Scutellaria baicalensis (Huang-Qin in Chinese medicine) produces bioactive 4′-deoxyflavones by a newly evolved metabolic pathway.
Keywords: Chinese Medicinal Plant, Flavone, Bioactive, Synthesis, Wogonin
Abstract
Wogonin and baicalein are bioactive flavones in the popular Chinese herbal remedy Huang-Qin (Scutellaria baicalensis Georgi). These specialized flavones lack a 4′-hydroxyl group on the B ring (4′-deoxyflavones) and induce apoptosis in a wide spectrum of human tumor cells in vitro and inhibit tumor growth in vivo in different mouse tumor models. Root-specific flavones (RSFs) from Scutellaria have a variety of reported additional beneficial effects including antioxidant and antiviral properties. We describe the characterization of a new pathway for the synthesis of these compounds, in which pinocembrin (a 4′-deoxyflavanone) serves as a key intermediate. Although two genes encoding flavone synthase II (FNSII) are expressed in the roots of S. baicalensis, FNSII-1 has broad specificity for flavanones as substrates, whereas FNSII-2 is specific for pinocembrin. FNSII-2 is responsible for the synthesis of 4′-deoxyRSFs, such as chrysin and wogonin, wogonoside, baicalein, and baicalin, which are synthesized from chrysin. A gene encoding a cinnamic acid–specific coenzyme A ligase (SbCLL-7), which is highly expressed in roots, is required for the synthesis of RSFs by FNSII-2, as demonstrated by gene silencing. A specific isoform of chalcone synthase (SbCHS-2) that is highly expressed in roots producing RSFs is also required for the synthesis of chrysin. Our studies reveal a recently evolved pathway for biosynthesis of specific, bioactive 4′-deoxyflavones in the roots of S. baicalensis.
INTRODUCTION
Scutellaria baicalensis Georgi is a species in the family Lamiaceae commonly used in traditional Chinese medicine, where it is known as Huang-Qin (Fig. 1, A and B). Huang-Qin has been used for more than 2000 years for the treatment of fever and lung and liver complaints and was first recorded in Shennong Bencaojing (written between 200 and 300 AD). The authoritative Materia Medica (Bencao Gangmu), written in 1593, describes the use of S. baicalensis for treatment of a wide range of disorders. Its author, Li Shizhen, reported successful self-administration to treat a severe lung infection (1). Modern day use of Huang-Qin has reported successful outcomes in combination therapies of non–small cell lung carcinomas (2–4). Huang-Qin has also been applied in the treatment of inflammation, respiratory tract infections, diarrhea, dysentery, liver disorders, hypertension, hemorrhaging, and insomnia (5).
Fig. 1. Specialized flavones found in S. baicalensis Georgi plant.
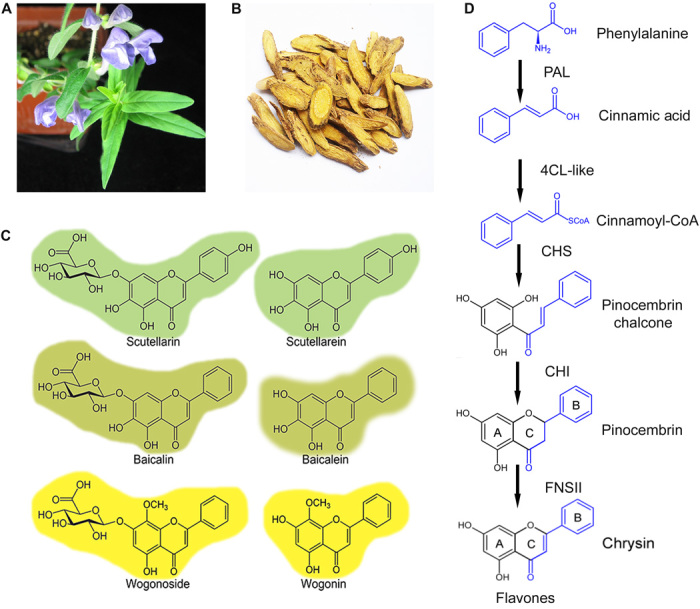
(A) S. baicalensis Georgi plant. (B) The dried roots of S. baicalensis Georgi used in traditional Chinese medicine. (C) Structures of its major flavones. (D) The proposed pathway responsible for biosynthesis of 4′-deoxyflavones in S. baicalensis.
Scutellaria is rich in flavones (Fig. 1, C and D), which are flavonoids widely distributed in the plant kingdom and most usually produced in flowers, where they serve as copigments with anthocyanins, giving bluer colors to flowers such as gentian. Dietary flavones have diverse beneficial properties for animal cells, including activities as free radical scavengers and anticancer properties (6, 7). Baicalin and wogonoside, and their respective aglycones baicalein and wogonin, are the major bioactive flavones produced in large amounts by the roots of S. baicalensis [the root-specific flavones (RSFs)]. RSFs lack a 4′-hydroxyl group on their B ring compared to the widely distributed “classic flavones” associated with aerial tissues such as flowers (Fig. 1C). The 4′-deoxyRSFs provide a variety of specific health benefits in Huang-Qin, such as antifibrotic activity in the liver, and antiviral and anticancer properties (8–13). Scutellaria RSFs specifically promote apoptosis in tumor cells but have low or no toxicity in healthy cells (13, 14). We are interested in elucidating the biosynthetic pathways for the RSFs for applications involving increased production of these bioactive compounds.
Flavones are synthesized by the flavonoid pathway, which is part of phenylpropanoid metabolism (15). Naringenin is a central intermediate in biosynthesis of normal 4′-hydroxyflavones (16). In the aerial parts of Scutellaria, the 4′-hydroxyflavones, scutellarin and scutellarein accumulates, derived from naringenin. However, Scutellaria roots accumulate large amounts of specialized RSFs lacking a 4′-OH group on their B rings (Fig. 1C) (17). These 4′-deoxyRSFs, which include baicalein and wogonin and their glycosides, are unlikely to be synthesized from naringenin because no dehydroxylase that removes hydroxyl groups from the B ring of flavonoids has been found in plants (Fig. 1C). This finding suggests that an alternative pathway recruits cinnamic acid to form cinnamoyl–coenzyme A (CoA) through a CoA ligase, which is then condensed with malonyl-CoA by chalcone synthase (CHS) to form a chalcone, and then isomerized by chalcone isomerase (CHI) to form pinocembrin, a 4′-deoxyflavanone. Pinocembrin could be converted by a flavone synthase (FNS) to form chrysin and subsequently decorated by hydroxylases, methyltransferases, and glycosyltransferases (GTs) to produce the different RSFs in S. baicalensis (Fig. 1C). To date, cDNAs encoding phenylalanine ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumaroyl–CoA ligase (4CL), CHS, and CHI have been reported from S. baicalensis (18, 19). However, biochemical and genetic evidence indicating which, if any, of these genes are involved in the biosynthesis of RSFs is still lacking. It is also possible that specific isoforms of CoA ligase are required for the formation of cinnamoyl-CoA (20–22), and isoforms of CHS and CHI for the formation of pinocembrin. In short, the entire pathway for RSF biosynthesis needs to be defined functionally.
FNS converts flavanones to flavones by introducing a double bond between the C2 and C3 positions. This reaction can be catalyzed by two different types of FNS (FNSI and FNSII). FNSI is a cytoplasmic 2-oxoglutarate– and Fe2+-dependent dioxygenase (23) and has been best characterized in members of the Apiaceae, particularly parsley (24), and in monocots (25). In contrast, FNSII is a membrane-associated cytochrome P450 (Cyt p450) monooxygenase (CYP93B) that requires the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) as cofactor and is widely distributed in angiosperms (16). Genes encoding FNSII have been isolated and characterized from a range of plants, and they all catalyze the conversion of naringenin or other flavanones with a 4′-OH group, such as eriodictyol or liquiritigenin, to flavones. No FNS that specifically converts pinocembrin (a 4′-deoxyflavanone) to chrysin (a 4′-deoxyflavone) (Fig. 1D) has yet been described at the molecular level.
The enzyme 4CL converts 4-coumaric acid and other substituted cinnamic acids such as caffeic acid and ferulic acid into the corresponding CoA esters, which are used for the biosynthesis of numerous phenylpropanoid-derived compounds including lignin, suberins, coumarins, wall-bound phenolics, and flavonoids (26). In Arabidopsis, there are four 4-CoA ligase isoforms that exhibit distinct substrate specificities and may participate in different flavonoid metabolic pathways (27, 28). It has been reported that 4CL-like (CLL) proteins may activate cinnamic, benzoic, or fatty acid derivatives, although specificity for cinnamic acid has not yet been demonstrated for any of these 4CL isoforms, which tend to show similar affinity for cinnamic acid and 4-coumaric acid as substrates in vitro (20–22, 29, 30).
Here, we describe a new isoform of FNSII, which is required for specialized 4′-deoxyflavone (RSF) biosynthesis in the roots of S. baicalensis. However, this activity alone is not sufficient for 4′-deoxyRSF synthesis in plants making conventional 4′-hydroxyflavonoids. We describe isoforms of two other enzymes involved in the RSF biosynthetic pathway (a CoA ligase and a CHS), which, together with CHI, are required for synthesis of 4′-deoxyflavones in nonspecialized host plants. We describe the discovery of these new enzymes in the pathway in the order in which we discovered them to illustrate the scientific steps whereby we identified the pathway, which runs parallel to that of classic flavone synthesis. The tools necessary for pathway identification, even in species such as S. baicalensis with very limited genetic and genomic resources, are relatively easy to establish (a complete transcriptome of the tissues synthesizing the metabolites and a rapid transformation system to test functionality), meaning that our approach in S. baicalensis could be applied to unravelling biosynthetic pathways of specialized metabolism even in recalcitrant species, such as many of those used in traditional Chinese medicine.
RESULTS
Identification of cDNAs encoding FNSII in S. baicalensis
To identify genes encoding enzymes that might be responsible for 4′-deoxyflavone biosynthesis in the roots of S. baicalensis, we performed RNA sequencing (RNA-seq) on RNA extracted from hairy root cultures that accumulated 4′-deoxyflavones (baicalein, baicalin, wogonin, and wogonoside) and screened for contigs, which were annotated as FNS or CYP93B from our Scutellaria RNA-seq database. We identified three putative FNSII cDNA fragments sharing 70 to 79% nucleotide identity with FNSII from Perilla frutescens, which also belongs to the mint family (Lamiaceae), like S. baicalensis. On the basis of the sequence of Unigene22612, we obtained its full-length cDNA by 3′ and 5′ rapid amplification of cDNA ends (RACE) (31) and designated it SbFNSII-1 (CYP93B24). The open reading frame of the SbFNSII-1 cDNA was 1509 bp long, encoding a predicted 502–amino acid protein of 56.77 kD. Subsequent analysis revealed that Unigene14383 belonged to another part of SbFNSII-1. The 1518-bp coding sequence of Unigene19446, obtained by reverse transcription polymerase chain reaction (RT-PCR), encoded a 505–amino acid protein of 57.36 kD, which we named SbFNSII-2 (CYP93B25). The two SbFNSII cDNAs were similar in their encoded proteins to FNS from closely related plants such as P. frutescens (CYP93B6), Ocimum basilicum (CYP93B23), and Antirrhinum majus (CYP93B3) (32–36).
A phylogenetic tree was constructed to assess the evolutionary relationship between SbFNSII-1 and SbFNSII-2, with other CYP93Bs (Fig. 2A). Both proteins grouped in the same clade as other FNSIIs from Lamiales, and were clearly separated from the group of CPY93B17, CPY93B2, and CPY93B5, which encode FNS from the family Asterales. The phylogenetic analysis also suggested that SbFNSII-2 diverged from SbFNSII-1 recently, after the divergence of the family Lamiaceae, and that either FNSII-1 or FNSII-2 may have undergone neofunctionalization and gained an activity different from its ancestors, exemplified by CYP93B24, CYP93B6, and CYP93B23. SbFNSII-1 shares 68% identity with SbFNSII-2 at the amino acid level, and the two proteins have 79 and 69% identity with FNSII (CYP93B6) from P. frutescens, respectively (fig. S1). These homologies suggested that SbFNSII-1 likely retained an activity similar to that of CYP93B6 from P. frutescens (32), whereas SbFNSII-2 could be an FNS with activity specific to S. baicalensis.
Fig. 2. Phylogenetic and RNA interference (RNAi) silencing of SbFNSII-1 and SbFNSII-2 in hairy root cultures of S. baicalensis.
