Summary
Dysregulation of sleep or feeding has enormous health consequences. In humans, acute sleep loss is associated with increased appetite and insulin insensitivity, while chronically sleep-deprived individuals are more likely to develop obesity, metabolic syndrome, type II diabetes, and cardiovascular disease. Conversely, metabolic state potently modulates sleep and circadian behavior; yet, the molecular basis for sleep-metabolism interactions remains poorly understood. Here, we describe the identification of translin (trsn), a highly conserved RNA/DNA binding protein, as essential for starvation-induced sleep suppression. Strikingly, trsn does not appear to regulate energy stores, free glucose levels, or feeding behavior suggesting the sleep phenotype of trsn mutant flies is not a consequence of general metabolic dysfunction or blunted response to starvation. While broadly expressed in all neurons, trsn is transcriptionally upregulated in the heads of flies in response to starvation. Spatially restricted rescue or targeted knockdown localizes trsn function to neurons that produce the tachykinin-family neuropeptide Leucokinin. Manipulation of neural activity in Leucokinin neurons revealed these neurons to be required for starvation-induced sleep suppression. Taken together, these findings establish trsn as an essential integrator of sleep and metabolic state, with implications for understanding the neural mechanism underlying sleep disruption in response to environmental perturbation.
Results and Discussion
In humans, sleep and feeding are tightly interconnected, and pathological disturbances of either process are associated with metabolism-related disorders. Acute sleep loss correlates with increased appetite and insulin insensitivity, while chronically sleep-deprived individuals are more likely to develop obesity, metabolic syndrome, type II diabetes, and cardiovascular disease [1–3]. Conversely, in humans and rodents internal metabolic state potently modulates sleep and circadian behavior [4–6]. Despite the widespread evidence for interactions between sleep loss and metabolic dysfunction, little is known about how these processes integrate within the brain.
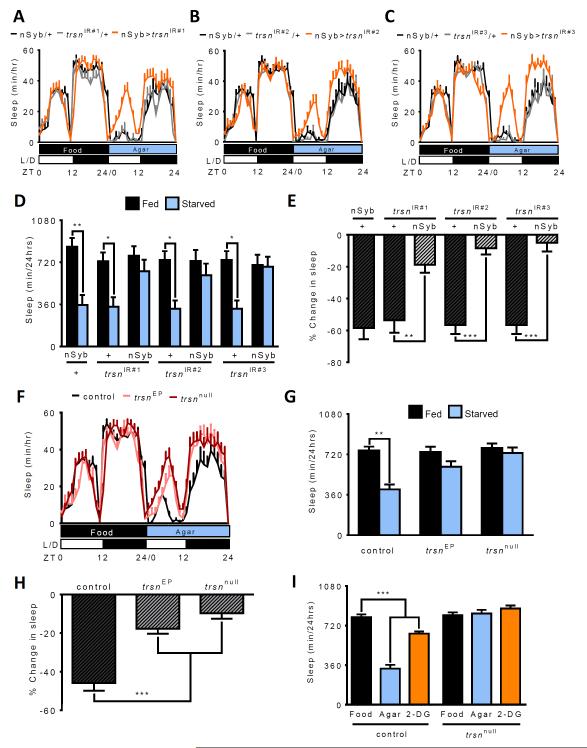
To address this question, we sought to identify integrators of sleep and metabolic state in the fruit fly, Drosophila melanogaster. Knockdown of genes from randomly selected RNAi lines was achieved by expression of UAS-RNAi under the control of the neuron-specific GAL4 driver, n-Synaptobrevin-GAL4 (nSyb-GAL4) [7, 8]. Following 24 hours of baseline sleep measurements on food, sleep was measured during 24 hours starvation on agar and the change in sleep was calculated as previously described [9]. Starvation-induced sleep suppression was reduced in flies with neuron-specific knockdown of the RNA/DNA binding protein translin (trsn) (Figure 1A). To confirm the effect of trsn-RNAi on sleep, we tested two additional RNAi transgenes. All three RNAi lines showed similar phenotypes; trsn knockdown flies slept similarly to control flies on food, while sleep loss resulting from starvation was reduced or absent (Figure 1B-E). Targeted knockdown of trsn in the fat body (yolk-GAL4) or muscle (24b-GAL4), two tissues involved in energy storage, showed normal sleep suppression in response to starvation (Figure S1A) supporting the notion that trsn functions primarily in neurons to regulate sleep.
Figure 1. trsn is required for metabolic regulation of sleep.
