Abstract
Mutations in CHD7, encoding ATP-dependent chromodomain-helicase-DNA-binding protein 7, in CHARGE syndrome leads to multiple congenital anomalies including craniofacial malformations, neurological dysfunction and growth delay. Currently, mechanisms underlying the CNS phenotypes remain poorly understood. Here, we show that Chd7 is a direct transcriptional target of oligodendrogenesis-promoting factors Olig2 and Smarca4/Brg1, and is required for proper onset of CNS myelination and remyelination. Genome-occupancy analyses, coupled with transcriptome profiling, reveal that Chd7 interacts with Sox10 and targets the enhancers of key myelinogenic genes, and identify novel Chd7 targets including bone formation regulators Osterix/Sp7 and Creb3l2, which are also critical for oligodendrocyte maturation. Thus, Chd7 coordinates with Sox10 to regulate the initiation of myelinogenesis and acts as a molecular nexus of regulatory networks that account for the development of a seemingly diverse array of lineages including oligodendrocytes and osteoblasts, pointing to the hitherto previously uncharacterized Chd7 functions in white matter pathogenesis in CHARGE syndrome.
INTRODUCTION
Chromatin remodeling control is essential to orchestrate cell growth and differentiation, and to maintain proper cell identity1, 2. Mutations in chromatin-modifying enzymes have been implicated in the pathogenesis of various human diseases, manifesting in growth/developmental defects, neurological disorders and tumorigenesis 3, 4. One class of chromatin-modifying enzymes, the chromodomain helicase DNA-binding (CHD) family of SNF2H-like ATP-dependent nucleosome remodeling enzymes, has emerged as an important regulator of multiple biological processes 5. Mutations in CHD7 are the major cause of human CHARGE syndrome (Coloboma of the eye, Heart defects, Atresia of the choanae, severe Retardation of growth/development, Genital abnormalities and Ear abnormalities), an autosomal dominant disorder characterized by a non-random association of multiple birth defects impairing normal development 5, 6. The life expectancy of CHARGE patients ranges from five days to approximately 46 years7. CHD7 modulates chromatin configurations to control cell-type specific transcriptional machinery, thereby controlling temporal and spatial gene expression 8–11. Most CHARGE patients exhibit some degree of intellectual disability, and many present with structural abnormalities of the corpus callosum and cerebellar vermis 5, 12. Recent reports of white matter defects in the brain of patients with CHARGE syndrome 13, 14 suggest a possible role for CHD7 in myelination in the central nervous system (CNS).
Oligodendrocytes (OLs) produce myelin sheaths that electrically insulate axons and promote rapid propagation of action potentials in the CNS. Failure of OLs to remyelinate demyelinated axons disrupts saltatory nerve conduction, which could lead to cognitive and motor function deficits or irreversible axonal degeneration in disorders such as multiple sclerosis (MS) and leukodystrophies 15–17. The onset and timing of CNS myelination and remyelination is tightly regulated by the balanced intrinsic and extrinsic cues 18–20. Differentiation of OL precursor cells (OPCs) into mature OLs requires precise coordination between epigenetic programming and transcriptional regulation. Chromatin reorganization is critical for OL differentiation processes 21. Recently, the SWI/SNF chromatin-remodeling enzyme Smarca4 (Brg1) has been demonstrated to complex with a pioneer transcription factor Olig2 to target active enhancer elements to initiate the differentiation of OL lineage cells 22.