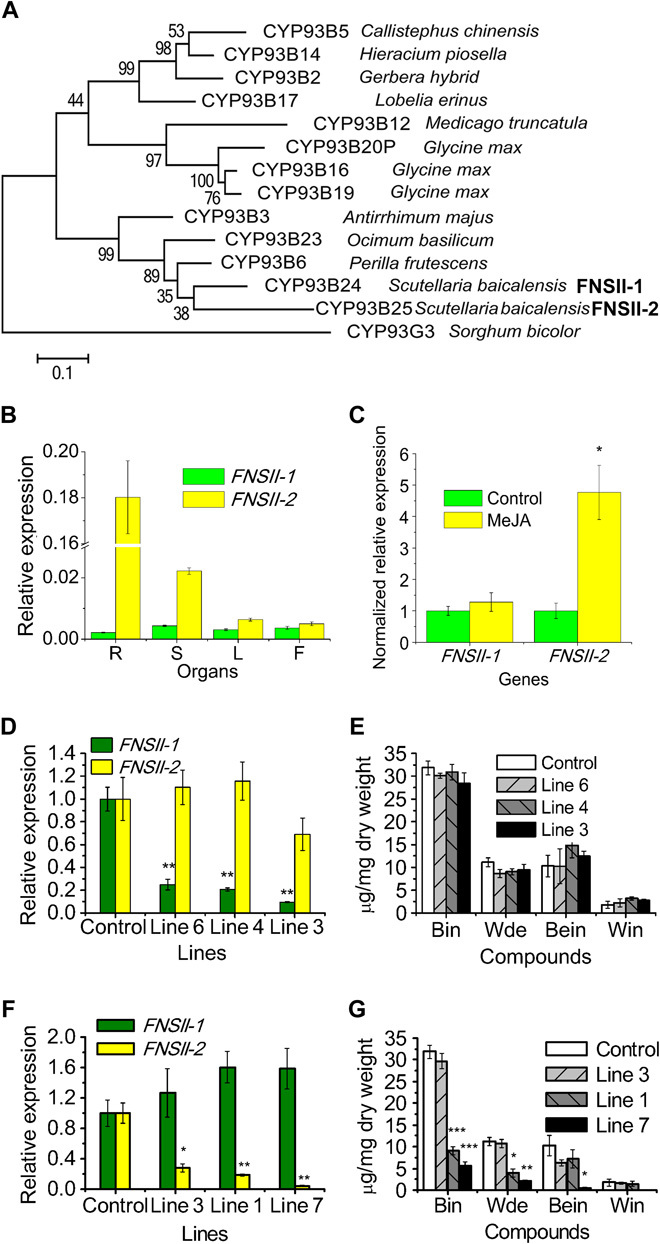
(A) Bootstrap consensus tree of the CYP93B subfamily. Maximum likelihood (ML) was used to construct this tree with 1000 replicate bootstrap support. The tree was rooted with Sorghum bicolor CYP93G. GenBank ID of the proteins used in the tree: CYP93B6, BAB59004.1; CYP93B23, AGF30365.1; CYP93B3, BAA84071.1; CYP93B17, BAF49323.1; CYP93B2, AAD39549.1; CYP93B5, AAF04115.1; CYP93B14, ACB56919.1; CYP93B12, ABC59104.2; CYP93B20P, KHN21998.1; CYP93B16, ACV65037.1; CYP93B19, NP_001241129.1; CYP93G3, XP_002461286.1. (B) Relative levels of SbFNSII-1 and SbFNSII-2 transcripts compared to β-actin were determined by qRT-PCR analyses performed on total RNA extracted from different organs. R, roots; S, stem; L, leaves; F, flowers. (C) Relative expression of SbFNSII-1 and SbFNSII-2 subjected to MeJA treatment for 24 hours. The expression levels were normalized to corresponding values from mock treatments. (D) Silencing of SbFNSII-1 was measured by monitoring relative transcript levels by qRT-PCR. The expression levels were measured relative to those obtained with empty vector as a control. (E) Measurements of RSFs from the SbFNSII-1 RNAi lines used for transcript analysis. (F) Silencing of SbFNSII-2 was measured by monitoring relative transcript levels by qRT-PCR. (G) Measurements of RSFs from the SbFNSII-2 RNAi lines used for transcript analysis. Bin, baicalin; Wde, wogonoside; Bein, baicalein; Win, wogonin. SEs were calculated from three biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Expression profiles of SbFNSII genes
The raw reads of the FNSIIs from the RNA-seq database offered clues to their expression patterns in the roots of S. baicalensis. FNSII-2 had 10,135 reads compared with 73 and 14 reads for the contigs encoding FNSII-2. This suggested that FNSII-2 is more highly expressed in the roots of S. baicalensis and may play a more important role in the synthesis of RSFs than FNSII-1 because hairy roots accumulate high levels of RSFs. The transcript levels of FNSII-1 and FNSII-2 were compared in different organs of S. baicalensis by quantitative RT-PCR (qRT-PCR) and with the levels of flavones present in each organ. The expression of FNSII-1 was relatively low and equally distributed in the four organs analyzed (Fig. 2B). Transcript levels of FNSII-2 were particularly high in roots, being 8-, 28-, and 36-fold higher than the levels detected in stems, leaves, and flowers, respectively. The expression patterns of FNSII-2 were very similar to the accumulation of baicalin and wogonoside, which were substantially higher in roots than in aerial parts of the plant (17) (fig. S2A).
Methyl jasmonate (MeJA) has been reported to enhance the expression of several genes in the phenylpropanoid pathway and induce production of RSFs in Scutellaria suspension culture cells (18). We observed the same effect of MeJA in hairy root cultures of S. baicalensis. After treatment of root cultures with 100 μM MeJA, baicalin and baicalein levels increased 2- and 3.5-fold, respectively, compared to untreated controls (fig. S2, B to F). Although no significant increase in wogonoside was detected, wogonin increased 5.8-fold. The transcript levels of FNSII-1 did not change in hairy roots following MeJA treatment (Fig. 2C), but FNSII-2 transcript levels increased 4.8-fold, emphasizing the correlation between FNSII-2 expression and the accumulation of RSFs.
RNAi silencing of FNSIIs in hairy roots of S. baicalensis
We used RNAi to confirm the different roles of the two FNSII genes in RSF biosynthesis in Scutellaria. Real-time qRT-PCR confirmed that the transcript levels of the FNSII-1 gene were significantly down-regulated in three independent hairy root lines (Fig. 2D). Silencing of FNSII-1 showed no effect on the accumulation of any of the four RSFs (Fig. 2E), even in line 3, which had only 9.6% of the levels of FNSII-1 transcripts compared to control roots.
Real-time qRT-PCR was used to measure FNSII-2 expression in RNAi lines relative to controls (Fig. 2F). The transcript levels of FNSII-2 declined by 82% in line 1, which resulted in reductions of 71 and 65% in baicalin and wogonoside, respectively, compared to controls (Fig. 2G). In RNAi line 7, RSFs levels were reduced to 17.9, 18.4, and 5.2% for baicalin, wogonoside, and baicalein, respectively, which had 4.6% of the FNSII-2 transcript levels of controls (Fig. 2, F and G, and fig. S3A). Extracts from the control lines showed consistently large peaks for baicalin, wogonoside, baicalein, and wogonin, whereas two FNSII-2 RNAi lines had substantially smaller peaks for these flavones, and instead, three new compounds not seen in controls (fig. S3, A to E) accumulated [peak I, m/z (mass/charge ratio) 449.04; peak II, m/z 433.17; peak III, m/z 287.30]. The m/z values of the new peaks were consistent with the protonated ions of dihydrobaicalin-O-glucuronide, pinocembrin-O-glucuronide, and dihydrowogonin, respectively, which are reduced derivatives or glycosylated versions of the putative substrate of FNSII-2. The identities of the new peaks were determined by tandem mass spectrometry (MS2) analysis (fig. S3, D and E). The m/z 273 of peak I is an in-source fragment from m/z 499 by losing a glucuronic acid, which was then fragmented into m/z 169 and 131, showing an essentially identical pattern to that previously reported for dihydrobaicalin (37). Peak II was further fragmented into m/z 257 by losing m/z 176 of glucuronic acid, and the MS2 had an identical spectrum to an authentic standard of pinocembrin (m/z 215, 153, and 131). Peak III was an aglycone, with MS2 of m/z 287, 183.1, and 131.0, which were identical to those reported for dihydrowogonin (38). Consequently, the three new compounds in the RNAi lines were identified as dihydrobaicalin-O-glucuronide, pinocembrin-O-glucuronide, and dihydrowogonin, respectively. These results offered direct evidence that it is FNSII-2 that functions in the biosynthesis of RSFs and is responsible for the synthesis of 4′-deoxyflavones in the roots of S. baicalensis.
Functional characterization of recombinant FNSII proteins
The coding sequences of FNSII-1 and FNSII-2 were individually expressed in S. cerevisiae WAT11, a yeast strain engineered for plant Cyt p450 protein studies by the coexpression of the NADPH Cyt p450 reductase 1 from Arabidopsis (39). Microsomes from the strain expressing each enzyme were assayed against pinocembrin, the proposed precursor of RSFs, as well as naringenin (with a 4′-OH group) and eriodictyol (with 4′- and 3′-OH groups), the classic substrates of FNSII (40) (see fig. S4A for the structures of the substrates). New peaks were detected from reactions with microsomes containing FNSII-1, in addition to the substrates pinocembrin, naringenin, or eriodictyol (fig. S4B). The peaks had the same retention time and MS spectra as the authentic standards of chrysin (m/z 255.3), apigenin (m/z 271.3), and luteolin (m/z 287.2), respectively. The m/z of the products was lower than their corresponding substrates by 2, indicating that FNSII-1 catalyzes a dehydrogenation reaction.
FNSII-2 could convert only pinocebrin to chrysin. Peaks for apigenin or luteolin were not detected following FNSII-2 incubation with naringenin or eriodictyol (fig. S4B), even with extended reaction times. No products were detected when NADPH was absent from the reactions, confirming that the dehydrogenation occurred in a NADPH-dependent manner, a property of Cyt p450 monooxygenases.
In vitro assays indicated that FNSII-2 cannot use substrates with a 4′-OH group. In vivo yeast assays were used to confirm this conclusion. Both pinocembrin (4′-deoxyflavanone) and naringenin (4′-hydroxyflavanone) were added to yeast medium, and the strains were grown overnight. Cells were collected and metabolites were extracted for analysis. A large peak corresponding to chrysin was detected in the yeast expressing FNSII-2 (fig. S4, C and D), but no apigenin was produced by this strain when it was incubated with naringenin. Control yeast cells carrying the empty vector did not produce any flavones.
Yeast microsomes enriched with either FNSII-1 or FNSII-2 were incubated with varying amounts of pinocembrin and naringenin to compare the kinetic parameters of the two enzymes (Table 1). FNSII-1 could convert both pinocembrin and naringenin to chrysin and eriodictyol, respectively, at high efficiency, with apparent Michaelis constant (Km) values of 0.24 and 0.28 μM, respectively, and apparent maximal velocity (Vmax) values of 27.65 and 60.93 pkat mg protein−1, respectively, giving a 1.9-fold higher Vmax/Km ratio for naringenin than for pinocembrin. These parameters suggested that the preferred substrate for FNSII-1 is naringenin. In contrast, FNSII-2 exhibited an apparent Km of 0.46 μM and a lower apparent Vmax of 9.02 pkat mg protein−1 with pinocembrin. Like classic CYP93Bs, SbFNSII-1 likely preferentially converts flavanones with a 4′-OH, such as naringenin and eriodictyol, to flavones, whereas SbFNSII-2 can use only 4′-deoxyflavanones, such as pinocembrin, as substrates. Phylogenetic analysis (Fig. 2A) suggested that SbFNSII-2 may have diverged from 4′-OH substrate–accepting enzymes.
Table 1. Kinetic parameters of FNSIIs toward pinocembrin and naringenin.