A-C. Sleep profile for hourly sleep averages over a 48 hour experiment. Flies are on food for day 1, then transferred to agar for day 2. Sleep does not differ between any of the groups for day 1. The trsn knockdown groups (nSyb>trsn; orange) sleep more than nSyb-GAL4/+ (black) and trsnIR/+ controls (grey) during day 2 (starved). D Control flies (nSyb-GAL4/+ and trsnIR/+) sleep significantly more on food (black) than when starved (blue, N≥36; P<0.001) while no signficant differences in sleep duration are observed in flies where nSyb-GAL4 drives expression of trsnIR#1 (N=45; P>0.98), trsnIR#2 (N=45; P>0.99), or trsnIR#3 (N=36; P>0.98). E. Quantifying the percentage change in sleep between fed (day 1) and starved (day 2) states reveals sigificantly greater sleep loss in nSyb-GAL4/+ controls (nSyb-Gal4/+ vs trsnIR#1/+, N≥38; P>0.95; nSyb-Gal4/+ vs trsnIR#2/+, N≥39; P>0.99; nSyb-Gal4/+ vs trsnIR#3/+, N≥37; P>0.99) compared to all three lines with neuronal expression of trsnIR#1(N≥38; P<0.01), trsnIR#2 (N≥39; P<0.001) and trsnIR#3 (N≥36; P<0.01). F. Sleep profile over 48 hours reveals that sleep in trsnEP and trsnnull does not differ from w1118 control flies on food. Both trsnEP and trsnnull mutant flies sleep more than control flies on agar. G. Sleep is significantly reduced in starved control flies (N≥54; P<0.001), while sleep differences are not significant in trsnEP (N=69; P>0.23) or trsnnull flies (N=58; P>0.98). H. Percentage sleep loss is also significantly reduced in trsnEP and trsnnull mutants compared to controls (N≥54; P<0.001). I) In control flies, sleep is significantly reduced in flies on agar (blue; N=44; P<0.001) or food laced with 2-deoxyglucose (2-DG; orange) (N≥64; P<0.001), compared to flies fed standard food (black). No differences are detected between flies fed standard food compared to agar or 2-DG in trsnnull mutants (N≥38; P>0.70). Bars for % change in sleep are mean ± SEM by one-way ANOVA. All other bars are mean ± SEM; P<0.01,**; P<0.001,*** by 2-way ANOVA. See also Figure S1.
In Drosophila, starvation induces hyperactivity in addition to sleep loss [10–12]. To determine whether trsn also regulates the hyperactivity response to starvation, we analyzed waking activity in fed and starved trsn knockdown flies. Neuronal knockdown of trsn had no effect on waking activity in fed flies, but reduced starvation-induced hyperactivity (Figure S1B). These findings are consistent with the notion that trsn does not modulate sleep or activity in the fed state, but is required for both sleep and locomotor changes that result from starvation.
To validate that the sleep phenotype in trsn knockdown flies was not due to off-target effects of RNAi, we measured sleep in flies with a mutation in the trsn locus. Both male and female flies with a P-element insertion in the trsn locus (trsnEP) or the excision allele (trsnnull) are viable [13] and exhibit reduced sleep suppression during starvation (Figure 1F-H and S1C-D), phenocopying flies with neuron-specific RNAi knockdown. The waking activity of trsnnull flies phenocopies RNAi knockdown flies under fed conditions, while starvation-induced hyperactivity is blunted or absent in trsn mutants (Figure S1E).
A number of systems have been developed for high-resolution video tracking that may provide a more accurate measure of sleep compared to infrared-based monitoring systems [14–17]. Tracking analysis revealed that w1118 control, but not trsnnull flies, suppress sleep during starvation, confirming that the results obtained using infrared tracking are not an artifact of the sleep acquisition system (Figure S1F). Taken together, these findings indicate starvation-induced sleep suppression and locomotor activity are reduced in trsn mutant flies.
Starved flies utilize glucose and fatty acids to maintain metabolic homeostasis, and the availability of these energy sources may regulate sleep. To determine the energy source required for normal sleep, we fed flies the glycolysis inhibitor 2-Deoxyglucose (2-DG) [18], or the carnitine palmitoyltransferase antagonist, etomoxir, an inhibitor of fatty acid β-oxidation [19]. Treatment with both of these drugs has been used extensively in mammals, and these inhibitors have similar effects on fly metabolism [20, 21]. Flies were fed standard food laced with 400mM 2-DG or 25μM etomoxir and monitored for sleep to determine whether the breakdown products of glucose or triglyceride stores (or both) contribute to reduced sleep during starvation. Flies fed 2-DG, but not etomoxir, significantly reduced sleep, suggesting that reduced glucose availability or the energy derived from its metabolism, rather than fatty acids, contribute to sleep suppression (Figure 1I; and data not shown). When trsn mutant flies were subjected to the same protocol, no changes in sleep were observed with 2-DG feeding (Figure 1I). The finding that trsn mutant flies are insensitive to sleep regulation in response to both acute food deprivation and pharmacological perturbation of energy utilization, suggests trsn is critical for the integration of sleep and metabolic state.
It is possible that the reduced ability of trsn mutants to suppress sleep during starvation stems from a general inability to modulate sleep in response to environmental or pharmacological disruption. To test this, sleep rebound was determined by mechanically shaking flies at 3-4 minute intervals for 12 hours during the night (ZT12-ZT24) and measuring sleep for 12 hours (ZT0-ZT12) the following day. Sleep-deprived trsnnull flies showed a significant increase in daytime sleep that was not present in undisturbed controls (Figure S1G). The sleep rebound in trsnnull flies was comparable to w1118 control flies indicating that trsn is dispensable for the homeostatic response to mechanical sleep deprivation (Figure S1G). In addition to mechanical deprivation, numerous pharmacological agents including the stimulant caffeine and free-radical inducing agent paraquat, disrupt sleep in flies [22, 23]. Both w1118 control and trsnnull flies significantly reduced sleep when fed food laced with caffeine (Figure S1H) or paraquat (Figure S1I), supporting the notion that the loss of starvation-induced sleep suppression in trsn mutant flies does not result from a generalized inability to suppress sleep.