Herein, we identify Chd7 as a downstream target of Brg1 and Olig2. We find that expression of Chd7 is highly enriched in OL lineage cells, with a peak of expression in differentiating OLs. Inactivation of Chd7 causes defects in OL differentiation and myelination while sparing OPC formation. We further show that Chd7 is required for OL remyelination after demyelinating injury. By genome-wide mapping of Chd7 targeting sites and co-immunoprecipitation, we demonstrate that Chd7 complexes with Sox10 and directly activates a distinct set of critical regulators for OL differentiation. Moreover, our studies identify the osteoblast-differentiation factor Osterix/Sp7 as an OL-specific Chd7 downstream target in the CNS, and demonstrate a critical requirement of Osterix for OL differentiation. Collectively, these data provide evidence that the chromatin remodeler Chd7 interacts with Sox10 to bridge Brg1/Olig2 activity during OL differentiation and controls the onset of OL myelination and remyelination via directly activating myelinogenic programs.
RESULTS
OL-enriched Chd7 is a direct target of Brg1/Olig2 complex
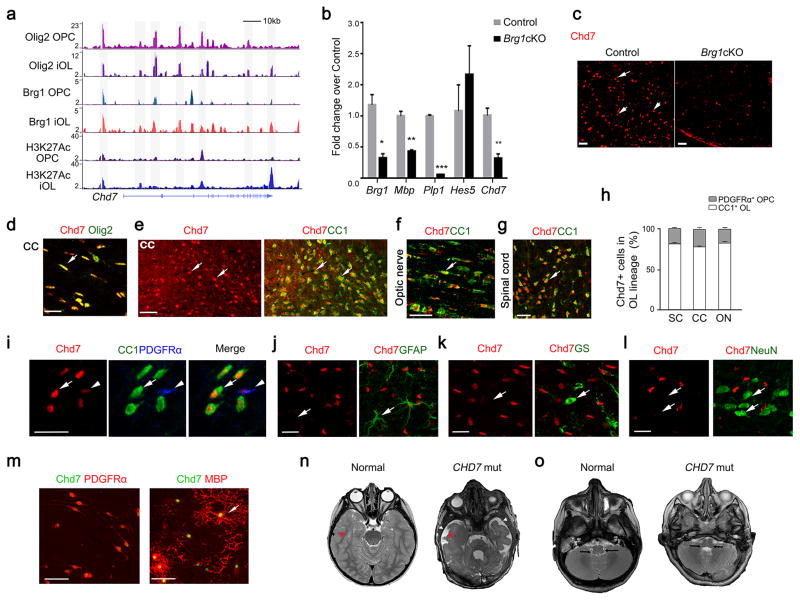
We have previously shown that Brg1 and Olig2 co-occupancy in the genome establishes a transcriptional program to initiate OPC differentiation 22. We integrated transcriptome profiling of the spinal cord from Brg1cKO mutants (Brg1flox/flox;Olig1-Cre+/−) by gene-chip microarray 22 and genome-wide Brg1/Olig2 occupancy at multiple phases of OL differentiation and identified Chd7 as a candidate target gene of Brg1/Olig2. Brg1 and Olig2 bound strongly at multiple sites both around the promoter region and within the Chd7 gene locus, accompanied by the presence of an activated histone acetylation mark H3K27Ac in OPCs and early differentiating immature OLs (iOLs) (Fig. 1a). Expression of Chd7 was significantly downregulated in Brg1cKO mutants (Fig. 1b). Consistently, Chd7-expressing cells appeared markedly reduced in the Brg1cKO spinal cord at P14 (Fig. 1c). These observations suggest that Chd7 is a downstream target of Brg1/Olig2.
Figure 1. Identification of OL-enriched Chd7 as a direct target of the Brg1/Olig2 complex.
(a) Track view of ChIP-Seq density profile for Olig2, Brg1 and H3K27Ac centered at the Chd7 locus in OPCs and OLs.
(b) qRT-PCR analysis of myelination-associated genes and Chd7 in spinal cords from control (Ctrl) and Brg1cKO animals at P14 (The data are presented as mean ± s.e.m. n = 3; for Brg1, * p = 0.024, t = 3.52; for Mbp, ** p = 0.0018, t = 7.40; for Plp1, *** p < 0.0001, t = 40.94; for Hes5, p = 0.448, t = 1.72; for Chd7, ** p = 0.007, t = 5.02; n, numbers of animals; Two-tailed unpaired Student’s t test).
