed to induce methylation and gene repression51. Endogenous targeting was also used to identify natural targets of enhancer regions52 (FIG. 2Bc). This was achieved by reducing the activities of enhancers with TALE–LSD1 (also known as KDM1A) fusions that demethylated H3K4, and identifying native targets of the enhancer by observing the resulting downregulation of proximal genes. Programmable DBDs also enable multiplex and combinatorial targeting, which has proved useful in several cases. For example, using ZFs, we recently co-recruited a library of 223 diverse chromatin-regulating proteins together with the commonly used transcriptional activator VP16, which provided a set of distinct, two-input gene expression logic53. Other groups have used multiple ZFs to increase the site specificity of synthetic DNA methylation by fusing two co-dependent parts of a DNA methyltransferase to two distinct ZFs that bind to adjacent genomic sites54–56. Future work is likely to take advantage of highly multiplexible CRISPR–Cas9 systems57,58 to increase the throughput and technical ease of introducing multiple site-specific chromatin modifications. Indeed, Cas9 could enable large-scale `rewiring' or reprogramming of the chromatin network for research in stem cells, development, genomic stability and evolution. Furthermore, libraries of multiplexed site-specific chromatin modifiers could be used to tune the expression of biosynthetic or endogenous genes without the need to alter gene sequences, which enable rapid screening and engineering of production organisms.
Site-specific chromatin modifications: heterochromatin and stable gene repression
Site-specific recruitment of chromatin modifiers also enables mechanistic studies of other complex behaviours such as gene expression `memory'. Indeed, chromatin has been extensively implicated in the establishment and maintenance of gene expression memory, so much so that the term epigenetic is often co-opted to refer to chromatin modifications in general. However, by a stricter definition, epigenetic refers only to gene expression states that are heritable through cell divisions. This is a crucial point, as it is clear that chromatin modifications alone do not transmit heritable `epigenetic' information. Instead, they must act in concert with RNA interference (RNAi) and protein factors through self-reinforcing feedback mechanisms to maintain memory59,60. As reviewed elsewhere59, much effort over the past several decades has focused on identifying these mechanisms and minimal components that give rise to heritable gene regulation, mainly in stable repressive regions called heterochromatin. Heterochromatin describes compact genomic regions that are often characterized by reduced histone acetylation, H3K9 trimethylation (H3K9me3) enrichment and occupancy of repressive proteins such as HP1.
Synthetic approaches have contributed to this area of research by demonstrating the causative functions of various chromatin regulators in the establishment of heterochromatic silencing. In particular, synthetically recruited heterochromatin proteins can demonstrate the requirement or sufficiency of DNA sequence elements, RNAi components, histone and DNA modifications, and regulatory proteins for memory, and such proteins can even induce heritable repression53,61–69. More recently, Ragunathan et al.70 showed that histone modifications and regulatory proteins are sufficient to establish stable heterochromatin in Schizosaccharomyces pombe, and that DNA sequence elements and RNAi are dispensable. They did so by transiently recruiting the S. pombe H3K9 methyltransferase Clr4 to a specific genomic location. This established a large domain of repressive H3K9me3 that was maintained over several mitotic and meiotic cell divisions in the absence of any DNA sequence elements, RNAi, DNA methylation or recruited Clr4. This supports a mechanism in which the histone modification, H3K9me3, and proteins, including Clr4, that both read and `write' H3K9me3 are able to propagate histone modifications over time and maintain heterochromatic memory. Interestingly, proteins that catalyse and remove H3K9me3 marks regulated the stability of this heterochromatic region, suggesting a potential way to tune this form of memory in engineering applications.
Recently, Hathaway et al.66 developed an inducible system that dynamically regulated (FIG. 2Bd) the recruitment of HP1 and the strong synthetic transcriptional activator VP64 to a reporter locus in mouse embryonic stem cells. The associations of HP1 with the synthetic ZFHD1 DBD and of VP64 with the Gal4 DBD were controlled by the inducible dimerization domains of the FKBP–FRB system and the PYL–ABI system, respectively. This approach provided unprecedented temporal resolution of the kinetics of heterochromatin formation and reactivation and, through an accompanying quantitative model, insights into molecular mechanisms of heterochromatin establishment. Notably, the authors showed that the recruited HP1 protein could induce 10-kb regions of heterochromatic H3K9me3 after 5 days, and that this region was stable for at least 8 days after HP1 recruitment was washed out. Their model and data showed that the stability and boundaries of the heterochromatic region were determined by a dynamic competition between placement of H3K9me3 marks and nucleosome turnover. An increase in DNA methylation was also measured; based on previous work that linked DNA methylation to stable heterochromatin69,71, this probably contributed to the observed gene repression memory. The continued development of this type of synthetic approach will be of particular interest to both chromatin biologists and synthetic biologists because memory and switch-like elements are of tremendous importance in cell biology, as well as in biomedical and industrial applications.
Site-specific chromatin modifications: ongoing challenges
Future work in site-specific chromatin modification should address the substrate specificity of synthetic chromatin modifiers. For example, a histone acetyltransferase may acetylate many lysine residues on a histone and recruit other regulatory proteins through binding interactions. These potential nonspecific modifications and interactions need to be fully characterized. The use of catalytically dead mutant forms of chromatin modifiers could reveal their residue specificity, whereas the truncation of chromatin modifiers to minimal catalytic domains could reduce the recruitment of other regulatory proteins51.
Defining the binding specificities of the various customizable DNA-targeting platforms is also of considerable interest in the field. Although gene regulation activities tend to be the strongest at intended target sites, ZFs, TALEs and Cas9 can all exhibit substantial off-target binding as observed by various methods, and even minor off-target activities could be oncogenic or toxic72–79. Some strategies to improve the specificity of binding (or of effector activity) may include obligate pairs of DNA-binding or effector domains54,55,80,81; weakening binding affinities to individual nucleosides while increasing the overall length of the target DNA sequence82; and altering the length of DNA-binding arrays for ZFs and TALEs or the guide RNA for Cas9 targeting77,83,84.
