Abstract
Purpose
This study used uncertainty and sensitivity analysis to evaluate a physiologically based pharmacokinetic (PBPK) model of the complex mechanisms of sorafenib and its two main metabolites, sorafenib glucuronide and sorafenib N-oxide in mice.
Methods
A PBPK model for sorafenib and its two main metabolites was developed to explain disposition in mice. It included relevant influx (Oatp) and efflux (Abcc2 and Abcc3) transporters, hepatic metabolic enzymes (CYP3A4 and UGT1A9), and intestinal β-glucuronidase. Parameterization of drug-specific processes was based on in vitro, ex vivo and in silico data along with plasma and liver pharmacokinetic data from single and multiple transporter knock-out mice.
Results
Uncertainty analysis demonstrated that the model structure and parameter values could explain the observed variability in the pharmacokinetic data. Global sensitivity analysis demonstrated the global effects of metabolizing enzymes on sorafenib and metabolite disposition and the local effects of transporters on their respective substrate exposures. In addition, though hypothesis testing, the model supported that the influx transporter Oatp is a weak substrate for sorafenib and a strong substrate for sorafenib glucuronide and that the efflux transporter Abcc2 is not the only transporter affected in the Abcc2 knock-out mouse.
Conclusions
Translation of the mouse model to humans for the purpose of explaining exceptionally high human pharmacokinetic variability and its relationship with exposure dependent dose-limiting toxicities will require delineation of the importance of these processes on disposition.
Keywords: Sorafenib, Physiologically Based Pharmacokinetics, Influx Transporters, Efflux Transporters, Sensitivity Analysis
Introduction
Sorafenib is a multikinase inhibitor of C-RAF, B-RAF, c-KIT, FLT-3, PDGR-β, and VEGFR 1–3 [1,2]. It has been approved for the treatment of advanced renal cell carcinoma, hepatocellular carcinoma, and advanced thyroid cancer. In addition, it is being evaluated in acute myeloid leukemia and other solid tumors in adults and pediatrics [3–5]. Due to its large interindividual variability and thus potentially higher than expected exposure, there is the possibility for exposure-related toxicities such as hand-foot skin reaction (HFSR) [6–8]. Therefore, developing a better understanding of factors related to this variability is important in determining sorafenib treatments that increase efficacy and decease toxicity [9].
Sorafenib is a small lipophilic molecule, has a high fraction absorbed, and high bioavailability (92% and 80% respectively, in mice) [10]. The primary route of elimination is hepatic metabolism and CYP3A4 is the primary enzyme responsible for sorafenib oxidative metabolism to an N-oxide metabolite in mice and humans [10,11]. Sorafenib also undergoes conjugation to sorafenib glucuronide (SG) by UGT1A9. This accounts for the clearance of approximately 15% of the administered sorafenib dose in humans [12]. There was no effect of renal dysfunction on sorafenib clearance in humans [13] and 19% of radioactivity following oral administration was present in urine, almost exclusively in the form of sorafenib metabolites [12]. Therefore, renal elimination is not considered a major factor in sorafenib clearance. The plasma/serum protein binding is high with an unbound fraction of 0.5% in mice and humans [10].
Transporters play a major role in the disposition of sorafenib and its metabolites and several involved in influx (Oatp1b2) and efflux (Abcc2, Abcc3) are localized to hepatocytes [14–16]. Sorafenib is an OATP [17,18] and OCT1 substrate [18] in vitro although in Oatp1b2(−/−) knockout mice, sorafenib and its N-oxide metabolite plasma exposure were not different from wildtype. However, SG concentrations were substantially increased in Oatp1b2(−/−) knockout mice [17] and this increase was not the result of altered glucuronidation capacity in the knockout mice [17]. This suggests that Oatp1 transport of the glucuronide into the hepatocytes is an important step in SG clearance. Using in vitro studies and in vivo studies with wildtype and knockout mice, it was determined that once SG is transported into hepatocytes, its disposition depends on Abcc2 (Mrp2) which shuttles SG into bile as well as Abcc3 (Mrp3) which shuttles SG back into circulation [19]. The fate of SG in the bile and intestinal lumen was assessed ex vivo with mouse luminal contents. It was demonstrated using neomycin, a non-systemic antimicrobial that eliminates gastrointestinal flora, that deconjugation was mediated by β-glucuronidase [19]. In vivo, sorafenib was found in plasma of mice following a single SG dose and absent when SG was co-administered with neomycin [19] demonstrating the importance of β-glucuronidase in sorafenib disposition in mice. In healthy volunteers treated with neomycin, the systemic exposure of a single 400 mg dose of sorafenib was reduced by 54% (Nexavar® package insert) and it has been estimated that the sorafenib exposure in humans is increased by 50% due to enterohepatic circulation (EHC) following oral administration [20].
The complexities of sorafenib disposition point to the need for data integration that allows for data interpretation. Physiologically Based Pharmacokinetic (PBPK) models integrate system- and drug-level information to estimate drug disposition. Specifically, PBPK models explicit definitions of anatomy and physiology allow for mechanistic modeling of transporters and drug metabolizing enzymes while their drug-specific information including transporter/enzyme affinity along with plasma/serum protein and tissue binding affinities defines how the drug interacts with the organism. Integration of these data ultimately leads to predictions of drug disposition in the absence of in vivo information. Next, PBPK model assessment with in vivo PK data is used for model refinement and further model evaluation. For example, hypothesis testing and subsequent hypothesis generation, benchmarked with in vitro and in vivo data, can guide us to further understand the importance of specific processes affecting overall drug disposition. For tyrosine kinase inhibitors (TKIs), PBPK models have the potential to provide insight into drug resistance mechanisms, drug-drug interactions, tumor uptake and ultimately drug efficacy [21]. For sorafenib, a previous PBPK model was used to demonstrate a lack of PK interaction of sorafenib with everolimus and was further used to generate the hypothesis that saturation of transporters is responsible for the nonlinear dose-tumor exposure relationship [22]. Owing to a lack of data, this model focused only on sorafenib and did not include EHC.
A full understanding of the important processes driving sorafenib and its metabolite disposition needs to be deciphered in a preclinical study as this is not possible in humans. Therefore, the objective of this study was to use uncertainty and sensitivity analysis to evaluate the complex mechanisms of a preclinical PBPK model of sorafenib and its metabolites. To accomplish this we defined a PBPK model that accounts for metabolism, active transport, and EHC and predicts sorafenib, sorafenib N-oxide, and SG in the plasma and liver of mice. In addition, the model was used to test and generate hypotheses on how metabolism, active transport, and EHC affect the disposition of sorafenib and its metabolites. This study will guide the future extrapolation to a human model that will support our understanding of sorafenib related efficacy and dose-limiting toxicities.