(c) Immunostaining showing Chd7 expression in spinal cords of control (Brg1flox/+;Olig1-Cre+/−) and Brg1cKO animals at P14. Arrows indicate Chd7+ cells. Scale bar, 50 μm.
(d) Co-immunolabeling of Chd7 and Olig2 on the corpus callosum (CC) of wildtype brain at P7. Scale bar, 20 μm.
(e) Co-immunolabeling of Chd7 and CC1 in the corpus callosum (CC) of P14 mice. Arrows indicate co-labeled cells. Scale bar, 50 μm.
(f,g) Immunolabeling of Chd7 and CC1 on longitudinal sections of optic nerves (f) and coronal sections of the spinal white matter (g) at P14. Arrows indicate co-labeled cells. Scale bars in (f), 25 μm; (g), 50 μm.
(h) The percentage of CC1 or PDGFRα+ cells among total Chd7+ cells in the cortex (Ctx), spinal cord (SC) and optic nerve (ON) from P14 wildtype mice (n = 3 animals).
(i) Immunostaining showing expression of CC1, PDGFRα and Chd7 in the cortical section of P14 mice. Arrows and arrowheads indicate CC1+Chd7hi OLs and PDGFRα+Chd7low OPCs, respectively. Scale bar, 30 μm.
(j–l) Immunostaining for Chd7 and GFAP (arrow in j) in the P24 cortex, glutamine synthetase (GS) (arrow in k) in the P7 cortex, and NeuN (arrow in l) in the P24 cortex. Arrow indicates GFAP+ astrocyte. Scale bars, 25 μm (j); 20 μm (k,l)
(m) Immunolabeling of Chd7 with PDGFRα in OPCs (left) or with MBP in OLs (right) in vitro. Scale bar, 50 μm.
(n,o) Representative T2-weighted magnetic resonance imaging (MRI) scans of cortices (n) and brainstem/cerebellar regions (o) in a normal and a 3-year old boy with CHARGE Syndrome carrying a CHD7 nonsense truncation mutation (7252C>T). In panel n, asterisks indicate the white matter, and arrowheads indicate CSF-filled fluid space. Arrows in o indicate white matter structures in the brainstem and cerebellum region that are dysmorphic in the patient.
To further characterize Chd7-expressing cell types in the developing cortex, we co-immunostained Chd7 with cell-type specific markers such as Olig2 for the OL lineage, and CC1 for OLs, glial fibrillary acidic protein (GFAP) and glutamine synthetase (GS) for astrocytes, and a pan-neuronal marker NeuN. Chd7 was detected in most Olig2-positive cells (Fig. 1d). The majority of Chd7+ cells in the OL lineage were CC1+ differentiated OLs in the corpus callosum, optic nerve and spinal cord at P14 (Fig. 1e–h). Intense Chd7 expression was detected in OLs but at a lower level in PDGFRα+ OPCs (Fig. 1i) in the P14 cortex. Similarly, in culture, Chd7 appeared more robustly expressed in MBP+ (myelin basic protein) mature OLs than PDGFRα+ OPCs (Fig. 1m), suggesting a potential role of Chd7 at the OL differentiation onset. In contrast, we did not observe Chd7 expression in GFAP or GS-expressing astrocytes in the corpus callosum (Fig. 1j,k). In addition, Chd7 was hardly detectable in NeuN+ neurons in the cortex at P24 (Fig. 1l). These data reveal that Chd7 is highly enriched in differentiating OLs in the developing CNS.