Most work has focused on writing chromatin modifications, but complementary technologies for reading these modifications85 will be equally important if conditional feedback of basal or altered chromatin states is desired, for example, to inform downstream actions such as the writing of additional modifications. The ability to sense aberrant chromatin states in live cells could also be useful in diagnosing disease states such as cancer.
These chromatin-editing and chromatin-sensing technologies will require creative, new strategies to enhance control over substrate modifications and sensing in diverse chromatin contexts; address typically modest protein–nucleosome binding energies86; improve genomic site specificity; and stabilize chromatin-based memory. For example, to address weak binding energies, inspiration could be drawn from natural systems and proteins that bind to chromatin through multiple domains or increased avidity87.
In addition to these improvements, the prospects for site-specific chromatin modifiers are bright and are likely to involve the incorporation of diverse molecular engineering techniques that will provide enhanced spatiotemporal resolution. In one recent example, Konermann et al.88 fused chromatin modifiers and TALEs to the Cry2 and CIB1 protein domains, respectively. Illumination with blue light dimerized the Cry2 and CIB1 domains, and dynamically localized chromatin modifiers to the DNA-bound TALE protein. These and other future systems may also adapt interesting biological systems that exhibit useful regulatory behaviours. For example, lowly expressed or non-expressed developmental genes have been observed to be `poised' for rapid activation by the presence of both active and repressive chromatin marks89. By engineering such bivalent states, this principle could be exploited for switch-like or adaptive responses in cellular engineering. Finally, although DBDs have mainly been used to effect changes in chromatin biochemistry, they can also recruit proteins that modulate many other useful aspects of chromatin biology (see below). In summary, site-specific chromatin modifications provide a powerful way to functionally study the roles of diverse chromatin-regulating activities and chromatin structures in gene expression.
Modulating nucleosome positioning
Beyond biochemical modifications, nucleosome positioning on DNA influences gene expression. Nucleosomes generally inhibit transcription by reducing accessibility of transcription factors and RNA polymerases to DNA90–92, thus creating a basally repressed state that contrasts with highly accessible bacterial DNA5. Both nucleosome-remodelling proteins and the underlying DNA sequence control nucleosome positioning93–95. Owing to the ease of manipulating genetic sequences, synthetic approaches have focused on the manipulation of DNA sequences to control nucleosome positioning and gene expression. For example, Raveh-Sadka et al.96 created a library of more than 70 promoter variants, each with different positioning and lengths of a poly(dA:dT) tract that is known to disfavour nucleosome binding. Using this system, the positioning of nucleosomes regulated the accessibility of transcription factors to DNA. This strategy provided a dynamic range of more than an order of magnitude in transcriptional activity. This highly tunable approach to design the strength of synthetic promoters could be useful in situations for which balancing the expression strength of different components is important for its behaviour, such as in genetic oscillators2.
In addition to controlling transcription levels, the positioning of nucleosomes can drive complex non-linear behaviours. For example, Lam et al.97 showed that, in yeast, nucleosomes can decouple the activation threshold of the PHO5 promoter from its dynamic range of expression. The activation threshold is controlled by the DNA-binding affinity of a protein that moves a nucleosome which originally blocks access of transcription factors to the promoter. When this nucleosome remodelling event is achieved, transcription factor binding sites uncovered in the promoter then determine the dynamic range. This type of regulation is reminiscent of a transistor in electrical engineering, in which the application of one signal allows the passage of a second signal, and could be used as a biological switch. Furthermore, the signal or protein expression level required to move the nucleosome and activate the PHO5 promoter could be engineered to be weaker than the protein expression level driven by the activated PHO5 promoter. This would essentially act as a signal amplifier that could be useful in circuit and cellular engineering. Future efforts could synthetically reposition nucleosomes by recruiting nucleosome remodellers (for example, SWI/SNF) to test other mechanisms that have been hypothesized to underlie complex behaviours, including the cooperative and switch-like activation or amplification of some genes98. These types of experiments demonstrate how nucleosome formation and repositioning could contribute to sophisticated and potentially useful gene expression programmes.
Spatial features of chromatin
Synthetic biology has primarily abstracted genomic material as a repository of regulatory components and genes that produce proteins and RNA. Complex behaviours are derived from the interactions between these components through temporally dynamic genetic and biochemical networks. The chromatin scaffold provides a unique opportunity to engineer cellular systems that also function in spatial dimensions. The repetitive array of nucleosomes on DNA and the polymeric nature of chromatin provide the physical properties needed to regulate genes at both small and large spatial scales in the nucleus. For example, domains of DNA and histone modifications (such as H3K9me3) often cover multiple tandem genes. At larger spatial scales, genomic loci interact within and between chromosomes and also localize to subnuclear structures such as the nuclear lamina. Synthetic approaches that control these spatial features are deepening our understanding of chromatin biology and revealing emergent behaviours that could be useful in cellular engineering (FIG. 3).
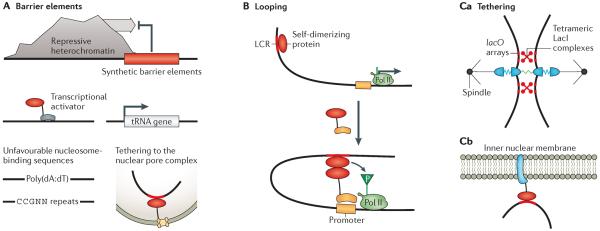
Figure 3. Synthetic spatial control of chromatin.