Flies with enhanced energy stores do not suppress sleep or increase activity in response to starvation [11, 21]. Drosophila primarily stores energy as triglycerides and glycogen, and prolonged food-deprivation results in depletion of both stores. To test the possibility that trsn mutant flies do not suppress sleep when fasted due to increased energy stores, we measured triglyceride and glycogen levels using colorimetric assays standardized to total protein level [24, 25]. No differences in glycogen, triglyceride or free glucose levels were observed between fed or 24 hour starved trsnnull flies and w1118 controls (Figure S2A-C), indicating that the loss of starvation-induced sleep suppression in trsn mutant flies is not due to an increase in energy stores.
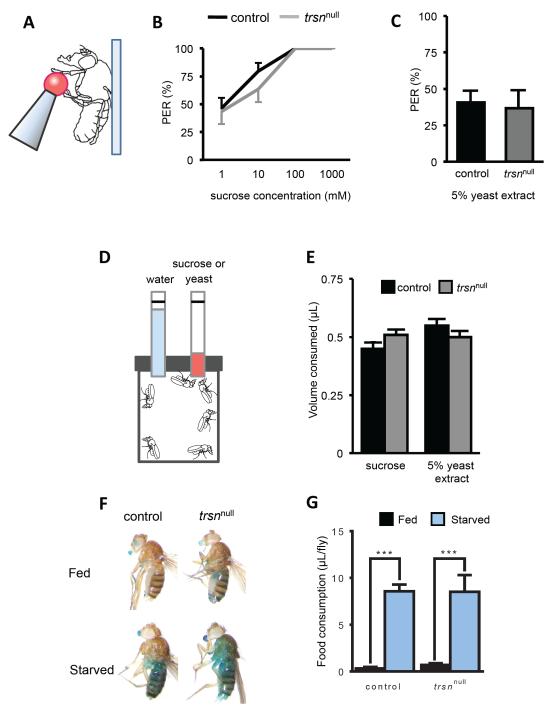
Many metabolism-related genes regulate both sleep and feeding [26], raising the possibility that trsn is generally required for hunger-dependent behaviors. To determine whether trsn modulates reflexive food acceptance response, we measured the Proboscis Extension Reflex (PER) of flies starved for 24 hours prior to behavioral testing (Figure 2A) [27, 28]). Total PER response did not differ between starved trsnnull and w1118 flies to sucrose concentrations of ranging from 1 to 1000 mM (Figure 2B), or 5% yeast extract (Figure 2C), indicating that trsn is dispensable for reflexive feeding. To measure food consumption, we provided flies with 100mM sucrose or 5% yeast extract in the Capillary Tube Feeding (CAFÉ) assay (Figure 2D) [29]. Flies were starved for 24 hours prior to the start of the assay and consumption was measured over 12 hours. No differences in total consumption of 100 mM sucrose or 5% yeast extract was detected between control and trsnnull flies (Figure 2E). To quantify feeding over a shorter timeframe, the blue-dye assay was used to determine the quantity of food consumed in fed and 24 hour starved flies over a 30 minute period [30]. No differences between control and trsnnull flies were detected in overall consumption in the fed or starved state, indicating that trsn does not regulate acute food consumption (Figure 2F-G). Taken together, three independent feeding assays indicate that trsn does not regulate feeding behavior during the starved state.
Figure 2. Starvation-induced feeding is normal in trsn mutant flies.
A. Diagram of the proboscis extension reflex (PER) assay. Tastant is supplied to the tarsi of a tethered female fly. B, C. No significant differences in PER are detected between control (black) and trsnnull mutants (grey) to increasing concentrations of sucrose (N≥10; 1mM, P>0.84; 10mM, P>0.21; 100mM and 1000mM P>0.95) (B) or 5% yeast extract (N=18; P>0.98) (C). D. Diagram of the Capillary Feeder Assay (CAFÉ) assay. Flies are presented with one capillary containing 100mM sugar or 5% yeast extract and a second containing water. E. No significant differences in sucrose (left bars, N=4; P>0.34) or yeast (right bars, N>4; P>0.18) were detected between control and trsnnull flies when presented with each tastant. F. Starved or fed flies are placed on food containing blue dye for 30 minutes and consumption is measured. Representative images of flies following the assay show increased consumption in starved control and trsnnull mutants compared to fed controls. G. Quantification of food intake reveals a significant increase in starved controls and trsnnull flies compared to fed flies from each genotype (N≥26; P<0.001). No differences were observed between genotypes in the fed (N≥29;P>0.99) or starved (N≥26;P>0.99) states All bars are mean ± SEM; P<0.001,*** by 2-way ANOVA. See also Figure S2.