To explore the potential function of Chd7 in human CNS myelination, we examined the white matter of a cohort of 17 patients with CHARGE syndrome carrying CHD7 mutations. MRI scans revealed severe cerebral white matter defects in 47% (8/17) of the CHARGE patients. Compared to the age-matched normal brain, these patients exhibited volumetric loss of cerebral white matter (Fig. 1n) and dysmorphic features in the white matter regions of the brainstem and cerebellum (Fig. 1o) that are commonly seen in patients with CHARGE Syndrome 13, indicating that white matter defects are a prominent feature of CHARGE syndrome with CHD7 mutations. These observations, together with Chd7 expression in the OL lineage, raise the possibility that CHD7 may be functionally required for CNS myelination in humans.
Chd7 is required for proper myelination in the CNS
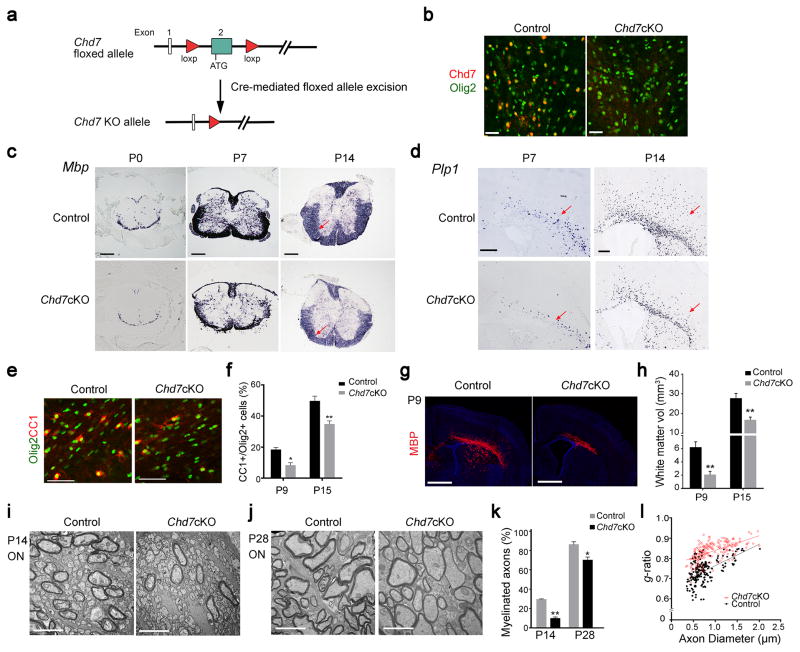
To investigate the function of Chd7 in CNS myelination in vivo, we selectively ablated Chd7 in OL lineage cells by breeding the floxed Chd7 allele with mice carrying an OL-lineage expressing Olig1-Cre line (Fig. 2a). Chd7 expression was substantially reduced in the corpus callosum of the Chd7 conditional knockout mice (Chd7flox/flox;Olig1-Cre+/−, hereafter referred to as Chd7cKO) at P14, suggesting that the floxed Chd7 allele was effectively recombined in the Chd7cKO brain as compared to the control (Chd7flox/+;Olig1-Cre+/−) (Fig. 2b). Chd7cKO animals were born at the expected Mendelian frequency and were of normal body size and weight. The analysis of appearance of MBP at embryonic stages and early postnatal stages indicated a substantial reduction of MBP-expressing OLs in the spinal cord of Chd7cKO animals (Supplementary Fig. 1a–c). Similarly, expression of Mbp and Plp1 (proteolipid protein 1) mRNAs was visibly diminished in both the spinal cord and corpus callosum of Chd7cKO mice at neonatal and perinatal stages (Fig. 2c,d). In addition, the numbers of CC1+ OLs and OL lineage cells marked by Olig1, Olig2, and Sox10 in Chd7cKO cortices were significantly decreased (Fig. 2e,f, Supplementary Fig. 2a–d). Consistently, MBP-expressing area assayed by immunohistochemistry was much less in the corpus callosum of Chd7cKO mice (Fig. 2g). Cerebral white matter volume assessed by total MBP+ area in the corpus callosum was significantly reduced in developing Chd7cKO mice (Fig. 2g,h; Supplementary Fig. 2e). These observations suggest that Chd7 is required for the onset of OL differentiation in the CNS.
