An epidermis control of plant shoot stem cells can explain the scaling and position of the niche expression domains.
Keywords: Stem cells, Shoot apical meristem, Arabidopsis thaliana, WUSCHEL, CLAVATA3, Computational Morphodynamics
Abstract
How molecular patterning scales to organ size is highly debated in developmental biology. We explore this question for the characteristic gene expression domains of the plant stem cell niche residing in the shoot apical meristem. We show that a combination of signals originating from the epidermal cell layer can correctly pattern the key gene expression domains and notably leads to adaptive scaling of these domains to the size of the tissue. Using live imaging, we experimentally confirm this prediction. The identified mechanism is also sufficient to explain de novo stem cell niches in emerging flowers. Our findings suggest that the deformation of the tissue transposes meristem geometry into an instructive scaling and positional input for the apical plant stem cell niche.
INTRODUCTION
Development requires coordination between molecular patterning and morphogenesis (1, 2). Several studies suggest how morphogens can drive gene patterning, growth, and shape generation in different systems (3, 4) and how mechanical constraints might feedback on growth (5, 6). Although adaptation to size is often seen in developmental tissues (1, 7), how tissue geometry and size feedback on gene expression domains is less understood. Using a combination of computational and experimental approaches, we address this question in the shoot apical meristem (SAM), the stem cell niche source of all aboveground plant organs (8).
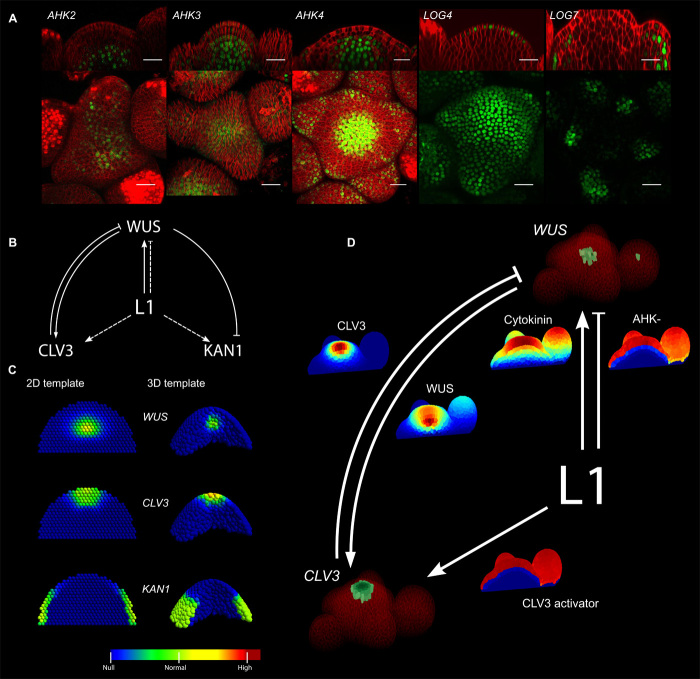
Considered as a main orchestrator of the SAM activity (9, 10), WUSCHEL (WUS) is expressed in a central domain, a few cell layers below the apex of the SAM. It encodes a transcription factor, moving between cells, and is involved in perpetuating stem cell activity and keeping differentiating cells at bay (11, 12). At the very tip of the SAM, the stem cells express the CLAVATA3 peptide (CLV3) that diffuses in the tissue and down-regulates WUS, closing the canonical CLV3/WUS feedback loop (13, 14), thought to be the main factor maintaining SAM homeostasis. On the flanks of the meristem, WUS also represses the differentiation program including the KANADI1 (KAN1) marker (12). Spatial aspects of WUS maintenance via cytokinin signaling have recently been elucidated: LONELY GUY (LOG) proteins, which catalyze cytokinin biosynthesis, are expressed in the epidermis (L1), and ARABIDOPSIS HISTIDINE KINASE (AHK) cytokinin receptors are expressed inside the tissue, a few cell layers away from the L1 (15, 16) (Fig. 1A). Along with previously published data (17, 18), this implies that the epidermis may provide a major positional cue for SAM maintenance.
Fig. 1. The epidermis-driven model reproduces the wild-type gene expression patterns.
(A) Expression domains of cytokinin receptors (AHK2, AHK3, AHK4), and cytokinin-activating enzymes (LOG4, LOG7) in green (fluorescent protein markers, Supplementary Materials) together with plasma membranes in red (FM4-64 dye). Scale bars, 20 μm. (B) Representation of the model. Solid lines indicate interactions supported by molecular evidence, whereas dashed lines show interactions supported by indirect evidence. (C) Optimized domains of WUS, CLV3, and KAN1 in two-dimensional (2D) (left) and 3D (right) abstract representations of the tissue. Gene expression intensity is color-coded. (D) Optimized domains of CLV3 and WUS in a segmented tissue obtained from confocal microscopy. Gene expression domains from the simulation are shown in green, and plasma membranes are shown in red (FM4-64 dye). The diffusive signal concentrations are displayed with the color map of (C) (minimum, blue, to maximum, red). Model equations are defined in the Supplementary Materials, and model parameter values are given in table S1.
RESULTS AND DISCUSSION
To investigate this hypothesis, we propose an integrated quantitative description of the processes regulating the SAM homeostasis where the L1 is central to the patterning of the stem cell regulatory genes (Fig. 1B). The spatial model integrates the SAM regulatory interactions described in a system of differential equations, using Hill formalism for transcription together with mass action and passive diffusion-like transport (Supplementary Materials) (11, 12, 19). It includes the CLV3/WUS feedback loop and the repression of KAN1 by WUS. In particular, the L1 produces four signals modeled as diffusive molecules. Of those, cytokinin acts as an activator of WUS by binding the AHK receptors. Given the expression pattern of the AHKs (Fig. 1A), we hypothesized a repressive L1 signal, keeping the AHKs away from the epidermis (termed AHK-). Cytokinin and AHK- form an incoherent feed-forward motif regulating WUS (20) (Fig. 1, A and B). Two additional signals produced by the epidermis also activate CLV3 (11, 12, 19) and KAN1 (12).
Different representations of the meristematic tissue are used throughout this study. Two abstract tissue templates, one 2D and one 3D, are composed of overlapping spherical cells constrained in a parabolic shape (Fig. 1C and fig. S1). A realistic tissue template obtained from segmented confocal data (20) is also used and includes cell volumes and cell contact surfaces (Fig. 1D). For all these templates, the bottom layer of cells implements a specific boundary condition (referred to as sink), abstracting the diffusion of molecules outside of the meristem and into the tissues below (stem and vasculature), where molecules are removed from simulations based on their diffusion rate. As such, the faster a molecule diffuses in the meristematic tissue, the faster it is carried out of the system at the bottom boundary. In contrast, epidermal cells have a nonflux boundary to the outside of the tissue.
Model parameters are optimized to obtain the wild-type expression domains of WUS, CLV3, and KAN1 using a dedicated strategy based on the Covariance Matrix Adaptation Evolution Strategy (CMA-ES) algorithm (21) (Supplementary Materials and fig. S2). At equilibrium, the model achieves a correct representation of the expression domains showing that the epidermis-driven model is sufficient for SAM patterning. This result is robust in the different representations of the meristematic tissue (Fig. 1, C and D). The model is also able to qualitatively reproduce a large set of experimentally observed perturbations including perturbations of the CLV3/WUS feedback loop and perturbations of cytokinin signaling (Supplementary Materials and fig. S3).