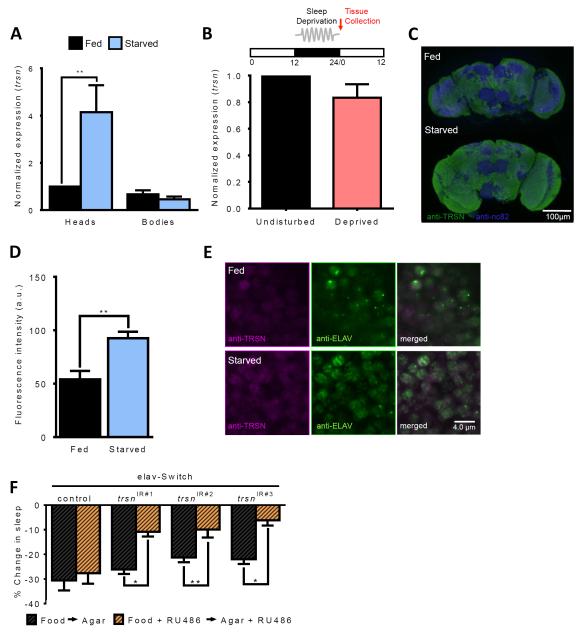
In Drosophila, trsn is expressed in the brain throughout development [31]. To determine whether trsn is acutely regulated in response to sleep or feeding state, we measured trsn transcript levels by quantitative PCR in flies that were previously starved or sleep-deprived. trsn was expressed at low-levels in the heads and bodies of fed flies and was specifically upregulated in the head following 24 hours of starvation (Figure 3A). No changes in trsn transcript were detected after 12 hours of mechanical sleep deprivation, suggesting the upregulation of trsn expression is not a generalized response to stress or environmental perturbation (Figure 3B). To confirm that TRSN protein is increased in response to starvation, we performed immunohistochemistry on brains immunostained with anti-TRSN. Quantification of whole-brain fluorescence confirmed that TRSN protein is increased in response to starvation (Figure 3C-D). In agreement with previous findings, TRSN signal is below detection in trsnnull mutants and dramatically reduced in nSyb-GAL4>trsn-IR flies confirming the antibody specifically labels TRSN (data not shown and [13]). Counterstaining with the neuronal marker Embryonic Lethal Abnormal Vision (ELAV) revealed that TRSN and ELAV are expressed in all neurons during the fed and starved states (Figure 3E), suggesting the observed changes in protein levels are not due to altered protein localization. Together, these data suggest that at the RNA and protein levels, trsn is increased in response to starvation.
Figure 3. Spatial and temporal localization of trsn function.
A. Expression of trsn is upregulated in the heads (N≥14; P<0.01) but not bodies of w1118 control flies (N≥14;P>0.99) following 24 hours of starvation. B. trsn transcript does not differ in heads between flies sleep-deprived for 12 hours from ZT12-ZT24 and undisturbed controls (N=3; P>0.17). Red arrow denotes point of tissue collection. C,D. Immunohistochemistry for whole-brain TRSN protein (B). Neuropils are labeled by NC82 for reference (magenta) and anti-TRSN (green) is observed throughout the brain. Whole-brain TRSN protein quantification of fluorescence intesity revealed TRSN is increased in starved flies compared to fed control (N≥6; P<0.002) by paired t-test. E. Immunostaining for anti-TRSN (magenta) and the neuronal marker anti-ELAV (green) reveals colocalization between TRSN and ELAV proteins in brains of fed (upper) and starved (lower) flies. Depicted is a representative section from the dorsomedial central brain, near the lateral horn region. Scale bar denotes 4 μm. F. Percentage sleep loss in experimental flies treated with RU486 (orange bars) or controls without drug treatment (black bars). Sleep suppression is significantly reduced in elav-Switch>trsnIR#1 flies (N≥36; P>0.031), elav-Switch>trsnIR#2 (N≥68; P<0.011) and elav-Switch>trsnIR#3 (N≥34; P<0.041) flies fed RU486 compared to non-RU486-fed controls. There is no effect of RU486 feeding in flies harboring the elav-Switch transgene alone (N≥39; P>0.99). All other bars are mean ± SEM; P<0.05,*; P<0.01,**; by 2-way ANOVA. See also Figure S3.
The finding that trsn is upregulated in response to starvation raises the possibility that it functions acutely to modulate sleep. RNAi targeted to trsn was acutely induced in three-day old animals using the GeneSwitch system. Flies were fed food laced with 0.25mM RU486, and sleep was measured on food and agar. Adult-specific pan-neuronal knockdown with all three RNAi lines under regulation of elav-Switch impaired sleep-suppression compared to genotype matched controls not fed RU486 or genetic controls lacking the trsnIRi transgene ([32, 33] Figure 3F and S3). These findings, coupled with the upregulation of trsn in response to starvation, provide evidence that trsn is required during adulthood for the integration of sleep and metabolic state.