Methods
Development of PBPK Model
The model describing sorafenib, sorafenib N-oxide, and SG was developed using PK-Sim® v5.3.2 and MoBi® v3.3.2 (Bayer Technology Services, Leverkusen, Germany). Unless otherwise described, the default anatomical and physiological parameters for mice defined in this software were used. Organ:plasma partition coefficients were predicted using the approach of Rodgers and Rowland [23,24]. The physicochemical parameters used in the model are presented in Supplemental Table S1.
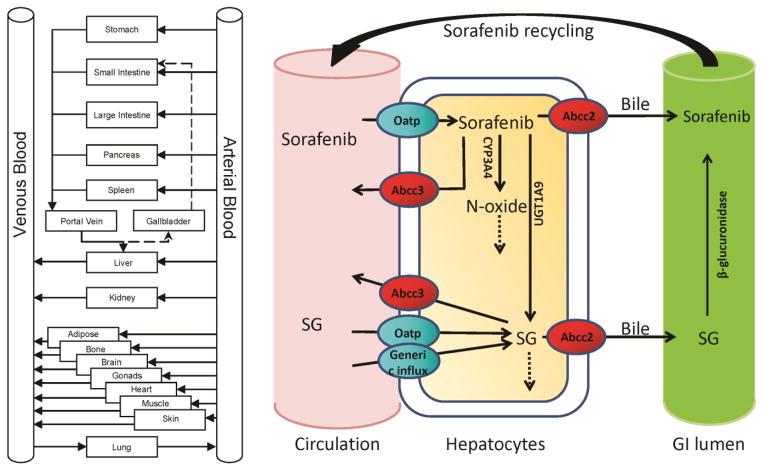
The active processes that may be responsible for metabolism and transport of sorafenib and its metabolites are presented in Figure 1. The hepatic enzymes CYP3A4 and UGT1A9 were modeled using a Michaelis-Menten process with literature-based Km values [11,25]. Metabolites were produced in the hepatic intracellular space and were available for metabolism, transport or diffusion as defined by the metabolite-specific processes. Influx and efflux transporters (sinusoidal and canalicular) were mostly modeled as linear due to limitations of available data, with the exception of the sorafenib Oatp transporter where an observed Km value was used [17]. The kinetics were determined using the plasma and liver concentration-time data from wild type (WT) and transporter knock-out (KO) mice and parameter estimation methods.
Figure 1.
Whole body PBPK model structure (left). Structure of Sorafenib and Sorafenib metabolite model including transporters (ovals) and metabolism processes in liver and gastrointestinal (GI) lumen (right).
Data supports the hypothesis that SG is converted back to sorafenib in the gut via β-glucuronidase. Therefore, the model included both EHC (via first-order kinetics) and β-glucuronidase activity in the gut. The recycling is accounted for by a continuous fraction of bile released into the gut (30% - default value) with subsequent bile dump every 8 hours. The β-glucuronidase activity was estimated ex-vivo in mouse cecal material treated with SG both with and without neomycin [19].
Experimental Data
Plasma and liver concentration-time data was obtained from Friend virus B-type (FVB) mice treated with 10 mg/kg of sorafenib, of the following genetic background:
WT; Oatp1a/1b KO [17] (used for parameter estimation)
WT; Oatp1a/1b KO; Abcc3 KO; Abcc2 KO; Oatp1a/1b and Abcc2 KO; Oatp1a/1b and Abcc3 KO; Oatp1a/1b, Abcc2 and Abcc3 KO [19] (used for simulations and hypothesis testing)
The activity of β-glucuronidase was estimated ex vivo using caecal material treated with SG. These experimental details have been previously reported [19]. Experiments were approved by the Institutional Animal Care and Use Committees of St. Jude Children’s Research Hospital.
Parameter Estimation
All of the organism-specific anatomical and physiological parameters were fixed at their default values and certain parameters associated with sorafenib or its metabolites were fixed to literature values (e.g. CYP3A4 Km, LogP, molecular weight). Model parameters that were less well-defined and that had supportive in vivo data were estimated using manual and numerical techniques and mouse WT and KO data. When estimating the parameters for the PBPK model we first exported the model to Matlab® (The Mathworks) using the MoBi® Toolbox for Matlab®. Then, within Matlab®, we used weighted least squares, where the weights were defined as 1/(n*concentration), where n is the number of samples in each group (e.g. plasma sorafenib, liver SG). Specifically, we used the plasma and liver concentration-time data from WT and Oatp1a\1b KO mice to estimate the following identifiable values: SG fraction unbound, SG Oatp1a/1b activity, UGT1A9 Vmax, and sorafenib N-oxide intrinsic clearance. These fixed and estimated parameters were set as the baseline values for all sensitivity and uncertainty analysis, hypothesis testing, and simulations in this study. Note that these parameters were defined for the purpose of understanding the observed differences in the experimental results and are not meant to be a unique set of values.
Sensitivity and Uncertainty Analysis
Uncertainty analysis was used to assess the prediction precision of the plasma and liver concentration-time profiles due to uncertainty in estimating the model parameters. We used Latin Hypercube Sampling (LHS) to simultaneously sample from selected inputs. In this process we used a uniform distribution where the parameters were varied ±two orders of magnitude about their nominal value. In addition, we assumed that the inputs were independent. A sensitivity analysis was completed that extended the uncertainty analysis by using Partial Rank Correlation Coefficients (PRCC) and the output from the uncertainty analysis to identify those parameters that contribute most to the prediction imprecision of model outputs [26,27]. The model parameters considered in the analysis were the Vmax for the drug-metabolizing enzymes, the rates for active transporters, non-specific hepatic intrinsic clearances, key parameters involved in EHC, and other physiological and physiochemical parameters where estimates were not well known. The model outputs considered were the Area Under the Curve (AUC0-8h) and the concentration at 8 hrs (e.g. a trough level or Cmin) for plasma and liver sorafenib, sorafenib N-oxide, and SG concentrations.
Simulations
Simulations were performed in Matlab® by exporting the PBPK model developed in PK-Sim® and MoBi® to Matlab® using the MoBi® Toolbox for Matlab®. In all simulations, only the parameters specified were altered while the remaining parameters were fixed to their nominal or estimated values (Supplemental Tables S1 and S2).