A | Several different types of barrier elements (red) can block the spreading of repressive heterochromatin. These include recruitment of transcriptional activators101, tethering to a nuclear pore complex102, insertion of actively transcribed tRNA genes or insertion of nucleosome-disfavouring DNA sequences99,100. B | The recruitment of a self-dimerizing protein can induce chromatin looping and localization of an enhancer or a locus control region (LCR) to a promoter107. This could localize factors that phosphorylate (P) and activate RNA polymerase II (Pol II). C | DNA-binding proteins can be used to answer biological questions, for example, whether the spindle checkpoint senses displacement between sister chromatids or within kinetochores116 (part Ca). Fusing DNA-binding domains to nuclear envelope proteins can localize (and often silence) specific genomic regions117 (part Cb). Part Ca adapted from REF. 116.
Controlling boundaries of genomic activity
The juxta-position of sequence elements (DNA) and non-sequence elements (nucleosomes and DNA methylation) is an intriguing property of chromatin with profound and broad implications for gene regulation. For example, nucleosomes and methylated DNA bases provide a repetitive scaffold to which regulatory proteins can bind and propagate over large spatial regions that contain multiple genes. However, these regions and their boundaries must ultimately be controlled by DNA sequence elements. Synthetic tools have provided insights into this crosstalk between sequence and non-sequence elements. In particular, many studies have focused on how the boundaries of large regions of silent heterochromatin, which are often marked by DNA methylation and H3K9 methylation, are specified. It is thought that these regions are established by self-reinforcing mechanisms in which protein complexes bind to heterochromatic modifications and then catalyse the same modifications on adjacent nucleosomes or DNA.
Synthetic approaches have revealed various protein and sequence elements that can control the spreading of heterochromatic regions. Telomeres have been used as a natural source of heterochromatin, with auxotrophic markers inserted nearby as sensors of heterochromatic spreading. Using similar systems, various sequence elements have been inserted between the markers and telomeres, and identified as barriers to heterochromatic spreading. These include nucleosome-disrupting sequences99,100, transcriptionally active tRNA genes, binding sequences for synthetic transcriptional activators such as SAS2 fused to the DBD of GAL4 (REF. 101), and sequences tethered to the nuclear pore complex102 (FIG. 3A).
There are also boundaries of transcriptional activity within individual genes that can and need to be controlled. For example, most promoters have a limited extent of divergent transcription of non-coding sequences. Marquadt et al.103 integrated a synthetic bidirectional promoter, which drives divergent transcription of two distinct fluorescent proteins, into a yeast knockout library and found that divergent transcription increased in CAF-I-knockout strains. As CAF-I regulates chromatin assembly, nucleosome positioning upstream of promoters probably limits divergent non-coding transcription. As more sophisticated synthetic gene expression systems are developed, both intragenic and intergenic boundary elements — such as CAF-I and nucleosome-disrupting sequences, respectively — will need to be used to avoid unintended spatial regulation.
Chromatin conformation and subnuclear interactions
Chromosomes exist in semi-restricted spatial domains within the nucleus called chromosome territories. In addition, the positioning of genomic loci within each chromosome is nonrandom, as is their positioning relative to subnuclear structures such as the nuclear lamina18,104. However, it is unclear whether these forms of spatial organization are just by-products of evolution or whether they have evolved for specific dynamic regulatory functions. Recent synthetic approaches have recapitulated some of these interactions, demonstrating that at least in some synthetic and physiological situations long-range interactions and chromatin conformation exhibit function.
Synthetic approaches have demonstrated that transcriptional effectors recruited to sites more than 1 kb downstream of promoters can both activate105 and repress106 yeast genes through intrachromosomal looping. There is also evidence that looping can regulate genes at much greater distances. One prominent example is the β-globin gene that is regulated by its locus control region (LCR) positioned 40 kb away. Deng et al.107 demonstrated that this looping was causative of gene activation. They designed a ZF that binds to the β-globin promoter and fused it to Ldb1, a protein that is present at the LCR. The synthetic tethering of Ldb1 to the β-globin promoter was enough to recruit the LCR region through Ldb1–Ldb1 binding and resulted in activation of β-globin transcription (FIG. 3B). The authors also used the same strategy to reactivate the developmentally silenced fetal γ-globin promoter in primary adult murine erythroblasts108. This strategy could yield therapeutic benefits in patients with sickle cell anaemia or β-thalassemia by using gene therapy approaches to introduce looping factors into cells, thus inducing transcription of non-sickling γ-globin to replace defective adult β-globin. Such potential therapeutics could be within reach in a decade or two, as there has been a recent resurgence in clinical trials of gene therapies109. This looping approach has several advantages, including the requirement of only a single ZF–Ldb1 factor for introduction into cells. Furthermore, the looping approach activated γ-globin expression by more than 1,000-fold, which is much stronger than the levels typically observed with synthetic activators. Thus, this strategy could lead to therapies that are more potent than the simple recruitment of transcriptional activators directly to the γ-globin promoter or the introduction of exogenous globin genes.
Chromatin–chromatin interactions also drive complex functions beyond gene activation. Noordermeer et al.110 tested whether the insertion of an artificial LCR onto a separate chromosome from the β-globin gene could activate β-globin transcription. They observed that interchromosomal interactions between the artificial LCR and the native β-globin gene were able to activate transcription, although this activation occurred only in rare `jackpot' cells. The authors hypothesized that rare long-range interactions might be a way for cells to stochastically generate phenotypic diversity and described this effect as `spatial effect variegation'. In addition to activating individual genes such as β-globin, chromatin–chromatin interactions can also temporally coordinate the expression of multiple genes, which could be useful in engineering the dynamic and ordered expression of different components in biosynthetic pathways. For example, active and inactive regions of the human genome tend to cluster together through predominantly intrachromosomal, as well as interchromosomal, contacts111. It is thought that regulatory factors, either activating112 or repressive113, are shared between spatially clustered genes, thus coordinating transcription regulation. Fanucchi et al.112 revealed an example of this coordination between the SAMD4A, TNFAIP2 and SLC6A5 genes, which are known to cluster in response to tumour necrosis factor-α (TNFα) exposure. They designed TALE nucleases to cleave each of the three loops that brought these genes together and measured changes in gene expression. They found that disrupting the chromatin contacts interfered with transcription of the other genes. Interestingly, this co-transcriptional effect was hierarchical. Some cleavage sites disrupted transcription of all three genes, whereas others affected only one or two genes.