Several mutants of Arabidopsis thaliana result in meristematic tissue defects (12, 22–24); an example is the clavata phenotype, causing plants with markedly enlarged and flattened meristems. In such tissues, the CLV3 domain forms a band a few cell layers thick capping the SAM, whereas WUS is expressed in another band-like domain below CLV3 (23) (Fig. 2A). Simulating clavata in the wild-type tissue geometry results in a limited enlargement of the CLV3 and WUS domains, as experimentally shown upon inducing the mutation: the first 24 hours shows an expansion of the CLV3 domain via dedifferentiation of a few cells, later leading to increased growth and major reorganization (25). When applied to an enlarged and flattened meristem, the model shows a substantial reorganization of the CLV3 and WUS domains in two superposed bands (Fig. 2A). This stresses the importance of geometrical changes to explain the shift in expression domains and is in strong agreement with experimental data. The pCLV3::WUS misexpression line (23) is another reproducible example of extreme geometrical rearrangement, resulting in both CLV3 and WUS overlapping in the external cell layers of a massively enlarged meristem (fig. S4).
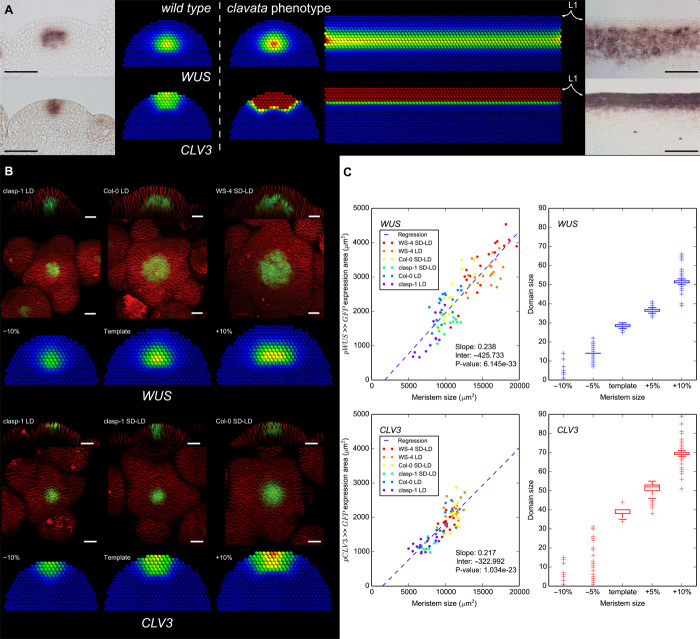
Fig. 2. WUS and CLV3 expression domains respond to the geometry of the meristem.
(A) From left to right: In situ hybridizations of WUS and CLV3 in wild-type meristems and their simulated counterparts, clavata phenotype simulated in a wild-type 2D meristem and in a geometry mimicking the enlarged tissue observed in mutant plants, and in situ hybridizations of WUS and CLV3 in clv1 loss of function meristems. Scale bars, 50 μm. (B) Expression domains of WUS and CLV3 for various meristem sizes. From top to bottom: WUS expression experimentally observed (lateral section and top projection) and simulated, and CLV3 expression experimentally observed and simulated. LD indicates plants grown under long-day conditions; SD-LD indicates growth in short days followed by long days. Scale bars, 20 μm. (B) shows representative examples of meristems presented in (C). (C) Correlation between meristem sizes and gene expression domain sizes: left, experimental data (from top projections); right, simulations (domain size is the number of expressing cells). Values obtained from linear regression are given for the experimental data (slope, interception, and P value; see the Supplementary Materials). Data for individual conditions are provided in figs. S5 and S6.
Wild-type meristems also show size variations, but contrary to the aforementioned mutants, the regulatory network is unaffected. To test the specific effect of geometry, we thus applied the model on tissue templates of various sizes. The simulations predict a robust positive correlation across optimized parameter sets between the size of WUS and CLV3 domains, measured by the number of expressing cells and the size of the tissue templates (Fig. 2, B and C, and Supplementary Materials). Hence, the model predicts an adaptive scaling behavior of the stem cell regulatory expression domains to the meristem size. We tested this hypothesis by growing different A. thaliana genotypes in different environmental conditions, resulting in a set of plants with various inflorescence SAM sizes (ranging from 5000 to 20,000 μm2 and from 5000 to 13,000 μm2 for pWUS >> GFP and pCLV3 >> GFP plants, respectively). Measured on top projections (Fig. 2B and Supplementary Materials), the area of the WUS domains ranged from 700 to 5000 μm2, and the size of the CLV3 domains ranged from 1000 to 3000 μm2, in both cases showing a significant correlation with the size of the tissue (linear regression P values <10−3), confirming the predicted adaptive scaling mechanism of the key regulatory components of stem cell regulation (Fig. 2C). Strikingly, tissue size variations seen in individual genotypes and growth conditions also correlate to the domain sizes (figs. S5 and S6). As opposed to the classical homeostatic view on the SAM domains, this result shows a built-in adaptability of the stem cell pool to changes in tissue size.
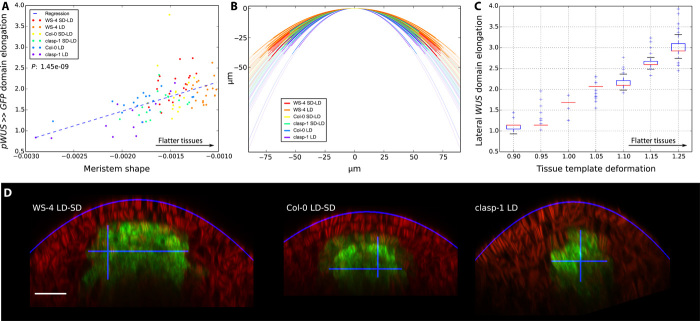
Next, we quantified the shape of the meristematic tissue across the data set (Supplementary Materials, Fig. 3, and figs. S7 to S19). We parameterized the surface of the meristems by parabolic functions. Notably, the curvature varies across samples, and this correlates with the shape of both pWUS >> GFP (Fig. 3, A, B, and D, and figs. S8 and S13) and pCLV3 >> GFP (figs. S7, A and B, and S14 to S19) expression domains; the flatter the tissue, the more laterally elongated are the domains. Simulating the model on a collection of tissue templates reproducing this geometrical variability shows that the model also captures this effect (Fig. 3C and figs. S7C and S20). Hence, the proposed regulatory network adapts the domains not only to the size of the tissue but also to its shape. Note that we observe a correlation between our size and shape measures (fig. S21), showing that larger meristems are also flatter.
Fig. 3. WUS expression domain is elongated for flatter meristems.