Results
Parameter Estimation
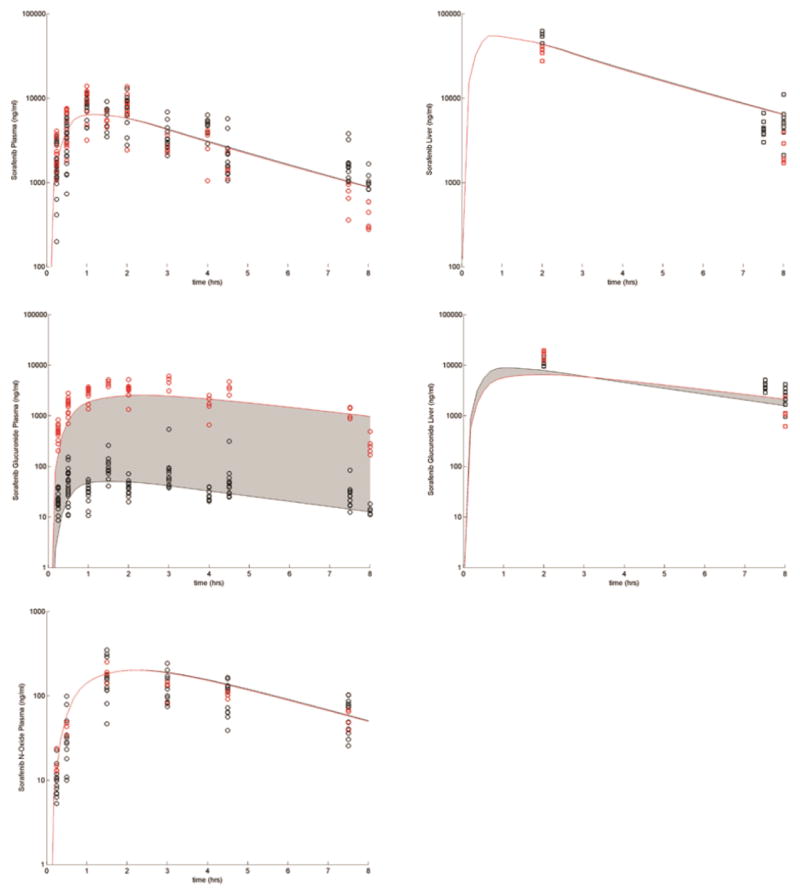
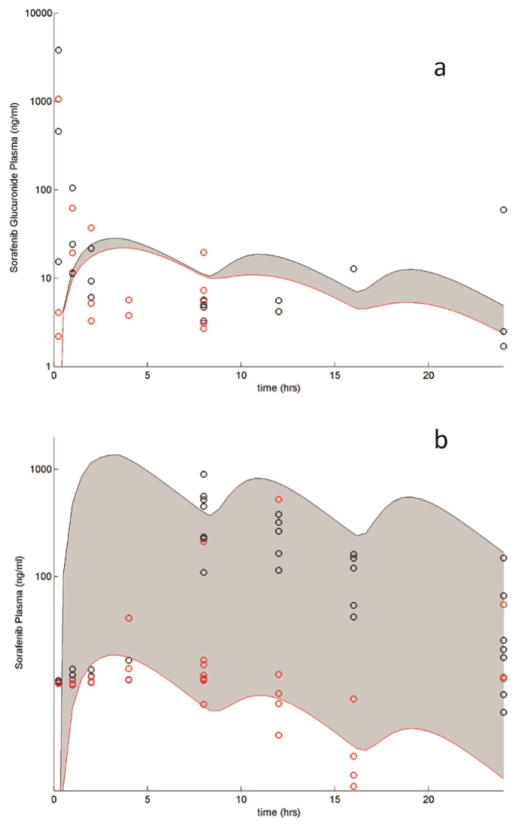
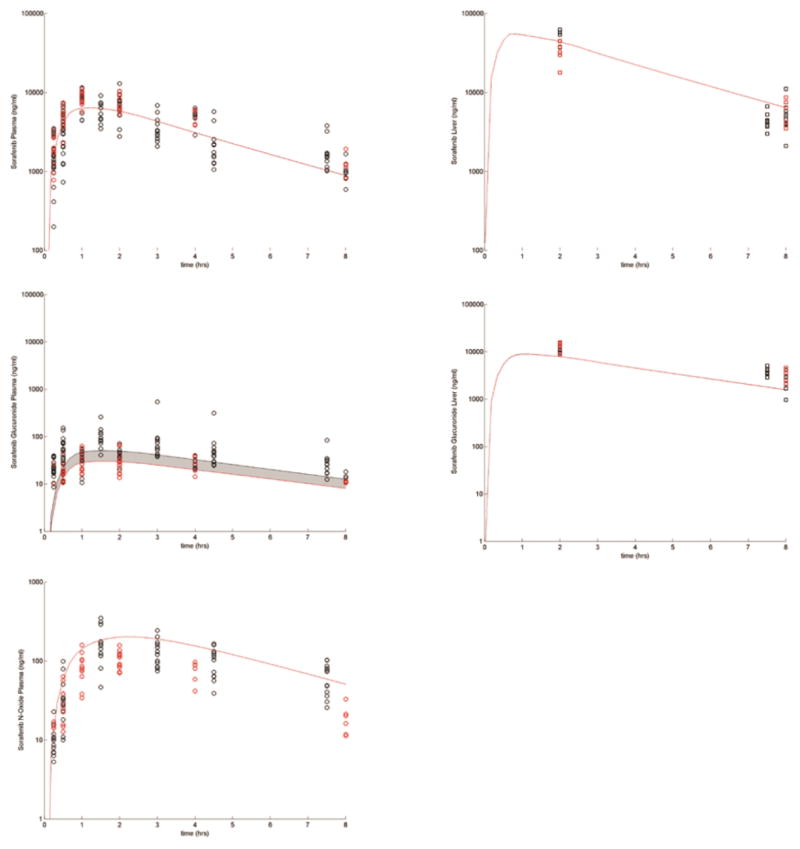
The drug-specific fixed and estimated parameters describing the data from WT and Oatp1a/1b influx transporter KO mice are presented in Supplemental Tables S1 and S2 and Figure 2. With the exception of the parameter(s) related to the activity of the KO transporter(s) (which were set equal to 0 in the KO cases), these parameters were fixed for all the simulations described in this study. Figure 3 presents sorafenib and SG plasma concentrations over 24 hours with and without neomycin co-administration after a dose of 10 mg/kg of SG. This simulation indicates the models ability to qualitatively account for the effects of EHC using the above described fixed and estimated parameters. In this example, the only parameter that was changed was the β-glucuronidase activity, which was decreased 2-fold from baseline for the neomycin-treated mice.
Figure 2.
Plasma (circles) and liver (squares) concentration time profiles following sorafenib administration to WT (black symbols) and Oatp1a/1b KO (red symbols) mice. The shaded region shows the variability in the disposition when SG Oatp1a/1b activity is decreased from the baseline value to 0.
Figure 3.
SG (a) and sorafenib (b) disposition given a 10 mg/kg dose of SG to WT mice either treated with saline (black circles) or neomycin (red circles). The shaded region shows the variability in the disposition when β-glucuronidase activity is decreased 2 fold from baseline.