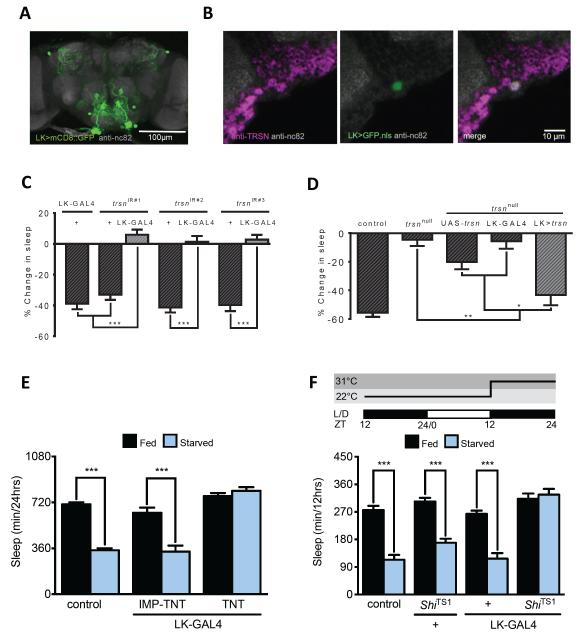
We next sought to identify neurons where trsn functions to modulate sleep. Peptidergic neurons are critical regulators of many behaviors, including sleep and feeding [34–36]; therefore, we screened GAL4 lines labeling defined populations of peptidergic neurons or neurons previously shown to regulate sleep. We identified the Leucokinin (LK) neurons, where knockdown of trsn reduced sleep modulation in response to starvation. LK has been implicated in a host of fly behaviors including feeding and water homeostasis, locomotion, and olfactory behavior [37, 38]. Driving membrane tethered CD8::GFP with LK-GAL4 labeled a single large neuron in the lateral horn and three pairs of neurons in the subesophageal zone ([38]; Figure 4A). Immunostaining brains of LK-GAL4 flies driving nuclear GFP (UAS-GFP.nls) revealed that the LK-GAL4 neurons that are co-labeled by TRSN antibody (Figure 4B). In addition, all three trsn-IR lines impaired starvation-induced sleep suppression when expressed under the control of LK-GAL4 (Figure 4C), whereas restoration of trsn specifically in LK-GAL4 neurons, or in all neurons with nSyb-GAL4, rescued starvation-induced sleep suppression to control levels (Figure 4D and S4A). Therefore, trsn function in LK neurons is essential for starvation-induced sleep loss.
Figure 4. trsn functions in Leucokinin neurons to regulate sleep.
A. Whole-brain confocal reconstruction of LK-GAL4>mCD8::GFP. GFP-expressing neurons (green) labeled the subeosphogeal zone and dorsal protocerebrum. The brain was counterstained with the neuropil marker nc82 (grey). Scale bar denotes 100 μm. B. Immunostaining for anti-TRSN (magenta) in the brain of LK-GAL4>UAS-GFP.nls reveals TRSN localizes to neurons labeled by LK-GAL4 (white). Depicted is a representative 2 μm section from the lateral horn region. Scale bar denotes 10 μm. The neuropil marker anti-nc82 (grey) is used as background. C. Knockdown of trsn in LK-GAL4 neurons alone reduces starvation-induced sleep suppression in all three trsnIR lines compared to control flies harboring a UAS-trsnIR transgene alone (N≥52; P<.001) or LK-GAL4 transgenes alone (N≥64; P<0.001). D. Expression of UAS-trsn under LK-GAL4 control in the background of a trsnnull mutation restores starvation-induced sleep suppression compared to flies harboring either UAS-trsn (N=87; P<0.05); or the GAL4 lines alone (N=79; P<0.01). No significant differences were detected between LK rescue and w1118 control flies (N≥38; P>0.10). E. Starvation-induced sleep suppression is abolished in flies expressing TNT in LK-GAL4 neurons (LK-GAL4>UAS-TNT, Fed vs Starved N=39; P=0.96) while controls expressing inactive UAS-IMP-TNT suppress sleep (Fed vs. Starved: LK-GAL4>UAS-IMP-TNT, N=33, P<0.001). Sleep duration on food does not differ significantly between LK-GAL4>UAS-TNT and UAS-IMP-TNT flies N≥34, P>0.06). F. Flies were transferred to agar at ZT9 then sleep was measured at 31°C on food (black) or agar (blue) over the 12hr night (ZT12-ZT24). Genetic silencing of LK-GAL4 abolished starvation-induced sleep suppression (LK-GAL4>UAS-ShiTS, Fed vs Starved, N≥40, P>0.98) while control flies robustly suppressed sleep (Fed vs Starved: control, N≥79, P<0.001; UAS- ShiTS1 /+, N≥42, P<0.0001; LK-GAL4/+, N≥51, P<0.002). No differences were observed between genotypes in the fed state (Fed vs Fed: control vs LK-GAL4>UAS-ShiTS , P>0.72; UAS-ShiTS/+ vs LK-GAL4>UAS-ShiTS , P>0.98, LK-GAL4/+ vs LK-GAL4>UAS-ShiTS, P=0.07). All columns are mean ± SEM; P<0.01,**; P<0.001,*** by 2-way ANOVA. See also Figure S4.
