Abstract
In the face of starvation animals will engage in high-risk behaviors that would normally be considered maladaptive. Starving rodents for example will forage in areas that are more susceptible to predators and will also modulate aggressive behavior within a territory of limited or depleted nutrients. The neural basis of these adaptive behaviors likely involves circuits that link innate feeding, aggression, and fear. Hypothalamic AgRP neurons are critically important for driving feeding and project axons to brain regions implicated in aggression and fear. Using circuit-mapping techniques, we define a disynaptic network originating from a subset of AgRP neurons that project to the medial nucleus of the amygdala and then to the principle bed nucleus of the stria terminalis, which plays a role in suppressing territorial aggression and reducing contextual fear. We propose that AgRP neurons serve as a master switch capable of coordinating behavioral decisions relative to internal state and environmental cues.
Ecological studies reveal that prey species display cost-benefit decision making when foraging for food. The costs of foraging include food-seeking energy demands along with environmental threats such as predation and thermal challenges. Many prey species forage within a familiar territory, in zones that are protected from predators and have moderate temperatures 1-3. However, when challenged with starvation, behavioral priorities adapt and prey species display higher-risk behavior to find food 4-7.
Orexigenic AgRP neurons are active in a starved state 8, 9 and elicit signals that are paramount to the sensation of hunger 10-12. Coined for the expression of agouti-related peptide (AgRP), AgRP neurons are inhibitory projections neurons; they are GABAergic and express two inhibitory neuropeptides, neuropeptide Y (NPY) and AgRP 8, 13-15. Somewhat paradoxically, AgRP neurons appear to stimulate hunger by inhibiting downstream brain regions involved in satiety. AgRP neurons are derived from at least two progenitors 16 and project (with minimal collaterals) to approximately 15 unique downstream brain regions 14, 17. Activation of distinct AgRP projections revealed a “parallel and redundant” signaling network, but interestingly, some AgRP target regions do not evoke a feeding response 17. We propose that the heterogeneous AgRP population functions to coordinate numerous behavioral and physiological adaptations that prioritize food seeking and energy conservation under conditions of starvation.
AgRP neurons may influence behavioral decisions by signaling to brain regions that are involved in sensory processing. For example, a subset of AgRP neurons project to the medial amygdala (MeA) 14, 18, 19, a brain region implicated in innate social behaviors including aggression 20. Chemosensory cues of conspecific mice activate cells in the MeA, as indicated by the expression of Fos 21, 22, and acute activation of GABAergic cells in the posterior dorsal MeA can induce attack behavior 23. Under conditions of starvation, AgRP signaling to the MeA may alter an animal's normal response to chemosensory cues, shifting behavior away from protecting an energy-depleted territory and toward exploratory, food-seeking behavior. To test this idea, we used a combination of viral and genetic tools to activate AgRP neurons, and compared the behavior of these mice to those in the fasted state. We describe a specific starved-state neural circuit that influences innate and learned behavioral responses (Supplementary Fig. 1).
RESULTS
AgRP circuits promote risk taking and reduce territorialism
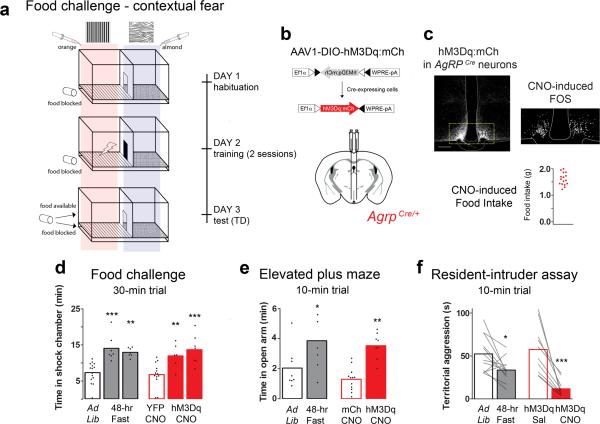
Starvation promotes higher-risk foraging behavior such that prey species become willing to search for food in exposed areas outside of their territorial safe zone 4. To model this behavioral shift in a laboratory setting, we designed an experiment that challenges mice to search for food in a chamber that they are conditioned to associate with a mild foot shock (Fig. 1a). We observed that, under normal conditions, mice avoided the shock-associated area, spending only 24.5 ± 2.5% of the trial in this chamber. Fasted animals, however, overcame the conditioned threat and spent more than 40% of the time in the shock-associated side. During habituation and training, food was present below the floor grid in the shock-associated chamber. On test day, the food either remained under the floor grid (food-blocked group), or was presented in the chamber and available for consumption (food-access group). The food-access group spent 46.9 ± 4.6% of the trial in the shock chamber; similarly, the food-blocked group spent 43.2 ± 1.6% of the trial in the shock chamber (Fig. 1d, black outlined bars; Supplemental STATISTCS provides details of all tests performed). We questioned whether the fasted state or the food cues biased the animals’ behavior. However, when the experiment was performed in the absence of food entirely, fasted mice behaved similar to fed controls, spending only 25.7 ± 4.7 % of the trial in the shock chamber (Supplementary Fig. 2a).
Figure 1.
AgRP stimulation recapitulates fasting-related foraging and reduced territorialism. (a) Schematic diagram of the Pavlovian food-challenge assay used to assess risk taking. (b) Viral construct: AAV1-DIO-hM3Dq:mCh, and diagram of the stereotaxic injection site in AgrpCre mice. (c) Viral expression of hM3Dq:mCh in AgRP neurons in conjunction with CNO-induced Fos immunoreactivity; scale bar, 100 μm. Animals included in the study (red dots) consumed >1.0 g of food 4 hr post CNO (1 mg/kg IP). Note, four animals consumed <1.0 g of food in this test and were excluded from the study. (d) Postconditioning (TD) quantification of time spent in the shock-associated chamber, compared using a 1-way ANOVA with Bonferroni's multiple comparison tests. Experimental and control animals were evaluated with either the food available, or blocked on test day. There was no difference between controls under these conditions, and they are plotted together. The experimental animals are separated by either condition: food available (left) and food blocked trial (right). (e) Time spent in the open arms of a plus maze, compared using a 1-way ANOVA with Bonferroni's multiple comparison tests. (f) Paired two-tailed Student's t-test analysis of home-cage aggressive behavior. (d-f) Bar outlines indicate mouse genotype: red indicates AgrpCre, while black indicates C57BL/6. The condition or treatments of the animals are indicated below the x-axis. (d,e) Conducted during the light cycle (5:00 – 17:00), (f) Conducted in the dark (17:00 – 19:00). See methods for detailed statistics.
We reasoned that AgRP neurons may promote high-risk exploration in the starved state. To test this idea, we made AgRP neurons excitable by transducing AgrpCre mice with a conditional virus containing the stimulatory DREADD (designer receptor exclusively activated by designer drugs), hM3Dq (Fig. 1b, c)24. The designer receptor ligand, clozapine-n-oxide (CNO), induces Gαq-mediated signal transduction and can be used to activate AgRP neurons via intraparietal delivery 11. We activated AgRP neurons in fed mice and asked whether this isolated circuit could recapitulate the foraging behavior of fasted mice in the food-challenge assay described above. Similar to fasted animals, we found that AgRP neuron-stimulated mice spent more time in the shock-associated chamber relative to controls. Both food-available and food-blocked groups spent 40.0 ± 5.6% and 45.6 ± 5.6% of the trial in the shock chamber, respectively; while control animals displayed an aversion to the shock-associated side and spent only 22.7 ± 3.2% of the trial in this chamber (Fig. 1d, red bars). These data support that, beyond promoting food intake, AgRP neurons can influence the behavioral response to environmental threats.
In a second test, we evaluated innate anxiety-like behavior by assessing the willingness of animals to enter an exposed platform on an elevated maze. In support of previous literature 25, both fasted and AgRP neuron-stimulated animals spent significantly more time in the exposed platforms compared to controls (Fig. 1e). AgRP neuron activation has been demonstrated to promote locomotor activity 11 and, consistent with this, stimulated mice moved an average total distance of 3148 ± 148.3 cm, while controls moved an average of 2024 ± 186.5 cm on the maze. The stimulated mice not only spent more time but also moved a greater distance in the open arms than the controls (stimulated, 48.2 ± 3.1%; controls, 30.7 ± 4.5%).
During starvation, the costs associated with foraging are not limited to environmental threats, but also include the threat of dwindling energy reserves. Consequently, organisms forage in a way that minimizes the energy costs associated with food seeking 1-3, 26. For example, territorial-defense behavior is not an efficient use of energy if a territory is depleted of resources 27. Experimentally, we evaluated territorial behavior using the resident-intruder assay and define territory as a defended area. Resident males were sexually experienced and territorialized to an isolated home cage. The intruder test evaluates aggressive territorial behaviors including: holding, fighting (boxing/attacking/mounting), high-speed chasing, and nudging when an intruder (younger, sexually naïve, group-housed littermates that do not display aggression toward residents) is placed in the cage. (Supplementary Fig. 2b). Compared to the fed state, fasted animals displayed less home-cage aggression toward an intruder (Fig. 1f, black bars). We observed that fasted residents spent significantly more time investigating the snout of the intruder—perhaps smelling food odorants on the intruder's snout—and displayed escape behaviors including rearing and jumping (Supplementary Fig. 2b-d). We used a 48 hr fast to maximally activate the feeding circuits; however, 24-hr fasted residents also displayed decreased home-cage aggression (Supplementary Fig. 2c).