Figure 2. Chd7 is required for proper CNS myelination.
(a) Diagram depicting Cre mediated excision of the floxed Chd7 exon 2.
(b) Immunolabeling of Chd7 and Olig2 in the corpus callosum of P14 brain from control and Chd7cKO mice. Scale bar, 40 μm.
(c) mRNA expression of Mbp in the spinal cord of control and Chd7cKO mice at P0, P7 and P14. Scale bars, 100 μm. Arrows point to the spinal white matter region.
(d) mRNA expression of Plp in P7 and P14 brain sections from control and Chd7cKO mice. Scale bar, 100 μm. Arrows point to the white matter.
(e) Immunostaining for CC1 and Olig2 in the corpus callosum from control and Chd7cKO mice at P7. Scale bar, 100 μm.
(f) Quantification of CC1+ OLs as a percentage of total Olig2+ cells in the corpus callosum of Control and Chd7cKO at indicated ages (The data are presented as mean ± s.e.m. For P9, n = 3 controls and 2 mutants, * p = 0.026, t = 4.12; for P15, n = 4 controls and 4 mutants, ** p = 0.009, t = 3.79; n, numbers of animals; Two-tailed unpaired Student’s t test).
(g) Immunolabeling of MBP on coronal brain sections from control and Chd7cKO mice at P9. Nuclei are counterstained with 4,6-diamidino-2-phenylindole (DAPI; blue). Scale bar, 1 mm.
(h) Quantification of total cerebral white matter volume (MBP+) of control and Chd7cKO mice at P9 and P15 (The data are presented as mean ± s.e.m. For P9, n = 3 controls and 3 mutants, ** p = 0.002, t = 7.22; for P15, n = 4 controls and 4 mutants, ** p = 0.009, t = 3.74; n, numbers of animals; Two-tailed unpaired Student’s t test).
(i,j) Electron micrographs of transverse optic nerve sections from P14 (i) and P28 (j) control and Chd7cKO mice. Scale bars, 4 μm.
(k) Quantification of myelinated axons as a percentage of total axons (The data are presented as mean ± s.e.m. For P14, n = 3 controls and 3 mutants, ** p = 0.0017, t = 10.73; for P28, n = 4 controls and 4 mutants, * p = 0.0217, t = 3.656; n, numbers of animals; Two-tailed unpaired Student’s t test).
(l) Quantification of myelin sheath thickness (g-ratio) in control and Chd7cKO optic nerves (p < 0.0001; for control, g-ratio = 0.7227 ± 0.007414, n = 196 axons from 3 control optic nerves; for Chd7cKO, g-ratio = 0.8325 ± 0.003235, n = 202 axons from 3 Chd7cKO optic nerves; Two-tailed unpaired Student’s t test).
Consistent with the reduction in mature OLs, ultrastructural analysis of optic nerves revealed compromised axonal ensheathment, with a significantly reduced number of myelinated axons in Chd7cKO animals at P14 (Fig. 2i,k). At P28, the number of myelinating axons increased but was still less than the control (Fig. 2j,k). Notably, the thickness of myelin sheaths measured by g-ratio (the ratio between the diameter of the axon itself and the axon plus its myelin sheath), was significantly thinner in Chd7-mutant optic nerves than controls (Fig. 2j,l). Intriguingly, this hypomyelination effect appears transient as the number of CC1+ OLs gradually increased during development and finally caught up in the spinal cord of adult mice (Supplementary Fig. 3a,b). By P60, the degree of myelination was essentially indistinguishable in the spinal cord between control and Chd7cKO mice (Supplementary Fig. 3c,d). Thus, these observations indicate that Chd7 loss causes a markedly delay in CNS myelination, and that increased numbers of differentiated OLs may account for the myelination recovery in adult Chd7cKO animals.