Each data set represents the mean ± SE from triplicate measurements.
| Protein and substrate | Km (μM) | Vmax (pkat mg protein−1 ) | Vmax/Km |
| FNSII-1 | |||
| Pinocembrin | 0.24 ± 0.07 | 27.65 ± 3.07 | 114.73 |
| Naringenin | 0.28 ± 0.06 | 60.93 ± 0.67 | 215.31 |
| FNSII-2 | |||
| Pinocembrin | 0.46 ± 0.06 | 9.02 ± 0.22 | 19.60 |
| Naringenin | — | — | — |
Heterologous expression of FNSII-2 in transgenic Arabidopsis
Flavones are made at very low levels, if at all, in most of the approximately 3000 Brassicaceae species, coincident with the absence of genes encoding FNSII in the genome of Arabidopsis thaliana (16). Consequently, Arabidopsis is an ideal plant to test the function of FNS genes, in the context of other enzymes of flavonoid metabolism. To examine whether FNSII-2 is functional in planta, the cDNA of the FNSII-2 gene was overexpressed in transgenic Arabidopsis under the control of the cauliflower mosaic virus (CaMV) 35S promoter. A number of primary transgenic lines were obtained, and the constitutive expression of FNSII-2 was confirmed by qRT-PCR analysis. T2 seedlings of five independent lines with high FNSII-2 expression levels (Fig. 3A), as well as empty vector controls, were grown on Murashige and Skoog medium with or without supplementation of pinocembrin at a concentration of 50 μM. The flavone chrysin was not detectable in the empty vector lines or in Arabidopsis transformed with SbFNSII-2 without supplementation of pinocembrin. However, when pinocembrin was present in the medium, a new peak corresponding to chrysin was detected in all five FNSII-2–expressing lines, but not in controls (fig. S5). Plants expressing FNSII-2 converted most of the pinocembrin they absorbed to chrysin, accumulating chrysin at 2.18 to 3.79 mg g−1 dry weight (DW) (Fig. 3B). No chrysin was detected in the empty vector line EV1. Chrysin was not formed when FNSII-2 was expressed in Arabidopsis except after feeding plants with pinocembrin. This clearly indicated that the formation of chrysin was dependent on the supply of pinocembrin from cinnamic acid, by a pathway absent in Arabidopsis.
Fig. 3. Overexpression of SbFNSII-2 in Arabidopsis.
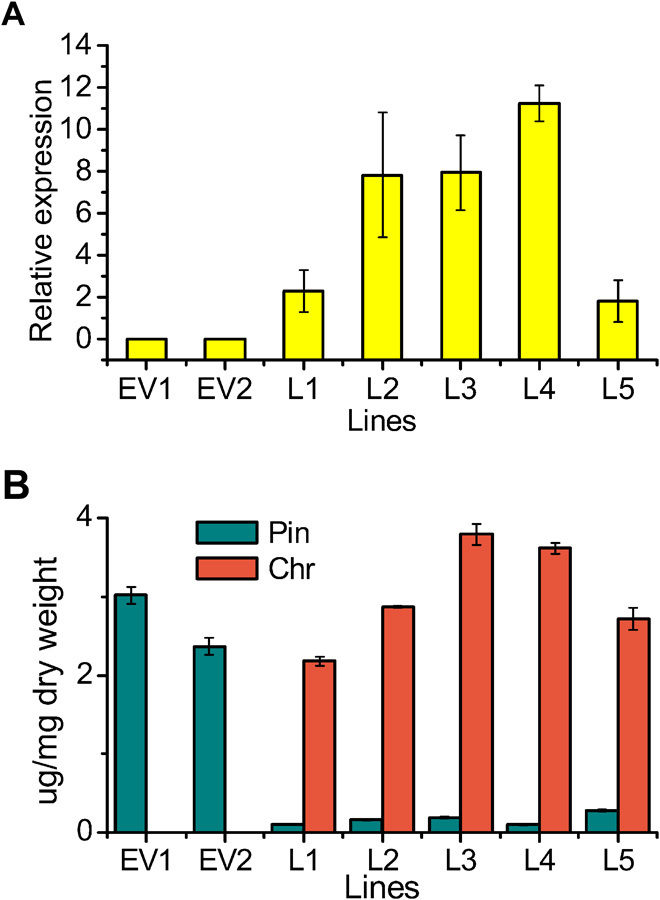
(A) Transcript levels of SbFNSII-2 relative to Arabidopsis UBI in two empty vector (EV) control lines and five transgenic lines determined by qRT-PCR. (B) Measurements of pinocembrin (pin) and chrysin (chr) from two empty vector lines and five transgenic lines grown on MS supplemented with pinocembrin. SEs were calculated from three biological replicates.
Identification of an SbCLL gene expressed preferentially in roots
We hypothesized that pinocembrin is the product of CHS and CHI and that a specific CoA ligase might be present in Scutellaria roots to convert cinnamic acid to cinnamoyl-CoA, the precursor of pinocembrin. We screened for contigs annotated as 4CL or CoA ligase-like (CLL) in the RNA-seq database. This identified cDNA fragments encoding eight putative CoA ligases, although no sequences highly homologous to the Arabidopsis and Petunia proteins encoding cinnamate–CoA ligase (AtCNL and PhCNL, respectively) (21, 22) were identified among the transcripts in S. baicalensis hairy roots. Full-length cDNAs of the five that showed >1000 raw reads in the root RNA-seq database were studied further.
A phylogenetic tree was constructed using protein sequences encoded by 4CLs and CLL genes expressed in the roots of S. baicalensis and from Arabidopsis (Fig. 4A). SbCLL-1 and SbCLL-5 were grouped in the same clade as Arabidopsis 4CL1, 4CL2, 4CL3, and 4CL5, all enzymes with traditional 4CL substrates [4-coumarate or its derivatives (22)]. The SbCLL-6, SbCLL-7, and SbCLL-8 proteins were clearly separated phylogenetically from the core group of SbCLL-1, SbCLL-2, SbCLL-3, and SbCLL-5. SbCLL-7 was most similar to At4CLL7 with a similarity of 67%, and SbCLL-8 was most similar to At4CLL10 with a similarity of 79%, suggesting that these two enzymes likely have catalytic specificities similar to their Arabidopsis counterparts.
Fig. 4. Phylogenetic tree of CLLs and qRT-PCR analysis of Sb4CLs genes.
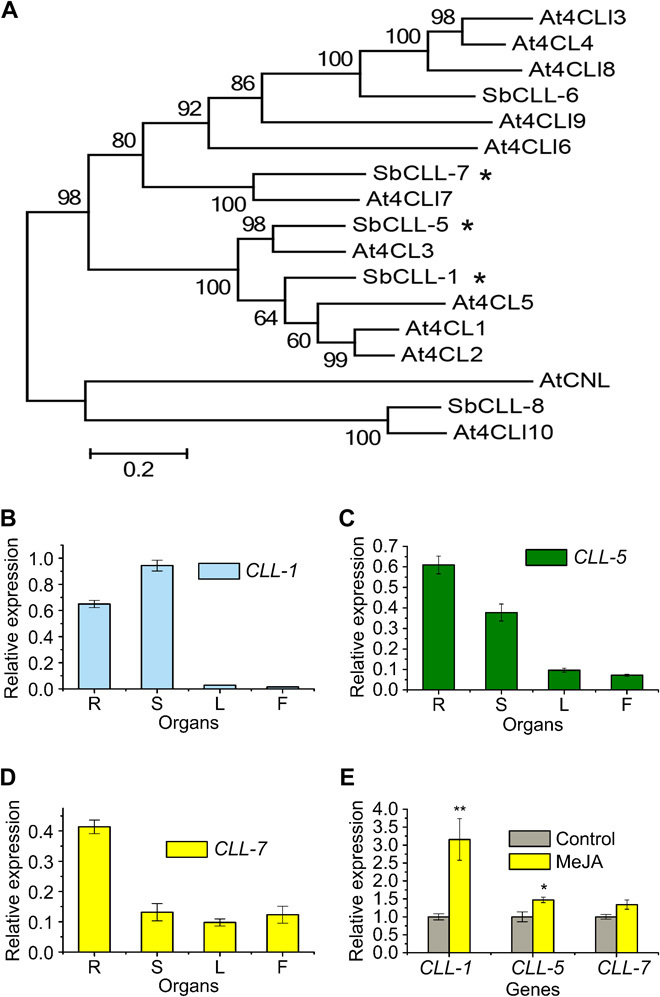
(A) Phylogenetic analysis of CLLs. ML method was used to construct this tree with 1000 replicates bootstrap support. TAIR (The Arabidopsis Information Resource) ID of the proteins used in the tree: At4CL1, AT1G51680; At4CL2, AT3G21240; At4CL3, AT1G65060; At4CLL3, AT1G20490; At4CL4, AT1G20500; At4CL5, AT3G21230; At4CLL6, AT4G19010; At4CLL7, AT4G05160; AT4CL8, AT5G38120; At4CLL9, AT5G63380; At4CLL10, AT3G48990; AtCNL, AT1G65880. (B) Relative SbCLL-1, (C) SbCLL-5, and (D) SbCLL-7 transcript levels to β-actin were determined by qRT-PCR analyses performed on total RNA extracted from different organs. (E) Relative expression of the three genes subjected to MeJA treatment for 24 hours. The expression levels were measured relative to those obtained from mock treatment as a control. SEs were calculated from three biological replicates. *P < 0.05, and **P < 0.01 (Student’s t test).
Analysis of the organ-specific expression patterns by qRT-PCR revealed that mRNA levels of SbCLL-1 were highest in stems and followed by root, whereas leaves and flowers contained only low levels (Fig. 4B). SbCLL-5 transcript levels were high in roots and stems but relatively low in other organs (Fig. 4C). SbCLL-7 was expressed most highly in roots. Its transcript levels in aerial parts were at least three times lower than those in roots (Fig. 4D) such that SbCLL-7 was expressed in a pattern coincident with baicalin and wogonoside accumulation. When treated with MeJA, SbCLL-1 and SbCLL-5 transcript levels were significantly enhanced; however, no significant difference was detected in levels of SbCLL-7 transcripts in response to MeJA (Fig. 4E).
Identification of SbCLL-7 encodes a cinnamic acid–specific CoA ligase
SbCLL proteins were expressed as hexahistidine-tagged fusions in E. coli (fig. S6A), and the soluble proteins were purified to apparent homogeneity by affinity chromatography under native conditions. Using a standard photometric 4CL assay, SbCLL-1 and SbCLL-5 could convert all three substrates tested (cinnamic acid, 4-coumaric acid, and caffeic acid). However, SbCLL-7 could convert only cinnamic acid, suggesting a unique role in the synthesis of RSFs in S. baicalensis. SbCLL-6 and SbCLL-8 did not show any activity toward any of the substrates tested. In extracts of bacterial strains harboring the empty expression vector (pQE), no CoA ligase protein or CoA ligase activity was detected.
The purified recombinant proteins were tested for their abilities to use different substrates. The Km values of SbCLL-1 and SbCLL-5 for 4-coumaric acid were similar to those reported for many purified plant 4CLs, and the apparent Km, apparent Vmax, and relative Vmax/Km values determined for different substrates tested are listed in Table 2. Cinnamic acid was also converted with low efficiency by both enzymes. However, SbCLL-7 had a substantially lower apparent Km value for cinnamic acid compared to SbCLL-1 and SbCLL-5. SbCLL-7 also had a considerably higher Vmax/Km value for cinnamic acid among the different enzymes tested, being 27 and 9 times higher than those of SbCLL-1 and SbCLL-5, respectively, and was specific for cinnamic acid with no activity with 4-coumaric acid or caffeic acid, indicating that SbCLL-7 is a cinnamate–CoA ligase.
Table 2. Kinetic parameters of CLLs toward different substrates.
Each data set represents the mean ± SE from triplicate measurements.
| Enzyme | Substrate | Km (μM) | Vmax (nkat mg protein−1) | Vmax /Km |
| SbCLL-1 | Cinnamic acid | 5300 ± 712 | 180 ± 16 | 0.03 |
| 4-Coumaric acid | 45 ± 3 | 536 ± 8 | 11.88 | |
| Caffeic acid | 18 ± 2 | 190 ± 9 | 10.58 | |
| SbCLL-5 | Cinnamic acid | 3060 ± 331 | 279 ± 23 | 0.09 |
| 4-Coumaric acid | 31 ± 3 | 687 ± 35 | 22.15 | |
| Caffeic acid | 260 ± 15 | 600 ± 10 | 2.31 | |
| SbCLL-7 | Cinnamic acid | 172 ± 17 | 168 ± 7 | 0.97 |
| 4-Coumaric acid | — | — | — | |
| Caffeic acid | — | — | — |
Silencing of SbCLL-7 in hairy roots of S. baicalensis
We used RNAi to study the role of SbCLL-7 in RSF biosynthesis in Scutellaria. We screened the RNAi hairy root lines using real-time qRT-PCR and identified three RNAi lines with considerably down-regulated SbCLL-7 transcript levels, with 48, 25, and 21% of the levels in controls, in lines 5, 8, and 2, respectively (Fig. 5A).
Fig. 5. RNAi of SbCLL-7 in hairy root cultures of S. baicalensis.