To test the role of AgRP neurons in fasting-related territorial behavior, we evaluated activated AgRP neurons in fed resident mice. Similar to fasting, AgRP neuron-stimulated mice displayed less home-cage aggression toward an intruder (Fig. 1f, red bars). If food was presented during the trial, AgRP neuron-stimulated residents spent the majority of the 10-min trial eating, consuming 0.34 ± 0.03 g (Supplementary Fig. 3).
AgRP → MeA signaling influences territorial behavior
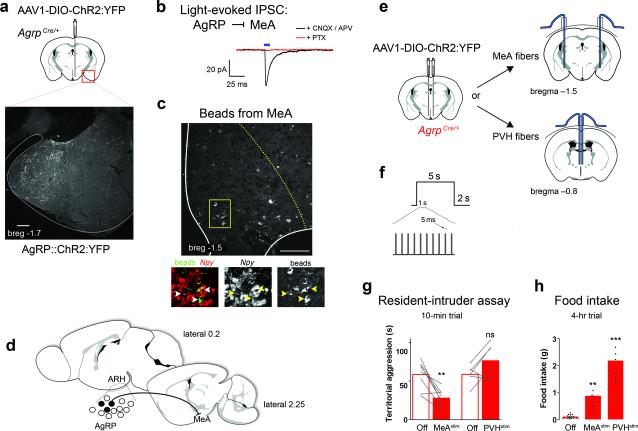
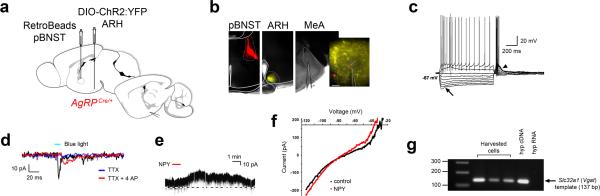
Because MeA neurons are involved in innate social behavior, including territorialism, we reasoned that the inhibitory AgRP → MeA circuit may be responsible for starved-state decreases in territorial aggression. To test whether AgRP fibers can directly inhibit cells in the MeA, we transduced AgrpCre mice with a conditional channelrhodopsin-2 (ChR2)-expressing virus (Fig. 2a)28. We photostimulated AgRP fibers in the MeA and performed whole-cell recordings in slice preparations. MeA soma in close proximity to fluorescent AgRP fibers were patched; 4 of 11 cells from 2 mice displayed a light-evoked inhibitory post-synaptic current (IPSC) that was blocked by the GABAA receptor antagonist picrotoxin (PTX) but not by glutamate receptor antagonists (Fig. 2b). We used retrograde tracing to quantify the subset of AgRPMeA-projecting neurons. Fluorescent RetroBeads injected into the MeA (Supplementary Fig. 4a) were retained in 7.1 ± 0.6 % of AgRP-expressing cells (167 ± 15 of 2356.3 ± 146 total hemisphere, n = 3; Fig. 2c, d). These data establish that a subset of AgRP neurons make direct inhibitory connections onto MeA neurons.
Figure 2.
MeA-projecting AgRP neurons evoke hunger and inhibit territorial aggression. (a) ChR2:YFP-expressing AgRP fibers observed in the MeA. Scale bar, 200 μm. (b) Whole-cell, patch clamp recordings (voltage-clamp, -70 mV) of MeA cells in proximity to AgRP::ChR2-expressing fibers. Light-evoked responses were measured with 10-ms light pulse (472 nm, 10 mW power at the tip) in the presence of CNQX and AP5 (black trace), followed by picrotoxin (red trace). Cells were recorded from three ChR2-transduced animals (tissue from the fourth animal was excluded due to lack of fluorescent reporter expression in the targeted AgRP population). (c) Green fluorescent RetroBeads injected into MeA were retained in 7.1 ± 0.6 % of Npy-expressing ARH cells; indicated by arrows. Scale bar, 100 μm. (d) Schematic demonstrating that a subset of AgRP neurons (black filled) project to the MeA. (e) Bilateral injection of AAV1-DIO-ChR2:YFP into the ARH with duel fiber optic cannulas implanted above either the MeA or PVH. (f) Stimulation paradigm for behavioral studies: 10 Hz with 5-ms pulses that continue for 5 s followed by 2 s light-off recovery. (g) Paired two-tailed Student's t-test analysis of home-cage aggressive behavior, comparing light on versus light off conditions; conducted during the first 2 hr of the dark cycle (17:00 – 19:00). (h) Cumulative food intake measured during the light cycle (10:00 – 14:00) compared using a 1-way ANOVA with Bonferroni's multiple comparison tests. See methods for detailed statistics.
We used optogenetic fiber stimulation to probe the behavioral impact of the AgRP → MeA circuit (Fig. 2e, f; Supplementary Fig. 4b). Similar to fasting, stimulation of AgRP axons in the MeA elicited less territorial aggression relative to a non-stimulated state (Fig. 2g). To gauge the specificity of the AgRP → MeA circuit on territorial behavior, we stimulated AgRP neurons that project to the PVH. Prior studies demonstrated that the AgRP → PVH circuit can induce food intake equivalent to that observed following a fast 29. If fasting-induced territorial behavior is a consequence of hunger, then this circuit should also modulate home-cage aggression. We did not observe a significant change in aggression upon AgRP → PVH stimulation (Fig. 2e, f; Supplementary Fig. 4c). Together, these data indicate that a distinct population AgRP neurons can mediate unique behaviors.
Along with social behavior, evidence suggests that the MeA is also involved in feeding behavior and body-weight regulation 30-32. We found that AgRP → MeA fiber-stimulated mice ate significantly more than non-stimulated, fiber-attached controls (0.87 ± 0.12 g vs 0.10 ± 0.02 g; Fig. 2h). To gauge the magnitude of this effect, we measured light-evoked food intake from AgRP → PVH fiber stimulation. The hyperphagia induced by AgRP → PVH fiber stimulation was 2.5-fold higher than AgRP → MeA fiber stimulation (Fig. 2h). These data add to evidence that there are redundant AgRP circuits that promote feeding 17.
Manipulating cells downstream of AgRP in the MeA
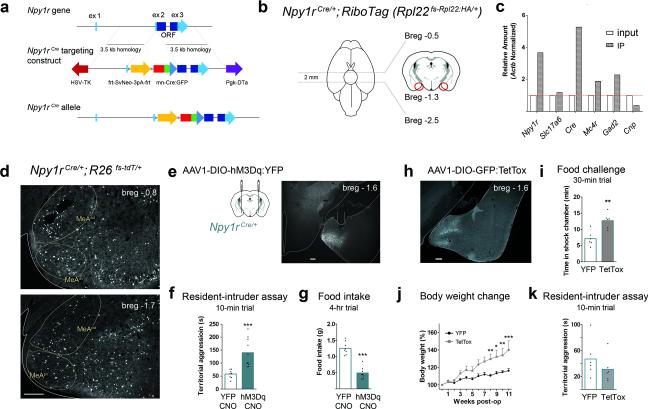
AgRP neurons co-express NPY8, 14; therefore, post-synaptic targets of AgRP neurons likely express NPY 1 or 5 receptors (Npy1R or Npy5R) 33 and the melanocortin 4 receptor (MC4R). To evaluate the function of MeA cells that receive information from AgRP neurons, we generated an Npy1rCre knock-in mouse line (Fig. 3a).
Figure 3.
Npy1R neurons in the MeA can evoke aggression and inhibit feeding. (a) Npy1rCre:GFP knock-in targeting construct (diagram not to scale; see methods for complete details). (b) RiboTag mice contain a cre-dependent epitope-taged polyribosome gene. MeA tissue was harvested (red circles) from Npy1rCre; RiboTag mice. (c) Comparison of precipitated transcripts versus input sample, demonstrated enrichment of Npy1r and Cre transcripts along with Gad2 and Mc4r. (d) Npy1R expression in the anterior-dorsal (AD) and anterior-ventral (AV) MeA; lower, posterior-dorsal (PD) and posterior-ventral (PV) MeA; three animals were evaluated demonstrating a similar expression pattern. Scale bar, 200 μm. (e) Bilateral injection of AAV1-DIO-hM3Dq:YFP into the MeA of Npy1rCre mice; histology demonstrating the injection site and Cre-dependent YFP fluorescence. Note, two animals were excluded from the study following histological analysis due to lack of reporter expression. Scale bar, 200 μm. (f) Stimulation of Npy1RMeA cells of resident animals evokes aggressive behavior (assessed during between 17:00 – 19:00) and (g) decreased food consumption during the dark cycle (17:00 – 21:00); compared using unpaired two-tailed Student's t-tests. (h) Bilateral injection of AAV1-DIO-GFP:TetTox into the MeA of Npy1rCre mice with histology to demonstrate the injection site. (i - k) Npy1RMeA neuron silencing resulted decreased threat avoidance behavior, compared using an unpaired two-tailed Student's t-test (i), and increased body weight, compared using a 2-way repeated measures ANOVA (j). (k) We did not observe a difference in home-cage aggression in Npy1RMeA silenced animals; compared using an unpaired two-tailed Student's t-test. (k). Conditioned avoidance, i, was measured by time spent in the shock-associated side on test day (detailed in Fig. 1a). See methods for detailed statistics.