In addition to intrachromatin interactions, chromatin also localizes with various subnuclear structures. Some subnuclear structures can be induced to form on chromatin by the tethering of specific proteins. For example, artificial kinetochores or centromeres can be created by the synthetic recruitment of specific proteins to DNA. When Ask1, a member of the Dam1–DASH microtubule complex in S. cerevisiae, was fused to LacI and recruited to eight copies of genomically integrated lacO operator sites, it was able to trigger assembly of a synthetic kinetochore114. This kinetochore carried out many native functions, including bi-orienting sister kinetochores at metaphase, segregating sister chromatids and repairing errors in chromosome attachment. In another example, synthetic recruitment of the Clr4 H3K9 methyltransferase to a euchromatic region in S. pombe promoted de novo centromere formation115. Synthetic control over centromere and kinetochore formation, coupled with knockouts of different centromere-related proteins, could reveal different roles of proteins in centromere establishment and function, including in regulating the fidelity and stability of the chromosome. Furthermore, the ability to engineer stable synthetic centromeres in synthetic chromosomes could be useful in expressing large cassettes that contain entire synthetic circuits, full biosynthetic pathways or large therapeutic genes that would be difficult to package in viruses for gene delivery (such as titin for various myopathies).
Synthetic tools can also be used to control the biophysical parameters of subnuclear structures, such as centromeres and kinetochores, to answer fundamental biological questions. In particular, it had been unclear how, during mitosis, the spindle checkpoint senses that paired chromosomes are attached to separate spindles. The general hypothesis was that when chromosomes were properly attached to separate spindles, the spindles would pull in opposite directions, which leads to a displacement either within kinetochore protein complexes or between the paired centromeres. To address this question, Nannas et al.116 synthetically restricted the ability of centromeres to stretch by integrating lacO sites on either end of the centromere of chromosome III in haploid S. cerevisiae (FIG. 3Ca). By expressing tetrameric LacI complexes during mitosis, the outside ends of the paired centromeres were tethered together and restricted from moving apart. Tethering did not alter the spindle checkpoint or chromatid segregation, suggesting the checkpoint senses intrachromosomal kinetochore strain rather than interchromosomal displacement between paired centromeres116.
Chromatin also localizes to other subnuclear structures that do not function as part of chromosomes. Some of these interactions are correlated with transcriptional activity. For example, the nuclear lamina is generally associated with repressed genes, whereas the nuclear pore is associated with active genes. Although the mechanisms underlying these correlations remain unclear, synthetic tethering of reporter genes to sub nuclear structures has been shown to alter gene expression61–64. In one example, Reddy et al.117 inserted an array of lacO operator sites downstream of a reporter gene. They also fused LacI to emerin, an inner nuclear membrane protein (FIG. 3Cb). Through interaction with LacI, the reporter was recruited to the nuclear membrane and silenced117. In a related experimental system, Kind et al.118 fused the nuclear lamina protein lamin B1 to the adenine methyltransferase dam. When they expressed the methyl-adenine-binding protein dpnI fused to the transcriptional activator VP16, the fusion protein was recruited and bound to methylated adenine bases located at the nuclear lamina. The dpnI–VP16 fusion protein then induced delamination of lamina-associated chromatin domains from the nuclear periphery. The authors hypothesized that activating transcription in lamina-associated chromatin domains induced their movement away from the nuclear periphery, which is consistent with the trend of a silent nuclear periphery117. Finally, in addition to controlling chromatin localization and conformations, synthetic DBDs such as LacI or Cas9 can also be used to recruit fluorescent proteins to observe the dynamic 3D structure of chromatin18,119,120. In summary, many rich behaviours arise from the conformations and subnuclear interactions of chromatin, and existing and future synthetic tools that control the spatial aspects of chromatin may reveal even more unique regulatory properties.
A bright future for synthetic chromatin biology
More than a decade ago, systems and synthetic biology enhanced our understanding of gene networks1,2,121,122. It is appealing to speculate how these fields may once again provide insights into the regulatory logic of chromatin5 and address the many open questions in chromatin and cell biology. The key advantages that synthetic approaches have provided are their ability to functionally perturb chromatin states with specificity in both histone residues and genomic locations. These attributes should continue to help researchers to determine the extent and validity of a histone code, as well as its functional role in the regulation of various molecular processes from gene expression activity and kinetics to DNA repair. Furthermore, although synthetic approaches often force the recruitment or manipulation of chromatin components in ways that are physiologically unnatural, their advantage is that they can identify the roles of context — for example, contexts in which regulation at one genomic site versus another or with specific combinations of components leads to distinct outcomes. Additionally, there has been recent and rapid development of technologies that expand our experimental capabilities, and it is exciting to speculate how the development and incorporation of these technologies could have substantial impacts on future research directions. For example, the synthetic construction of entire chromosomes123 could lend itself to studies that interrogate the relationships between regulatory DNA sequences, distances between genes and spatial chromosome conformation. Moreover, CRISPR–Cas9 technologies57,58,124 could enable tractable experiments that induce genome-wide rewiring of chromatin states and subsequent analyses of their effects on many biological processes, including cellular differentiation, proliferation and oncogenesis. Finally, optical tools125 could probe chromatin dynamics at unprecedented spatiotemporal resolution, thus revealing the kinetics of gene activation and repression, memory formation and the relative order of action of different components in executing biological processes such as transcription.