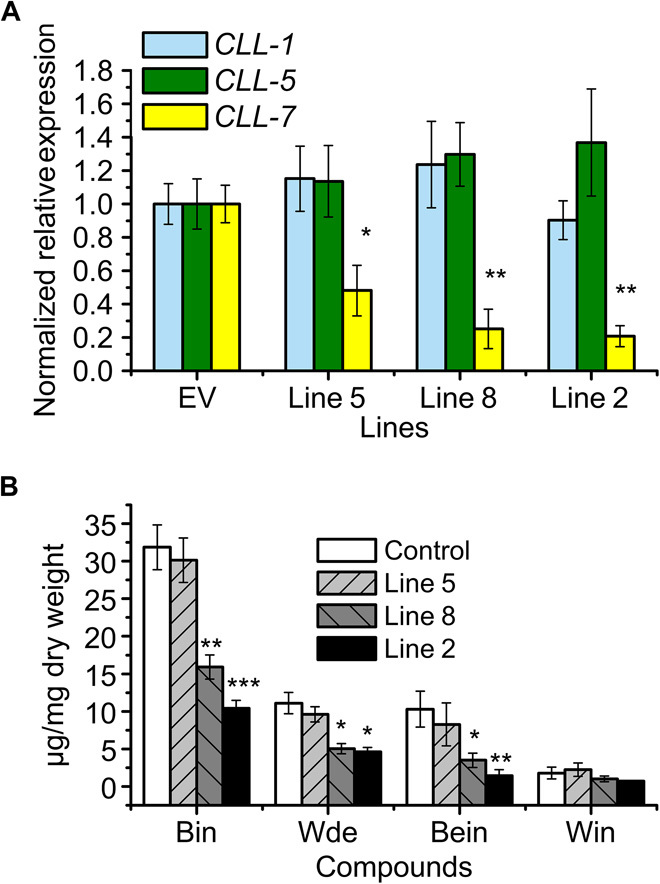
(A) Silencing of SbCLL-7 was measured by monitoring relative transcript levels by qRT-PCR. The expression levels were measured relative to those obtained from an empty vector line as a control. (B) Measurements of RSFs from SbCLL-7 RNAi lines used for transcript analysis. SEs were calculated from three biological replicates. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test).
Although no significant reduction in flavone levels was detected in line 5, the levels of the four major root flavones were reduced in lines 8 and 2, which had only 50 and 30% baicalin, respectively, compared to empty vector controls. Wogonoside was also reduced from 11.10 μg mg−1 DW in the control to 5.05 and 4.62 μg mg−1 in lines 8 and 2, respectively. The levels of baicalein were reduced from 10.30 μg mg−1 DW in the control to 3.51 μg mg−1 in line 8 and 1.43 μg mg−1 in line 2 (Fig. 5B and fig. S7).
To uncover more clues about the specialized flavone pathway in roots, we transformed Arabidopsis with the SbCLL-7 gene driven by the 35S promoter. The T2 generation of the transgenic plants was grown on Murashige and Skoog medium supplemented with cinnamic acid. However, we did not detect any pinocembrin peak from these seedlings, indicating that Arabidopsis CHS cannot use cinnamoyl-CoA as a substrate in vivo. This implied that genes specific to S. baicalensis (and close relatives making RSFs), which encode CHS and/or CHI and can preferentially use cinnamic acid, are active in the pathway for synthesis of RSFs.
Identification of genes encoding CHS and CHI from S. baicalensis
We isolated two full-length genes coding for CHS from our deep sequencing databases (from hairy roots and flowers) and named them SbCHS-1 and SbCHS-2. SbCHS-1 is expressed specifically in flowers and has not been reported by previous studies. It was represented by 4038 raw reads in RNA-seq data of flowers but only by 45 reads in RNA-seq of hairy roots. SbCHS-2 had abundant raw reads (40887) in RNA-seq of hairy roots but was represented by a much lower number of reads in RNA-seq of flowers (145 raw reads). This information suggested that SbCHS-2 might be responsible for the synthesis of RSFs. We compared the encoded protein sequences to those in the databases for CHS from S. baicalensis. SbCHS-2, which was highly expressed in roots, was 98% identical to SbCHS2a/SbCHS-C (BAB03471.1) (41, 42) and 99% identical to another CHS from S. baicalensis, SbCHS2b (BAA23373.1) (43), suggesting that all three sequences represent alleles of the same gene. SbCHS-2 was 94% identical to SbCHS-P (AAB88208.1), identified by Morita et al. (42). SbCHS-2 was 98% identical to a gene encoding CHS from S. vestidula, SvCHS (ACC68839.1), a close relative of S. baicalensis that also makes RSFs (44). However, SbCHS-2 has only 83% identity with the SbCHS-1 gene highly expressed in flowers of S. baicalensis. A phylogenetic tree was constructed to assess the evolutionary relationship between SbCHS-1, SbCHS-2, and other CHS (Fig. 6A). This analysis suggested that CHS-1 and CHS-2 separated relatively recently, after the divergence of the family Lamiaceae.
Fig. 6. Phylogenetic analysis of Scutellaria CHS isoforms and the expression patterns of their genes.
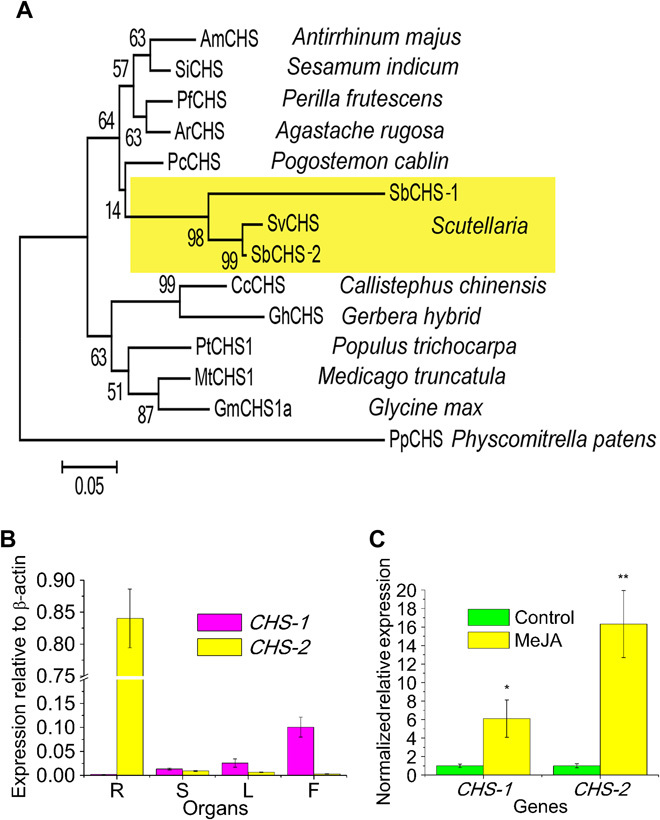
(A) Phylogenetic tree of CHS proteins. ML was used to construct this tree with 1000 replicate bootstrap support. The tree was rooted with Physcomitrella patens CHS. GenBank ID of the proteins used in the tree: AmCHS, CAA27338.1; SiCHS, XP_011091402.1 ; PfCHS, O04111.1; ArCHS, CAA27338.1; PcCHS, AJO53275.1; SvCHS, ACC68839.1; CcCHS, P48385.2; GhCHS, CAA86220.1; PtCHS, XP_002303821.2; MtCHS1, XP_003601647.1; GmCHS1a, AAB01004.1; PpCHS, ABB84527.1. (B) Relative levels of SbCHS-1 and SbCHS-2 transcripts compared to β-actin were determined by qRT-PCR analyses performed on total RNA extracted from different Scutellaria organs. (C) Relative expression of SbCHS-1 and SbCHS-2 subjected to MeJA treatment for 24 hours. The expression levels were measured relative to those obtained from mock treatment as a control. SEs were calculated from three biological replicates. *P < 0.05 and **P < 0.01 (Student′s t test).
The expression patterns of the two genes encoding CHS from S. baicalensis were further confirmed by qPCR analysis (Fig. 6B). The expression of SbCHS-1 was relatively low in roots but very high in flowers (Fig. 6B). Transcript levels of SbCHS-2 were particularly high in roots but very low in flowers. When treated with MeJA, transcript levels of both of the SbCHS genes were enhanced. Their expression levels were elevated 6- and 16-fold compared with the control for SbCHS-1 and SbCHS-2, respectively (Fig. 6C).
Transcripts encoding CHI in the roots of S. baicalensis were sought in the RNA-seq database, but the single transcript identified encoded a protein comparable to two sequences in National Center for Biotechnology Information (NCBI) (ADQ13184.1 and AJR10104.1), which themselves differ by one amino acid (position 160 in ADQ13184.1 and position 31 in AJR10104.1) in their encoded protein sequences and are therefore likely allelic. This finding suggested that there was not a novel isoform of CHI involved in the synthesis of RSFs and that pinocembrin is formed from pinocembrin chalcone either spontaneously or through the CHI that is also active in aerial parts of the plant (Fig. 1D) (19).
Reconstruction of the 4′-deoxyRSF pathway in tobacco leaves
It is reasonable to propose that SbCLL-7 synthesizes cinnamoyl-CoA and that at least one of the CHS genes could efficiently use cinnamoyl-CoA as a substrate and channel this precursor into the pinocembrin pool, which would be converted to chrysin by FNSII-2 and then decorated by a flavone-6-hydroxylase, a flavone-8-hydroxylase, and GTs to form the different 4′-deoxyRSFs of S. baicalensis.
To test this idea, we expressed SbCLL-7, SbCHS-2, SbCHI, and SbFNSII-2 under the control of the 35S promoter in the HyperTrans transient expression system in the leaves of Nicotiana benthamiana (45), with or without supplementation with cinnamic acid. Inoculation with the control vector expressing green fluorescent protein (GFP) gave no new products as established by comparison to authentic standards of pinocembrin and chrysin (fig. S8). Inoculation with a vector expressing SbCLL-7 also did not result in pinocembrin or chrysin formation, confirming our previous observations in Arabidopsis (Table 3). Inoculation with SbCLL-7 and SbCHS-2 resulted in the formation of detectable levels of pinocembrin both in unsupplemented leaves and in leaves supplemented with cinnamic acid (Table 3). This result established that SbCHS-2 has a specific activity in the formation of pinocembrin that cannot be complemented by the standard CHS active in the leaves of N. benthamiana. These results also showed that an S. baicalensis–specific CHI activity was not required and that the CHI gene from N. benthamiana could function in pinocembrin formation, a conclusion verified by inoculation with SbCLL-7 plus SbCHS-2 and SbCHI, which gave rise to similar production of pinocembrin as SbCLL-7 plus SbCHS-2, with or without cinnamic acid supplementation (Table 3 and fig. S8). The production of pinocembrin by SbCLL-7 and SbCHS-2 in N. benthamiana showed that SbCLL-7 can compete with the endogenous C4H activity for cinnamic acid, an activity hypothesized, but not found, in old man’s cactus (Cephalcereus senilis) by Liu et al. (30). Indeed, transcript levels of C4H were particularly high in the roots and stems of Scutellaria (fig. S9), suggesting that the complete selectivity of SbCLL-7 for cinnamic acid might be particularly important for root-specific production of 4′-deoxyflavones. Finally, either inoculation with genes encoding four proteins (SbCLL-7, SbCHS-2, SbCHI, and SbFNSII-2) or encoding three proteins (SbCLL-7, SbCHS-2, and SbFNSII-2) gave rise to chrysin production, even in the absence of supplementation with cinnamic acid (Table 3 and fig. S8). Therefore, reconstruction of this specialized pathway in a novel host confirmed that a new pathway for RSF biosynthesis has evolved relatively recently in S. baicalensis and its close relatives, as evidenced by the phylogenetic relationships between SbCHS-1 and SbCHS-2 and those between SbFNSII-1 and SbFNSII-2. The complexity of the CLL family makes the point of recruitment of SbCLL-7 for RSF synthesis difficult to ascertain with confidence. The lack of requirement for a specific CHI activity may reflect the fact that isomerization of chalcones can occur spontaneously as well as catalytically (46).