(A) Meristem and expression domain shapes. The x axis is the value of the a parameter of the parabola outlying the meristem; the higher the value, the flatter the meristem (see the Supplementary Materials). The y axis is the ratio between the longest vertical axis of pWUS >> GFP expression domain and its longest horizontal axis; the higher the value, the more horizontally elongated the domain is. (B) Recorded meristem shapes plotted as parabolas. The horizontal span of a plain parabola corresponds to the width of the treated image. (C) Modeled meristem and expression domain shapes. The x axis shows the different tissue templates displayed in fig. S12. For the parameter sets optimized for the 2D template, the y axis shows WUS expression domain elongation (longest vertical axis over longest horizontal axis). (D) Examples of processed images. The automatically defined parabolas and expression domains axes are plotted in blue. Scale bar, 20 μm.
Strikingly, the data also show a variability of the position of the WUS domain relative to the tip of the meristem: for the same genotype and similar growth conditions, the highest expression point of WUS may be either the third or the fourth cell layer (figs. S8 to S13). Although the model was optimized for a specific position and size of the expression domains, variability in positioning of WUS would easily be achieved by minor changes in the model parameter values or noise in the reaction rates.
A recurring morphological change in the SAM is the initiation of floral buds at its periphery. Following the tissue deformation marking the forming organ, new WUS and stem cell domains are established in domains resembling the SAM domains (Fig. 4); the developmental program leading to a fully differentiated flower then proceeds. As of now, no mechanism explaining the formation of the flower CLV3 and WUS domains has been identified. The model applied to a tissue template altered to comprise a dome-shaped SAM and a growing primordium-like lateral structure shows that the geometrical alteration is sufficient to generate new WUS and CLV3 domains in the primordium, along with maintaining the domains in the SAM (Fig. 4, movie S1, and Supplementary Materials). Hence, our model can discern between general size changes in the meristem, adapting the expression domains, and can account for more precise morphological changes, such as bud formation where it can promote the induction of new stem cell domains. Experimentally proving that the geometrical changes in the tissue upon flower formation cause the appearance of new stem cell domain is a difficult task. However, as the live image acquisition and processing steadily improve, it should be possible within the next few years to test this hypothesis using single cell resolution time courses, encompassing the meristematic tissue and the precise morphological changes associated with flower development (including the cryptic bract and multiple stages of flower development).
Fig. 4. A localized tissue deformation is sufficient to explain the formation of new stem cell domains in flower primordia.
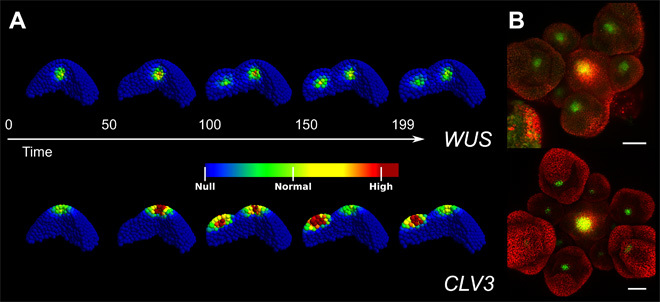
(A) Model equilibria in a set of different tissues mimicking the growth of a flower primordium. As the deformation expands, new WUS and CLV3 domains appear in the expanding organ. (B) Top-view projections of WUS and CLV3 expression domains in a SAM surrounded by flower primordia at different stages. Older (larger) primordia express the constructs whereas the younger ones do not. Scale bars, 50 μm.
To understand the source of the adaptability of the system, we explored the parameter values from multiple model optimizations. The analysis reveals that two types of epidermal signals are required to achieve the wild-type expression domains (Fig. 5A and fig. S22). Indeed, in all cases, cytokinin is a long-range signal, with its diffusion rate much larger than its degradation rate. This observation is coherent with the small size of cytokinin and its involvement in organism-scale signaling (26). The three other signals produced in the L1 are short range and predicted to form similar gradients within the tissue.
Fig. 5. Scaling mechanism: Two types of epidermal signals form an incoherent feed-forward motif.
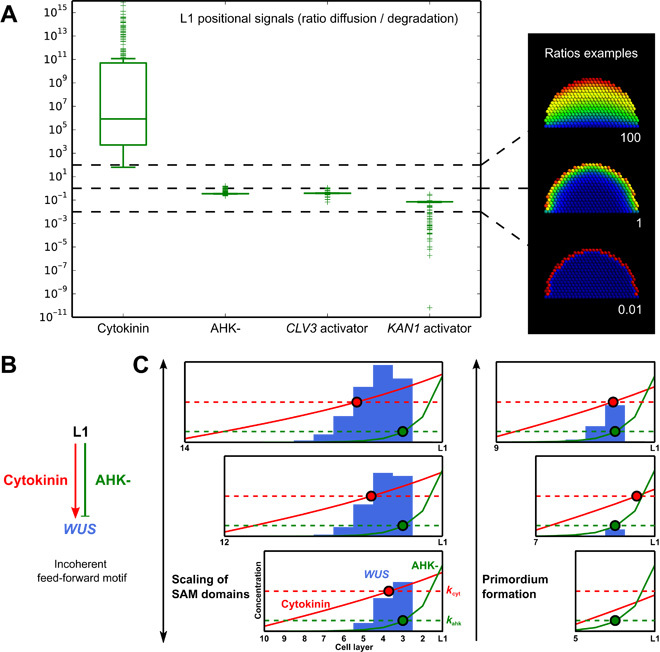
(A) Ratios between diffusion and degradation rates for the L1 signals obtained after multiple optimizations of the 2D template (left). Dashed lines mark the shape of the resulting gradients (right). The bottom two examples represent gradients from short-range signals, whereas the top one represents a gradient from a long-range signal. (B) The incoherent feed-forward network motif activating WUS. (C) Illustration of the mechanism within a simplified model [shown in (B)] simulated on a 1D tissue of varying size (Supplementary Materials). Lines show signal concentrations (red, cytokinin; green, AHK-), and dashed lines are activity thresholds (kcyt, kahk); the point where the concentration of a signal meets its threshold is represented by a large dot. WUS expression is shown in blue. WUS is activated to the right of the red dot and repressed to the right of the green dot. When the tissue size varies, the distance between the red and the green dots changes, causing the WUS expression domain to scale with the tissue (left). As the tissue expands, a de novo WUS expression domain is formed when the red dot passes the green dot (right). Note that the regulations are not step functions and some “leakage” of expression appears outside the activity thresholds.
The identified long-range and short-range epidermal signals are combined into an incoherent feed-forward regulation of WUS expression (Fig. 5B). This motif by itself accounts for the positioning and scaling of the pattern (Fig. 5C). The short-range AHK- signal remains largely unaffected by size changes, whereas the long-range cytokinin signal scales with tissue size. As the tissue expands, cytokinin meets its concentration threshold further away from the L1 and the AHK- threshold, leading to an increase of the WUS domain. The same effect can explain the appearance of WUS in primordia as they grow (Fig. 5C) and the lateral expansion of the domains in flatter tissues by juxtaposing cell layers (cf. Figs. 2A and 3).
We use a quantitative representation of some of the most studied expression domains and molecular interactions participating in the homeostasis of the SAM and show that this regulatory network is sufficient to describe domains in wild-type and perturbed plants. We propose the interactions between external cell layers and internal tissue as a mechanism to understand the stem cell regulation in the SAM. In particular, we propose that the combination of long- and short-range epidermal signals for the activation of WUS expression provides a mechanism for adaptive scaling and for the emergence of stem cell domains in flower primordia. This regulation of the gene expression domains by the geometry of the tissue offers a straightforward way of adapting the size of the stem cell domains to the host tissue, possibly allowing plants to fit their need for new cells to their environmental conditions and providing a mechanism allowing for the wide variety of SAM sizes in plant species to develop.