We validated the correct targeting of this knock-in using multiple approaches. Prior to injection, neomycin-resistant ES cell colonies were screened for the proper insertion of Cre in the targeted Npy1r allele by Southern blot analysis. We also evaluated transcripts expressed in Npy1rCre cells by crossing this line to a Cre-dependent RiboTag mouse that expresses an epitope-tagged ribosomal protein (RPL22:HA)34. The conditional expression of the hemagglutinin (HA)-tagged ribosomes in Cre-positive cells allows us to isolate mRNA transcripts from these cells. Npy1rCre-RiboTagged cells in the MeA (Fig. 3b) were enriched in both Npy1r and Cre transcripts relative to transcripts expressed in all cells within the same region (Fig. 3c).
To further profile the Npy1RMeA cells, we mined the RiboTag-isolated transcriptome, probing for genes characteristic of excitatory, inhibitory and glial cells. We found that Npy1R-RiboTagged cells were enriched for both Mc4r and Gad2 transcripts. The glutamate transporter, Slc17a6 (Vglut2) was not enriched and the glial cell marker, Cnp was de-enriched (Fig. 3c). These data suggest that Npy1R cells in the MeA may be inhibitory neurons.
Npy1RMeA cells are anatomically distributed throughout the MeA, with a slightly biased distribution in the anteroventral subdivision (Fig. 3d and Supplementary Fig. 5), a pattern that resembles Mc4r expression 18, 19. To investigate whether Npy1RMeA cells are involved in feeding behavior or aggression, we bilaterally transduced the MeA of Npy1rCre mice with AAV1-DIO-hM3Dq:YFP (Fig. 3e). Because inhibitory AgRP → MeA fiber stimulation decreased territorial aggression, we predicted that activation of target Npy1R neurons would have the opposite effect. We found that CNO-induced Npy1RMeA neuron activation significantly increased territorial aggression (Fig. 3f and Supplementary Movie 1). Consistent with previous work using DREADDs to manipulate social behavior 23, we observed a scale of aggressive phenotypes upon CNO-induced activation. Four of nine stimulated animals displayed overt attack behavior, while the rest engaged in other aggressive behaviors including, nudging, aggressive grooming, and holding. To determine the degree of aggression evoked, we evaluated the overtly aggressive males in the presence of an anesthetized intruder; all four mice attacked the anesthetized conspecific within 59.0 ± 5.9 s (Supplementary Movie 2), a behavior never observed in non-stimulated mice. Along with changes in aggression, activation of the Npy1RMeA neurons significantly decreased food consumption within the first 4 h of the dark cycle (Fig. 3g).
To determine the necessity of the Npy1RMeA population for satiety, territorial aggression, and high-risk exploration, we used a viral approach to chronically inhibit Npy1rCre-expressing cells in the MeA. Mice were transduced with a conditional virus containing the light-chain of the tetanus toxin (AAV1-DIOGFP:TetTox) which can silence neurons by preventing synaptic transmission 35, 36 (Fig. 3h). In the food-challenge assay, TetTox-silenced Npy1RMeA mice spent more time in the shock-associated chamber postconditioning (Fig. 3i). They also gained significantly more weight after viral transduction relative to controls (Fig. 3j), but there was no statistical difference between TetTox-silenced mice relative to controls in territorial aggression (Fig. 3k). Because acute inhibition from AgRP → MeA fibers was sufficient to suppress territorial aggression, this finding was surprising but may be the result of compensatory phenomena. We also evaluated anxiety using the elevated plus maze. There was no difference in open-arm exploration between TetTox-silenced mice relative to YFP controls (17.2 4.2% for TetTox versus 21.2 3.1% for YFP controls, P = 0.5; Supplementary Fig. 6).
A disynaptic network: AgRP → Npy1RMeA → pBNST
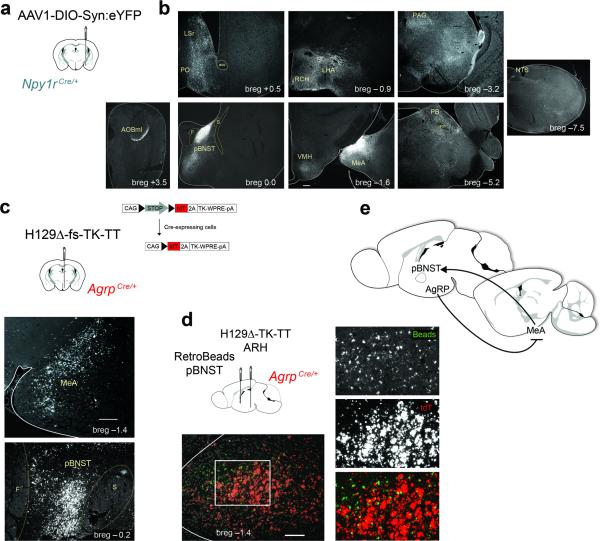
To identify candidate secondary targets of the AgRP → MeA circuit, we mapped the projection field of Npy1RMeA cells using a virus expressing a Cre-dependent, synapse-specific reporter, synaptophysin:YFP(Fig. 4a). We observed dense reporter expression in the posterior principle region of the bed nucleus of the stria terminalis (pBNST), along with several other brain regions including the lateral hypothalamic area, periaqueductal grey, parabrachial nucleus, ventromedial hypothalamus and anterior olfactory bulb (Fig. 4b). To determine whether these were secondary targets of AgRP neurons, we injected a Cre-dependent and trans-synaptic anterograde tracing virus, H129Δ-fs-TK-TT 37, into the ARH of AgrpCre animals (Fig. 4c). TdTomato fluorescence was observed in many sites throughout the brain, notably the MeA and the pBNST (Fig. 4c).
Figure 4.
The posterior BNST is a secondary target of AgRP and receives direct input from Npy1RMeA neurons. (a) Injection of AAV1-DIO-Synaptophysin:YFP into the MeA of Npy1rCre mice. (b) Immuno-reactive YFP fibers in the: AOBmi (accessory olfactory bulb, mitral layer), LSr (lateral septal nucleus, rostroventral part), PO (preoptic area), pBNST (bed nucleus of the stria terminalis, posterior division, principle nucleus), RCH (retrochiasmatic area), LHA (lateral hypothalamic area), VMH (ventromedial hypothalamus), MeA (medial amygdala), PAG (periaqueductal gray), PB (parabrachial nucleus), NTS (nucleus of the solitary tract). This finding was similarly observed in 3 animals with properly targeted viral injections. All images were scaled equally; scale bar, 200 μm (see breg –1.3). (c) AgRP neurons relay information to the pBNST and MeA. The trans-synaptic virus, H129Δ-fs-TK-TT, was injected into the ARH of AgrpCre mice (top panel). Immunoreactive DsRed cells were present in the MeA (middle panel) and pBNST (bottom panel). Scale bar, 200 μm. (d) The MeA relays signals from AgRP neurons to the pBNST. Diagram shows co-injection of green RetroBeads into the pBNST and H129Δ-fs-TK-TT into the ARH of AgrpCre mice. DsRed immune-reactive cell bodies and retrobead-positive cells are present in the MeA. Scale bar, 100 μm. Two animals were evaluated with proper targeting of both the ARH and pBNST demonstrating a similar expression profile in the MeA. (e) Model of the AgRP → MeA circuit.
The pBNST is an established target of the MeA 38, 39 and while AgRP fibers have been found in the anterior BNST 14, 17, they have not been observed in the pBNST. Likewise, we observed few, if any, fluorescent cell bodies in the ARH following injection of fluorescent RetroBeads into the pBNST (Supplementary Fig. 7a, b). To test the idea that the pBNST is a secondary target of AgRP neurons via the MeA, we co-injected RetroBeads into the pBNST and H129Δ-fs-TK-TT into the ARH of AgrpCre mice (Supplementary Fig. 7c, d). We observed expression of both reporters in the MeA (Fig. 4d), consistent with the idea that the pBNST is a secondary target of the AgRP → MeA circuit (Fig. 4e). Due to the nature of H129 infection, the cells were not healthy enough to quantify the overlap of these reporters in the MeA. Instead, we used an alternative approach to characterize the cells in the MeA that receive input from AgRP neurons and project to the pBNST.