Table 3. Tobacco leaves infiltrated with different combinations of flavone biosynthetic genes.
| Scutellaria genes used | Cinnamic acid (supplemented) | Pinocembrin (detected) | Chrysin (detected) |
| GFP | − | No | No |
| + | No | No | |
| CLL-7 | − | No | No |
| + | No | No | |
| CLL-7, CHS-2 | − | Yes | No |
| + | Yes | No | |
| CLL-7, CHS-2, CHI | − | Yes | No |
| + | Yes | No | |
| CLL-7, CHS-2, CHI, FNSII-2 | − | No | Yes |
| + | No | Yes | |
| CLL-7, CHS-2, FNSII-2 | − | No | Yes |
| + | No | Yes |
DISCUSSION
S. baicalensis is noted for its high-level production of bioactive 4′-deoxyflavones in roots. We wanted to establish whether there is a specific biosynthetic pathway that uses pinocembrin rather than naringenin to produce these flavones. We first isolated two candidate FNSII genes that encode proteins that are homologous to previously reported CYP93B proteins and named them CYP93B24 (FNSII-1) and CYP93B25 (FNSII-2). Phylogenetic analysis suggested that CYP93B25 may have diverged from CYP93B24 following a recent gene duplication event, after the divergence of Scutellaria from other members of the family Lamiaceae (Fig. 2A). The elongated branch length of CYP93B25 suggests accelerated evolution, which could be the result of positive selection or neutral drift following release from the evolutionary constraints imposed on the ancestral gene (Fig. 2A) (47). A similar event has been observed in Arabidopsis, where a recently duplicated gene encoding CYP84A4 is involved in the biosynthesis of α-pyrone (47). Multiple isoforms of FNS have not been reported for other members of the order Lamiales, perhaps because of a lack of genome sequence information. However, both Medicago truncatula and Glycine max have genes encoding three FNSII proteins (CYPB93B) (40), supporting divergent roles in the synthesis of flavones and isoflavones in the legume family.
All the other FNS genes identified in species of the order Lamiales have been identified from RNA from aerial organs or have a higher expression in aerial parts of the plants; CYP93B3, CYP93B4, and CYP93B13 were isolated from petals of Antirrhinum, Torenia, and Gentiana, respectively (32–36), and therefore are likely involved in synthesis of flavones derived from naringenin. FNSII (CYP93B6) from P. frutescens is highly expressed in leaves and is responsive to light (32). Coincidently, the leaves of Perilla (mint family, Lamiaceae) have been reported to contain scutellarein and its derivatives (48). Several species of the genus Scutellaria produce baicalein and wogonin and their glycosides, although these are not always accumulated predominantly in roots (14). To our knowledge, 4′-deoxyflavones such as baicalein and wogonin have been reported only in Anodendron affine and Cephalocereus senilis (49, 50) outside the order Lamiales. A broad-specificity 4CL activity supporting deoxyflavonoid biosynthesis was described in C. senilis (30), suggesting that a pathway different from the one we have characterized in S. baicalensis operates in this widely diverged species. Baicalein or wogonin and their derivatives have been reported in species such as Oroxylum indicum vent (51) and Plantago major L. (52), which are members of the order Lamiales but belong to families outside the mint family, Lamiaceae. It would be interesting to identify the enzymes that synthesize these flavones, in O. indicum vent, P. major L., and A. affine, which likely acquired their functions by convergent evolution (53).
Multiple isoforms of FNSII with different expression patterns have also been observed in Medicago, a species relatively distant to S. baicalensis. The activity of these isoforms results in different product profiles in vitro, although these were interpreted to be the result of different rates of transition through dihydroxyflavanone intermediates in the formation of the flavone apigenin (54). It could be that the products of these two FNSII genes in Medicago are distinct in vivo, in a manner analogous to the situation for the FNSIIs producing 4′-deoxyRSFs in roots and the 4′-hydroxyflavones produced predominantly in aerial parts of S. baicalensis. Further functional diversity of CYP93 genes in legumes may reflect activity of the closely related iso-FNS genes (CYP93C proteins), which catalyze an aryl ring migration in addition to the creation of the double bond between C2 and C3 that is catalyzed by FNSII enzymes (55).
Pinocembrin is the likely substrate of FNSII-2 in S. baicalensis roots. When SbFNSII-2 was silenced, this precursor was converted by a GT to produce pinocembrin-O-glucuronide. A 7-O-glucosyltransferase activity has been detected in S. baicalensis that is active with a broad range of substrates including flavanones (56), although this activity was not able to transfer glucuronic acid to flavones. Pinocembrin might also serve as a substrate for flavone-6-hydroxylase, flavone-8-hydroxylase, and O-methyltransferase to produce dihydrobaicalin and dihydrowogonin (fig. S3). A flavone-6-hydroxylase was recently reported from sweet basil, a member of the family Lamiaceae. This enzyme was able to use the flavonone sakuranetin as a substrate, although with relatively lower activity than the flavone genkwanin (36).
The enzymatic reaction of FNSII is stereoselective (57), and both FNS from Scutellaria favored the (S) over the (R) enantiomer. In our assays of FNSII enzyme kinetics, we used pure (S) enantiomer, explaining why we found somewhat lower apparent Km and higher apparent Vmax values than previously reported for FNSII enzymes (32, 57). Similar to other reports for FNSII, SbFNSII-1 is a relatively promiscuous enzyme and has high catalytic efficiency for naringenin. However, SbFNSII-2 is specific for pinocembrin, with a lower apparent Vmax than SbFNSII-1 (Table 1). Phylogenetic analysis suggested that SbFNSII-2 originated from a recent gene duplication event of the common ancestor of SbFNSII-1 and SbFNSII-2. Mutation of the ancestor of SbFNSII-2 likely leads to neofunctionalization, such that SbFNSII-2 exclusively uses pinocembrin as a substrate, albeit at the price of lower catalytic activity than SbFNSII-1.
The results of ectopic expression of FNSII-2 in Arabidopsis showed that there must be specific enzymes supplying pinocembrin to FNSII-2 because chrysin was produced only upon feeding FNSII-2 plants with pinocembrin. The intermediate pinocembrin-glucuronide accumulated when FNSII-2 was silenced in S. baicalensis roots, suggesting that a specific isoform of CoA ligase could activate cinnamic acid and that specific isoforms of CHS and CHI are also required for the formation of pinocembrin. Five CLL genes are expressed in S. baicalensis hairy roots. SbCLL-1 and SbCLL-5 aligned with the Arabidopsis 4CL enzymes that activate 4-coumarate and its derivatives (Fig. 4A). No CLLs expressed in roots aligned with the CNL proteins from Arabidopsis or Petunia. Both SbCLL-1 and SbCLL-5 have high expression in roots and stems, the organs with relatively large amounts of vascular tissues. SbCLL-1 showed high specificity for 4-coumaric acid and caffeic acid as substrates in vitro, suggesting functions in the biosynthesis of lignin (Fig. 4B), as shown for At4CL1 (28). SbCLL-5 is most closely related structurally to At4CL3 and showed similar enzyme characteristics to At4CL3; both of which have high affinity for 4-coumarate (Table 2).
SbCLL-7 showed three times higher expression levels in roots than in aerial organs (Fig. 4D). SbCLL-7 protein had improved catalytic characteristics with cinnamic acid compared with all other SbCLLs. This enzyme worked only with cinnamic acid among three substrates tested. On the basis of the three-dimensional model of the At4CL2 active site and mutation analysis, 12 amino acid residues have been identified in the active site that form a signature motif determining 4CL substrate specificity (20). Multiple sequence alignments identified candidate amino acids that might determine the substrate preferences of SbCLL-7. SbCLL-7 carries a noncharged residue, Ala, at position 249, which in At4CL2 is Asn (Asn256), which forms a hydrogen bond with the 4-hydroxyl group of 4-coumaric acid (20). Correspondingly, the SbCLL-7 protein is unlikely to recognize 4-hydroxycinnamic acid derivatives because of this change from a charged to an uncharged residue (fig. S6, B and C). The predicted active sites of the two closely related proteins, At4CLL7 and SbCLL-7, are exclusively made up of hydrophobic amino acids (fig. S6, B and C) and are therefore predicted to bind hydrophobic substrates preferentially. By increasing the hydrophobicity of its active site, At4CL2 can be converted to a cinnamic acid–utilizing enzyme (20); therefore, our analysis supports the idea that a CLL protein with a hydrophobic active site may function as a cinnamate–CoA ligase. However, SbCLL-7 and At4CLL7 share the same 12 amino acid residues in their active sites (fig. S6, B and C), but At4CLL7 cannot ligate CoA to cinnamic acid (58). This finding suggests that amino acids additional to the 12 previously reported determine the specificity of SbCLL-7 as a cinnamate–CoA ligase.
SbCLL-7 aligns phylogenetically with At4CLL7 and shares 67% amino acid identity with this protein. In Arabidopsis, At4CLL7 activates medium-chain fatty acids, medium-chain fatty acids carrying a phenyl substitution, and long-chain fatty acids, as well as the jasmonic acid precursors 12-oxo-phytodienoic acid and 3-oxo-2-(2-pentenyl)-cyclopentane–1-hexanoic acid, and has been suggested to be an enzyme in jasmonic acid biosynthesis (58). SbCLL-7 appears to have been recruited from a CLL ancestor and not by duplication of a gene encoding 4CL and subsequent neofunctionalization.
SbCLL-7 was aligned with BZO (AtCNL) from Arabidopsis (Fig. 4A). BZO encodes a cinnamate–CoA ligase (21) that is structurally closely related to PhCNL from Petunia hybrida (22). PhCNL has a somewhat higher affinity for cinnamic acid than 4-coumaric acid (22), and it is likely that AtCNL has similar substrate specificities to PhCNL, although acceptor specificities beyond cinnamic acid were not reported for AtCNL (21). The phylogenetic alignment showed independent origins for SbCLL-7 and AtCNL despite the activities of their encoded proteins being similar, to the extent that they will both accept cinnamic acid as a substrate. The broad specificity of CNLs for both cinnamic acid and 4′-hydroxycinnamic acids (22) means that this activity is likely relatively ineffectual at producing cinnamoyl-CoA for pinocembrin formation when C4H is also highly active, as in the roots of Scutellaria (fig. S9). The peroxisomal location of CNL activity (22) might limit its ability to supply cinnamoyl-CoA for cytoplasmic production of 4′-deoxyflavones. However, SbCLL-7 also has a peroxisome localization motif (SKL at its C terminus), perhaps reflecting its origins in genes encoding fatty acid–metabolizing CLL enzymes. The abundant supply of CoA from β oxidation and import of cinnamic acid into peroxisomes (22) might promote the production of cinnamoyl-CoA by SbCLL-7, despite concomitant C4H activity competing for cinnamic acid. Recruitment of a cinnamic acid–specific CoA ligase with high catalytic efficiency induced in roots likely paved the way for high-level production in Scutellaria, involving subsequent recruitment of other enzymes (CHS-2 and FNSII-2) for the synthesis of 4′-deoxyflavones. These comments emphasize the importance of convergence in the evolution of specialized metabolism with related enzyme activities being derived from different ancestral genes to provide specialized features suitable for different pathways (Fig. 4A) (53).
Working with sequences from NCBI and our own RNA-seq database, we identified two CHS transcripts (with variations likely to be the result of single-nucleotide polymorphisms in the same gene in different accessions) for S. baicalensis. SbCHS-1 was expressed at very low levels in roots, whereas SbCHS-2 [also called SbCHS-C (41, 42)] was expressed at high levels in roots. A CHS sequence from the roots of Scutellaria viscidula (SvCHS), which also makes wogonin and baicalin, is most closely related to SbCHS-2 (Fig. 6A) (44). SbCHS-2 appears to have diverged from SbCHS-1 after the divergence of the Lamiales, similar to that of SbFNSII-2 from SbFNSII-1 (Fig. 6A). This suggests that the pathway specific for RSF synthesis evolved relatively recently in S. baicalensis and its close relatives, following duplication of genes active in the standard flavone pathway and subsequent neofunctionalization, and by recruitment of a gene (SbCLL-7) whose ancestor was likely involved in fatty acid metabolism. This type of convergent mechanism for the evolution of specialized pathways in plants is gaining considerable experimental support (53).