We have shown the ability of our model to describe a large amount of previously reported and novel behaviors of the system. However, the description relies on hypotheses, one of the major ones being the epidermis-based cytokinin gradient activating WUS expression. Because cytokinins have not been precisely measured in the SAM, we formulated the hypothesis on the basis of convergent clues, with attention to parsimony. First of all, cytokinins have been known for many years to be diffusive molecules (26) and have been shown to form a concentration gradient in the root meristem (27). Second, the connection between cytokinin and shoot development has been reported several times: multiple LOG mutant induces dwarf development in Arabidopsis (28), LOG mutant induces meristem termination in rice (29), and external application has been shown to increase WUS expression (15, 30). Third, we could only detect LOG4 and LOG7 in the SAM, both expressed along the epidermis. The L1 was thus used as the source of cytokinin in the model. A localized source of cytokinin is sufficient to create a cytokinin gradient, and this gradient in combination with the AHK- signal is sufficient to explain WUS expression domain position and shape in relation to tissue geometry. Given the current data, the cytokinin gradient control of WUS is the most parsimonious hypothesis we could formulate. Future work will need to also explore hypotheses involving additional factors. For instance, the cytokinin-degrading CKXs enzymes are expressed in the SAM and procambium and affect WUS expression and meristem size (31). CKXs are also induced by cytokinin signaling (32). Whether their role in the meristem is to reinforce the robustness of WUS expression to cytokinin fluctuations via this negative feedback or to have a defining role on WUS expression remains to be fully assessed.
The second major hypothesis of the model is the AHK- signal. The formulation of this hypothesis is based on the expression pattern of the AHK genes in the SAM (Fig. 1). As pointed out, they are expressed away from the epidermis. Again, we formulated the hypothesis following Occam’s razor and introduced a repressive signal keeping the AHKs away from the L1. This hypothesis is in line with the L1 originating signals used in previous efforts to model CLV3 dynamics across various phenotypes [notably the pCLV3::WUS phenotype (22)] and offers a straightforward explanation for the specific band-like expression domains observed in the large and flat clv3 meristems (Fig. 2A). The three predicted short-range signals form similar types of gradients, all falling sharply within a few cell layers of the L1, and thus following the L1 very closely. This suggests that the three overlapping signals could be mediated by similar means. miR394, produced in the L1 of the SAM and diffusing over a few cell layers, has been shown to participate in defining the stem cell domain (18). In line with this observation, microRNAs offer interesting molecular candidates to mediate the hypothesized short-range signals. The sharpness of the short-range signals resulting from the optimization also indicates that the model could accommodate “step-like” nondiffusing signals, as long as they follow the L1. For instance, lineage difference between tunica and corpus (33) would likely be sufficient as a positional cue. Another interesting candidate would be mechanical stresses as they have been suggested to be stronger in the epidermis and shown to regulate molecular behavior (6, 34, 35).
Although the CLV3/WUS feedback does provide robustness to the system (figs. S23 and S24), it does not enforce invariable expression domains and shows a remarkable adaptability to the shape of the meristematic tissue. The epidermis has a strong role in meristem growth and guiding mechanical properties (6, 36), therefore affecting tissue geometry. Our work fills the gap between geometry and gene expression via an epidermis-driven mechanism. The next major challenge is to understand how gene regulation interplays with mechanics and tissue geometry to balance the generation of new tissue and the control of the stem cell niche.
MATERIALS AND METHODS
The following describes the techniques used to compute the stable state of the models describing the gene expression patterns in the SAM. Two cases are presented:
1) Abstract tissue. In the simplest scenario, the tissue is described with spherical cells. Molecules can diffuse between overlapping cells. No topological information other than cell neighborhood is taken into account.
2) Realistic tissue. In the second scenario, the tissue geometry is drawn from segmented confocal images. In this case, computations make use of the realistic topology of the tissue, taking into account cell volumes and contact surfaces between cells.
Meristem topology
In what is defined as the abstract tissue, the SAM is represented by a set of spheres corresponding to cells. Overlapping cells are considered in contact and can exchange diffusing molecules.
The complete abstract tissue is composed of cells enclosed in a 2D or a 3D parabola. The 3D tissue can be extended to account for a primordium; this is modeled by adding a half-sphere on the side of the parabola. The methods used to acquire the realistic template are described in the Realistic template section.
The abstract tissues count 369 and 2567 cells for the 2D and the 3D cases, respectively. The realistic tissue counts 3965 cells.
The expression domains of the genes of interest (WUS, CLV3, and KAN1) are defined on the template from experimental data (fig. S1). The epidermis (L1) is defined as the cells in contact with the parabola.A boundary acting as a sink is also defined as the cells at the bottom of the tissue; in those cells, diffusing molecules are eliminated from the system at a rate proportional to their diffusion. This boundary represents the forming vasculature and the connection to the rest of the plant.
Gene expression
RNA production is modeled using Hill functions; RNAs undergo a degradation g. For a set of NA activators Aa and NI inhibitors Ib, the concentration of an RNA X varies as
with V being the maximal rate of RNA production. The Hill constants k set the required concentration of activators or inhibitors to switch a gene between its active and inactive states. The Hill coefficients n control the slope of the transition between states. The equilibrium of X is found for
Molecular transport
Molecular movement between cells is modeled by a passive diffusion-like transport across cell membranes. Such diffusing molecules are produced by a gene expression domain (WUS, CLV3) or the L1. This domain is referred to as P, a vector of cell RNA concentrations for gene expression or a vector of 1 or 0 indicating that a cell belongs to the L1 or not. The domain is associated to a production rate p.
The bottom cell layer of the tissue represents the sink, S. As for the L1, it is a vector of 1 and 0. In those cells, diffusing molecules undergo degradation equal to their diffusion rate D, approximating a continued flux into the nonmodeled tissue below the meristem. Diffusing molecules also undergo a passive degradation of rate g.
For a vector of concentrations of a diffusing molecule x, we have
where Δ is the Laplace operator; transport in the model is assumed to be passive.
For the abstract templates and a cell i with ni neighbors j, the diffusion of x follows
For the realistic template and a cell i of volume Vi and surface contact with its neighbors Ci, having ni neighbors j sharing a contact area Cij with i
Finally, the equilibrium state of the considered molecule is found by solving
Here, this is achieved using the linalg.solve function of the python scientific package SciPy (www.scipy.org).
Model
The model is described by a set of nine differential equations for each cell. It comprises the dynamics of three gene RNAs: WUS (W), CLV3 (C), and KAN1 (K) and the modeled protein/peptides WUS (w) and CLV3 (c). It also describes the dynamics of four signals produced by the external cell layer: Lc, La, LC, and LK, which stand for cytokinin, AHK-, CLV3 activator, and KAN1 activator, respectively. The L1 cell layer and the sink are L and S, respectively.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
The full system is 3321, 23,103, and 35,685 equations in the 2D abstract, 3D abstract, and realistic tissues, respectively. Given that the exact signaling pathways controlling the gene expressions are not fully known, we simplified RNA production in the model to single Hill equations combining input factors from activators and repressors. WUS regulation is controlled by the incoherent feed-forward regulation from the L1 where the activating cytokinin (Lc) is combined with the repressing AHK- (La). We have abstracted the repression from the L1 of the cytokinin receptors (AHKs) and the receptors’ part in activating WUS in the single AHK- signal. An additional factor represents CLV3 repression (c). Following the same principle, we have combined activating L1 signals with WUS activation (repression) for regulation of CLV3 (KAN1).