Numerous cell types in the posterior MeA have been defined based on electrophysiological and morphological properties 40-42. To investigate the properties of pBNST-projecting MeA neurons we injected RetroBeads into the pBNST and performed whole-cell recordings on bead-labeled cells in the MeA (Fig. 5). When subjected to current-step injections, we observed a prominent hyperpolarization-activated voltage sag (h-current, denoted by the arrow, in 10 of 13 cells recorded), which has been described in type 1 GABAergic projections neurons in the posterior MeA 42. In addition, we discovered that that a subset of these neurons expressed a T-type calcium current (denoted by the arrow head, in 7 of 13 cells; Fig. 5c).
Figure 5.
Identification of Vgat-expressing NPY-responsive cells in the MeA that received direct input from AgRP neurons and project to the pBNST. (a) Sagittal diagram of injection and recording sites. (b) Coronal sections confirming targeted injections of RetroBeads into the pBNST (left) and DIO-ChR2:YFP into the ARH (center). Recordings were performed on bead-labeled cells in the MeA (right); zoom image (right) represents a red cell in proximity to YFP fibers. Note, three of eight animals injected with both beads and ChR2 contained the correct ipsilateral targeting of both the ARH and pBNST. Scale bar, 50 μm. (c) Voltage response to current step injections. The majority of bead-positive neurons exhibited a prominent hyperpolarization-activated voltage sag (h-current; n = 10 cells; arrow) and a low threshold potential (a T type calcium current, n = 7 cells; arrow head). (d) Light-responsive, bead-positive cells in the MeA (n = 3 cells); blue light-evoked fast IPSP (black trace) was abolished in the presence of TTX (blue trace), and rescued with the addition of the K+ channel blocker 4-AP (red trace). (e) Bead-positive neurons that displayed an outward current in response to bath application of NPY (1 μM; red line) (14.5 ± 4.5 pA; n = 4 cells from 4 sections across 3 animals with proper targeting of the retrograde label) when held at –60 mV (note, gaps in the trace indicate the voltage ramp interval in f). (f) IV relationship during a voltage ramp performed on bead-positive, light-responsive cells, before (black) and after (red) NPY application. (g) PCR detection of Slc32a1 (Vgat) cDNA in cell lysates harvested from recorded cells. Reverse-transcribed cDNA from the hypothalamus was used as a positive control, while hypothalamic RNA was tested as the negative control, all light-responsive and bead-positive cells were harvested for post-hoc analysis.
We determined that AgRP neurons can evoke GABA-mediated IPSCs in MeA neurons. Using the RetroBead labeling described here, we sought to determine whether ChR2-expressing AgRP neurons could evoke light-induced responses in pBNST-projecting MeA cells (Fig. 5a, b). Bead-positive MeA soma in proximity to YFP fibers demonstrated light-evoked IPSPs in 3 of 11 recorded cells (Fig. 5d). Similar to the recordings in Fig 2b, the light-evoked inhibitory response occurred with a short latency to the photostimulation, suggesting a direct connection. To support this idea, we identified a shifted (smaller and longer latency) light-evoked IPSP in the presence of the action-potential blocker, TTX along with a potassium channel blocker, 4-aminopyridine (4-AP)—a property indicative of monosynaptic connections in ChR2-assisted circuit mapping (Fig. 5d, red trace) 43.
The light-responsive, bead-positive MeA cells also respond to NPY. Bath application of NPY in the presence of TTX resulted in an outward current when held at a membrane potential of −60 mV in 4 out of 4 cells tested (Fig. 5e). To investigate the NPY-induced current, voltage ramps were performed in the presence and absence of NPY (Fig. 5f). Consistent with the idea that NPY-induced current is mediated by G protein-coupled, inwardly-rectifying potassium channel activation 44, 45, we found that the reversal potential for the outward current was at -85 mV, close to the Nernst equilibrium potential for potassium.
Based on the firing properties recorded in bead-positive cells (Fig. 5c) along with enrichment of Gad2 in Npy1R-RiboTagged cells (Fig. 3c), we predicted that these neurons were GABAergic42. Following recording, we harvested the cytosol of the cells 46 and performed single-cell RT-PCR to test for Slc32a1 (Vgat) expression. The majority of cells were Vgat-positive—out of 11 successfully harvested Actb-positive cells, 7 were positive for Vgat expression, while the remaining 4 did not amplify either Vgat or Vglut templates (Fig. 5g). These data are consistent with the idea that a subset of AgRP neurons synapse on a population of inhibitory, NPY-responsive cells in the MeA that project to the pBNST.
Npy1RMeA → pBNST signaling influences territorial behavior
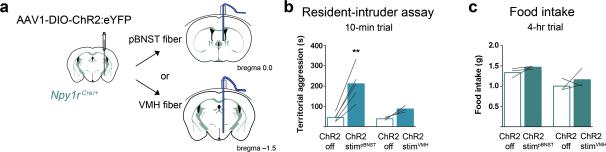
The Npy1RMeA population projects to numerous efferent targets, some of which have been implicated in aggressive behavior, including the VMH and PAG 22, 47. Because AgRP neurons make a disynaptic connection to the pBNST via the MeA, we hypothesized that this circuit may be involved in aggressive/territorial behaviors. We virally transduced Npy1rCre mice unilaterally with ChR2:YFP virus and placed a fiber optic cannula above either the ipsilateral VMH or pBNST (Fig. 6a). Optogenetic stimulation of Nyp1RMeA fibers in the pBNST evoked significantly more territorial defensive behavior (Fig. 6b). However, rather than overt attack behavior, the pBNST-fiber stimulation increased nudging activity; the resident mouse followed the intruder for the majority of the assay, constantly nudging the intruder into the wall of the cage (Supplementary Movie 3). As opposed to violent aggression, a nudging threat display may be adequate for territorial defense from most competitors (Supplementary Fig. 7e). We activated Npy1RMeA soma using metabotropic hM3Dq DREADD receptors while we stimulated the Npy1RMeA fibers in the pBNST using ionotropic photostimulation; hence, we cannot exclude the possibility that this difference accounts for the behavioral difference.
Figure 6.
Npy1RMeA neurons that project to the pBNST evoke territorialism. (a) Unilateral injection of AAV-DIO-ChR2:YFP into the MeA of Npy1rCre mice, with ipsilateral optic cannulas implanted above the pBNST or the VMH. (b) Stimulation of NpyRMeA fibers over pBNST increased aggression in a home-cage intruder assay (left panel), while stimulation of fibers over VMH does not (right panel); assessed during the dark cycle (17:00 – 19:00) and compared using paired two-tailed Student's t-tests. (c) Neither VMH nor pBNST-projecting Npy1RMeA cells decreased feeding (recorded from 17:00 – 21:00); compared using paired two-tailed Student's t-tests. Stimulation paradigm (472 nm, 10 mW power at the tip): 10 Hz with 5-ms pulses that continue for 5 s followed by 2 s light-off recovery. See methods for detailed statistics.
Optogenetic stimulation of Npy1RMeA fibers in the VMH did not result in significant differences in territorial aggression (Fig. 6b). We also measured the effect of photostimulating Npy1RMeA axons in the pBNST or VMH on food intake, but did not observe a significant effect of stimulating either of these projections (Fig. 6c). Because Npy1R neurons project to numerous downstream targets, it is likely that the overt/violent aggression observed following Npy1RMeA neuron stimulation (Fig. 3f and Supplementary Movie 1,2), and also feeding behavior (Fig. 3g) are orchestrated by projections to targets other than the pBNST or VMH. The modulatory effect of AgRPMeA fiber stimulation on territorial behavior likely involves Npy1R neurons that project to the pBNST, a circuit that can be modulated under physiological conditions of negative energy balance.
DISCUSSION
Risk assessment and territorialism require sensory processing of environmental cues. Rodents select a territorial domain for nesting and foraging with respect to the risk of predation and will defend the limited resources of this area from conspecific intruders. However, under conditions of starvation, mice forage in more exposed or threatening areas and are less willing to defend a territory that is depleted of resources, exemplifying a behavioral adaptation that is associated with an internal state change 48.
Hungry animals will aggressively defend limited food resources from competitors. However, if food is depleted, starving mice try to escape from their territorialized home cage (Supplementary Fig. 1a, c), display less aggression toward an intruding conspecific, are less anxious, and engage in risky exploration to seek food. This shift in behavior is accompanied by a coincident change in AgRP neuron activity. In the absence of food, AgRP neurons are activated by interoceptive cues of negative energy balance, but when food or food-related cues are present, AgRP neuron activity is rapidly silenced 49, 50. We demonstrate that a subset of AgRP neurons can evoke GABA-mediated inhibition of the MeA and argue that this circuit is responsible for modulating aggressive territorial behavior when food is limited. If food is discovered during foraging, this cue should rapidly relieve GABA-mediated AgRP inhibition of the MeA, providing a switch to adjust behavior for food acquisition. This behavioral switch is difficult to model in the confines of a small arena and isolated housing conditions. When food was presented to hungry residents (artificially induced by AgRP neuron-stimulation) during an intruder trial, they choose to eat rather than interact with the intruder (Supplementary Fig. 3). Fluctuations in territorial aggression levels with respect to a limited or depleted food source could be evaluated with the development of techniques to study and track individual animals in a large, group-housed arena.