When SbCHS-2 was expressed transiently in leaves of N. benthamiana together with SbCLL-7 and SbFNSII-2, the accumulation of chrysin was detected, indicating that the core steps in the pathway for synthesizing 4′-deoxyRSFs in S. baicalensis had been identified. The new pathway, which has evolved for the synthesis of 4′-deoxyRSFs in the roots of S. baicalensis, uses pinocembrin rather than naringenin as an intermediate to produce baicalein and wogonin and their glycosides (Fig. 7). The major difference in the new root-specific pathway is that C4H is bypassed to provide cinnamic acid rather than 4-coumaric acid for activation by addition of CoA. SbCLL-7 has high affinity for cinnamic acid, meaning that this enzyme should be able to compete effectively with C4H for substrate in roots. The ability of SbCLL-7 to direct pinocembrin production in N. benthamiana (in combination with SbCHS-2) indicates that, indeed, SbCLL-7 can compete effectively with C4H for cinnamic acid, presumably on the basis of its specific catalytic properties. Specific isoforms of CoA ligase (SbCLL-7), CHS (SbCHS-2), and FNSII (SbFNSII-2) are responsible for the synthesis of bioactive 4′-deoxyRSFs, which are then further decorated by flavone-6-hydroxylases, flavone-8-hydroxylases, and GTs, which likely work on all types of flavones produced in S. baicalensis, as judged by the different types that are found in this species (Fig. 1C). The roots of S. baicalensis produce particularly high levels of RSFs, compared to other sources of bioactive flavones such as parsley and celery (59). The capacity to accumulate high levels of specialized flavones and the consequent ethnobotanical use of Huang-Qin might have been a consequence of the evolution of a specialized flavone biosynthetic pathway with its regulation independent of standard flavone (scutellarin) production, dedicated to the production of these specific bioactives in the genus Scutellaria.
Fig. 7. Proposed pathways for synthesis of flavones in S. baicalensis.
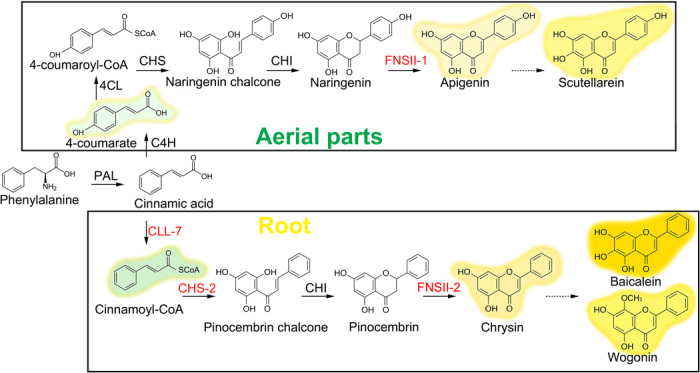
The proteins labeled in red are those for which genes encoding specific isoforms are reported in this study.
MATERIALS AND METHODS
Plant materials and induction of Scutellaria hairy roots
S. baicalensis Georgi seeds were purchased from Anguo County, Hebei Province, China. Single colonies of the Agrobacterium rhigogenes A4 strain carrying the pK7WG2R plasmid with the dsRed marker gene (60) were inoculated into 5 ml of tryptone yeast broth supplemented with spectinomycin (50 mg liter−1) and kanamycin (50 mg liter−1), and cultured overnight at 28°C with shaking (180 rpm). The cultures were centrifuged for 15 min at 3000g, 4°C, and resuspended in Murashige and Skoog liquid medium containing 50 μM acetosyringone.
Leaf explants were collected from 3-month-old S. baicalensis Georgi plants grown in a green house. The leaves were first treated with 75% ethanol for 30 s, followed by surface sterilization using 10% bleach for 10 min, and then washed five times with sterile water. The leaf explants were scratched using a knife dipped with A. rhigogenes infection solution. All explants were blotted dry on sterile filter paper and cocultured on Murashige and Skoog medium containing 50 μM acetosyringone at 25°C in the dark for 3 days. The explants were then transferred into B5 medium containing cefotaxime (500 mg liter−1; Sigma). The first hairy roots developed on cut ends after about 2 weeks of cocultivation and were screened for expression of dsRed under a fluorescence microscope. Hairy roots with red fluorescence were excised from explants and cultured in fresh B5 medium containing cefotaxime (500 mg liter−1) at 23°C in the dark. Hairy root cultures were transferred to fresh medium every 3 weeks and maintained as separate independent clones.
To obtain liquid cultures, the elongated root tips were cut and transferred to flasks containing 50 ml of B5 liquid with cefotaxime (400 mg liter−1) and maintained in the dark at 25 ± 1°C with shaking at 90 rpm. Roots were harvested for further analysis at about 50 days after inoculation
Analysis and identification of flavonoids
Baicalin, baicalein, scutellarein (C98%), scutellarin (C98%), wogonin, pinocembrin, chrysin, narigenin, apigenin, cinnamic acid, 4-coumaric acid, and caffeic acid were purchased from Sigma-Aldrich or Extrasynthese. Wogonoside was purchased from Carbosynth Ltd. Standard stock solutions (1 mg ml−1) were obtained by dissolving the compounds in methanol. Stock solutions were serially diluted with 70% methanol to obtain working standard solutions of various concentrations.
Hairy root tissue (50 days old), was harvested, frozen, and ground into a fine powder with liquid N2 before freeze-drying. The freeze-dried samples were extracted with 70% methanol at a concentration of 1 mg ml−1 in a sonicator bath for 2 hours. Plant debris was removed by centrifugation. Samples were filtered through 0.2-μm filters before injection. High-performance liquid chromatography (HPLC) was performed using a Waters 2659 HPLC system. Separation used a 100 × 2 mm Luna 3 μm C18(2) column with the following gradient: acetonitrile/MeOH (1:1) + 0.1% formic acid (A) versus 0.1% formic acid in water (B), run at 260 μl min−1 and column 35°C (0 to 3 min, 20% B; 20 min, 50% B; 20 to 30 min, 50% B; 36 min, 30% B; 37 min, 20% B; and 37 to 43 min, 20% B). Absorption was measured at 280 nm with a diode array detector (Waters). Flavonoids were quantified by calculating the area of each individual peak and comparing this to standard curves obtained from the pure compounds.
Liquid chromatography–MS/MS (LC-MS/MS) was carried out on a Surveyor HPLC system attached to a Deca XP Plus ion trap MS (Thermo). Separation was on a 100 × 2 mm Luna 3 μm C18(2) column using the same gradient previously described. Detection was by absorbance and by positive-mode electrospray MS. For light absorbance, the instrument collected full scans from 200 to 600 nm but also from a single channel at 280 nm (bandwidth, 9 nm). For MS, the instrument collected full spectra from m/z 200 to 2000 and data-dependent MS2 of the most abundant precursor ions, at a collision energy of 35% and an isolation width of m/z 4.0. Dynamic exclusion was used to ensure that after an ion had been selected for fragmentation twice, it would be ignored for 0.5 min in favor of the next most abundant ion. This maximized the range of precursors for which we collected MS2 data. Spray chamber conditions were 50 U of sheath gas, 5 U of aux gas, 350°C capillary temperature, and 3.8-kV spray voltage in positive mode, and were conducted using a steel needle kit.
RNA sequencing
Fifty-day-old hairy roots and young flowers from 6-month-old plants were used for transcriptome analysis. Library construction and sequencing was carried out by the Beijing Genomics Institute (Shenzhen, China).
Total RNA was obtained using the RNeasy Plant Mini Kit (Qiagen). After deoxyribonuclease I treatment, magnetic beads with oligo(dT) were used for isolation of mRNA. Mixed with fragmentation buffer, mRNA was broken into short fragments. Then, cDNA was synthesized using the mRNA fragments as templates. Short fragments were purified and resolved with elution buffer for end repair and single nucleotide A (adenine) addition. After that, the short fragments were ligated with adapters. Suitable fragments were selected for PCR amplification as templates. During the quality control steps, an Agilent 2100 Bioanalyzer and an ABI StepOnePlus Real-Time PCR System were used in quantification and quality control of the sample library. The libraries were sequenced using Illumina HiSeq 2000.
Image data output from sequencing was transformed by base calling into sequence data, called raw reads, and stored in FASTQ format. Problematic raw reads were discarded to remove reads with adaptors, reads with unknown nucleotides amounting to more than 5% of the sequence, and low-quality reads in which the percentage of low-quality bases (base quality ≤ 10) was more than 20%, to leave clean reads. The de novo transcriptome assembly was carried out with the clean reads assembly program Trinity (55) to produce Unigenes.
Unigene sequences were first aligned by blastx against protein databases NR, Swiss-Prot, KEGG, and COG (e value < 0.00001), and then aligned by blastn to nucleotide databases NT (e value <0.00001), retrieving proteins with the highest sequence similarity to the given Unigenes along with their functional protein annotations. Using those annotations, we used the Blast2GO program (61) to obtain GO (Gene Ontology) annotation for the Unigenes. After getting GO annotation for every Unigene, we used the WEGO software (62) for GO functional classification of Unigenes and to understand the distribution of gene functions within the species.
Calculation of Unigene expression used the FPKM (fragments per kilobase per million) method (63), with the formula FPKM = 106C/NL/103, where FPKM was the expression of Unigene A, C was the number of fragments that aligned uniquely to Unigene A, N was the total number of fragments that aligned uniquely to all Unigenes, and L was the base number in the coding DNA sequence (CDS) of Unigene A. The FPKM method eliminated the influence of different gene lengths and sequencing levels on the calculation of gene expression. Therefore, the calculated gene expression could be used directly for comparing the differences in gene expression between samples. The formulas of FPKM and RPKM (reads per kilobase per million) were the same. The only difference between them was the method used to compute the parameters of N and C. If both pairs of reads aligned to a gene, they were treated as one fragment with FPKM, but treated as two reads with RPKM. Both algorithms were rational.
The RNA-seq data for this work have been deposited in the SRA (Sequence Read Archive) database under BioProject ID PRJNA300475, with the following accession numbers: study ID, SRP068883; sample ID, SRS1263237; experiment ID, SRX1546377; run ID, SRR3123399.
Characterization of genes in the flavone pathway
The Scutellaria deep sequencing database was searched for sequences homologous to the genes in flavone biosynthesis pathway. SbFNSII-1 contig Unigene22612 was used to identify a full-length cDNA from Scutellaria hairy root cDNA using 3′ and 5′ RACE PCR (31). Total RNA was isolated, and first-strand cDNA was synthesized (64) using the primer AP. All the primers used for cloning are listed in table S1. The 3′ end of the cDNA was amplified using the primer AUAP and gene-specific primers (GSPs). The amplified sequence was cloned into the pGEM-T Easy vector system (Promega) and sequenced. The 5′ RACE PCR was conducted using a Roche 5′ RACE Kit. First-strand cDNA was synthesized using the primer SP1. The 5′ end of the cDNA was amplified using the anchored oligo(dT) primer and SP primers. The full-length cDNA was then reamplified using the SbFNSII-1 CDS primer pairs based on the 3′ and 5′ sequence amplified by RACE.
The genes with full-length sequences in the Scutellaria RNA-seq database (SbFNSII-2, SbCLL-1, SbCLL-5, SbCLL-6, SbCLL-7, SbCLL-8, SbCHS-1, SbCHS-2, and SbCHI) were amplified with the primers listed in table S1. The PCR products were subcloned into plasmid pDONR207 using the Gateway BP Clonase II enzyme mix (Thermo Fisher) according to the protocol offered and verified by complete gene sequencing. The sequences for these genes have been submitted to the NCBI database with the following accession numbers: SbFNSII-1, KT963453; SbFNSII-2, KT963454; SbCLL-1, KT963455; SbCLL-5, KT963456; SbCLL-6, KT963457; SbCLL-7, KT963458; SbCLL-8, KT963459; SbCHS-1, KT963460; SbCHS-2, KT963461; SbCHI, KT963462.
Full-length cDNAs of SbFNSII-1 and SbFNSII-2 were cloned into plasmid pYesdest52 for yeast expression. SbFNSII-2 cDNA was cloned into plasmid pK7WG2R (60, 65) for Arabidopsis transformation. SbCLL-1, SbCLL-5, SbCLL-6, SbCLL-7, and SbCLL-8 cDNAs were cloned into plasmid pDest17 for expression in E. coli. The full-length cDNAs of SbCLL-7, SbCHS-2, SbCHI, and SbFNSII-2 were cloned into pEAQ-HT-DEST1 for tobacco leaf infiltration (45). All constructs were made using the Gateway LR Clonase II enzyme mix (Thermo Fisher) according to the manufacturer’s protocols.
Phylogenetic analysis
Amino acid sequences were aligned using the Clustal X2 program (66). The ML trees were built using MEGA6 (Molecular Evolutionary Genetics Analysis version 6.0) (67) with the following options: 1000 bootstrap replications, Poisson substitution model, uniform rates, partial deletion for gaps/missing data, 95% site coverage cutoff, and a strong branch swap filter.