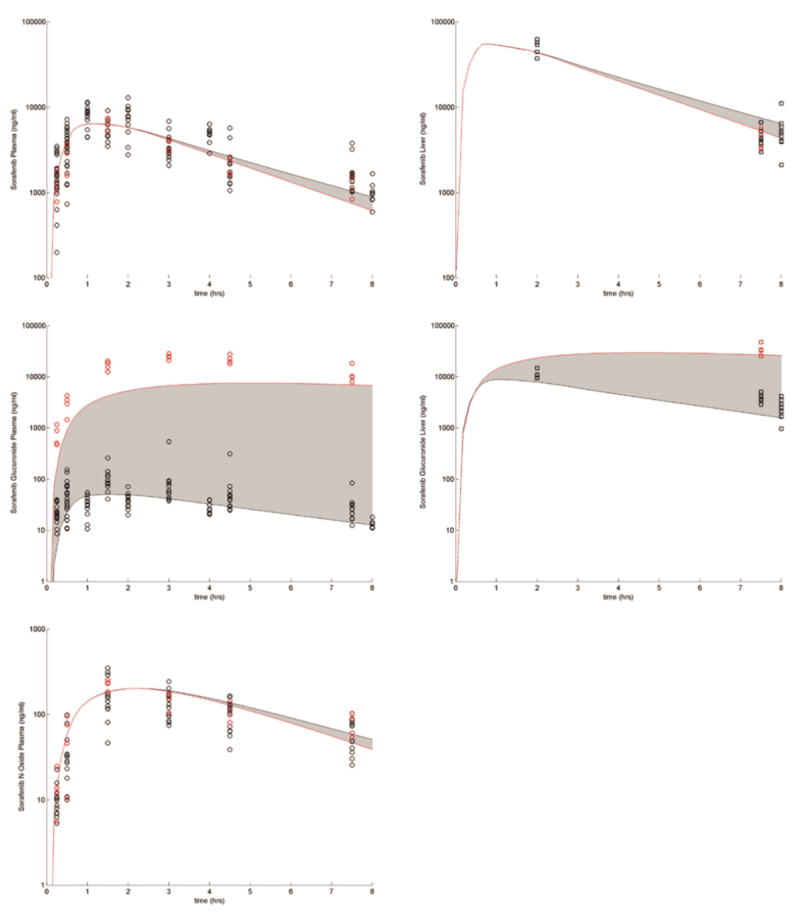
Uncertainty Analysis
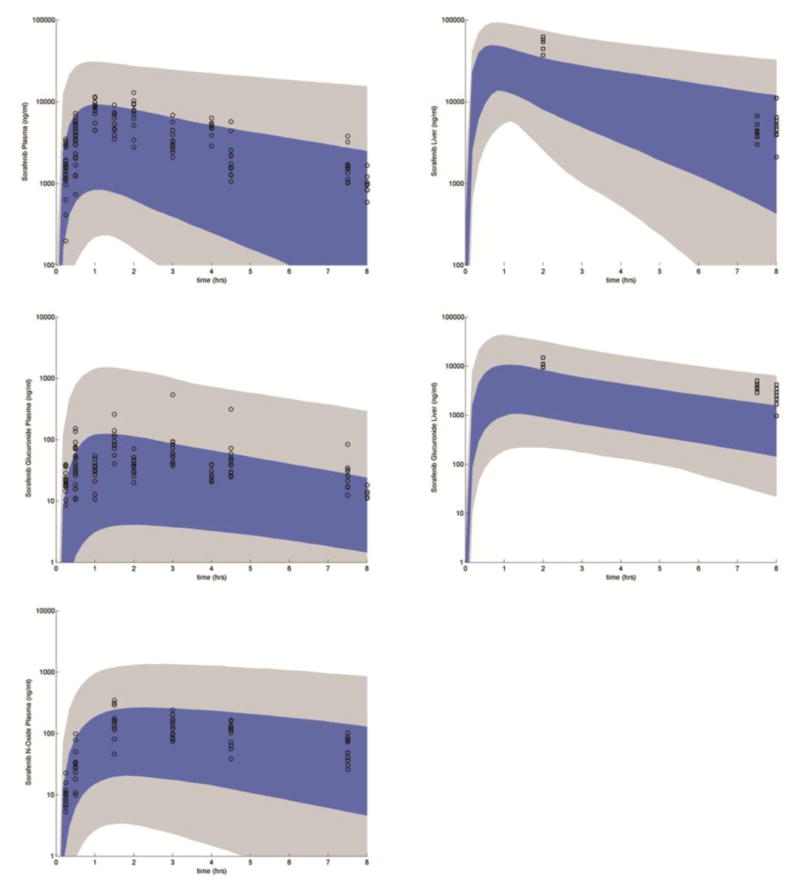
The observed variability in disposition in WT mice was explained by varying the 16 model parameters listed in Table 1 and is presented in Figure 4. The range of the output variability resulting from the variability in the parameters covered the observed concentrations. This suggests that the model has the ability to describe the variability in the data and that the hypothesis that model structure is adequate is not rejected.
Table 1. Results from the sensitivity analysis.
The values are the Partial Ranked Correlation Coefficients (PRCC) for the given model parameters. Red shading indicates PRCC≤−0.4 and green shading indicates PRCC≥0.4. Results are based on 5000 LHS with the parameters varied ±100×baseline. Model outputs include AUC (0 to 8 hrs) and Cmin (concentration at 8 hrs).