To further examine the role of LK neurons in sleep regulation we blocked synaptic release from LK neurons and measured sleep in fed and starved flies [38, 39]. Chronic blockade of synaptic release in LK neurons with tetanus toxin (TNT) impaired starvation-induced sleep suppression compared to control flies expressing an inactive form of TNT (UAS-IMP-TNT) or genetic controls harboring only a single transgene ([40] Figure 4E and S4B). In fed conditions, silencing of LK neurons increased sleep compared to controls that approached significance, raising the possibility that these neurons are wake promoting (Figure S4B). To examine the effects of acutely silencing LK-GAL4 neurons, the dominant-negative form of the GTPase Shibire (ShiTS1) was expressed in LK neurons and sleep was measured in both fed and starved flies during the night period [41]. Flies expressing ShiTS1 in LK labeled neurons failed to suppress sleep at the non-permissive temperature of 31°C (Figure 4F and S4C-E). Control and experimental groups did not suppress sleep at 22°C due to the lower temperature and shortened duration of the assay (Figure S4C,D). Therefore, LK neurons are acutely required for modulation of sleep in response to starvation, supporting the notion that trsn function in LK neurons is essential for the integration of sleep and metabolic state.
Taken together, we have identified trsn as an essential regulator of sleep-metabolism interactions. While many genes have been identified as genetic regulators of sleep or metabolic state, multiple lines of evidence indicate that trsn functions as a unique integrator of these processes. trsn is not required for the homeostatic increase in sleep following mechanical deprivation or response to stimulants, suggesting trsn is not generally required for acute modulation of sleep. Further, trsn-deficient flies display normal feeding behavior, indicating that it is not required for modulation of behavior in response to food deprivation. Finally, energy stores in trsn mutant flies are normal indicating that the starvation-induced sleep suppression phenotype is not due to increased nutrient storage. These results provide evidence that trsn is not required for the perception of starvation or the general induction of hunger-related behaviors, but is required for the induction of wakefulness in the absence of food.
While trsn is broadly expressed in the Drosophila nervous system, we localize the function of trsn in metabolic regulation of sleep to LK-expressing neurons. Targeted knockdown of trsn in LK neurons disrupts metabolic control of sleep, while restoring trsn to LK neurons rescues sleep regulation in trsn mutants. In addition to regulating sleep, ablation of LK neurons reduces meal number, while increasing consumption during individual feeding bouts, suggesting a role in feeding behavior [38]. LK is expressed in the subesophageal zone, the insect taste center, and in modulatory neurons within the lateral horn, raising the possibility that the sleep and feeding phenotypes associated with LK mutations or manipulation of LK neurons may localize to distinct brain regions [38]. It is possible that the same populations of LK neurons regulate meal frequency and sleep or distinct neurons modulate each process. Combinatorial genetic approaches to manipulate subsets of GAL4-labeled neurons in combination with recent advances in behavioral analysis of meal frequency may allow for the localization of LK neurons involved in each behavioral process [42–44].
In addition to its known role in the synthesis of non-coding RNA, TRSN physically associates with Translin- Associated Protein X (TRAX) [45, 46]. TRSN and TRAX are essential components for the RNA induced silencing complex (RISC), suggesting a role in post-transcriptional gene silencing through the generation of small RNAs. trsn knockout mice have diminished forebrain monoamine levels, indicating that trsn may serve to regulate neurotransmitter synthesis [47]. Further investigation of the mechanistic relationship between trsn and neural regulation of sleep will provide a framework to study the molecular properties and neural networks that are associated with interactions between sleep and metabolic state.
Experimental Procedure
Drosophila maintenance and fly stocks
Flies were grown and maintained on standard food (New Horizon Jazz Mix, Fisher Scientific). Flies were maintained in incubators (Powers Scientific; Dros52) at 25°C on a 12:12 LD cycle, with humidity set at 55-65%. The background control line used in this study is the w1118 fly strain, and all experimental fly strains including trsnEP and nSyb-GAL4 were outcrossed for 5-6 generations into this background. The nSyb-GAL4 line was a generous gift from Dr. Julie Simpson (UCSB). For further genotype information see Supplemental Methods.
Sleep and feeding analysis
Unless otherwise noted, fly activity was monitored using Drosophila Activity Monitors (DAM2; Trikinetics, Waltham, MA) as previously described [48]. Female flies were briefly anesthetized using CO2 within 1 hour of lights on at Zeitgeber Time on at (ZT0) and placed into plastic tubes containing standard fly food. All flies were given at least 22 hours to recover from anesthesia prior to behavior experiments. For detailed description of all behavioral paradigms, see Supplemental Methods
Pharmacological manipulation
For pharmacological manipulation of glucose and fatty acid utilization, flies were loaded into tubes containing standard fly food. Following a 24 hour acclimation period, flies were transferred at ZT0 into tubes containing standard fly food (control), food laced with either 400mM 2-DG, 25μM etomoxir, or 400mM 2-DG and 25μM etomoxir and sleep was measured for an additional 24 hours. For GeneSwitch experiments, a 100mM stock solution of RU486 (Sigma, St. Louis) was added to fly food or 1% agar solution to a final concentration of 0.25mM RU486. For further details, see Supplemental Methods.