Chd7 regulates the transition from OPC to OL
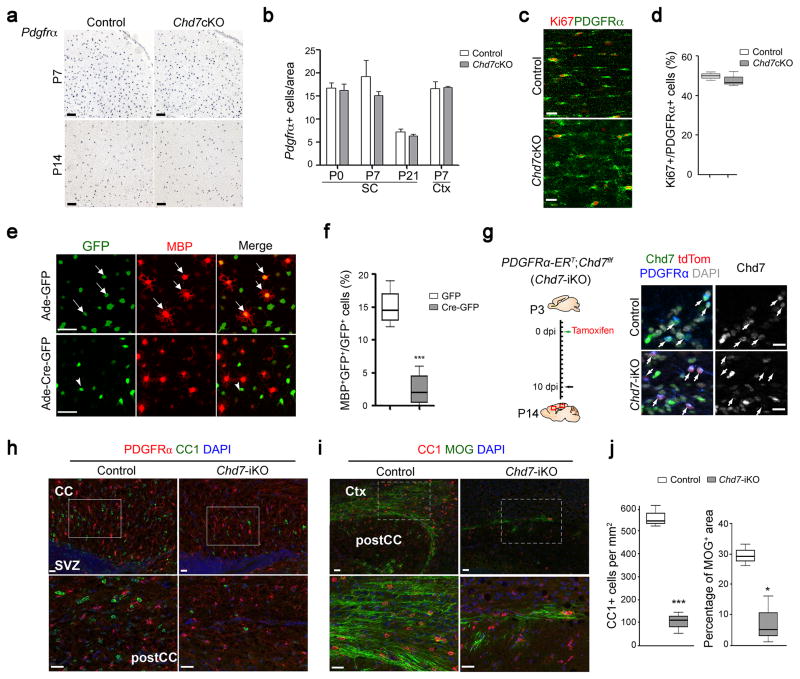
Given the possibility that the dysmyelinating phenotype in Chd7cKO mice could be due to a failure to generate OPCs, we assessed OPC development in the brain. Expression of the OPC marker PDGFRα was comparable between control and mutant cortices at P7 and P14 (Fig. 3a). Quantification of PDGFRα+ cells at postnatal stages (P0, P7 and P21) indicated that deletion of Chd7 did not significantly alter OPC formation (Fig. 3b). Supporting this, we found similar OPC proliferation rate (percentage of Ki67+ proliferative OPCs, Fig. 3c,d) and survival (absence of activated Caspase 3, Supplementary Fig. 4) between mutant and control mice. It is also noteworthy that Chd7 inactivation in the OL lineage did not lead to notable alterations in astrocytes and neurons marked by GFAP and NeuN, respectively (Supplementary Fig. 4).
Figure 3. Chd7 deletion does not affect OPC development but impairs their differentiation in a cell-autonomous manner.
(a) In situ hybridization for Pdgfrα on forebrain sections of control and Chd7cKO mice at P7 and P14, as indicated. Scale bars, 100 μm.
(b) Quantification of the number of Pdgfrα+ cells per area (0.04 mm2) in the cortex (Ctx) and spinal cord (SC) of control and Chd7cKO (The data are presented as mean ± s.e.m. For P0 SC, n = 4 controls and 3 mutants, p = 0.741, t = 0.348; for P7 SC, n = 3 controls and 3 mutants, p = 0.417, t = 0.905; for P21 SC, n = 4 controls and 3 mutants, p = 0.434, t = 0.849; for P7 Ctx, n = 3 controls and 3 mutants, p = 0.873, t = 0.17; n, numbers of animals; Two-tailed unpaired Student’s t test).
(c) Immunostaining for Ki67 and PDGFRα in the corpus callosum from P7 control and Chd7cKO mice. Scale bar, 20 μm.