| Parameter | Plasma | Liver | |||
|---|---|---|---|---|---|
| Sorafenib | Sorafenib N-oxide | Sorafenib Glucuronide | Sorafenib | Sorafenib Glucuronide | |
| AUC | |||||
| Sorafenib | |||||
| Intestinal permeability | 0.66 | 0.63 | 0.50 | 0.71 | 0.64 |
| Fraction unbound in plasma | −0.94 | 0.14 | 0.10 | 0.10 | 0.12 |
| Influx Activity | −0.87 | 0.34 | 0.22 | 0.34 | 0.31 |
| Abcc2 Activity | −0.03 | −0.04 | −0.02 | −0.05 | −0.04 |
| Abcc3 Activity | 0.03 | 0.01 | 0.00 | 0.02 | 0.02 |
| CYP3A4 Vmax | −0.36 | 0.92 | −0.20 | −0.44 | −0.33 |
| UGT1A9 Vmax | −0.84 | −0.81 | 0.65 | −0.89 | 0.80 |
| Sorafenib Glucuronide | |||||
| Intestinal permeability | −0.03 | −0.02 | 0.25 | −0.01 | 0.08 |
| Fraction unbound in plasma | 0.00 | 0.01 | −0.85 | 0.00 | −0.02 |
| Abcc2 Activity | 0.01 | 0.00 | −0.80 | 0.01 | −0.93 |
| Abcc3 Activity | 0.01 | 0.00 | 0.55 | 0.00 | 0.00 |
| Oatp1a/1b Activity | 0.01 | 0.01 | −0.84 | 0.01 | 0.02 |
| Non-specific hepatic loss | −0.01 | −0.01 | −0.25 | −0.01 | −0.38 |
| β-glucuronidase | 0.03 | 0.02 | −0.03 | 0.04 | 0.01 |
| Sorafenib N-oxide | |||||
| Non-specific hepatic loss | 0.01 | −0.92 | −0.02 | 0.01 | 0.00 |
| Other | |||||
| EHC continuous fraction | 0.05 | 0.04 | 0.23 | 0.07 | 0.15 |
| Parameter | Plasma | Liver | |||
|---|---|---|---|---|---|
| Sorafenib | Sorafenib N-oxide | Sorafenib Glucuronide | Sorafenib | Sorafenib Glucuronide | |
| Cmin | |||||
| Sorafenib | |||||
| Intestinal permeability | −0.07 | 0.16 | 0.08 | −0.01 | −0.03 |
| Fraction unbound in plasma | −0.79 | −0.01 | 0.00 | −0.11 | −0.05 |
| Influx Activity | −0.82 | −0.18 | −0.16 | −0.42 | −0.31 |
| Abcc2 Activity | −0.03 | −0.05 | −0.02 | −0.05 | −0.04 |
| Abcc3 Activity | 0.03 | 0.00 | 0.01 | 0.01 | 0.02 |
| CYP3A4 Vmax | −0.45 | 0.78 | −0.32 | −0.57 | −0.43 |
| UGT1A9 Vmax | −0.87 | −0.86 | −0.03 | −0.91 | −0.27 |
| Sorafenib Glucuronide | |||||
| Intestinal permeability | −0.06 | −0.04 | 0.20 | −0.06 | 0.06 |
| Fraction unbound in plasma | 0.01 | 0.00 | −0.59 | 0.00 | 0.00 |
| Abcc2 Activity | 0.03 | 0.02 | −0.69 | 0.03 | −0.82 |
| Abcc3 Activity | 0.01 | −0.01 | 0.41 | −0.01 | 0.04 |
| Oatp1a/1b Activity | −0.01 | 0.01 | −0.73 | 0.00 | −0.04 |
| Non-specific hepatic loss | −0.05 | −0.03 | −0.26 | −0.05 | −0.32 |
| β-glucuronidase | 0.09 | 0.07 | −0.06 | 0.09 | 0.01 |
| Sorafenib N-oxide | |||||
| Non-specific hepatic loss | 0.02 | −0.92 | −0.03 | 0.02 | 0.00 |
| Other | |||||
| EHC continuous fraction | 0.16 | 0.11 | 0.37 | 0.18 | 0.31 |
Figure 4.
Uncertainty analysis: Symbols present the WT FVB mouse plasma (circles) and liver (squares) concentration time data. The blue shaded region represents 25th–75th percentile in the variability of the concentrations and the gray shaded region represents the range.
Sensitivity Analysis
We used sensitivity analysis to describe which model parameters used in the uncertainty analysis were most correlated with changes in sorafenib, sorafenib N-oxide, and SG plasma and liver disposition. The PRCC of each model parameter and output measure are summarized in Table 1.
Sorafenib metabolizing enzymes have global effects. For example, an increase in UGT1A9 Vmax increases plasma and liver SG AUC0-8h (ρ = 0.65 & 0.80) and decreases plasma and liver sorafenib (ρ = −0.84 & −0.89) and sorafenib N-oxide AUC0-8h (ρ = −0.81). Sorafenib intestinal permeability also had global effects, where increases in this parameter correlated with increases in the plasma and liver AUC0-8h of sorafenib and its metabolites.
Drug transporters have local effects. For example, sorafenib influx primarily affects plasma sorafenib AUC0-8h (ρ = −0.87). Meanwhile, the system is not sensitive to sorafenib efflux to either the circulation (Abcc3) or bile (Abcc2) (|ρ| < 0.05 for all). In addition, SG Oatp1a/1b influx and Abcc3 efflux to circulation only affects SG AUC0-8h (ρ = −0.84 & 0.55, respectively) while Abcc2 efflux to the bile only affects plasma and liver SG AUC0-8h (ρ = −0.80 & −0.93).
Variation in EHC continuous fraction and β-glucuronidase activity are only minor contributors to variability in the exposure to sorafenib and its metabolites (Table 1).
Other parameters considered such as the fraction unbound in plasma, SG intestinal permeability, and non-sorafenib hepatic intrinsic clearance all have local effects only significantly altering one or two outputs directly affected by the parameter. For example sorafenib N-oxide intrinsic clearance only has a local effect on sorafenib N-oxide disposition (Table 1).
Similar relationships were also observed when comparing inputs to the 8 hr concentrations (Cmin) of sorafenib and its metabolites (Table 1). The main differences in parameter sensitivity between the output measure of AUC0-8h and Cmin involved intestinal permeability and UGT1A9 Vmax. These differences are due to the possibility of two concentration-time curves having different AUCs over the interval (0–8 hrs) but similar Cmin at 8 hrs.
Influx Transporters
We tested via simulation whether sorafenib active influx accounts for differences observed in the Oatp1a/1b KO mice. We observed that decreasing sorafenib influx activity to 0 while keeping the SG Oatp1a/1b activity fixed did not explain the increased exposure of SG in the KO mice. This result was predicted by the sensitivity analysis (Table 1); the PRCC for sorafenib influx activity was not highly correlated with SG plasma (ρ = 0.22) or liver disposition (ρ = 0.31). This suggests that sorafenib is, at best, a weak substrate for Oatp1a/1b. This assertion is based on the sensitivity analysis (Table 1), which shows decreases in sorafenib influx Vmax cause increases in sorafenib exposure (ρ = −0.87)---an assertion that is not supported by the experimental data due to the lack of change in the observed sorafenib exposure in Oatp1a/1b KO mice compared to the WT mice. Next, we tested whether SG influx accounted for differences observed in the Oatp1a/1b KO mice. We observed that decreasing the SG activity of Oatp1a/1b to 0 was sufficient to describe the observed changes in SG in the Oatp1a/1b KO mice (Figure 2). Again, this result was predicted by the sensitivity analysis (Table 1); the PRCC for SG Oatp1a/1b activity was strongly negatively correlated with SG disposition (ρ = −0.84).
Efflux Transporters
We tested via simulations whether efflux of sorafenib and SG via Abcc3 would account for differences in sorafenib disposition in Abcc3 KO mice. We observed that decreasing the sorafenib and SG activity of Abcc3 to 0 was sufficient to describe the observed changes in sorafenib and SG in the Abcc3 KO mice (Figure 5). It was observed that the effects of Abcc3 on the disposition of sorafenib and the two metabolites were small, which was consistent with the sensitivity analysis (Table 1); the PRCC for SG Abcc3 efflux activity is only weakly correlated with SG disposition (ρ = 0.55) and the correlation between sorafenib activity and disposition of each of the components was very small (|ρ| < 0.05).