Paraquat dichloride (Sigma, St. Louis) was dissolved directly into 1% agar with 5% sucrose and poured into plates to obtain a 1mM concentration of paraquat. To test the effect of caffeine on sleep, caffeine (Sigma, St. Louis) was dissolved in melted fly food and poured into plates to a concentration of 4mg/mL. Further details are provided in Supplemental Methods.
Protein, glycogen, and triglyceride measurements
Assays for quantifying triglyceride, glycogen and protein content of flies were performed as previously described [24, 25]. Further details are provided in Supplemental Methods.
qPCR and immunohistochemistry
Flies were collected 5-7 days after eclosion. Ten or more flies were separated into fed and starved groups and were flash frozen. Total RNA was extracted from fly heads using the QIAGEN RNeasy Tissue Mini kit according to the manufacturer’s protocol. RNA samples were reverse transcribed using iScript (Biorad), and the generated cDNA was used for real-time PCR (Biorad CFX96™, SsoAdvanced™ Universal SYBR® Green Supermix qPCR Mastermix Plus for SYBRGreen I) using 1.7ng of cDNA template per well and a primer concentration of approximately 300nM. Specific primer details are provided in Supplemental Methods.
Statistical Analysis
Statistical analyses were performed using InStat software (GraphPad Software 5.0 Inc.) or IBM SPSS 22.0 software (IBM, Somers, NY, USA). For analysis of sleep, we employed a one or 2-way ANOVA followed by a Tukey’s posthoc test. For PER experiments each fly was sampled three times with the same stimulus. The response was binary (PER yes/no), and these three responses were pooled for values ranging from 0-3. The Kruskal-Wallis test (nonparametric ANOVA) was performed on the raw data from single flies and Dunn's Multiple Comparisons test was used to compare different groups. For the capillary feeding assay, 30-60 flies were used per tube and 4-20 tubes per group were tested. The Wilcoxon signed rank test (non-parametric) with two-tailed p-value was used to test significance on single groups.
Supplementary Material
Highlights.
- Flies deficient for translin fail to integrate sleep and metabolic state.
-translin does not regulate stress response, metabolic function, or feeding.
- translin functions in Leucokinin neurons to regulate sleep.
- Silencing of Leucokinin neurons abolishes starvation-induced sleep suppression.
Acknowlegements
We are grateful to Ellen Scully (UNR) and Nicolai Oh (Davidson Academy, Reno) for technical support. This work was supported by NIH Grants 1R01 NS085252 to ACK and R15NS080155 to ACK and JRD. The authors are grateful to Erik Duboue (Carnegie Institution) for feedback on this manuscript.
Footnotes
Author Contributions:
Conceptualization: P.M,, J.R.D, A.C.K; Methadology: J.R.D, A.C.K; Investigation: K.M., M.E.Y, B.A.S., P.M., A.M., R.H., W.B., R.M.G.; Writing original draft: B.S., J.R.D., A.C.K.; Writing review and editing: All authors contributed; Funding Acquisition: J.R.D and A.C.K; Resources: Y.J.K.,, B.S., and W.W.J.; Supervision: P.M., J.R.D., and A.C.K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knutson KL, Van Cauter E. Associations between sleep loss and increased risk of obesity and diabetes. Ann. N. Y. Acad. Sci. 2008;1129:287–304. doi: 10.1196/annals.1417.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peppard PE, Young T, Palta M, Dempsey J, Skatrud J. Longitudinal study of moderate weight change and sleep-disordered breathing. JAMA. 2000;284:3015–3021. doi: 10.1001/jama.284.23.3015. [DOI] [PubMed] [Google Scholar]
- 3.Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:210–217. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danguir J, Nicolaidis S. Dependence of sleep on nutrient’s availability. Physiol. Behav. 1979;22:735–740. doi: 10.1016/0031-9384(79)90240-3. [DOI] [PubMed] [Google Scholar]
- 5.Macfadyen U, Oswald I, Lewis S. Starvation and human slow-wave sleep. J Appl Physiol. 1973;35:391–4. doi: 10.1152/jappl.1973.35.3.391. [DOI] [PubMed] [Google Scholar]
- 6.Green CB, Takahashi JS, Bass J. The Meter of Metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 8.Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J. Neurosci. 2009;29:1948–1961. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB, Waddell S, Blau J. Clock and cycle limit starvation-induced sleep loss in drosophila. Curr. Biol. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB, Waddell S, Blau J. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr. Biol. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattaliano MD, Montana ES, Parisky KM, Littleton JT, Griffith LC. The Drosophila ARC homolog regulates behavioral responses to starvation. Mol. Cell. Neurosci. 2007;36:211–221. doi: 10.1016/j.mcn.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claußen M, Koch R, Jin ZY, Suter B. Functional characterization of Drosophila Translin and Trax. Genetics. 2006;174:1337–1347. doi: 10.1534/genetics.106.063016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI. A video method to study Drosophila sleep. Sleep. 2008;31:1587–1598. doi: 10.1093/sleep/31.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donelson N, Kim EZ, Slawson JB, Vecsey CG, Huber R, Griffith LC. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat. Protoc. 2012;7:995–1007. doi: 10.1038/nprot.2012.041. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22538850. [DOI] [PubMed] [Google Scholar]