RNAi
Nonhomologous DNA regions of SbFNSII-1, SbFNSII-2, and SbCLL-7 were amplified with the primers listed in table S1. The PCR products were first subcloned into plasmid pDONR207 as previously described, and then cloned into plasmid pK7WGIGW2R (65) using the Gateway LR Clonase II enzyme mix (Thermo Fisher) according to the manufacturer’s instructions. The vectors for RNAi were introduced into A. rhigogenes A4 by electroporation. Successful transformants were screened on LB solid medium supplemented with spectinomycin (50 mg liter−1) and kanamycin (50 mg liter−1).
Quantitative reverse transcription polymerase chain reaction
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized from 2 μg of total RNA using the adaptor oligo(dT)17 primer plus random primers (31) (Sigma) and SuperScript III (Invitrogen). qRT-PCR was performed using GSPs as shown in table S1, using procedures described previously (64).
Yeast strains, growth, and in vivo assays for enzyme activity
S. cerevisiae WAT11 (39, 68) was used as the host strain for the expression of candidate Cyt p450 genes FNSII-1 and FNSII-2 in pYesdest52 Gateway Vector. The constructs, as well as an empty vector, were transformed into yeast, and the proteins were induced as described in the pYesdest52 instruction manual (Thermo Fisher). Transformants were selected on synthetic drop-out medium –Ura (SD–Ura) containing glucose (20 g liter−1) and grown at 28°C for 48 hours. The resulting recombinant strains were initially grown in SD−His liquid medium with glucose (20 g liter−1) at 28°C for about 24 hours to an OD600 (optical density at 600 nm) of 2 to 3. Cells were centrifuged and washed with sterile water to remove residual glucose. The cells were resuspended in the SD-Ura containing galactose (20 g liter−1) to induce expression of the target proteins. Two hours later, cultures were supplemented with 100 μM pinocembrin or narigenin. After 24 hours, the cells were harvested by centrifugation, ice-dried, and extracted with MeOH for metabolite analysis.
FNSII enzyme assays and kinetics
Yeast strains were grown, and target proteins were induced as described above. Microsomal proteins were isolated using the procedure described by Truan et al. (68). Microsomal proteins were suspended in storage buffer containing 50 mM tris-HCl (pH 7.5), 1 mM EDTA, and 20% (v/v) glycerol, and were adjusted to a final concentration of 10 to 20 mg ml−1 using the Bradford assay (69).
For the enzyme assay for FNSII (70), the incubation mixture (final volume, 200 ml) contained 100 mM tris-HCl (pH, 7.9), 50 μM substrate (pinocembrin, naringenin, or eriodictyol), 0.5 mM reduced glutathione, and 2.5 mg of crude protein extract. The reactions were initiated by addition of NADPH. The assays were incubated for 6 hours at 28°C, and the reactions were quenched by addition of MeOH (to a final concentration of 70%). Extracts were filtered and analyzed by HPLC. Assays without NADPH or with microsomes from yeast harboring the empty vector were used as controls.
For kinetics measurements, pinocembrin or naringenin were used at concentrations ranging from 0.1 to 30 μM. Reaction time was reduced to 30 min. Km and Vmax values were calculated from the Eadie-Hofstee plot.
CLL protein expression and enzyme assays
All Scutellaria CLL cDNAs were cloned into pDest17 and expressed in E. coli strain BL21 using the expression vector pDEST17 and thus contained a His6 tag at their N termini. E. coli transformants were grown overnight in 5 ml of LB medium with ampicillin (100 mg liter−1) at 37°C. The cultures were then grown in 200 ml of fresh medium for about 3 hours. To induce expression, cells were grown for 3 hours in the presence of 1 mM isopropyl-1-thio-β-d-galactopyranoside and collected by centrifugation.
For protein purification, pelleted cells were suspended in 5 ml of 50 mM sodium phosphate (pH, 7.8) containing 300 mM NaCl (buffer A) plus 2 mM β mercaptoethanol, 20% glycerol, and 10 mM imidazole, and lysed by sonification; insoluble components were removed by centrifugation (20,000g for 20 min at 4°C). Ni–nitrilotriacetic acid (NTA) agarose (1 ml) (Qiagen) equilibrated with buffer A was added to the supernatant, and for adsorption of the His6-tagged proteins, the suspension was stirred on ice for 1 hour. The Ni-NTA agarose was washed with 30 ml of buffer B [50 mM sodium phosphate (pH, 7.8) containing 300 mM NaCl, 20 mM imidazole, 15% glycerol, and 2 mM β mercaptoethanol] and washed three times with 40 ml of 50 mM sodium phosphate (pH, 7.5), 300 mM NaCl, 30 mM imidazole, and 15% glycerol (buffer B). Subsequently, the agarose was packed to a column, and the protein was eluted by buffer C, 50 mM sodium phosphate (pH, 7.8) containing 300 mM NaCl, 250 mM imidazole, 20% glycerol, and 2 mM β mercaptoethanol. Fractions (1 ml) were collected, and imidazole was removed by ultrafiltration. Protein concentrations were determined using the Bradford assay with bovine serum albumin as a standard (69). The samples were analyzed by SDS–polyacrylamide gel electrophoresis and Western blots (71).
4CL activity was determined with the spectrophotometric assay as previously described (28). The change in absorbance was monitored at 311, 333, and 363 nm according to the absorption maxima for cinnamoyl-CoA, 4-coumaroyl–CoA, and caffeoyl-CoA, respectively (72). Enzyme assays were tested in a system of 0.3 μM CoA, 5 μM MgCl2, 50 mM tris-HCl (pH, 7.8), 5 μM adenosine 5′-triphosphate, and protein. Phenolic substrates were used at concentrations ranging from 0.1 to 4 mM (cinnamic acid) and from 0.005 to 0.8 mM (all other substrates). The kinetic constants Km and Vmax for the phenolic substrates were determined at fixed concentrations of all other substrates by linear regression of v against v/s (Eadie-Hofstee plot). Similar values were obtained from Hanes plots (v/s against s). Each plot contained at least six points.
Arabidopsis transformation and metabolite assays
A. thaliana ecotype Columbia was used in transformation. Seeds were sown on soil and stratified at 4°C for 2 days before transfer to a growth chamber maintained under long-day (16-hour light, 8-hour dark) conditions. Temperature and humidity were maintained at 23°C and 50%.
Binary vector pK7WG2R, for expression of FNSII-2 under the control of the CaMV 35S promoter or the empty vector, was introduced into Agrobacterium tumefaciens strain GV3101 pMP90 by electroporation, and transformation of A. thaliana by the resulting bacteria was performed by in planta infiltration (73). Transformed seedlings (T1) were identified by selection on Murashige and Skoog solid medium containing kanamycin (50 mg liter−1) and were transferred to soil. Positive transgenic plants were confirmed by genomic PCR. Homozygous T3 plants were used in further studies.
For substrate feeding experiments, 2-week-old sterile Arabidopsis seedlings were transferred to 100 ml of Murashige and Skoog solid medium, with or without pinocembrin or naringenin, and cultured under long-day conditions for 3 weeks. The resulting seedlings were washed with distilled water three times, freeze-dried, and ground to a fine powder. Arabidopsis samples of 10 mg DW were extracted for metabolites in 1 ml of 70% methanol, and then sonicated in an ultrasonic water bath at room temperature for 1 hour. The resulting extract was centrifuged at 12,000g for 5 min at 4°C; the supernatant was used for acid hydrolysis. An equal volume of 2 N HCl was added to the samples for incubation at 90°C for 1 hour. Filtered samples (10 μl) were used for LC-MS analysis.
Pathway reconstitution in N. benthamiana
Full-length cDNAs of GFP (as a control), SbCLL-7, SbCHS-2, SbCHI, and SbFNSII-2 were cloned into the CPMV-HT (HyperTrans) plasmid pEAQ-HT-DEST1 (45) and transformed into A. tumefaciens strain GV3101 pMP90. Leaves of N. benthamiana plants were infiltrated as previously described (74). Infiltrated leaves were harvested 7 days after infiltration, and metabolites were extracted and analyzed as described for Arabidopsis samples.
Statistics
All experiments were repeated using at least three biological replicates. Data are presented as means ± SEM unless otherwise stated. To compare group differences, paired or unpaired, two-tailed Student’s t tests were used. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
Funding:This work was supported by a China Scholarship Council (CSC) award to Q.Z.; Chinese Academy of Sciences (CAS)/John Innes Centre (JIC) and Centre of Excellence for Plant and Microbial Sciences (CEPAMS) joint foundation support to Q.Z., H.X., X.-Y.C., and C.M. Q.Z., C.M., L.H., and Y.Z. were also supported by the Institute Strategic Program, “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004596/1), from the Biotechnology and Biological Sciences Research Council (BBSRC) to JIC. Y.Z. was supported in part by the BBSRC Synthetic Biology Research Centre “OpenPlant” award (BB/L014130/1). Author contributions: C.M. and Q.Z. designed the research; Q.Z., Y.Z., and L.H. performed the research; C.M., Q.Z., Y.Z., G.W., L.H., J.-K.W., X.-Y.C., and H.X. analyzed and interpreted the data; Q.Z. and C.M. wrote the paper with significant input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The RNA-seq data for this work have been deposited in the SRA database under BioProject ID: PRJNA300475, with the accession numbers: study ID, SRP068883; sample ID, SRS1263237; experiment ID, SRX1546377; run ID, SRR3123399. The sequences for all the genes described in this paper have been submitted to the NCBI database with the accession numbers: SbFNSII-1, KT963453; SbFNSII-2, KT963454; SbCLL-1, KT963455; SbCLL-5, KT963456; SbCLL-6, KT963457; SbCLL-7, KT963458; SbCLL-8, KT963459; SbCHS-1, KT963460; SbCHS-2, KT963461; SbCHI, KT963462. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Any additional data related to this paper may be requested from C.M. (cathie.martin@jic.ac.uk).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1501780/DC1
Fig. S1. Multiple alignment of CPY93B6, CPY93B24, and CPY93B25.
Fig. S2. Flavone accumulation patterns in S. baicalensis.
Fig. S3. RNAi of SbFNSII-2 in hairy root cultures of S. baicalensis.
Fig. S4. In vitro assay of SbFNSII-1 and SbFNSII-2 and in vivo assay of SbFNSII-2.
Fig. S5. HPLC metabolite profiles of Arabidopsis plants carrying empty vector or a representative SbFNSII-2 line, grown on MS with or without supplementation of pinocembrin.
Fig. S6. Western blot analysis of the recombinant SbCLLs and SPB domain analysis of At4CLs and SbCLL-7.
Fig. S7. Metabolite profiles by HPLC from empty vector line and a representative SbCLL-7 RNAi line.
Fig. S8. Metabolite profiles of HPLC analysis of infiltrated N. benthamiana leaves.
Fig. S9. Transcript levels of SbC4H relative to actin.
Table S1. Primers used in this study. Underlined sequences mean recombination cites for Gateway cloning.