Figure 5.
Sorafenib and SG efflux via Abcc3. Black circles and curve: WT; Red circles curve: Abcc3 KO. The shaded region shows the variability in the disposition when sorafenib and SG Abcc3 activity are decreased from baseline value to 0.
Next, we tested whether the efflux of sorafenib and/or SG via Abcc2 could account for differences in sorafenib disposition in Abcc2 KO mice. We observed that decreasing the activity of Abcc2 to 0 relative to sorafenib and SG was not sufficient to describe the observed changes in the Abcc2 KO mice (Supplemental Figure S1). Specifically, while the simulation adequately described the minimal effects of Abcc2 on the disposition of sorafenib, which was consistent with the sensitivity analysis (Table 1), it did not fully account for the large increase in the disposition of plasma SG. This suggests that there are changes in the Abcc2 KO mice other than an elimination of Abcc2 activity. Other studies have suggested that the activity of transporters other than Abcc2 is altered in Abcc2 KO mice. For example one study observed a 6-fold increase in Abcc4 mRNA and protein in Abcc2 KO mice [15]. While the Abcc3 and Abcc4 expression in liver of mice similar to those used in this study did not show significant changes between WT and Abcc2 KO mice (unpublished data), there could be another, currently unknown transporter that is altered in the Abcc2 KO mouse relative to the WT. The sensitivity analysis gives us an approach to form a hypothesis on what may be altered to adequately describe the observed changes in the disposition of plasma SG in the Abcc2 KO mice. Specifically, either increases in the activity of an efflux transporter with the same orientation as Abcc3/4 (sinusoidal membrane efflux transporter) or decreases in the activity of an influx transporter would cause a local increase to SG plasma disposition (Table 1). Simulating a simultaneous decrease in Abcc2 activity to 0 and an increase in the activity of an unknown sinusoidal membrane efflux transporter by 2-fold showed that this hypothesis was sufficient to explain the changes in plasma SG in Abcc2 KO mice (Figure 6).
Figure 6.
Sorafenib and SG efflux via Abcc2. Black circles and curve: WT; Red circles and curve: Abcc2 KO. The shaded region shows the variability in the disposition when SG Abcc2 activity is decreased from baseline value to 0 and another undefined sinusoidal membrane efflux transporter’s activity is increased 2 fold.
Double and triple KO of transporters
Finally we simulated the effects of Oatp1a/1b, Abcc3 double KO; Oatp1a/1b, Abcc2 double KO; and Oatp1a/1b, Abcc2, Abcc3 triple KO. In all cases altering the transporter activity parameters in the same manner as in the single KO simulations while continuing to fix all remaining parameters at their nominal levels, we are able to adequately describe the disposition of the plasma and liver sorafenib, sorafenib N-oxide, and SG disposition (Supplemental Figures S2–S4).
Discussion
A PBPK model of sorafenib, sorafenib N-oxide, and SG was developed to explain its disposition in mice. All physiological and anatomical inputs specific to the mouse were fixed at reasonable values. Parameters that were dependent on the molecule(s) were either fixed to literature-based values or set based on preliminary assessment and numerical estimation. These parameter values and model structure described the WT and Oatp1a/1b KO data well (Figure 2). Due to the availability of sorafenib and sorafenib metabolite plasma and liver concentration time data in transporter KO mice, there was the ability to explicitly define transport processes. This approach differed from a previous sorafenib PBPK model that did not include efflux or influx transporters and focused on sorafenib alone [22]; a structure and focus that was sufficient for the question being addressed in that study. Because the purpose for our PBPK model was to determine those processes responsible for sorafenib and metabolite disposition with potential for later translation to humans, explicit delineation of important metabolic and transport processes was required.
Following the PBPK model development, it was initially evaluated using PK data resulting from SG administration to mice in the presence and absence of neomycin. While the presence of neomycin did not greatly affect SG concentrations, it significantly decreased sorafenib plasma concentrations both in the observed and simulated data. The simulation did not characterize the sorafenib curve shape in the first few hours without neomycin where the sorafenib concentrations were similar in the presence or absence of neomycin. In order to adequately simulate the early points, we would need to prevent β-glucuronidase from cleaving SG in the cecum and large intestine during the first few hours post administration. To accomplish this the small intestinal transit time would have needed to be extended to non-physiological values. One possible approach to address this discrepancy would be to include a time lag in the model. Considering that neomycin inactivation of β-glucuronidase was not an important process to understanding other mechanisms of disposition, we did not include a time lag and this deviation did not invalidate the model.
While the parameter values in Supplemental Table S2 were those that were moved forward, it was understood that these values were not unique. Determining a unique set of parameter values in mice was not a goal of this study nor was it possible to achieve given the complexities of the model and the lack of identifiability of some parameters.
Using the values from the above model in WT mice, uncertainty analysis allowed for an assessment of the ability of the model to explain the variability in the observed data. The variation in each parameter was set a priori to ± two orders of magnitude following a uniform distribution (a conservative choice). The observed data was covered by the uncertainty analysis (Figure 4), suggesting that the model structure and parameter values were reasonable. Note that while the uncertainty analysis was used to examine if our model structure was sufficient enough to account for observed variability, it did so in the absence of anatomical and physiological variability not related to drug metabolism and transport. Since the mice used in the experiments were homogenous the addition of anthropometric variability is most likely far less than the variability imposed by drug transport and metabolism.
Global sensitivity analysis was then performed to identify those parameters that contributed most to the prediction of sorafenib disposition. The model output of AUC0-8 was selected as a metric of overall exposure within a typical human dosing interval and the model output Cmin was relevant since troughs are commonly used for therapeutic drug monitoring. The results of this analysis showed that sorafenib metabolism had global effects such that all output metrics were highly correlated to metabolism parameters. However, in the case of transporters, output metrics specific to the transporter substrate were highly correlated only to its transporter activity. Sensitivity analysis provided an expectation of the PK results observed in KO mice and thus an expectation of the importance of each process on sorafenib disposition.
The availability of data sets that included various transporter KOs provided further opportunity for understanding the mechanisms important in sorafenib disposition as well as to evaluate model structure and parameter values. For the Abcc3 KO where all Abcc3 activities were set to 0, simulated profiles well described the observed data and reflected the sensitivity analysis, such that sorafenib was unaffected by the KO and SG concentrations slightly decreased in plasma but not liver (Figure 5). From the sensitivity analysis we expected that the Abcc2 KO would not affect sorafenib concentrations and would lead to increased SG concentrations; a result mirrored in the observed data (Figure 6) and previous studies [28]. While this was qualitatively captured in the simulation, the magnitude of the change in simulated plasma SG was not as great as in the observed data. To account for this difference, an additional increase in efflux to systemic circulation was required.