We propose that the AgRP → MeA circuit is involved in territorial adaptations during starvation, but questioned whether hunger itself could influence territorialism. In this experiment we targeted the AgRP → PVH circuit which has been demonstrated to evoke robust feeding behavior, equivalent to that following a fast. Unlike activating AgRP → MeA fibers, stimulation of the AgRP → PVH circuit did not reduce territorial aggression (Fig. 2g). These data support the idea that hunger itself does not change territorial behavior. We define an MeANpy1R → pBNST circuit that is downstream of AgRP and can alter territorial behavior. There are, however, many other AgRP targets throughout the brain, and similar to feeding, they may play redundant roles in territorial adaptations.
The behavioral adaptations that occur during starvation facilitate food acquisition and minimize unnecessary energy expenditure, a complex state-change likely attributed to the broad projection profile AgRP neurons throughout the brain. Not all anatomically distinct AgRP subsets contribute equally to food-intake behavior. Optogenetic stimulation of AgRP fibers in the PVH evokes maximal food consumption, while stimulation of AgRP fibers in the paraventricular thalamus and parabrachial nucleus evoke lesser, if any, influence on food consumption 17, 29. Similarly, we find that AgRP fibers in the MeA can evoke food intake, but the magnitude of this effect is much less compared to equivalent fiber stimulation in the PVH. The long-term consequence of inhibiting cells in the MeA is a significant increase in body weight (our data and ref 31). One idea to resolve the differing degrees of food consumption observed by activating distinct AgRP neuron target regions is that their contributions are additive; however, because PVH stimulation is equivalent to stimulating AgRP cell bodies, that explanation is unlikely. Instead, brain regions where AgRP-axon stimulation promotes less food consumption may help coordinate non-feeding behaviors with hunger. For example, AgRP-mediated inhibition of the MeA induces feeding and suppresses territorialism, providing a circuit that can function independently to coordinate two behaviors. It is also possible that under some conditions, select populations of AgRP neurons become activated. The potential for AgRP neurons to orchestrate a complex behavioral response is broad. Future studies detailing the behavioral and physiological contribution of other AgRP targets will provide a complete profile of the starved-state behavioral response.
Online Methods
Animals
Adult male mice (2-6 months old) were bred onto a C57BL/6 background and housed on a 12-hr light cycle (5:00 – 17:00). All experiments were approved by The Animal Care and Use Committee at the University of Washington, and in accordance with NIH guidelines. AgrpCre:GFP and RiboTag (Rpl22fs-Rpl22:HA) knock-in mice have been characterized 34, 51. The Cre-dependent reporter strain, Rosa26Sor fs-tdT, was acquired from Jackson Laboratories (stock# 007914).
Npy1rCre:GFP knock-in mice were generated by gene targeting in ES cells. A Cre:GFP fusion gene was inserted just 5’ of the normal initiation codon for Npy1r. The targeting construct had 3.5 kb 5’ and 3’ arms that were prepared by PCR (Npy1r gene) and inserted into a targeting vector with SvNeo for positive selection and HSV-TK and PGK-DTa for negative selection (Npy1rCre targeting construct, Fig. 3a). After targeting, Cre:GFP expression is under control of Npy1r regulatory elements. The Sv40-Neo selectable gene can be removed by action of FLP recombinase.
Stereotaxic injections and tissue preparation
Stereotaxic surgery and injection coordinates
Mice were anesthetized and positioned on a stereotaxic alignment device (David Kopf instruments). During the procedure, body temperature was maintained with a heating pad and a nose cone delivered isoflurane (1.5 – 2%). Either a Hamilton syringe (88000) or pulled glass capillary was used to inject the target brain regions; ARH / bregma −1.25, lateral ±0.25, ventral −5.8; MeA (viral injections) / bregma −1.0, lateral ±2.25, ventral −5.55 and pBNST / bregma 0.4, lateral ±0.9, ventral −4.25. Cannulas were implanted 0.5 mm above the position of the injection with the exception of the MeA in which the cannula was implanted caudal to the injection at bregma −1.5.
Fiber placement
MeA and PVH tracks were evaluated at bregma −1.5 mm and −0.8 mm, respectively. Terminal placement is represented in Supplementary Fig. 4. It is likely that some of the MeA-targeted fibers terminated in the lateral ventricle, as we did not find terminals for every implant.
Viruses and neuronal tracers
AAV serotype 1 viruses (AAV1-Ef1α-DIO-hM3Dq:YFP, AAV1-Ef1α-DIO-hM3Dq-mCh, AAV1-CAG-DIO-GFP:TeTx, AAV1-Ef1α-DIO-ChR2:YFP, AAV1-Ef1α-DIO-Synaptophysin:YFP, AAV1-Ef1α-DIO-YFP) were generated at the University of Washington as described 52. We did not observe side effects in animals injected with any of the AAV1 viruses. AAV1 virus (500 nL) was injected at a titer of ~109 viral particles per μl. Behavioral tests began following a minimum 2-wk, post-operative incubation. For viral tracing using Syn:GFP, the tissue was harvested after a minimum of 5 days after viral injection. H129Δfs-TkTT was generously provided by David Anderson and Liching Lo 37. H129 virus (300 nL) was diluted and injected at ~106 viral particles per μl. Following a 5-day incubation, H129-injected animals were euthanized and the tissue collected for analysis. To label afferents, we injected 300 nL of green RetroBeads (Lumafluor, RetroBeads™ IX) into the desired target location.
Histology
Mice were anesthetized and transcardially perfused with saline, followed by 4% (w/v) paraformaldehyde in 0.1 M phosphate buffer (PB). Brains were post-fixed (6 hr), washed, cryoprotected in 0.1 M PB with 30% sucrose (w/v), embedded in OCT and frozen at −80 C. For immunohistochemistry, 30-μm floating sections were stained for YFP or tdTomato with the primary antibodies: rabbit anti-GFP (Life Technology A11122; diluted 1:2000) or rabbit anti-DsRed (Clonetech 632496; diluted 1:1000). Antibodies were diluted in 0.1M PB with 0.1% Triton and 2% donkey serum and developed overnight at 4 C.
CNO administration for Fos analysis
CNO was injected at a dose of 1.0 mg/kg body weight 2 hr prior to euthanizing the animals. The tissue was stained with goat anti-Fos (Santa Cruz 48869, diluted 1:300).
Electrophysiology
Figure 2b
Whole-cell recordings were made using an Axopatch 700B amplifier (Molecular Devices) with filtering at 1 KHz using 4-6 MΩ electrodes. Coronal brain slices (250 μm) were prepared in an ice slush solution containing (in mM): 250 sucrose, 3 KCl, 2 MgSO4, 1.2 NaH2PO4, 10 D-glucose, 25 NaHCO3, 0.1 CaCl2. Slices recovered for 1 h at 34°C in artificial cerebral spinal fluid (ACSF) continually bubbled with O2/CO2 and containing (in mM): 126 NaCl, 2.5 KCl, 1.2 NaH2PO4, 1.2 MgCl2 11 D-glucose, 18 NaHCO3, 2.4 CaCl2. Patch electrodes were filled with an internal solution containing (in mM): 130 CsCl, 2 MgCl2, 0.5 EGTA, 10 HEPES, 0.25 Na-GTP, and 2.5 Mg-ATP, pH 7.2-7.4, 280 mOsm. ACSF at 32°C was continually perfused over slices at a rate of ~2 ml/min during recording. Drugs were purchased from Abcam and were applied to the bath where indicated at the following concentrations: CNQX: 10 μM; APV: 100 μM; picrotoxin: 100 μM. For light-evoked responses the fiber optic cable was lowered into the bath and 10-ms light pulses (10 mW) were delivered at a rate of 0.1 Hz while cells were held in voltage-clamp mode at −70 mV. Example traces are averages of 15 sweeps.
Figure 5
Coronal brain slices were prepared as described 53. Whole-cell patch recordings were performed in voltage clamp and current clamp using an Olympus BX51W1 upright microscope equipped with video-enhanced, infrared-differential interference contrast (IR-DIC) and an Exfo X-Cite 120 Series fluorescence light source. Electrodes were fabricated from borosilicate glass (1.5 mm outer diameter; World Precision Instruments, Sarasota, FL) and filled with a normal internal solution (in mM): 128 potassium gluconate, 10 NaCl, 1 MgCl2, 11 EGTA, 10 HEPES, 2 ATP, and 0.25 GTP (pH was adjusted to 7.3-7.4 with 1N KOH, 290-300 mOsm); for measurement of IPSCs, patch pipettes were filled with a high chloride solution (in mM): 140 mM KCl, 10 mM Hepes, 0.1 mM EGTA, 5 mM MgCl2, 0.3 mM Na-GTP, and 5 mM K2-ATP, pH was adjusted to 7.3-7.4 with 1N KOH, 290-300 mOsm. Pipette resistances ranged from 3–5 MΩ. In whole cell configuration, access resistance was less than 20 MΩ; access resistance was 80% compensated. For optogenetic stimulation, a light-induced response was evoked using a light-emitting diode (LED) 470 blue light source controlled by a variable 2A driver (ThorLabs, Newton, NJ) with the light path directly delivered through an Olympus 40× water-immersion lens. Example traces are averages of 10 sweeps. Electrophysiological signals were digitized with Digidata 1322A (Molecular Devices, Foster City, CA), and the data were analyzed using p-Clamp software (version 9.2, Molecular Devices). The liquid junction potential was corrected for all data analysis.