In addition to expanding our fundamental understanding of biology, an ongoing achievement of synthetic biologists is the abstraction of biology into modular components with defined properties, which greatly streamlines design cycles for cellular engineering and applications. An intriguing question that remains is whether an analogous design framework is possible at the chromatin level and, if so, what form this framework might take. The most effective synthetic approaches will continue to benefit from the wealth of knowledge and conceptual thinking developed by chromatin biologists over the past 50 years. In fact, our fundamental understanding of chromatin has already had a substantial impact in identifying regulatory properties that could be harnessed in artificially engineered cellular systems for bioindustrial (FIG. 4A) and biomedical (FIG. 4B) applications. For example, spatially coordinated gene expression and multigene silencing53,70 could be used by metabolic and microbial engineers to efficiently regulate large biosynthetic cassettes in a manner that cannot be achieved by bacterial systems (FIG. 4Aa). The many barrier elements that have been identified99–102 could also be used to control these regions of long-range regulation and prevent the spreading of regulation to endogenous genes. The direct role of chromatin in gene expression memory has been synthetically recapitulated53,66,70 and could be used to record and respond to environmental events to which cells are subjected, such as exposure to antibiotics, heat shock or toxins, or changes in cell density in a bioreactor (FIG. 4Ab). Finally, given the many ways to now target, modulate and modify chromatin, these approaches are likely to be used to correct disease-associated chromatin states in cancer, neurodegenerative diseases and many other conditions, or to artificially alter chromatin changes that are involved in developmental and differentiation pathways (FIG. 4B). Given that several epigenetic therapeutics, typically small-molecule inhibitors of chromatin-modifying proteins, have already been used in the clinic86, more-specific strategies derived from synthetic biology approaches to intervene in epigenetic states could become promising next-generation therapeutic avenues in the near future. All of these types of applications are theoretically only as far from the clinic or the pilot plant as our ability to integrate them into existing synthetic biology frameworks. Initial applications of chromatin-based regulation in industry are likely to augment and enhance existing synthetic systems and organisms, and they could be closer to implementation than biomedical applications. Applications in biomedically relevant cell types will be slower to realize, as they are limited by our understanding of the role of chromatin in cellular regulation, as well as by the higher standard for off-target effects amid worries of the oncogenic potential of artificial factors. However, synthetic approaches will probably contribute substantially to our understanding of biomedical problems and present novels ways to address them.
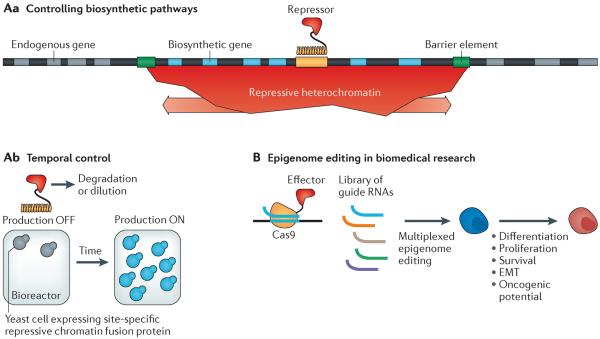
Figure 4. Potential applications of synthetic chromatin biology.
A | Eukaryotic organisms such as yeast are widely used in industry to produce diverse molecules that range from food flavourings to pharmaceuticals. Often, multiple biosynthetic genes need to be introduced into an organism's genome. Regulating many genes can be difficult, especially with many distinct promoters and other regulatory elements. Aa | In chromatin-based regulation, chromatin states such as heterochromatin can exert regulation over several kilobases of DNA through the simple recruitment of a repressor to a single location in the genome53,70. Barrier elements can be used to prevent spreading of repressive regulation into endogenous genes99–102. Ab | Repression of biosynthetic genes when cell densities are low promotes cell growth and fitness by avoiding taxing of cell resources. When cell densities in a bioreactor are optimal, the repressor can be degraded or diluted out, thus turning on expression of the biosynthetic genes. B | Chromatin has been widely implicated in many areas relevant to human health, including cell differentiation, cell proliferation, cell survival in different environmental conditions, epithelial-to-mesenchymal transition (EMT, which is relevant to cancer metastasis) and oncogenic potential6–12. Functional tools are lacking to directly perturb chromatin states at specific locations in the genome and to test hypotheses of their roles in these biomedical processes. Type II CRISPR–Cas9 DNA targeting technologies57,58 could be used to edit chromatin states at multiple genomic locations by fusing chromatin regulators to the Cas9 protein and expressing a library of guide RNAs that are complementary to genomic target sites.
Farther down the road, perhaps in the next two decades, we may truly begin to benefit from the regulatory sophistication of chromatin. We are only beginning to uncover the vast regulatory and `computational' potential of chromatin, including switch-like and amplification behaviours at promoters through the positioning and modification of nucleosomes66,89,97,98. The combination of these behaviours with genetic networks could lay the foundation for the next generation of gene expression systems and circuits for programming cells. Synthetic circuits that incorporate chromatin-based components and regulatory functions could give rise to a new layer of sophistication and complexity in synthetic circuits and could be useful in programming `smart' and adaptive industrial organisms or long-term sentinel and therapeutic cells introduced into the human body.
In conclusion, synthetic biology has great potential to advance two broad goals: first, in providing tools and approaches to complement studies that address fundamental questions in biology; second, in developing an engineering framework to harness chromatin components for cellular engineering applications9,11,48,126,127. Our path to both goals will be accelerated by the continued collaboration between scientists and engineers. Given the broad importance of chromatin in most cellular processes and the many diseases with epigenetic components in their aetiology, collaborative efforts could have profound effects on our understanding of cell biology and our treatment of a vast number of diseases.