Parameter values
Because parameter values are unknown, they are inferred via an optimization strategy. To maintain the computations linear, we propose a workaround for the CLV3/WUS feedback loop by optimizing the system in several steps, where subparts of the network are optimized and the resulting optimized parameters are carried forward to the following steps until the full model is optimized. All optimizations are carried out using the CMA-ES algorithm (21).
The strategy can be summarized as follows: (step 1) optimize the WUS domain using the static CLV3 domain (Ct) as defined in the template (fig. S1), (step 2) optimize the CLV3 domain keeping the optimized WUS domain fixed, and (step 3) optimize the CLV3 gradient such that the optimized CLV3 domain produces a gradient as similar as possible to the one resulting from the first step. This is followed by a confirmation step (step 4), where the equilibrium of the complete optimized network is computed. Finally, relevant KAN1 expression domain is computed in step 5. For steps 1, 2, and 5, the optimization proceeds to the next step if parameters are found, for which the cost function falls below a defined threshold. The step is repeated otherwise. If step 4 fails to achieve equilibrium, the optimization restarts at step 1. A schematic of the optimization strategy is provided in fig. S2.
In the following, diff(P) will refer to computing the equilibrium concentration of a diffusing molecule across the tissue, given a production domain P. P is a cell vector containing either 1s (L1) or 0s (non-L1) if the production domain is the L1. P is a cell vector of RNA concentrations if it refers to the expression domains of WUS or CLV3 (the equilibrium computation is described in the Molecular transport section). Similarly, will refer to computing the equilibrium of a gene expression regulated by NA activators A and NI inhibitors I (both activators and inhibitors are cell vectors of diffusing molecule concentrations; the equilibrium computation is described in the Gene expression section).
The function mes(X,Y,[p,γ]) measures the difference between two vectors X and Y following and is the main part of the cost functions used in the different steps of the optimization strategy. It allows one to differently weight some indices in the final sum, allowing, for instance, the computation of the difference only on subelements of the vectors or a different weight (p) to be attributed to cells for which the condition γ is met. This is used to increase the weight in the cost function for cells within the expression domains of target genes because these are fewer than cells outside the domains. For the 3D abstract template and the realistic template, this function is computed only on what is defined as the meristem (see fig. S1). As such, for these two templates, the expression domains present in the primordia are considered to be predictions of the model.
Step 1: WUS expression domain
The optimization first minimizes the difference between WUS as defined in the template (Wt) and WUS (W) at equilibrium [where cells expressing WUS (Wt = 1) have an increased weight p0]. A second component of the cost function ensures that the WUS domain (W2) is enlarged in clavata compared to wild-type (which is added as a sigmoidal contribution defined by p2, p3 with maximal weight p1). Finally, a third component limits the Hill coefficient values by weighting them with a factor p4, ensuring that the resulting expression domains are smoother than the binary domains defined in the template. The cost function, EW, is defined by
The computation of WUS at equilibrium for the wild-type and the clavata phenotype follows
Step 2: CLV3 expression domain
Following the same formalism as in the previous step, the second optimization minimizes the difference between CLV3 as defined in the template (Ct) and CLV3 at equilibrium (C). A second component ensures that the Hill coefficient values are limited. The cost function, EC, is defined by
CLV3 at equilibrium is computed following
Step 3: CLV3 peptide gradient
The first step of the optimization used a hypothetical CLV3 signal (ch) produced by the manually defined CLV3 expression domain. As an equilibrium CLV3 domain has been computed, one can compute the corresponding equilibrium for the CLV3 peptide. Optimizing CLV3 aims at minimizing its difference from the previously computed ch.
Equilibrium for CLV3 follows
and the cost function is
The parameters of Eq. 4 are reoptimized (the parameters of Eq. 4 optimized in step 1 are discarded).
Step 4: CLV3/WUS feedback loop: Equilibrium of the core model
As the previous steps of the optimization independently found equilibria for components of the model, one has to ensure that an adequate stable equilibrium state exists for the complete model as well. The feedback between WUS and CLV3 tends to induce dampened oscillations of the two genes. We propose an algorithm making use of that aspect and designed to quickly reach the equilibrium of the system using the parameter values obtained in the previous steps
whereas Ed > T0
else
At this point in the optimization, the eigenvalues of the system are computed to verify whether the equilibrium is an attractor. If they are all negative, the optimization proceeds. We also used numerical ordinary differential equation integrations to confirm equilibria for specific parameter values.
Step 5: KAN1 expression domain
The equilibrium of KAN1 can be computed after the equilibrium of the core model is obtained. This part of the optimization minimizes the difference between KAN1 at equilibrium and the manually defined KAN1 domain (Kt). The cost function also contains a component limiting the value of the Hill coefficients involved. For the abstract 3D template, the mes element of the cost function is only evaluated in the half of the template that does not contain the primordium.
The equilibrium for KAN1 is obtained via
and the cost function is
Optimization results
For the 2D abstract, the 3D abstract, and the realistic tissue templates, the optimizations resulted in 255, 40, and 6 parameter sets, respectively, correctly describing the wild-type gene expression patterns and exhibiting an increase of WUS expression for the clavata phenotype. Example parameter sets are given in table S1.
Computing equilibrium for wild type, mutants, and sensitivity analysis
Using the conventions defined in the previous section, the equilibrium of the L1 model can be found using optimized parameters. The wild-type cases use the parameters obtained from optimization and the tissue templates.
Using the parameters of the equations for L1 signals (cytokinin, AHK-, and CLV3, respectively)
On the basis of those signals, WUS and WUS are computed, followed by CLV3 and CLV3. They correspond to a clv3 loss of function scenario
The CLV3/WUS feedback loop algorithm from the optimization section is then applied to obtain the stable states WUS, WUS, CLV3, and CLV3.
Finally, for the abstract templates, KAN1 is computed
The mutants presented in fig. S3 are computed by changing either the parameter values or the equations
The same procedure is used to compute the sensitivity analysis presented in the figures. Each parameter of each parameter set optimized for the 2D abstract tissue is increased by 1%; the corresponding stable states for CLV3 and WUS are computed and compared to their wild-type counterpart. The y axis of both figures displays , where p is the parameter value, Δp is the modification applied, m is the measure (either C or W summed over all cells), and Δm is the modification of the measure. Following this convention, the closer a result is to 0, the less sensitive a model is to variations of the considered parameter (37). Results are presented in figs. S23 and S24; the difference between the values obtained for the full model and the clavata phenotype shows the effect of the CLV3/WUS feedback loop in improving the robustness of the model.