Behavior
To minimize the hierarchical dominance behavior observed in adult group-housed male mice, our behavior studies were performed on singly-housed mice. The behavior results were scored blindly.
AgRP neuron-stimulated cohort: prescreening
Numerous studies using viral targeting as a means of activating AgRP neurons have found that the efficiency (number of cells transduced) correlates with the magnitude of the feeding response when activated 11, 12. Based on this, we used a prescreening criteria to select animals that consumed at least 1 g of food 4 hr after administration of CNO (1mg/kg).
Food-challenge assay
Rooms of a two-chamber arena were distinguished by visual (vertical striped or wood grain wall paper), olfactory (Nestlet below the floor containing 3 drops of either almond or orange extract) and tactile (wire mesh versus metal bar flooring) cues (Fig. 1a). Animals were exposed to this context on 3 consecutive days during the light cycle (5:00 – 17:00). On habituation Day 1, mice were placed in the almond side of the cage and allowed to explore both chambers of the arena for a 30-min session. Mice were excluded from the test if they displayed ≥70% preference for either room. On training Day 2, mice were again placed in the almond side; upon movement into the orange chamber, the door dividing the chambers was closed and a shock paradigm (2 s, 0.3-mA shock, every 3 min over the course of 30 min) was initiated. Immediately following the last shock interval, mice were removed and placed in their home cage. Following a 2 hr inter-trial interval, mice were returned to the almond chamber, the dividing door was shut and the mice did not receive shocks during this 30-min session. Food was present below the floor grid, on the orange side on Days 1 and 2. On testing Day 3, mice were placed on the almond side with the chamber door open and their movement was tracked for a 30-min session. Depending on the experimental conditions, food was either available above the floor grid (food-available cohort) or was again placed below the floor grid (food-blocked cohort) in the orange/shock chamber. Food-available mice were excluded from the test if they moved the food pellet into the almond side during the test session. Video recordings on Days 1 and 3 were evaluated manually in a blinded manner. Group 1, fed versus fasted: all animals were food deprived between Day 1 and Day 2 training. Food was returned to “fed” animals following training, while “fasted” animals were continuously deprived. Group 2, AgRP stimulated (hM3Dq + CNO) versus AgRP controls (YFP + CNO): on test day, food was removed from the home-cage hopper 1 h prior to testing and animals were injected with CNO (1 mg/kg body weight). Group 3, Npy1R silenced (TetTox) versus Npy1R controls (YFP): food was provided ad libitum throughout the trial. Note, we did not repeat this test on individual subjects, each animal was evaluated once.
Elevated Plus Maze
Movement within the arms of a plus maze was recorded and scored for time spent in the open arms. The trial was conducted during the light cycle (5:00 – 17:00). The open-arm score is the cumulative time the animal spent in either open arm during the 10-min session. The center of the maze was excluded from scoring. The animal's movement was video recorded and scored using Ethovision XT (Noldus) video-tracking software. Mice were excluded from the study if they failed to stay on the maze for the duration of the trial. Group 1, fed versus fasted: fasted animals were food deprived for 48 h prior to testing. A separate cohort of age-matched mice, given ad libitum access to food was used as controls. Group 2, AgRP-stimulated (hM3Dq + CNO) versus AgRP controls (YFP + CNO): on test day, food was removed from the home-cage hopper 1 h prior to testing and animals were injected with CNO (1mg/kg). Group 3, Npy1R silenced (TetTox) versus Npy1R controls (YFP): food was provided ad libitum throughout the trial. We found that a second exposure to this maze will bias the animals’ movement. Thus, we evaluated only one trial for each animal.
Resident-intruder assay
Resident males were sexually experienced and territorialized to an isolated home cage for a period of two weeks prior to testing. Intruder males were group-housed littermates between 7 to 10 weeks of age and weighed less than resident animals (Supplementary Fig. 2a). The test consisted of exposing a resident animal to an unfamiliar intruder for a 10-min session. In order to expose the cage arena for video recording, the wire rack (containing food and water) was removed from the cage 1 h prior to testing. CNO versus saline control trials: CNO (Tocris ®, diluted in saline and administered intraperitoneally at 1 mg/kg body weight); saline (administered intraperitoneally at 10 uL/g body weight). Injections were performed 30 min prior to testing. Optogenetic trials: light stimulation (pulse paradigm Fig 2f) versus tethered controls (attached to dummy patch cords with no light stimulation). Stimulated animals were exposed to light 30 min prior to, and during testing. Anesthetized intruders were injected with ketamine (0.75 mg/kg; Ketaset® Pfizer). All trials were conducted within the first 2 h of the dark cycle (17:00 – 19:00) and video recorded with infrared lighting. The videos were scored in a blinded manner using the manual settings of Ethovision XI. Aggressive behavior included: holding, fighting (boxing/attacking/mounting), high-speed chasing, and nudging (Supplementary Fig. 7e). We did not observe aggressive behavior initiated by intruders. In some cases aggression was tested twice on the same animal (paired tests). Because aggression levels may change with repeated testing, we designed a randomized crossover study with respect to the experimental and control conditions. The tests were separated by a week and unique intruders were used in the second trail.
Optogenetics
Fiber-optic cannulas were assembled as described 54. A 0.22 NA, 220-μm coated multimode fiber (Thorlabs, FG200LEA) was fit into a 2.5-mm ferrule (custom bore hole of 230 μm; Precision Fiber). The fiber lengths for targeted sites were: MeA, 5.0; pBNST, 3.4; VMH, 5.0 mm. Custom duel-fiber-optic cannulas (fibers 1 mm apart and 4.5 mm long, Doric Lenses) and patch cords were used for PVH fiber stimulation. Before implantation, each cannula was evaluated for power loss at the fiber ending. Fiber-optic cannulas were permanently fixed to the skull using a layer of C&B Metabond (Parkell), followed by a top coat of Jet Acrylic cement (Lang Dental). Cannulas were connected to a blue (473 nm) laser (LaserGlow, LSR-0473) via a patch cord (0.22 NA, Doric Lenses), and set to deliver light at 10 mW from the end of the implanted fiber. Pulses (5 ms) were delivered at 10 Hz for 5 s, followed by a 2 s no-light recovery. The laser waveform was generated by Master-8 pulse stimulator (AMPI).
RiboTag
The MeA was isolated from a 2-mm-thick coronal section. Tissue punches from 6 male animals (Npy1rCre;RiboTag) were pooled and homogenized in buffer, as described previously 34. An antibody against HA (mouse-α-HA; Covance MMS-101R) was used to precipitate the epitope-tagged ribosomes and associated transcripts while the remaining lysate was used as the input sample as described 34. Transcript expression levels were quantified by qRT-PCR.
PCR analysis
RiboTag qRT-PCR analysis
Transcript expression levels were quantified using either SYBR Green or TaqMan PCR assays from Agilent Technologies as described 51. The following primer sets were used for SYBR Green primer-mediated amplification: (Npy1r) 5′-TGATCTCCACCTGCGTCAAC/ 5′-ATGGCTATGGTCTCGTAGTCAT; (Slc17a6) 5’- AGAGAGCGCAAATCTGCTAGGT/ 5’-GCGTAGACGGGCATGGAT; (Cre) 5’-CTGCCACCAGCCAGCTAT/ 5’- GGGCACTGTGTCCAGACC; (Actb) 5’-AGTGTGACGTTGACATCCGTA/ 5’- GCCAGAGCAGTAATCTCCTTCT. The following probes were used for TaqMan amplification: Mc4r, ID Mm00457483_s1; Gad2, ID Mm00484623_m1; Cnp, ID Mm01306641_m1
Post-recording single-cell PCR
Following whole-cell recordings, the content of the patched cell was harvested by applying negative pressure to gently aspirate the cell content into the tip of the recording pipette. The cell content was expelled into a 500 μl harvesting tube containing 5 μl of RT solution and stored at −80 C until further processing. RNA transcripts in the harvested lysate were reverse transcribed and PCR was performed as previously described 46 using the following primer sets: Vglut2 (Slc17a6) 5’-ATCTGCTAGGTGCAATGG/ 5’- TAAGCTGGCTGACTGATG; (Slc32a1) 5’- GTCACGACAAACCCAAGATCAC/ 5’- GGCGAAGATGATGAGGAACAAC; (Actb) 5’- AAGGCCAACCGTGAAAAGAT; 5’-GTGGTACGACCAGAGGCATAC
STATISTICS
Statistical analysis was performed with GraphPad's Prism software. A P value of < 0.05 was the threshold for significance (*P < 0.05, **P < 0.01, ***P < 0.001), and all error bars represent standard errors of the mean. Unless otherwise indicated, all n values represent individual mice. No statistical methods were used to pre-determine sample sizes because many of the outcomes were unknown. When possible, sample sizes were based on previous studies 11, 17, 23, but all data was gathered from at least three independent experiments. The data distribution was assumed to be normal, but this was not formally tested. Please note, a supplementary methods checklist is also available through Nature Neuroscience.