Box 1 | Synthetic non-coding RNAs regulate chromatin structure.
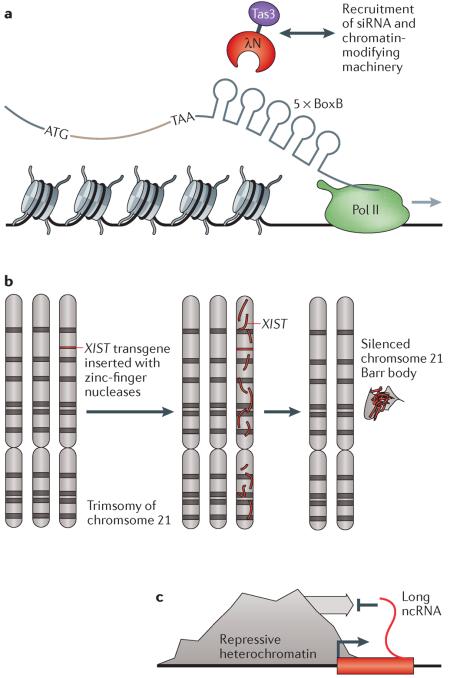
The terms chromatin and epigenetics evoke images of histones and DNA methylation. However, in order to direct chromatin components and modifications to specific genomic regions, it is clear that elements are required to provide sequence specificity. Some of this specificity arises from a bias in nucleosome occupancy based on the underlying DNA sequence and from transcription factors that bind to DNA motifs. More recently, non-coding RNAs (ncRNAs) have been shown to be specificity elements as well. Synthetic approaches have provided evidence that these ncRNAs can act in diverse ways to regulate chromatin. In Schizosaccharomyces pombe, the RNA-induced transcriptional silencing (RITS) complex was hypothesized to target heterochromatin silencing components to DNA via small interfering RNA (siRNA) base-pairing. Buhler et al.128 showed that recruitment of RITS to a ura4 gene — through fusion of ura4 to BoxB mRNA hairpin sequences and fusion of the RITS component Tas3 to λN protein (which binds to BoxB) — induced silencing of ura4 (see the figure, part a). Additionally, siRNAs that targeted ura4 were generated and silenced an additional copy of ura4 in trans128.
In human females, the X chromosome is silenced in a phenomenon known as dosage compensation. The silent X chromosome expresses the long ncRNA X inactive-specific transcript (XIST) that coats the entire chromosome and induces heterochromatic silencing. Recently, using zinc-finger nucleases, the XIST cDNA was integrated into one of the three copies of chromosome 21 in induced pluripotent stem cells (iPSCs) derived from an individual with Down syndrome (see the figure, part b). This silenced the third chromosome 21 and even rescued defects in proliferation and rosette formation in the iPSCs129. This type of synthetic approach could lead to `chromosome therapies' for other trisomy diseases. A recent study has demonstrated that ncRNAs can also function as barriers to heterochromatin spreading (see the figure, part c). Keller et al.130 discovered a long ncRNA termed BORDERLINE that prevented the spreading of pericentromeric heterochromatin on chromosome 1 in S. pombe. Interestingly, deleting BORDERLINE and synthetically replacing it with a ura3 cassette from Candida albicans maintained barrier activity, indicating that active expression of any RNA sequence is likely to be sufficient to block heterochromatin spreading.
Pol II, RNA polymerase II. Reprinted from Cell, 125, Buhler, M., Verdel, A. & Moazed, D., Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing, 873–886, Copyright (2006), with permission from Elsevier.
Acknowledgements
This work was supported by a US National Institutes of Health (NIH)-National Institute of General Medical Sciences (NIGMS) Ruth L. Kirschstein Postdoctoral Fellowship (A.J.K.), an NIH Director's Pioneer Award (J.K.J.), a Defense Advanced Research Projects Agency grant (J.K.J., A.S.K., and J.J.C.), start-up funds from the Department of Biomedical Engineering at Boston University (A.S.K.), a National Science Foundation CAREER Award (A.S.K.), a NIH R24 (J.J.C.), the Wyss Institute for Biologically Inspired Engineering (J.J.C.) and the Howard Hughes Medical Institute (J.J.C.). Owing to space limitations, the authors regret that many important publications were not able to be discussed in this Review.
Glossary
- Nucleosomes
Octamer protein complexes in which an octamer is comprised of two copies each of H3, H4, H2A and H2B histone proteins.
- Non-coding RNAs
Functional RNA molecules that are not translated into proteins.
- Lysine acetylation
A post-translational modification in which an acetyl group reacts with the primary amine on the side chain of a lysine residue.
- Correlation
A relationship between two or more variables. The correlation between the occupancy of a chromatin modification and transcriptional activity does not directly prove that the modification causes transcriptional activity, or vice versa.
- Pleiotropy
The phenomenon whereby one gene influences multiple other seemingly unrelated genes or traits.
- Unnatural amino acid
An amino acid that is not naturally encoded or found in the genetic code of an organism.
- Chemical ligation
A chemical reaction that links a fully chemically derived peptide to the end of a recombinant protein.
- Zinc-finger
(ZF). A small protein structural motif coordinated to one or more zinc ions that stabilize its fold.
- Transcription activator-like effector
(TALE). A bacterial protein with a variable number of 34-amino-acid repeats, of which 2 residues specify binding to a DNA base.
- CRISPR
(Clustered regularly interspaced short palindromic repeat). An important part of a prokaryotic adaptive immune system that uses short RNAs to guide the CRISPR-associated 9 (Cas9) nuclease to specific targets, which cleaves foreign DNA elements such as plasmids and phage genomes.
- RNA interference
(RNAi). A biological process that inhibits gene expression through RNA molecules interacting and interfering with specific mRNA molecules.
- FKBP–FRB
A protein pair consisting of FKBP and FRB, which dimerize by mutually binding to the small molecule rapamycin.