Live imaging (Fig. 1A)
Reporter cloning
pLOG4::2xYPET-N7 and pLOG7::2xYPET-N7 were previously described by Chickarmane et al. (16). pAHK2, pAHK3, and pAHK4 transcriptional reporter transgenes were constructed using standard molecular biology techniques. A tandem fusion of the YPET cDNA sequence with an N7 nuclear localization signal sequence (2xYPET-N7) was inserted between the translation start site and the stop codon of each AHK gene by gateway-mediated recombination. pAHK2::2xYPET-N7 and pAHK3::2xYPET-N7 sequences were subcloned into the pMOA34 vector. The pAHK4::2xYPET-N7 sequence was subcloned into the pMOA33 vector. Transgenic lines were established using the floral dip method. T1 plants were selected on the basis of the appropriate selection, hygromycin for pAHK2::2xYPET-N7 and pAHK3::2xYPET-N7 or kanamycin for pAHK4::2xYPET-N7. Subsequent resistant T1 individuals were further screened for the presence of signal in both the root tissue and the SAM tissue.
Growth and imaging conditions
T3 generation homozygous plants were grown under continuous light in a growth chamber on soil at 19°C. Inflorescence SAMs were dissected from plants at a shoot height of approximately 1 cm. Floral primordia were removed, and the SAM was imaged underwater. All images were obtained on a Zeiss 780 upright confocal laser-scanning microscope. For each transgene reporter, conditions were standardized using a 40× 1.0 numerical aperture water-dipping lens. Samples were imaged using the 514-nm laser with the output attenuated to 5%. The digital gain was set between 720 and 800 depending on the transcriptional reporter transgene. For the pAHK2 reporter, the laser output was attenuated to 10% total output to generate an image comparable to the other pAHK reporter transgenes. The digital offset was adjusted for each image such that all background pixels were not underexposed.
Image processing
Each of the images was processed using ImageJ for background subtraction and adjustment of the brightness and contrast. These adjustments did not alter the interpretation of the original image data for the extraction of expression domains in the parameterization of the model.
Realistic template (Fig. 1D)
Wild-type Arabidopsis Columbia (Col.0) plants were grown on soil under short-day conditions (8 hours light) for a period of 3 weeks and were then subjected under long-day conditions (16 hours light) and allowed to bolt. After reaching a size of approximately 5 cm, the shoot apices were dissected under a stereo microscope. After carefully removing older flowers and floral primordia, the apices were stained with FM4-64 and imaged with a Zeiss LSM 780 confocal microscope. Confocal Z sections of the apical meristem with 0.5-μm depth were acquired at a neutral angle and two additional stacks of the same apical meristem were captured after tilting the sample in two different orientations of 20° to 30°. The three individual Z stacks were then fused to create a single sample. The merged 3D image is segmented. We used MARS (20) for the fusion and the segmentation. The segmented image can then be viewed as a 3D image in which the voxels (volumetric pixels) belonging to a given cell are marked by the same label. The labels can be considered as cell identifiers. From the segmented image, quantitative data on cell volumes, cell centers, and contact surfaces between neighboring cells were obtained. Apart from this, an adjacency graph in which vertices correspond to cells was also generated. The edges of the adjacency graph correspond to cell walls and connect cells to their neighbors. The output of the model was visualized using a 3D viewer implemented using VTK, python, NumPy, and SciPy packages.
In situ hybridization (Fig. 2)
Plant material and treatments
Col.0 plants were grown at 23°C under constant light condition. The dominant negative clv1 allele corresponds to clv1-8, which was originally named fully fasciated (38).
In situ hybridization
In situ hybridizations were performed in accordance with standard protocols (39). Polyvinyl alcohol was added to the nitro blue tetrazolium–bromochloroindolyl phosphate staining solution at a final concentration of 3%. RNA probes were in vitro transcribed from full-length coding regions.
Meristem size variations (Fig. 2)
Plant lines and growth conditions
Seeds of A. thaliana, ecotypes Col.0 and Wassilewskija (WS), were provided by the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Clasp-1 was provided by G. Wasteneys and has been described previously (40). The ethanol-inducible transcriptional reporters for WUS and CLAVATA3 were provided by P. Laufs and have also been described (41, 42). Plants were grown on soil in a phytotron under constant light or, if stated otherwise, under long-day conditions directly (16 hours/8 hours light/dark period) or under short-day conditions (8 hours/16 hours light/dark period) for 1 month and then under long-day conditions.
Observation of SAMs
To access the inflorescence meristem, flowers and older floral buds [up to stage 3 as defined by Smyth et al. (43)] were dissected out. Then, the meristem was cut and kept in an apex culture medium for imaging (20). To induce the expression of green fluorescent protein (GFP) in the pWUS >> GFP and pCLV3 >> GFP lines, plants were kept in a box overnight in a wet atmosphere with ethanol vapors before they were dissected. The meristems were imaged using either a Zeiss LSM 700 or an LSM 780 upright confocal microscope: optical sections were obtained every 1 μm, and full projections of the stack of images and orthogonal sections were realized using the ImageJ software.
Meristem quantifications
The measurements of meristem size were performed as described by Landrein et al. (44). To measure the size of the WUS and CLV3 expression domains, 3D maximum intensity z projections of the confocal stacks were used, ensuring that the meristem was always oriented perpendicular to the z axis. Then, an ellipse was drawn manually around the projected domain and the surface of this ellipse was quantified using the ImageJ software. Note that as the pWUS >> GFP and pCLV3 >> GFP lines were generated in a WS-4 background, the measurements of meristem size and expression domains in clasp-1 and Col.0 were performed on an F2 segregation of the inter-ecotype crosses explaining the larger variation of meristem size observed in these genotypes [see Landrein et al. (44)].
For simulated data on the templates of different sizes, the domain sizes are counts of cells having the expression of either WUS or CLV3 above a threshold. For Fig. 2, the threshold is set to 0.5.
Regression analysis
The regressions were performed using the stats.linregress function of the SciPy scientific package for python. The given values are the slope and intercept of the regression line. A two-tailed P value testing the slope of the regression line being null as a null hypothesis is also given. Data from individual conditions are presented in figs. S5 and S6.
Meristem and expression domain geometrical quantifications (Fig. 3)
Not only do SAMs show size variations, the shape of the dome they form also varies. We measured that aspect on the pool of meristems used for the size analysis of Fig. 2.
The first part of the image processing was done with the Fiji software (45). For each meristem confocal stack (composite colors), 5-μm-thick transversal sections cutting through the center of the meristem were extracted and further projected following maximum intensity. When necessary, images were rotated so that the meristem would be displayed vertically. The images were then cropped to contain only the SAMs. Finally, each image was converted to png format and analyzed using the python PIL library (www.pythonware.com/products/pil/).
In the following, we consider a reference frame with an origin at the bottom left corner of each image where discrete x and y values correspond to pixel positions in the image. To measure the shape of a meristem, for each row of pixels, we extracted the topmost pixel registering FM4-64 fluorescence (for each x: maximum y with red channel value >50). This results in a collection of x,y coordinates outlying the shape of the meristem. The parameters of a second-degree polynomial (describing a parabola) were then fit to the extracted coordinates using the numpy.polyfit function. The parameter a of a parabola described by ax2 + bx + c = 0 controls how narrow the domain contained within the parabola is (where ax2 + bx + c < 0). In the case of the parabolas outlying the meristem shape, the higher the value of the a parameter, the flatter the meristem. This value was computed for each meristem. The parabolas and a parameters are displayed on the meristem images in figs. S8 to S19.