REFERENCES AND NOTES
- 1.S. Li, in Compendium of Materia Medica (Bencao Gangmu) (Huaxia Press, China, 2012), pp 543–546. [Google Scholar]
- 2.Pan M. Q., Li Y. H., Liu J. A., Tan Y. X., Reports for 80 patients with bronchial lung squamous carcinoma (mid or late stage) treated by Lung FuFang and chemotherapy. Journal of Traditional Chinese Medicine and Pharmacy 5, 19–21 (1990). [Google Scholar]
- 3.Pan M. Q., Li Y. H., Jiang Y. L., Clinical observation of old people NSCLC (mid or late stage) treated by Lung FuFang combined with chemotherapy. Shanxi Traditional Chinese Medicine 31, 389–390 (2000). [Google Scholar]
- 4.Duan X., Jia C. F., Duan M., Treatmenf of non-small-cell lung cancer by FuZheng anti-cancer prescription combined with chemotherapy. Shanxi Traditional Chinese Medicine 35, 311–312 (2014). [Google Scholar]
- 5.Li-Weber M., New therapeutic aspects of flavones: The anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treat. Rev. 35, 57–68 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Wang X., Wang X., Xu Z., Liu Z., Ni Q., Chu X., Qiu M., Zhao A., Jia W., Protective effect of flavonoids from Scutellaria baicalensis Georgi on cerebral ischemia injury. J. Ethnopharmacol. 108, 355–360 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Sak K., Cytotoxicity of dietary flavonoids on different human cancer types. Pharmacogn. Rev. 8, 122–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang M. D., Chiang Y.-M., Higashiyama R., Asahina K., Mann D. A., Mann J., Wang C. C. C., Tsukamoto H., Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor γ in hepatic stellate cells for their antifibrotic effect. Hepatology 55, 1271–1281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox J. T., Sakamuru S., Huang R. L., Teneva N., Simmons S. O., Xia M. H., Tice R. R., Austin C. P., Myung K., High-throughput genotoxicity assay identifies antioxidants as inducers of DNA damage response and cell death. Proc. Natl. Acad. Sci. U.S.A. 109, 5423–5428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himeji M., Ohtsuki T., Fukazawa H., Tanaka M., Yazaki S.-i., Ui S., Nishio K., Yamamoto H., Tasaka K., Mimura A., Difference of growth-inhibitory effect of Scutellaria baicalensis-producing flavonoid wogonin among human cancer cells and normal diploid cell. Cancer Lett. 245, 269–274 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Huang S.-T., Wang C.-Y., Yang R.-C., Chu C.-J., Wu H.-T., Pang J.-H. S., Wogonin, an active compound in Scutellaria baicalensis, induces apoptosis and reduces telomerase activity in the HL-60 leukemia cells. Phytomedicine 17, 47–54 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Nayak M. K., Agrawal A. S., Bose S., Naskar S., Bhowmick R., Chakrabarti S., Sarkar S., Chawla-Sarkar M., Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 69, 1298–1310 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Baumann S., Fas S. C., Giaisi M., Müller W. W., Merling A., Gülow K., Edler L., Krammer P. H., Li-Weber M., Wogonin preferentially kills malignant lymphocytes and suppresses T-cell tumor growth by inducing PLCγ1- and Ca2+-dependent apoptosis. Blood 111, 2354–2363 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Parajuli P., Joshee N., Rimando A. M., Mittal S., Yadav A. K., In vitro antitumor mechanisms of various Scutellaria extracts and constituent flavonoids. Planta Med. 75, 41–48 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Ferreyra M. L. F., Rius S. P., Casati P., Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens S., Mithöfer A., Flavones and flavone synthases. Phytochemistry 66, 2399–2407 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Islam M. N., Downey F., Ng C. K. Y., Comparative analysis of bioactive phytochemicals from Scutellaria baicalensis, Scutellaria lateriflora, Scutellaria racemosa, Scutellaria tomentosa and Scutellaria wrightii by LC-DAD-MS. Metabolomics 7, 446–453 (2011). [Google Scholar]
- 18.Xu H., Park N. I., Li X. H., Kim Y. K., Lee S. Y., Park S. U., Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 101, 9715–9722 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Park N. I., Xu H., Li X., Kim S.-J., Park S. U., Enhancement of flavone levels through overexpression of chalcone isomerase in hairy root cultures of Scutellaria baicalensis. Funct. Integr. Genomics 11, 491–496 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Schneider K., Hovel K., Witzel K., Hamberger B., Schomburg D., Kombrink E., Stuible H. P., The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. U.S.A. 100, 8601–8606 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S., Kaminaga Y., Cooper B., Pichersky E., Dudareva N., Chapple C., Benzoylation and sinapoylation of glucosinolate R-groups in Arabidopsis. Plant J. 72, 411–422 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Klempien A., Kaminaga Y., Qualley A., Nagegowda D. A., Widhalm J. R., Orlova I., Shasany A. K., Taguchi G., Kish C. M., Cooper B. R., D’Auria J. C., Rhodes D., Pichersky E., Dudareva N., Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell 24, 2015–2030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britsch L., Purification and characterization of flavone synthase I, a 2-oxoglutarate-dependent desaturase. Arch. Biochem. Biophys. 282, 152–160 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Martens S., Forkmann G., Matern U., Lukačin R., Cloning of parsley flavone synthase I. Phytochemistry 58, 43–46 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Lee Y. J., Kim J. H., Kim B. G., Lim Y., Ahn J.-H., Characterization of flavone synthase I from rice. BMB Rep. 41, 68–71 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Dixon R. A., Paiva N. L., Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Kim J. I., Pysh L., Chapple C., Four isoforms of Arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehlting J., Büttner D., Wang Q., Douglas C. J., Somssich I. E., Kombrink E., Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19, 9–20 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Shockey J. M., Fulda M. S., Browse J., Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 132, 1065–1076 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q., Bonness M. S., Liu M., Seradge E., Dixon R. A., Mabry T. J., Enzymes of B-ring-deoxy flavonoid biosynthesis in elicited cell cultures of “old man” cactus (Cephalocereus senilis). Arch. Biochem. Biophys. 321, 397–404 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Frohman M. A., Dush M. K., Martin G. R., Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. U.S.A. 85, 8998–9002 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitada C., Gong Z., Tanaka Y., Yamazaki M., Saito K., Differential expression of two cytochrome P450s involved in the biosynthesis of flavones and anthocyanins in chemo-varietal forms of Perilla frutescens. Plant Cell Physiol. 42, 1338–1344 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Nakatsuka T., Nishihara M., Mishiba K., Yamamura S., Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol. Breeding 17, 91–99 (2006). [Google Scholar]
- 34.Akashi T., Fukuchi-Mizutani M., Aoki T., Ueyama Y., Yonekura-Sakakibara K., Tanaka Y., Kusumi T., Ayabe S.-i., Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol. 40, 1182–1186 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Nakatsuka T., Nishihara M., Mishiba K., Yamamura S., Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci. 168, 1309–1318 (2005). [Google Scholar]
- 36.Berim A., Gang D. R., The roles of a flavone-6-hydroxylase and 7-O-demethylation in the flavone biosynthetic network of sweet basil. J. Biol. Chem. 288, 1795–1805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gafner S., Bergeron C., Batcha L. L., Angerhofer C. K., Sudberg S., Sudberg E. M., Guinaudreau H., Gauthier R., Analysis of Scutellaria lateriflora and its adulterants Teucrium canadense and Teucrium chamaedrys by LC–UV/MS, TLC, and digital photomicroscopy. J. AOAC Int. 86, 453–460 (2003). [PubMed] [Google Scholar]
- 38.Lin L. Z., Harnly J. M., Upton R., Comparison of the phenolic component profiles of skullcap (Scutellaria lateriflora) and germander (Teucrium canadense and T. chamaedrys), a potentially hepatotoxic adulterant. Phytochem Anal. 20, 298–306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pompon D., Louerat B., Bronine A., Urban P., Yeast expression of animal and plant P450s in optimized redox environments. Method Enzymol. 272, 51–64 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Li L., Cheng H., Gai J., Yu D., Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 226, 109–123 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Morita H., Takahashi Y., Noguchi H., Abe I., Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochem. Biophys. Res. Commun. 279, 190–195 (2000). [DOI] [PubMed] [Google Scholar]
- 42.H. Morita, H. Noguchi, T. Akiyama, M. Shibuya, Y. Ebizuka, Chalcone synthase from Scutellaria baicalensis, in Towards Natural Medicine Research in the 21st Century (Elsevier, Tokyo, 1998), pp. 401–409. [Google Scholar]
- 43.Zhou Y., Nagashima S., Hirotani M., Suzuki H., Yoshikawa T., Expression of the chalcone synthase gene in Scutellaria baicalensis hairy root cultures was unusually reduced by environmental stresses. Plant Biotechnol. 20, 207–214 (2003). [Google Scholar]
- 44.Lei W., Tang S.-H., Luo K.-M., Sun M., Molecular cloning and expression profiling of a chalcone synthase gene from hairy root cultures of Scutellaria viscidula Bunge. Genet. Mol. Biol. 33, 285–291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peyret H., Lomonossoff G. P., The pEAQ vector series: The easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83, 51–58 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Jez J. M., Bowman M. E., Dixon R. A., Noel J. P., Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7, 786–791 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Weng J.-K., Li Y., Mo H., Chapple C., Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337, 960–964 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Meng L., Lozano Y., Bombarda I., Gaydou E., Li B., Anthocyanin and flavonoid production from Perilla frutescens: Pilot plant scale processing including cross-flow microfiltration and reverse osmosis. J. Agr. Food Chem. 54, 4297–4303 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Tai M. C., Tsang S. Y., Chang L. Y. F., Xue H., Therapeutic potential of wogonin: A naturally occurring flavonoid. CNS Drug Rev. 11, 141–150 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin L., Markham K. R., Paré P. W., Dixon R. A., Mabry T. J., Flavonoids from elicitor-treated cell suspension cultures of Cephalocereus senilis. Phytochemistry 32, 925–928 (1993). [Google Scholar]
- 51.Chen L.-J., Games D. E., Jones J., Isolation and identification of four flavonoid constituents from the seeds of Oroxylum indicum by high-speed counter-current chromatography. J. Chromatogr. A 988, 95–105 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Samuelsen A. B., The traditional uses, chemical constituents and biological activities of Plantago major L.: A review. J. Ethnopharmacol. 71, 1–21 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pichersky E., Lewinsohn E., Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Zhang J. A., Subramanian S., Zhang Y., Yu O., Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 144, 741–751 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ayabe S.-i., Akashi T., Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 5, 271–282 (2006). [Google Scholar]
- 56.Hirotani M., Kuroda R., Suzuki H., Yoshikawa T., Cloning and expression of UDP-glucose: Flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210, 1006–1013 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Fliegmann J., Furtwängler K., Malterer G., Cantarello C., Schüler G., Ebel J., Mithöfer A., Flavone synthase II (CYP93B16) from soybean (Glycine max L.). Phytochemistry 71, 508–514 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Schneider K., Kienow L., Schmelzer E., Colby T., Bartsch M., Miersch O., Wasternack C., Kombrink E., Stuible H.-P., A new type of peroxisomal acyl-coenzyme a synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 280, 13962–13972 (2005). [DOI] [PubMed] [Google Scholar]
- 59.D. B. Haytowitz, S. Bhagwat, J. Harnly, J. M. Holden, S. E. Gebhardt. Sources of Flavonoids in the U.S. Diet Using USDA’s Updated Database on the Flavonoid Content of Selected Foods, www.ars.usda.gov/SP2UserFiles/Place/12354500/Articles/AICR06_flav.pdf [accessed 18 October 2015].
- 60.Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J. F., Harris J. M., Oldroyd G. E. D., Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20, 2681–2695 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M., Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J., WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Luo J., Nishiyama Y., Fuell C., Taguchi G., Elliott K., Hill L., Tanaka Y., Kitayama M., Yamazaki M., Bailey P., Parr A., Michael A. J., Saito K., Martin C., Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50, 678–695 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J., Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Truan G., Cullin C., Reisdorf P., Urban P., Pompon D., Enhanced in vivo monooxygenase activities of mammalian P450s in engineered yeast cells producing high levels of NADPH-P450 reductase and human cytochrome b5. Gene 125, 49–55 (1993). [DOI] [PubMed] [Google Scholar]
- 69.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 70.Martens S., Forkmann G., Genetic control of flavone synthase II activity in flowers of Gerbera hybrids. Phytochemistry 49, 1953–1958 (1998). [Google Scholar]
- 71.Burnette W. N., “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 (1981). [DOI] [PubMed] [Google Scholar]
- 72.J. Stockigt, M. H. Zenk, in Zeitschrift fur Naturforschung. Section C: Biosciences (Walter de Gruyter, Berlin, 1975), vol. 30, pp. 352–358. [DOI] [PubMed] [Google Scholar]
- 73.Bent A. F., Arabidopsis in planta transformation. Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 124, 1540–1547 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geisler K., Hughes R. K., Sainsbury F., Lomonossoff G. P., Rejzek M., Fairhurst S., Olsen C.-E., Motawia M. S., Melton R. E., Hemmings A. M., Bak S., Osbourn A., Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc. Natl. Acad. Sci. U.S.A. 110, E3360–E3367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/4/e1501780/DC1
Fig. S1. Multiple alignment of CPY93B6, CPY93B24, and CPY93B25.
Fig. S2. Flavone accumulation patterns in S. baicalensis.
Fig. S3. RNAi of SbFNSII-2 in hairy root cultures of S. baicalensis.
Fig. S4. In vitro assay of SbFNSII-1 and SbFNSII-2 and in vivo assay of SbFNSII-2.
Fig. S5. HPLC metabolite profiles of Arabidopsis plants carrying empty vector or a representative SbFNSII-2 line, grown on MS with or without supplementation of pinocembrin.
Fig. S6. Western blot analysis of the recombinant SbCLLs and SPB domain analysis of At4CLs and SbCLL-7.
Fig. S7. Metabolite profiles by HPLC from empty vector line and a representative SbCLL-7 RNAi line.
Fig. S8. Metabolite profiles of HPLC analysis of infiltrated N. benthamiana leaves.
Fig. S9. Transcript levels of SbC4H relative to actin.
Table S1. Primers used in this study. Underlined sequences mean recombination cites for Gateway cloning.