- PYL–ABI
A protein pair consisting of PYL and ABI, which dimerize by mutually binding to abscisic acid.
- Avidity
The accumulated strength of multiple affinities from multivalent non-covalent binding interactions.
- Cry2 and CIB1
(Cryptochrome 2 and cryptochrome-interacting basic helix–loop–helix 1). A pair of proteins that dimerize at the subsecond timescale upon blue-light exposure and that dissociate on the minute timescale.
- Nuclear lamina
A dense fibrillar network of intermediate filament proteins at the periphery of the nucleus.
- Telomeres
The regions at the ends of chromosomes comprised of repetitive nucleotide sequences that are typically repressed by heterochromatin.
- Auxotrophic markers
Genes absent in an organism that normally produce organic compounds required for survival of the organism.
- Yeast knockout library
A collection of yeast strains, each of which harbour a knockout allele for a single gene. Strains are either haploid and have a non-essential gene knocked out, or diploid and have the knockout allele in a heterozygous state.
- β-globin
A subunit of the major haemoglobin complex found in adult mammals.
- Locus control region
(LCR). A genomic region that enhances the expression of genes from a distance.
- Kinetochores
The protein structures assembled on the centromere to which spindle fibres attach during cell division to pull sister chromatids apart.
- Centromeres
The genetic loci on chromosomes that link sister chromatids during mitosis and on which kinetochores assemble.
Footnotes
Competing interests statement The authors declare competing interests: see Web version for details.
References
- 1.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl Acad. Sci. USA. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye H, Aubel D, Fussenegger M. Synthetic mammalian gene circuits for biomedical applications. Curr. Opin. Chem. Biol. 2013;17:910–917. doi: 10.1016/j.cbpa.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 6.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol. Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 7.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nature Rev. Mol. Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rheinbay E, Louis, David N, Bernstein, Bradley E, Suvà, Mario L. A. Tell-tail sign of chromatin: histone mutations drive pediatric glioblastoma. Cancer Cell. 2012;21:329–331. doi: 10.1016/j.ccr.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jost J, Saluz H. DNA Methylation: Molecular Biology and Biological Significance. Birkhäuser Basel: 2011. [Google Scholar]
- 14.Ballare C, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell. 2013;49:67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 16.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 17.Benveniste D, Sonntag HJ, Sanguinetti G, Sproul D. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl Acad. Sci. USA. 2014;111:13367–13372. doi: 10.1073/pnas.1412081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 19.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ram O, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 22.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 23.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidali G, Gershey EL, Allfrey VG. Chemical studies of histone acetylation. The distribution of ε-N-acetyllysine in calf thymus histones. J. Biol. Chem. 1968;243:6361–6366. [PubMed] [Google Scholar]
- 25.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Turner BM. The adjustable nucleosome: an epigenetic signaling module. Trends Genet. 2012;28:436–444. doi: 10.1016/j.tig.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Pick H, Kilic S, Fierz B. Engineering chromatin states: chemical and synthetic biology approaches to investigate histone modification function. Biochim. Biophys. Acta. 2014;1839:644–656. doi: 10.1016/j.bbagrm.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 30.He S, et al. Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis. Proc. Natl Acad. Sci. USA. 2003;100:12033–12038. doi: 10.1073/pnas.2035256100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimko JC, Howard CJ, Poirier MG, Ottesen JJ. Preparing semisynthetic and fully synthetic histones H3 and H4 to modify the nucleosome core. Methods Mol. Biol. 2013;981:177–192. doi: 10.1007/978-1-62703-305-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen UT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nature Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]; By reconstituting chemically synthesized histones with barcoded DNA molecules, this study was able to pool diverse histone–DNA complexes, thus greatly reducing the number of biochemical assays needed when exploring a large combinatorial histone modification space.
- 33.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nature Rev. Mol. Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. Genetically encoding Nε-methyl-l-lysine in recombinant histones. J. Am. Chem. Soc. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen DP, Garcia Alai MM, Virdee S, Chin JW. Genetically directing ε-N, N-dimethyl-l-lysine in recombinant histones. Chem. Biol. 2010;17:1072–1076. doi: 10.1016/j.chembiol.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 38.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JA, Hsu JY, Smith MM, Allis CD. Mutagenesis of pairwise combinations of histone amino-terminal tails reveals functional redundancy in budding yeast. Proc. Natl Acad. Sci. USA. 2012;109:5779–5784. doi: 10.1073/pnas.1203453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai J, et al. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddique AN, et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3A–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 2013;425:479–491. doi: 10.1016/j.jmb.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 42.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 43.de Groote ML, Verschure PJ, Rots MG. Epigenetic editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012;40:10596–10613. doi: 10.1093/nar/gks863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeder ML, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE–TET1 fusion proteins. Nature Biotech. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, et al. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2014;42:1563–1574. doi: 10.1093/nar/gkt1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvin CD, Parr RD, Kladde MP. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 2003;31:6493–6501. doi: 10.1093/nar/gkg853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivenbark AG, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2012;7:350–360. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nunna S, Reinhardt R, Ragozin S, Jeltsch A. Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PLoS ONE. 2014;9:e87703. doi: 10.1371/journal.pone.0087703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Falahi F, et al. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol. Cancer Res. 2013;11:1029–1039. doi: 10.1158/1541-7786.MCR-12-0567. [DOI] [PubMed] [Google Scholar]; References 48–50 show the potential advantage, over other gene repression technologies, of site-specific DNA methyltransferases in stably inhibiting oncogenes.
- 51.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]; The combined use of site-specific recruitment, minimal histone-modifying catalytic domains and catalytically dead mutants in this study provides strong evidence for the causative repressive nature of H3K9 methylation.
- 52.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nature Biotech. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, TALEs are used to alter the epigenetic state of endogenous enhancers, which enables the functional identification of their target genes in a highly native and physiological context.