Fig. 1d Sample size: ad lib, n = 15; fasted food-available, n = 7; fasted food-blocked, n = 7; YFP + CNO, n = 13; hM3Dq + CNO food-available, n = 6; hM3Dq + CNO food-blocked, n = 7. 1-Way ANOVA: F (5, 49) = 10.35, ***P < 0.0001. Bonferroni's multiple comparison tests: ad lib versus fasted food available, 95% CI (−10.67 to −2.78) ***P; ad lib versus fasted food blocked, 95% CI (−9.58 to −1.69) **P; YFP + CNO versus hM3Dq + CNO food available, 95% CI (−9.44 to −9.32) **P; YFP + CNO versus hM3Dq + CNO food blocked, 95% CI (-10.93 to -2.85) ***P.
Fig. 1e Sample size: ad lib, n = 8; fasted, n = 7; YFP + CNO, n = 12; hM3Dq + CNO, n = 8. 1-Way ANOVA: F (3, 31) = 8.58, ***P = 0.0003. Bonferroni's multiple comparison tests: ad lib versus fasted, 95% CI (−3.58 to −0.05) *P; YFP + CNO versus hM3Dq + CNO, 95% CI (−3.79 to −0.68) **P < 0.01
Fig. 1f Sample size: ad lib vs fasted, n = 12; hM3Dq + saline vs hM3Dq + CNO, n = 10. Paired two-tailed Student's t-test: ad lib (M = 52.42, SD = 21.10) versus fasted (M = 33.58, SD = 13.60): t (11) = 2.87, *P = 0.015; hM3Dq + saline (M = 57.61, SD = 28.16) versus hM3Dq + CNO (M = 11.73, SD = 9.76): t (9) = 5.16, ***P = 0.0006
Fig. 2g Sample size: ChR2 off vs ChR2 stimMeA, n = 8; ChR2 off vs ChR2 stimPVH, n = 6. Paired two-tailed Student's t-test: ChR2 off (M = 58.62, SD = 17.97) versus ChR2 stimMeA (M = 27.93, SD = 11.27): t (7) = 4.67, **P = 0.0023; ChR2 off (M = 58.73, SD = 12.54) versus ChR2 stimPVH (M = 76.37, SD = 22.82): t (5) = 2.02, not significant P = 0.0999
Fig. 2h Sample size: ChR2 off n = 18; ChR2 stimMeA, n = 8; ChR2 stimPVH, n = 10. 1-Way ANOVA: F (2, 33) = 58.68, ***P < 0.0001. Bonferroni's multiple comparison tests: ChR2 off versus ChR2 stimMeA, 95% CI (−1.29 to −0.25) **P; ChR2 off versus ChR2 stimPVH, 95% CI (−2.56 to −1.59) ***P.
Fig. 3f Sample size: YFP + CNO, n = 8 vs hM3Dq + CNO, n = 10. Unpaired two-tailed Student's t-test: YFP + CNO (M = 57.88, SD = 16.42) versus hM3Dq +CNO (M = 140.0, SD = 54.23): t (16) = 4.11, ***P = 0.0008
Fig. 3g Sample size: YFP + CNO vs n = 8; hM3Dq + CNO, n = 8. Unpaired two-tailed Student's t-test: YFP + CNO (M = 1.25, SD = 0.19) versus hM3Dq + CNO (M = 0.49, SD = 0.18): t (14) = 8.15, ***P < 0.0001
Fig. 3i Sample size: YFP, n = 6 vs TetTox, n = 6. Unpaired two-tailed Student's t-test: YFP (M = 7.16, SD = 2.26) versus TetTox (M = 12.71, SD = 2.31): t (10) = 4.21, **P = 0.0018
Fig. 3j Sample size: TetTox, n = 6; YFP, n = 6. 2-Way repeated measures ANOVA. Bonferroni's multiple comparisons: week 8, **P < 0.01; week 9 *P < 0.05; week 10 **P < 0.01; week 11 ***P < 0.001
Fig. 3k Sample sizeYFP, n = 6 vs TetTox, n = 6. Unpaired two-tailed Student's t-test: YFP (M = 46.83, SD = 28.32) versus TetTox (M = 31.05, SD = 20.66): t (10) = 1.10, not significant P = 0.296
Fig. 6b Sample size: ChR2-off vs ChR2 stimpBNST, n = 5; ChR2-off vs ChR2 stimVMH n = 3. Paired two-tailed Student's t-test: ChR2-off (M = 45.40, SD = 20.45) versus ChR2 stimpBNST (M = 211.0, SD = 82.79): t (4) = 5.86, **P = 0.004; ChR2-off (M = 38.33, SD = 8.02) versus ChR2 stimVMH not significant (M = 86.67, SD = 16.17): t (2) = 3.46, not significant P = 0.074
Fig. 6c Sample size: ChR2-off vs ChR2 stimpBNST, n = 5; ChR2-off vs ChR2 stimVMH n = 3. Paired two-tailed Student's t-test: ChR2-off (M = 1.34, SD = 0.12) versus ChR2 stimpBNST (M = 1.47, SD = 0.04): t (2) = 1.60, not significant P = 0.251; ChR2-off (M = 1.00, SD = 0.58) versus ChR2 stimVMH (M = 1.16, SD = 0.56): t (2) = 0.73, not significant P = 0.540
Supplementary Fig. 2a Sample size: ad lib (n = 6) vs 48-hr fasted (n = 6). Unpaired two-tailed Student's t-test: ad lib (M = 7.65, SD = 3.09) versus 48-hr fasted (M = 7.71, SD = 3.45): t (10) = 0.03, not significant P = 0.976
Supplementary Fig. 2b Sample size: intruders (n = 23); residents, ad lib (n = 12); residents, fasted (n = 12). 1-Way ANOVA: F (2, 44) = 46.64, ***P < 0.0001. Bonferroni's multiple comparison tests: intruder versus resident, ad lib, 95% CI (−9.17 to −5.61) ***P, intruder versus resident, fasted, 95% CI (−4.85 to −1.29) ***P.
Supplementary Fig. 2c Sample size: ad lib vs 24-hr fasted (n = 6). Paired two-tailed Student's t-test: ad lib (M = 59.27, SD = 27.92) versus 24-hr fasted (M = 27.62, SD = 18.09): t (5) = 3.41, *P = 0.019
Supplementary Fig. 2e Sample size: ad lib vs 48-hr fasted (n = 9). Paired two-tailed Student's t-test: ad lib (M = 17.54, SD = 4.15) versus fasted (M = 57.82, SD = 13.53): t (8) = 8.90, *** P = 0.0001.
Supplementary Fig. 2f Sample size: ad lib vs 48-hr fasted (n = 9). Paired two-tailed Student's t-test: ad lib (M = 7.29, SD = 4.10) versus fasted, (M = 3.75, SD = 22.17): t (8) = 3.63, **P = 0.007
Supplementary Fig. 3a Sample size: hM3Dq + saline or hM3Dq + CNO (no food), n = 10; hM3Dq + CNO (with food), n = 7. 1-Way ANOVA: F (2,24) = 20.90, ***P < 0.0001. Bonferroni's multiple comparison tests: saline versus CNO (no food), 95% CI (25.91 to 65.85) ***P; saline versus CNO (with food), 95% CI (28.89 to 72.90) ***P.