The same images were used to quantify the shape of the expression domains of the pWUS >> GFP and pCLV3 >> GFP constructs. We chose to measure the elongation of the domains along the axis of the reference frame. To do so, we find the maximal distance between two pixels registering GFP fluorescence (green channel value >50) and having the same y coordinate, giving us the longest horizontal axis of the GFP expression domain. We repeat the procedure using the same x coordinate and find the longest vertical axis. As a measure of the GFP domain elongation, we use the ratio of the longest vertical axis over the longest horizontal axis. Thus, the higher the value, the more the domain is horizontally elongated. Axis and elongation values are displayed on their corresponding meristem images in figs. S8 to S19.
Figure 3A and fig. S7A show a correlation between the shape of the meristematic tissue and the shape of the expression domains of both WUS and CLV3. The flatter the SAM, the more horizontally elongated the domains of expression are. Figure S21 shows the correlation between the size of the meristems from Fig. 2 and the shape of the meristems. We observe a strong correlation between the shape of the tissue and its size, with the larger meristems also being the flatter ones. Note that the number of meristems plotted in fig. S21 may differ from the number of meristems displayed in figs. S8 to S19, because only the meristems that could also be processed for Fig. 2 are displayed. Regression analysis is performed as described in the previous section.
To assess if the L1 model can capture this effect, we generated a set of new 2D templates. The optimization template was first scaled by a factor and then scaled horizontally again by the same factor. This generated a set of smaller and narrower tissue templates and a set of larger and flatter ones. The factors displayed in fig. S20 are 0.9, 0.95, 1, 1.05, 1.1, 1.15, and 1.2. As exemplified, the larger and flatter meristems show elongated domains of expression for both WUS and CLV3, whereas the domains tend to narrow down for the smaller and narrower meristems. The data obtained for all parameter sets optimized for the 2D template are shown in Fig. 3C and fig. S7C.
Primordium growth (Fig. 4 and movie S1)
Cells are allowed to grow by increasing the cell radii and are allowed to divide into two daughter cells when a threshold size is attained. A set of spring forces between cell centers moves cells apart when overlapping and back inside the lid when pushed out of it. Cells falling below the lower boundary of the lid are removed from the simulation (12).
As the simulation runs, a sphere moves from the inside of the parabola, exiting laterally, and stopping when its center reaches the parabola. As the sphere moves, the lid is updated to comprise the parabola and the part of the sphere outside of the parabola. This results in an increase of the space allocated to cells. The growth simulation is performed using the organism software (http://dev.thep.lu.se/organism).
For 200 time points evenly spaced over the length of the simulation, the cell positions and radii are recorded and the cells belonging to the boundaries (L1 and Sink) are found, allowing us to find the equilibrium of the L1 model as previously described. The final movie is obtained by assembling the equilibrium variables of the model for each time point.
It is worth noting that the growth and cell division algorithm used to generate the movie is noisy, likely more than what could be expected from the actual meristematic tissue. This generates large unrealistic fluctuations of the expression domains of WUS and CLV3 as the simulation runs. Keeping such an algorithm, the fluctuations could however be reduced by extending the model to add additional feedback loops known to be present in the system, such as the negative feedback of cytokinin of its signaling mediated by type B Arabidopsis response regulators. Another plausible approach is to trade some of the expression domain pattern quality for more robustness of the patterns by adding additional components to the cost functions used in the optimization of the parameters.
WUS and CLV3 live imaging (Fig. 4)
pWUS::GFP-ER and pCLV3::dsRED-N7 plants were grown under conditions described in the Realistic template section. For the live imaging, plants were transferred immediately after bolting to square boxes containing lukewarm MS media with 1% agar and allowed to solidify. The plants were then stained with FM4-64 and imaged with a Zeiss LSM 780 confocal microscope.
1D simulations (Fig. 5)
To illustrate the scaling effect driven by the incoherent feed-forward loop controlling WUS expression, simulations were performed on a set of 1D tissue templates (ranging from 5 to 14 cells), each having an L1 cell at one extremity and a Sink cell at the other. The simplified model comprises the activation of WUS mediated by cytokinin and its repression mediated by AHK4-.
The 10-cell template was chosen to manually fit the parameters of the system to obtain a central WUS domain. The chosen parameters are
Supplementary Material
Acknowledgments
We thank L. Guignard, G. Malandain, and C. Godin for their help in using MARS for the meristem segmentation. We also thank J. Locke for comments on the manuscript. Funding: This work was funded by grants from the Gatsby Charitable Foundation (GAT3395/PR4) and the Swedish Research Council (VR2013-4632) to H.J. and by Gatsby Charitable Foundation grants GAT3272/C and GAT3273-PR1, the U.S. NIH (R01 GM104244), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation (GBMF3406) (to E.M.M.). Author contributions: J.G. designed the study, performed modeling, collected and analyzed data, and prepared the manuscript. B.L. and P.T. equally contributed in collecting and analyzing data. C.S., Y.R., and A.S. collected and analyzed data. C.S. commented on the manuscript. O.H. and E.M.M. contributed in designing the study and commented on the manuscript. H.J. designed the study and prepared the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding author.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/1/e1500989/DC1
Fig. S1. Templates.
Fig. S2. Optimization strategy.
Fig. S3. The model is able to represent a large collection of perturbations.
Fig. S4. pCLV3::WUS.
Fig. S5. WUS domain size variation.
Fig. S6. CLV3 domain size variation.
Fig. S7. pCLV3 >> GFP meristem shapes and expression domains.
Fig. S8. clasp1 pWUS >> GFP meristems, grown in long days.
Fig. S9. clasp1 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S10. Col.0 pWUS >> GFP meristems, grown in long days.
Fig. S11. Col.0 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S12. WS-4 pWUS >> GFP meristems, grown in long days.
Fig. S13. WS-4 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S14. clasp1 pCLV3 >> GFP meristems, grown in long days.
Fig. S15. clasp1 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S16. Col.0 pCLV3 >> GFP meristems, grown in long days.
Fig. S17. Col.0 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S18. WS-4 pCLV3 >> GFP meristems, grown in long days.
Fig. S19. WS-4 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S20. Example of expression domain variations upon tissue shape changes.
Fig. S21. Meristem size measure versus shape measure.
Fig. S22. Long-range and short-range signals.
Fig. S23. WUS sensitivity analysis.
Fig. S24. CLV3 sensitivity analysis.
Table S1. Example parameter sets.
Movie S1. Primordium growth.