(d) Quantification of Ki67+ cells as a percentage of PDGFRα+ OPCs within the corpus callosum of P7 control and Chd7cKO mice (n = 3 control and 3 mutant animals, p = 0.709, t = 0.41; whiskers show minimum and maximum, box limits are first and third quartile and center lines are median; Two-tailed unpaired Student’s t test).
(e) Immunolabeling of MBP after 2 days of differentiation in Chd7f/f OPCs transduced with control GFP or Cre-GFP expressing adenoviruses. Arrows and arrowheads indicate the GFP or Cre-GFP virus transduced cells, respectively. Scale bar, 50 μm.
(f) Quantification of MBP+ OLs as a percentage of total GFP+ cells after 2 days of differentiation (n = 4 independent experiments, *** p < 0.001, t = 5.937; whiskers show minimum and maximum, box limits are first and third quartile and center lines are median; Two-tailed unpaired Student’s t test).
(g) Left: Diagram showing tamoxifen (TAM) administration to Chd7-iKO mice from P3 to P10 followed by tissue collection at P14. Right: Immunolabeling for Chd7, PDGFRα and tdTomato on the corpus callosum of wildtype and Chd7-iKO:Rosa26Tom mice. Nuclei are counterstained with DAPI. Scale bar, 20 μm.
(h,i) Immunolabeling for CC1 and PDGFRα (h) or MOG (i) on sagittal corpus callosum (CC) sections of TAM-treated control and Chd7-iKO mice at P14. Ctx, Cortex; SVZ, subventricular zone. Scale bars, 20 μm. Boxed areas in upper panels were shown at a high magnification in corresponding lower panels.
(j) Quantification of CC1+ OL density (left) and percentage of MOG+ area (right) in the corpus callosum of TAM-treated control and Chd7-iKO mice at P14 (n = 3 controls and 3 Chd7iKO animals; for CC1, *** p < 0.0001, t = 32.609; for MOG, * p = 0.0123, t = 4.331; whiskers show minimum and maximum, box limits are first and third quartile and center lines are median; Two-tailed unpaired Student’s t test).
To further determine if defects in OL differentiation are a cell-autonomous effect of Chd7 ablation, we purified OPCs from the neonatal cortex of Chd7flox/flox pups, transduced them with adenoviral vectors expressing either eGFP as a control (Ade-GFP) or Cre-IRES-eGFP (Ade-Cre-GFP), and analyzed their ability to differentiate in vitro after withdrawal of mitogens PDGFAA and NT3. Chd7flox/flox OPCs transduced with Ade-GFP or wildtype OPCs transduced with Ade-Cre-GFP were able to differentiate into mature MBP+ OLs. Chd7flox/flox cells transduced with Ade-Cre-GFP, however, essentially failed to differentiate into MBP+ OLs (Fig. 3e,f), suggesting that Chd7-deleted OPCs are intrinsically defective in maturation.
To examine the effects of inactivation of Chd7 on OPC differentiation during postnatal development in a time-controlled manner, we generated OPC-inducible Chd7 mutants by breeding Chd7flox/flox mice with PDGFRα-CreERT, an OPC-specific tamoxifen-inducible Cre 23 and a Rosa26tdTomato reporter. Tamoxifen administration in PDGFRαCreERT:Chd7flox/flox mutants (referred as Chd7-iKO) pups at P3 induced effective recombination of both Chd7 and Rosa26 loci with approximately 90% PDGFRα+ OPCs being tdTomato+ and negative for Chd7 expression after 11 days post-induction (Fig. 3g). Loss of Chd7 in OPCs led to a dramatic (~ 76%) reduction in differentiating OLs (CC1high expressing cells) in the cortical regions of Chd7-iKO mice compared to control littermates (PDGFRαCreERT:Chd7flox/+) (Fig. 3h,j). Similarly, expression of MOG myelin proteins in mature OLs was reduced compared to robust expression in controls (Fig. 3i,j). Collectively, these observations indicate that the primary role of Chd7 lies in the genesis of differentiated OLs from OPCs rather than OPC generation and expansion.