Supplementary Fig. 6 Sample size:YFP vs n = 4; TetTox, n = 4. Unpaired two-tailed Student's t-test: YFP (M = 2.11, SD = 0.62) versus TetTox (M = 1.72, SD = 0.84): t (6) = 0.75, P = 0.479.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Kafer and M. Chiang for technical assistance generating the new line of mice and maintaining the mouse colonies. We thank M.A. Patterson, and B.C. Jarvie for careful reading of this paper and the entire Palmiter laboratory for helpful discussions and critiques. We thank D.J. Anderson and L. Lo for generously providing H129Δ-fs-TK-TT. We thank E. Strakbein at U.W. Scientific Instruments for the development of tools and apparatuses used in this manuscript. This work was supported by funds from the Hilda Preston Davis Foundation (SLP); the National Institutes of Health (R01DK068098; MJK&OKR), (R01MH094536; LSZ) and (R01DA024908; RDP); Marie Sklodowska-Curie award (H2020-MSCA-IF-2014-658352; ES); AQ is a Ramón y Cajal fellow (RyC-2012-11873) and is funded by European Research Council Starting grant NEUROMITO (ERC-2014-StG-638106) and MINECO Proyectos I+D de Excelencia (SAF2014-57981P)
Footnotes
AUTHOR CONTRIBUTIONS
SLP designed the study under the guidance of RDP; JQ performed and analyzed electrophysiological data in the laboratories of MJK and OKR; MES performed and analyzed electrophysiology data in the laboratory of LSZ; ES performed the RiboTag pulldown and qPCR in the laboratory of AQ; CCN performed single-cell PCR on harvested cells post whole-cell recordings; RDP generated the AgRPCre and Npy1rCre lines of mice; FDB assisted with behavior experiments and blind scoring; RDP, MJK, OKR and LSZ provided laboratory space and resources to conduct the experiments; SLP and RDP wrote the manuscript with revisions and input from all contributing authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Sih A. Optimal Behavior - Can Foragers Balance 2 Conflicting Demands. Science. 1980;210:1041–1043. doi: 10.1126/science.210.4473.1041. [DOI] [PubMed] [Google Scholar]
- 2.Krebs JR. Optimal Foraging, Predation Risk and Territory Defense. Ardea. 1980;68:83–90. [Google Scholar]
- 3.Brown JL. Citation Classic - the Evolution of Diversity in Avian Territorial Systems. Cc/Agr Biol Environ. 1981:16–16. [Google Scholar]
- 4.Anderson PK. Foraging Range in Mice and Voles - the Role of Risk. Can J Zool. 1986;64:2645–2653. [Google Scholar]
- 5.Magnhagen C. Predation Risk and Foraging in Juvenile Pink (Oncorhynchus-Gorbuscha) and Chum Salmon (O-Keta). Can J Fish Aquat Sci. 1988;45:592–596. [Google Scholar]
- 6.Kohler SL, Mcpeek MA. Predation Risk and the Foraging Behavior of Competing Stream Insects. Ecology. 1989;70:1811–1825. [Google Scholar]
- 7.Whitham J, Mathis A. Effects of hunger and predation risk on foraging behavior of graybelly salamanders, Eurycea multiplicata. J Chem Ecol. 2000;26:1659–1665. [Google Scholar]
- 8.Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- 9.Krashes MJ, Shah BP, Koda S, Lowell BB. Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab. 2013;18:588–595. doi: 10.1016/j.cmet.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luquet S, Perez FA, Hnasko TS, Palmiter RD. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310:683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 11.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011 doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath TL, Bechmann I, Naftolin F, Kalra SP, Leranth C. Heterogeneity in the neuropeptide Y-containing neurons of the rat arcuate nucleus: GABAergic and non-GABAergic subpopulations. Brain Res. 1997;756:283–286. doi: 10.1016/s0006-8993(97)00184-4. [DOI] [PubMed] [Google Scholar]
- 14.Broberger C, Johansen J, Johansson C, Schalling M, Hokfelt T. The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proc Natl Acad Sci U S A. 1998;95:15043–15048. doi: 10.1073/pnas.95.25.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 16.Padilla SL, Carmody JS, Zeltser LM. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med. 2010;16:403–405. doi: 10.1038/nm.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betley JN, Cao ZF, Ritola KD, Sternson SM. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell. 2013;155:1337–1350. doi: 10.1016/j.cell.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23:7143–7154. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishi T, et al. Expression of melanocortin 4 receptor mRNA in the central nervous system of the rat. J Comp Neurol. 2003;457:213–235. doi: 10.1002/cne.10454. [DOI] [PubMed] [Google Scholar]
- 20.Stowers L, Cameron P, Keller JA. Ominous odors: olfactory control of instinctive fear and aggression in mice. Curr Opin Neurobiol. 2013;23:339–345. doi: 10.1016/j.conb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereno GL, Balaszczuk V, Beltramino CA. Detection of conspecific pheromones elicits fos expression in GABA and calcium-binding cells of the rat vomeronasal system-medial extended amygdala. J Physiol Biochem. 2011;67:71–85. doi: 10.1007/s13105-010-0051-5. [DOI] [PubMed] [Google Scholar]
- 22.Lin D, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong W, Kim DW, Anderson DJ. Antagonistic Control of Social versus Repetitive Self- Grooming Behaviors by Separable Amygdala Neuronal Subsets. Cell. 2014;158:1348–1361. doi: 10.1016/j.cell.2014.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich MO, Zimmer MR, Bober J, Horvath TL. Hypothalamic Agrp neurons drive stereotypic behaviors beyond feeding. Cell. 2015;160:1222–1232. doi: 10.1016/j.cell.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laundre JW. How large predators manage the cost of hunting. Science. 2014;346:33–34. doi: 10.1126/science.1260170. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Anderson DJ. Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature. 2010;463:227–231. doi: 10.1038/nature08678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 29.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Garza JC, Li W, Lu XY. Melanocortin-4 receptor in the medial amygdala regulates emotional stress-induced anxiety-like behaviour, anorexia and corticosterone secretion. Int J Neuropsychopharmacol. 2013;16:105–120. doi: 10.1017/S146114571100174X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King BM, Cook JT, Rossiter KN, Rollins BL. Obesity-inducing amygdala lesions: examination of anterograde degeneration and retrograde transport. Am J Physiol Regul Integr Comp Physiol. 2003;284:R965–982. doi: 10.1152/ajpregu.00249.2002. [DOI] [PubMed] [Google Scholar]
- 32.Xu P, et al. Estrogen receptor-alpha in medial amygdala neurons regulates body weight. J Clin Invest. 2015;125:2861–2876. doi: 10.1172/JCI80941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S, Boey D, Herzog H. NPY and Y receptors: lessons from transgenic and knockout models. Neuropeptides. 2004;38:189–200. doi: 10.1016/j.npep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heimer-McGinn V, Murphy AC, Kim JC, Dymecki SM, Young PW. Decreased dendritic spine density as a consequence of tetanus toxin light chain expression in single neurons in vivo. Neurosci Lett. 2013;555:36–41. doi: 10.1016/j.neulet.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, Soleiman M, Soden ME, Zweifel LS, Palmiter RD. A neural circuit for aversive teaching signal during associative learning. Cell. 2015 in press. [Google Scholar]
- 37.Lo L, Anderson DJ. A cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- 39.Canteras NS, Simerly RB, Swanson LW. Organization of projections from the medial nucleus of the amygdala: a PHAL study in the rat. J Comp Neurol. 1995;360:213–245. doi: 10.1002/cne.903600203. [DOI] [PubMed] [Google Scholar]
- 40.Niimi K, et al. Heterogeneous electrophysiological and morphological properties of neurons in the mouse medial amygdala in vitro. Brain Res. 2012;1480:41–52. doi: 10.1016/j.brainres.2012.08.050. [DOI] [PubMed] [Google Scholar]
- 41.Bian X. Physiological and morphological characterization of GABAergic neurons in the medial amygdala. Brain Res. 2013;1509:8–19. doi: 10.1016/j.brainres.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Keshavarzi S, Sullivan RK, Ianno DJ, Sah P. Functional properties and projections of neurons in the medial amygdala. J Neurosci. 2014;34:8699–8715. doi: 10.1523/JNEUROSCI.1176-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cruikshank SJ, Urabe H, Nurmikko AV, Connors BW. Pathway-specific feedforward circuits between thalamus and neocortex revealed by selective optical stimulation of axons. Neuron. 2010;65:230–245. doi: 10.1016/j.neuron.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun QQ, Akk G, Huguenard JR, Prince DA. Differential regulation of GABA release and neuronal excitability mediated by neuropeptide Y1 and Y2 receptors in rat thalamic neurons. J Physiol. 2001;531:81–94. doi: 10.1111/j.1469-7793.2001.0081j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun QQ, Huguenard JR, Prince DA. Neuropeptide Y receptors differentially modulate G- protein-activated inwardly rectifying K+ channels and high-voltage-activated Ca2+ channels in rat thalamic neurons. J Physiol. 2001;531:67–79. doi: 10.1111/j.1469-7793.2001.0067j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch MA, Tonsfeldt KJ, Ronnekleiv OK. mRNA expression of ion channels in GnRH neurons: subtype-specific regulation by 17beta-estradiol. Mol Cell Endocrinol. 2013;367:85–97. doi: 10.1016/j.mce.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behbehani MM. Functional characteristics of the midbrain periaqueductal gray. Prog Neurobiol. 1995;46:575–605. doi: 10.1016/0301-0082(95)00009-k. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy A, et al. Internal States and Behavioral Decision-Making: Toward an Integration of Emotion and Cognition. Cold Spring Harb Symp Quant Biol. 2014;79:199–210. doi: 10.1101/sqb.2014.79.024984. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Lin YC, Kuo TW, Knight ZA. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell. 2015;160:829–841. doi: 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Betley JN, et al. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature. 2015 doi: 10.1038/nature14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanz E, et al. Fertility-regulating kiss1 neurons arise from hypothalamic pomc-expressing progenitors. J Neurosci. 2015;35:5549–5556. doi: 10.1523/JNEUROSCI.3614-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gore BB, Soden ME, Zweifel LS. Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Curr Protoc Neurosci. 2013;4:4, 35, 31–34, 35, 20. doi: 10.1002/0471142301.ns0435s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu J, et al. Rapid signaling of estrogen in hypothalamic neurons involves a novel G-protein- coupled estrogen receptor that activates protein kinase C. J Neurosci. 2003;23:9529–9540. doi: 10.1523/JNEUROSCI.23-29-09529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sparta DR, et al. Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nat Protoc. 2012;7:12–23. doi: 10.1038/nprot.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.