REFERENCES AND NOTES
- 1.Nahmad M., Lander A. D., Spatiotemporal mechanisms of morphogen gradient interpretation. Curr. Opin. Genet. Dev. 21, 726–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeger J., Irons D., Monk N., Regulative feedback in pattern formation: Towards a general relativistic theory of positional information. Development 135, 3175–3183 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Coen E., Rolland-Lagan A.-G., Matthews M., Bangham J. A., Prusinkiewicz P., The genetics of geometry. Proc. Natl. Acad. Sci. U.S.A. 101, 4728–4735 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauret-Güeto S., Schiessl K., Bangham A., Sablowski R., Coen E., JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLOS Biol. 11, e1001550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shraiman B. I., Mechanical feedback as a possible regulator of tissue growth. Proc. Natl. Acad. Sci. U.S.A. 102, 3318–3323 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamant O., Heisler M. G., Jönsson H., Krupinski P., Uyttewaal M., Bokov P., Corson F., Sahlin P., Boudaoud A., Meyerowitz E. M., Couder Y., Traas J., Developmental patterning by mechanical signals in Arabidopsis. Science 322, 1650–1655 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Ben-Zvi D., Barkai N., Scaling of morphogen gradients by an expansion-repression integral feedback control. Proc. Natl. Acad. Sci. U.S.A. 107, 6924–6929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T. A. Steeves, I. M. Sussex, Patterns in Plant Development (Cambridge Univ. Press, New York, 1989). [Google Scholar]
- 9.Mayer K. F. X., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T., Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Schoof H., Lenhard M., Haecker A., Mayer K. F. X., Jürgens G., Laux T., The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Yadav R. K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G. V., WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav R. K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G. V., Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9, 654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark S. E., Williams R. W., Meyerowitz E. M., The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y., Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Gordon S. P., Chickarmane V. S., Ohno C., Meyerowitz E. M., Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106, 16529–16534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chickarmane V. S., Gordon S. P., Tarr P. T., Heisler M. G., Meyerowitz E. M., Cytokinin signaling as a positional cue for patterning the apical–basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 109, 4002–4007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhardt D., Frenz M., Mandel T., Kuhlemeier C., Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130, 4073–4083 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Knauer S., Holt A. L., Rubio-Somoza I., Tucker E. J., Hinze A., Pisch M., Javelle M., Timmermans M. C., Tucker M. R., Laux T., A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132 (2013). [DOI] [PubMed] [Google Scholar]
- 19.H. Jönsson, B. E. Shapiro, E. Meyerowitz, E. Mjolsness, in On Growth, Form, and Computers, S. Kumar, P. Bentley, Eds. (Academic Press, London, 2003), pp. 156–161. [Google Scholar]
- 20.Fernandez R., Das P., Mirabet V., Moscardi E., Traas J., Verdeil J.-L., Malandain G., Godin C., Imaging plant growth in 4D: Robust tissue reconstruction and lineaging at cell resolution. Nat. Methods 7, 547–553 (2010). [DOI] [PubMed] [Google Scholar]
- 21.N. Hansen. The CMA evolution strategy: A comparing review, in Towards a New Evolutionary Computation. Advances on Estimation of Distribution Algorithms, J. A. Lozano, P. Larranaga, I. Inza, E. Bengoetxea, Eds. (Springer-Verlag, Heidelberg, Germany, 2006), pp. 75–102. [Google Scholar]
- 22.Brand U., Grünewald M., Hobe M., Simon R., Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R., Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Wahl V., Brand L. H., Guo Y.-L., Schmid M., The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 10, 285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy G. V., Meyerowitz E. M., Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310, 663–667 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Kieber J. J., Schaller G. E., Cytokinins. Arabidopsis Book 12, e0168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniadi I., Plačková L., Simonovik B., Doležal K., Turnbull C., Ljunga K., Novák O., Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 27, 1955–1967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., Fukuda H., Sugimoto K., Sakakibaraa H., Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21, 3152–3169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J., Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Lindsay D. L., Sawhney V. K., Bonham-Smith P. C., Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. 170, 1111–1117 (2006). [Google Scholar]
- 31.Bartrina I., Otto E., Strnad M., Werner T., Schmülling T., Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhargava A., Clabaugh I., To J. P., Maxwell B. B., Chiang Y.-H., Schaller G. E., Loraine A., Kieber J. J., Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 162, 272–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. Schmidt, Histologische Studien an phanerogamen Vegetationspunkten. Bot. Arch. 9, 345–404 (1924). [Google Scholar]
- 34.Bozorg B., Krupinski P., Jönsson H., Stress and strain provide positional and directional cues in development. PLOS Comput. Biol. 10, e1003410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampathkumar A., Krupinski P., Wightman R., Milani P., Berquand A., Boudaoud A., Hamant O., Jönsson H., Meyerowitz E. M., Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3, e01967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savaldi-Goldstein S., Peto C., Chory J., The epidermis both drives and restricts plant shoot growth. Nature 446, 199–202 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Savageau M. A., Parameter sensitivity as a criterion for evaluating and comparing the performance of biochemical systems. Nature 229, 542–544 (1971). [DOI] [PubMed] [Google Scholar]
- 38.Medford J. I., Behringer F. J., Callos J. D., Feldmann K. A., Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4, 631–643 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D. Weigel, J. Glazebrook, Arabidopsis: A Laboratory Manual (Cold Spring Harbor Laboratory Press, New York, 2002). [Google Scholar]
- 40.Ambrose J. C., Shoji T., Kotzer A. M., Pighin J. A., Wasteneys G. O., The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19, 2763–2775 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grandjean O., Vernoux T., Laufs P., Belcram K., Mizukami Y., Traas J., In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16, 74–87 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deveaux Y., Peaucelle A., Roberts G. R., Coen E., Simon R., Mizukami Y., Traas J., Murray J. A. H., Doonan J. H., Laufs P., The ethanol switch: A tool for tissue-specific gene induction during plant development. Plant J. 36, 918–930 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Smyth D. R., Bowman J. L., Meyerowitz E. M., Early flower development in Arabidopsis. Plant Cell 2, 755–767 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landrein B., Refahi Y., Besnard F., Hervieux N., Mirabet V., Boudaoud A., Vernoux T., Hamant O., Meristem size contributes to the robustness of phyllotaxis in Arabidopsis. J. Exp. Bot. 66, 1317–1324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/1/e1500989/DC1
Fig. S1. Templates.
Fig. S2. Optimization strategy.
Fig. S3. The model is able to represent a large collection of perturbations.
Fig. S4. pCLV3::WUS.
Fig. S5. WUS domain size variation.
Fig. S6. CLV3 domain size variation.
Fig. S7. pCLV3 >> GFP meristem shapes and expression domains.
Fig. S8. clasp1 pWUS >> GFP meristems, grown in long days.
Fig. S9. clasp1 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S10. Col.0 pWUS >> GFP meristems, grown in long days.
Fig. S11. Col.0 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S12. WS-4 pWUS >> GFP meristems, grown in long days.
Fig. S13. WS-4 pWUS >> GFP meristems, grown in long days followed by short days.
Fig. S14. clasp1 pCLV3 >> GFP meristems, grown in long days.
Fig. S15. clasp1 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S16. Col.0 pCLV3 >> GFP meristems, grown in long days.
Fig. S17. Col.0 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S18. WS-4 pCLV3 >> GFP meristems, grown in long days.
Fig. S19. WS-4 pCLV3 >> GFP meristems, grown in long days followed by short days.
Fig. S20. Example of expression domain variations upon tissue shape changes.
Fig. S21. Meristem size measure versus shape measure.
Fig. S22. Long-range and short-range signals.
Fig. S23. WUS sensitivity analysis.
Fig. S24. CLV3 sensitivity analysis.
Table S1. Example parameter sets.
Movie S1. Primordium growth.