In this study, the transporters have been named based on available KO mouse data (e.g. Abcc2, Abcc3) but may not be unique in their activity. For example, it is possible that both Abcc3 and Abcc4 work to transport SG into systemic circulation from the liver, however unique resolution of these two processes is not possible given the available KO data. In addition, we assumed that the KO transporter had no activity, but there is no guarantee that other similar, but unknown, transporters are upregulated relative to WT. This has been demonstrated in another study where an Abcc2 KO had a 6-fold increase in Abcc4 mRNA and protein compared to WT [15]. We observed that the Abcc2 KO SG data could not be explained by simply knocking out Abcc2 activity in the model. Only when efflux to the systemic circulation (e.g. Abcc3- or Abcc4-like) was also doubled could the SG data be reproduced by the model. This is not necessarily a deficiency in the model but a failure to fully understand the transporter changes as a function of the KO process.
Moving forward to simulate multiple KOs, we assumed that the processes that were modified for the single KOs carried over to the multiple KOs. By setting all relevant transporter activities to 0, the PK data for sorafenib and its metabolites in plasma and liver were well described. This indicates that the process of knocking out multiple transporters did not cause changes beyond what the single KO may have altered. It also suggests that the flux of molecule movement through the liver has been well described even though the parameter values are not necessarily unique.
One of the primary goals of this model building and evaluation process was to test and generate hypotheses within an in silico system. Because of their mechanistic basis a PBPK model allows us to test hypotheses, with extrapolation being the most commonly applied hypothesis testing process. This has been used to assess the relevance of a drug-drug interaction [29] or the first-in-pediatric dose using extrapolation from an adult model [30]. Hypothesis generation on the other hand is a process to help us understand mechanisms. In the context of the sorafenib model, we were able to test the importance of specific parameters on plasma and liver concentrations (hypothesis testing) as well as develop questions about mechanisms relating to the integrity of the KO development process (hypothesis generating). In addition, the model could be used to generate and test hypotheses to help explain the large exposure variability observed in the clinical use of sorafenib [6].
Delineating the important processes that drive disposition may aid in understanding high exposure variability, particularly in the context of exposure dependent dose-limiting toxicities. For example, HFSR is related to sorafenib exposure where it has been observed that greater than 61% of adults treated with sorafenib developed this adverse event with the incidence of higher grades of toxicity linked to higher sorafenib plasma concentrations [6,8,7]. Further evaluation of this exposure-toxicity relationship, including exposure to sorafenib metabolites, may allow for correlation between HFSR, sorafenib dose, and transporter or metabolizing enzyme single nucleotide polymorphisms (SNPs). Individualization of sorafenib dosing may be possible by targeted SNP assessment such that the activities of those proteins most responsible for relevant compound disposition are accounted for in the dosing algorithm. For sorafenib, this PBPK model, once translated to adults and children, will aid in determining those SNPs that should be accounted for in the context of efficacy and dose-limiting toxicities. The use of TKI PBPK models coupled with genomic data for the purposes dose optimization is an area of current and future interest [21].
Supplementary Material
Acknowledgments
This work was supported, in part, by the American Lebanese Syrian Associated Charities (ALSAC), USPHS Cancer Center Support Grant 3P30CA021765 (S.D. Baker), and NCI Grants 5R01CA138744 (S.D. Baker)
Footnotes
Conflict of Interest/Disclosure
The authors have no conflicts of interest to declare.
Author Contributions
ANE, EIZ, AV, SDB, JCP wrote the manuscript
ANE, SDB, JCP designed the research
EIZ, AV performed experimental studies
ANE and JCP performed the research and analyzed the data
References
- 1.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23(3):329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 2.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5(10):835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 3.Mori S, Cortes J, Kantarjian H, Zhang W, Andreef M, Ravandi F. Potential role of sorafenib in the treatment of acute myeloid leukemia. Leuk Lymphoma. 2008;49(12):2246–2255. doi: 10.1080/10428190802510349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inaba H, Rubnitz JE, Coustan-Smith E, Li L, Furmanski BD, Mascara GP, Heym KM, Christensen R, Onciu M, Shurtleff SA, Pounds SB, Pui CH, Ribeiro RC, Campana D, Baker SD. Phase I pharmacokinetic and pharmacodynamic study of the multikinase inhibitor sorafenib in combination with clofarabine and cytarabine in pediatric relapsed/refractory leukemia. J Clin Oncol. 2011;29(24):3293–3300. doi: 10.1200/JCO.2011.34.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadaleta-Caldarola G, Infusino S, Divella R, Ferraro E, Mazzocca A, De Rose F, Filippelli G, Abbate I, Brandi M. Sorafenib: 10 years after the first pivotal trial. Future Oncol. 2015;11(13):1863–1880. doi: 10.2217/fon.15.85. [DOI] [PubMed] [Google Scholar]
- 6.Fukudo M, Ito T, Mizuno T, Shinsako K, Hatano E, Uemoto S, Kamba T, Yamasaki T, Ogawa O, Seno H, Chiba T, Matsubara K. Exposure-toxicity relationship of sorafenib in Japanese patients with renal cell carcinoma and hepatocellular carcinoma. Clin Pharmacokinet. 2014;53(2):185–196. doi: 10.1007/s40262-013-0108-z. [DOI] [PubMed] [Google Scholar]
- 7.Henin E, Blanchet B, Boudou-Rouquette P, Thomas-Schoemann A, Freyer G, Vidal M, Goldwasser F, Tod M. Fractionation of daily dose increases the predicted risk of severe sorafenib-induced hand-foot syndrome (HFS) Cancer Chemother Pharmacol. 2014;73(2):287–297. doi: 10.1007/s00280-013-2352-1. [DOI] [PubMed] [Google Scholar]