- 17.Garbe D, Bollinger W, Vigderman A, Masek P, Gertowski J, Sehgal A, Keene A. Context-specific comparison of sleep acquisition systems in Drosophila. Biol Open. 2015;4:1558–68. doi: 10.1242/bio.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puschner B, Schacht J. Energy metabolism in cochlear outer hair cells in vitro. Hear. Res. 1997;114:102–106. doi: 10.1016/s0378-5955(97)00163-9. [DOI] [PubMed] [Google Scholar]
- 19.Lopaschuk GD, Wall SR, Olley PM, Davies NJ. Etomoxir, a carnitine palmitoyltransferase I inhibitor, protects hearts from fatty acid-induced ischemic injury independent of changes in long chain acylcarnitine. Circ. Res. 1988;63:1036–1043. doi: 10.1161/01.res.63.6.1036. [DOI] [PubMed] [Google Scholar]
- 20.Dus M, Min S, Keene AC, Lee GY, Suh GSB. Taste-independent detection of the caloric content of sugar in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11644–11649. doi: 10.1073/pnas.1017096108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J. Neurosci. 2009;29:11029–11037. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassu ED, McDermott JE, Keys BJ, Esmaeili M, Keene AC, Birnbaum MJ, DiAngelo JR. Mio/dChREBP coordinately increases fat mass by regulating lipid synthesis and feeding behavior in Drosophila. Biochem. Biophys. Res. Commun. 2012;426:43–48. doi: 10.1016/j.bbrc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingras RM, Warren ME, Nagengast AA, Diangelo JR. The control of lipid metabolism by mRNA splicing in Drosophila. Biochem. Biophys. Res. Commun. 2014;443:672–676. doi: 10.1016/j.bbrc.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurgel M, Masek P, DiAngelo J, Keene A. Genetic dissection of sleep-metabolism interactions in the fruit fly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2014 doi: 10.1007/s00359-014-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dethier VG. The Hungry Fly: A Physiological Study of the Behavior Associated with Feeding. Harvard University Press; 1976. [Google Scholar]
- 28.Masek P, Scott K. Limited taste discrimination in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14833–14838. doi: 10.1073/pnas.1009318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong R, Piper MDW, Wertheim B, Partridge L. Quantification of food intake in Drosophila. PLoS One. 2009;4:e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suseendranathan K, Sengupta K, Rikhy R, D'Souza JS, Kokkanti M, Kulkarni MG, Kamdar R, Changede R, Sinha R, Subramanian L, et al. Expression pattern of Drosophila translin and behavioral analyses of the mutant. Eur. J. Cell Biol. 2007;86:173–186. doi: 10.1016/j.ejcb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffith LC. Neuromodulatory control of sleep in Drosophila melanogaster: Integration of competing and complementary behaviors. Curr. Opin. Neurobiol. 2013;23:819–823. doi: 10.1016/j.conb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghert PH, Nitabach MN. Peptide Neuromodulation in Invertebrate Model Systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nässel DR, Winther ÅME. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 37.De Haro M, Al-Ramahi I, Benito-Sipos J, López-Arias B, Dorado B, Veenstra JA, Herrero P. Detailed analysis of leucokinin-expressing neurons and their candidate functions in the Drosophila nervous system. Cell Tissue Res. 2010;339:321–336. doi: 10.1007/s00441-009-0890-y. [DOI] [PubMed] [Google Scholar]
- 38.Al-Anzi B, Armand E, Nagamei P, Olszewski M, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr. Biol. 2010;20:969–978. doi: 10.1016/j.cub.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 41.Kitamoto T. Conditional modification of behavior in drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 42.Itskov PM, Moreira J-M, Vinnik E, Lopes G, Safarik S, Dickinson MH, Ribeiro C. Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat. Commun. 2014;5:4560. doi: 10.1038/ncomms5560. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4143931&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ro J, Harvanek ZM, Pletcher SD. FLIC: high-throughput, continuous analysis of feeding behaviors in Drosophila. PLoS One. 2014;9:e101107. doi: 10.1371/journal.pone.0101107. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4076220&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–36. doi: 10.1016/j.neuron.2006.08.028. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1713190&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–6. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 46.Wu R, Osatomi K, Terada L, Uyeda K. Identification of Translin/Trax complex as a glucose response element binding protein in liver. Biochim. Biophys. Acta. 2003;1624:29–35. doi: 10.1016/j.bbagen.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Stein JM, Bergman W, Fang Y, Davison L, Brensinger C, Robinson MB, Hecht NB, Abel T. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J. Neurosci. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila activity monitoring (DAM) system. Cold Spring Harb. Protoc. 2010;5 doi: 10.1101/pdb.prot5518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.