- 53.Keung AJ, Bashor CJ, Kiriakov S, Collins JJ, Khalil AS. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell. 2014;158:110–120. doi: 10.1016/j.cell.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]; A library of more than 200 chromatin-modifying proteins were fused to ZFs and screened against diverse synthetic reporter architectures to reveal chromatin-based combinatorial and spatiotemporal regulatory behaviours.
- 54.Meister GE, Chandrasegaran S, Ostermeier M. Heterodimeric DNA methyltransferases as a platform for creating designer zinc finger methyltransferases for targeted DNA methylation in cells. Nucleic Acids Res. 2010;38:1749–1759. doi: 10.1093/nar/gkp1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaikind B, Kilambi KP, Gray JJ, Ostermeier M. Targeted DNA methylation using an artificially bisected M.HhaI fused to zinc fingers. PLoS ONE. 2012;7:e44852. doi: 10.1371/journal.pone.0044852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nomura W, Barbas CF., 3rd In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 57.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nature Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nature Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ptashne M. Epigenetics: core misconcept. Proc. Natl Acad. Sci. USA. 2013;110:7101–7103. doi: 10.1073/pnas.1305399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 62.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 63.Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 64.Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 65.Lustig AJ, Liu C, Zhang C, Hanish JP. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes an experimental system to dynamically recruit HP1 to specific genomic sites, which allowed measurement and modelling of the kinetics of heterochromatin establishment and memory.
- 67.Laurent BC, Treitel MA, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol. Cell. Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li F, et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–112. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J. Biol. Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- 70.Ragunathan K, Jih G, Moazed D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2014 doi: 10.1126/science.1258699. http://dx.doi.org/10.1126/science.1258699. [DOI] [PMC free article] [PubMed]; Using transient site-specific induction of ectopic H3K9 methylation, this study shows that histone modification can itself act epigenetically and be inherited in the absence of sequence-specific factors, DNA methylation or RNAi.
- 71.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 72.Grimmer MR, et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 2014;42:10856–10868. doi: 10.1093/nar/gku708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotech. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 74.Smith C, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X, Kriz AJ, Sharp PA. Target specificity of the CRISPR–Cas9 system. Quantitative Biol. 2014;2:59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotech. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 77.Guilinger JP, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nature Methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nature Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nature Biotech. 2014 doi: 10.1038/nbt.3117. http://dx.doi.org/10.1038/nbt.3117. [DOI] [PMC free article] [PubMed]
- 80.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotech. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotech. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR–Cas nuclease specificity using truncated guide RNAs. Nature Biotech. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan S, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc. Natl Acad. Sci. USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haynes KA, Silver PA. Synthetic reversal of epigenetic silencing. J. Biol. Chem. 2011;286:27176–27182. doi: 10.1074/jbc.C111.229567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fierz B, Muir TW. Chromatin as an expansive canvas for chemical biology. Nature Chem. Biol. 2012;8:417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes light-activated induction of site-specific transcriptional activation and histone modifications.
- 89.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 90.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 91.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 92.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 93.Hughes AL, Jin Y, Rando OJ, Struhl K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol. Cell. 2012;48:5–15. doi: 10.1016/j.molcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shrader TE, Crothers DM. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanaka S, Zatchej M, Thoma F. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 1992;11:1187–1193. doi: 10.1002/j.1460-2075.1992.tb05159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nature Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]; This study generates a library of synthetic promoters with fine-tuned activity by altering the number and location of nucleosome-disfavouring sequences.
- 97.Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 103.Marquardt S, et al. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014;157:1712–1723. doi: 10.1016/j.cell.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; By integrating a bidirectional fluorescent protein reporter into each strain of the non-essential yeast deletion library, the authors identified the CAF-I complex as an important regulator of divergent transcription.
- 104.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 105.de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 106.Zaman Z, Heid C, Ptashne M. Telomere looping permits repression “at a distance” in yeast. Curr. Biol. 2002;12:930–933. doi: 10.1016/s0960-9822(02)00865-5. [DOI] [PubMed] [Google Scholar]
- 107.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 107 and 108 describe how the synthetically induced looping of chromatin between the promoters and LCRs of the β- and γ-globin loci can activate these genes.
- 109.Brennan TA, Wilson JM. The special case of gene therapy pricing. Nature Biotech. 2014;32:874–876. doi: 10.1038/nbt.3003. [DOI] [PubMed] [Google Scholar]
- 110.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nature Cell Biol. 2011;13:944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155:606–620. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- 113.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lacefield S, Lau DT, Murray AW. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nature Cell Biol. 2009;11:1116–1120. doi: 10.1038/ncb1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kagansky A, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nannas NJ, Murray AW. Tethering sister centromeres to each other suggests the spindle checkpoint detects stretch within the kinetochore. PLoS Genet. 2014;10:e1004492. doi: 10.1371/journal.pgen.1004492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 118.Kind J, et al. Single-cell dynamics of genome–nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 119.Aragon-Alcaide L, Strunnikov AV. Functional dissection of in vivo interchromosome association in Saccharomyces cerevisiae. Nature Cell Biol. 2000;2:812–818. doi: 10.1038/35041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 122.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 123.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sander JD, Joung JK. CRISPR–Cas systems for editing, regulating and targeting genomes. Nature Biotech. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nature Rev. Mol. Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotech. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 128.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 129.Jiang J, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]; By inserting the long non-coding RNA XIST into one of three copies of chromosome 21 in Down syndrome pluripotent stem cells, this study was able to completely silence the extra chromosome, suggesting a potential therapeutic avenue for many polyploidy diseases.
- 130.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Buhler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nature Struct. Mol. Biol. 2013;20:1340. doi: 10.1038/nsmb1113-1340. [DOI] [PubMed] [Google Scholar]