- 8.Boudou-Rouquette P, Ropert S, Mir O, Coriat R, Billemont B, Tod M, Cabanes L, Franck N, Blanchet B, Goldwasser F. Variability of sorafenib toxicity and exposure over time: a pharmacokinetic/pharmacodynamic analysis. Oncologist. 2012;17(9):1204–1212. doi: 10.1634/theoncologist.2011-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drenberg CD, Baker SD, Sparreboom A. Integrating clinical pharmacology concepts in individualized therapy with tyrosine kinase inhibitors. Clin Pharmacol Ther. 2013;93(3):215–219. doi: 10.1038/clpt.2012.247. [DOI] [PubMed] [Google Scholar]
- 10.Saber-Mahloogi H, Morse DE. Pharmacology review-Sorafenib. Center for Drug Evaluation and Research; Rockville: 2005. [Google Scholar]
- 11.Ghassabian S, Rawling T, Zhou F, Doddareddy MR, Tattam BN, Hibbs DE, Edwards RJ, Cui PH, Murray M. Role of human CYP3A4 in the biotransformation of sorafenib to its major oxidized metabolites. BiochemPharmacol. 2012;84(2):215–223. doi: 10.1016/j.bcp.2012.04.001. S0006-2952(12)00259-6 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Lathia C, Lettieri J, Cihon F, Gallentine M, Radtke M, Sundaresan P. Lack of effect of ketoconazole-mediated CYP3A inhibition on sorafenib clinical pharmacokinetics. Cancer ChemotherPharmacol. 2006;57(5):685–692. doi: 10.1007/s00280-005-0068-6. [DOI] [PubMed] [Google Scholar]
- 13.Miller AA, Murry DJ, Owzar K, Hollis DR, Kennedy EB, Abou-Alfa G, Desai A, Hwang J, Villalona-Calero MA, Dees EC, Lewis LD, Fakih MG, Edelman MJ, Millard F, Frank RC, Hohl RJ, Ratain MJ. Phase I and pharmacokinetic study of sorafenib in patients with hepatic or renal dysfunction: CALGB 60301. JClinOncol. 2009;27(11):1800–1805. doi: 10.1200/JCO.2008.20.0931. JCO.2008.20.0931 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaher H, Meyer zu Schwabedissen HE, Tirona RG, Cox ML, Obert LA, Agrawal N, Palandra J, Stock JL, Kim RB, Ware JA. Targeted disruption of murine organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) significantly alters disposition of prototypical drug substrates pravastatin and rifampin. Mol Pharmacol. 2008;74(2):320–329. doi: 10.1124/mol.108.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu XY, Strauss JR, Mariano MA, Li J, Newton DJ, Cai X, Wang RW, Yabut J, Hartley DP, Evans DC, Evers R. Characterization of mice lacking the multidrug resistance protein MRP2 (ABCC2) J Pharmacol Exp Ther. 2006;317(2):579–589. doi: 10.1124/jpet.105.098665. [DOI] [PubMed] [Google Scholar]
- 16.Vlaming ML, Pala Z, van Esch A, Wagenaar E, van Tellingen O, de Waart DR, Oude Elferink RP, van de Wetering K, Schinkel AH. Impact of Abcc2 (Mrp2) and Abcc3 (Mrp3) on the in vivo elimination of methotrexate and its main toxic metabolite 7-hydroxymethotrexate. Clin Cancer Res. 2008;14(24):8152–8160. doi: 10.1158/1078-0432.CCR-08-1609. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman EI, Hu S, Roberts JL, Gibson AA, Orwick SJ, Li L, Sparreboom A, Baker SD. Contribution of OATP1B1 and OATP1B3 to the disposition of sorafenib and sorafenib-glucuronide. ClinCancer Res. 2013;19(6):1458–1466. doi: 10.1158/1078-0432.CCR-12-3306. 1078-0432.CCR-12-3306 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swift B, Nebot N, Lee JK, Han T, Proctor WR, Thakker DR, Lang D, Radtke M, Gnoth MJ, Brouwer KL. Sorafenib hepatobiliary disposition: mechanisms of hepatic uptake and disposition of generated metabolites. Drug Metab Dispos. 2013;41(6):1179–1186. doi: 10.1124/dmd.112.048181. dmd.112.048181 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasilyeva A, Durmus S, Li L, Wagenaar E, Hu S, Gibson AA, Panetta JC, Mani S, Sparreboom A, Baker SD, Schinkel AH. Hepatocellular Shuttling and Recirculation of Sorafenib-Glucuronide is Dependent on Abcc2, Abcc3, and Oatp1a/1b. Cancer Res. 2015 doi: 10.1158/0008-5472.can-15-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain L, Woo S, Gardner ER, Dahut WL, Kohn EC, Kummar S, Mould DR, Giaccone G, Yarchoan R, Venitz J, Figg WD. Population pharmacokinetic analysis of sorafenib in patients with solid tumours. BrJClinPharmacol. 2011;72(2):294–305. doi: 10.1111/j.1365-2125.2011.03963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo JM. Physiologically based pharmacokinetic models of tyrosine kinase inhibitors: a systems pharmacological approach to drug disposition. ClinPharmacolTher. 2013;93(3):236–238. doi: 10.1038/clpt.2012.244. clpt2012244 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Pawaskar DK, Straubinger RM, Fetterly GJ, Hylander BH, Repasky EA, Ma WW, Jusko WJ. Physiologically based pharmacokinetic models for everolimus and sorafenib in mice. Cancer ChemotherPharmacol. 2013;71(5):1219–1229. doi: 10.1007/s00280-013-2116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodgers T, Leahy D, Rowland M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases. JPharmSci. 2005;94(6):1259–1276. doi: 10.1002/jps.20322. [DOI] [PubMed] [Google Scholar]
- 24.Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. JPharmSci. 2006;95(6):1238–1257. doi: 10.1002/jps.20502. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerman EI, Roberts JL, Li L, Finkelstein D, Gibson A, Chaudhry AS, Schuetz EG, Rubnitz JE, Inaba H, Baker SD. Ontogeny and sorafenib metabolism. ClinCancer Res. 2012;18(20):5788–5795. doi: 10.1158/1078-0432.CCR-12-1967. 1078-0432.CCR-12-1967 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blower SM, Dowlatabadi H. Sensitivity and Uncertainty Analysis of Complex-Models of Disease Transmission - an Hiv Model, as an Example. Int Stat Rev. 1994;62(2):229–243. doi: 10.2307/1403510. [DOI] [Google Scholar]
- 27.Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254(1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu S, Chen Z, Franke R, Orwick S, Zhao M, Rudek MA, Sparreboom A, Baker SD. Interaction of the multikinase inhibitors sorafenib and sunitinib with solute carriers and ATP-binding cassette transporters. ClinCancer Res. 2009;15(19):6062–6069. doi: 10.1158/1078-0432.CCR-09-0048. 1078-0432.CCR-09-0048 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner C, Pan Y, Hsu V, Sinha V, Zhao P. Predicting the Effect of CYP3A Inducers on the Pharmacokinetics of Substrate Drugs Using Physiologically Based Pharmacokinetic (PBPK) Modeling: An Analysis of PBPK Submissions to the US FDA. Clin Pharmacokinet. 2015 doi: 10.1007/s40262-015-0330-y. [DOI] [PubMed] [Google Scholar]
- 30.Maharaj AR, Barrett JS, Edginton AN. A workflow example of PBPK modeling to support pediatric research and development: case study with lorazepam. AAPS J. 2013;15(2):455–464. doi: 10.1208/s12248-013-9451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.