Chd7 is critical for OL remyelination after demyelination
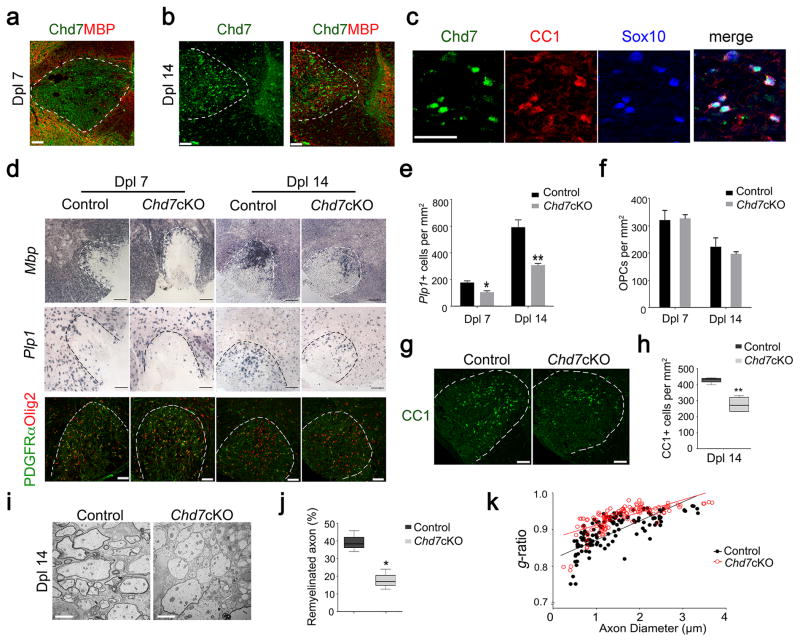
Given that myelination was normal in adult Chd7cKO animals, i.e. the numbers of myelinated axons and myelin morphology were comparable between Chd7cKO and control mice (Supplementary Fig. 3c,d), we then assessed the function of Chd7 in the remyelination process following lysolecithin-induced demyelination. Local injection of lysolecithin in the white matter induces rapid myelin breakdown followed by myelin regeneration through an OPC recruitment phase at 7 days post-lesion (Dpl 7) and a remyelinating phase at Dpl 14 24. In normal adult spinal white matter, expression of Chd7 was hardly detectable; however, Chd7 was re-expressed following demyelination induced by lysolecithin beginning at Dpl 7 (Fig. 4a), while Chd7+ cells increased substantially at Dpl 14 within the lesion (Fig. 4b). Chd7 was mainly expressed in Sox10+ OLs and predominantly confined to CC1+ differentiating OLs at Dpl 14 (Fig. 4c).
Figure 4. Chd7 is required for timely CNS remyelination.
(a,b) Immunostaining showing Chd7 and MBP at Dpl 7 and 14 in spinal cord LPC lesions from 8-week old mice. Scale bar, 50 μm.
(c) Immunostaining showing Chd7, CC1, and Sox10 at Dpl 14 in spinal cord LPC lesions. Scale bar, 50 μm.
(d) In situ hybridization for Mbp and Plp and immunolabeling for PDGFRα and Olig2 in spinal LPC lesions of control and Chd7cKO mutants at Dpl 7 and 14. Scale bars, 100 μm.
(e,f) Quantification of Plp1+ OLs and PDGFRα+ OPCs in LPC lesion sites at Dpl 7 and 14 (The data are presented as mean ± s.e.m. For Plp1 at Dpl 7, n = 4 controls and 4 mutant animals, * p = 0.022, t = 3.079; for Plp1 at Dpl 14, n = 4 controls and 4 mutant animals, ** p = 0.0098, t = 3.726; for PDGFRα+ OPCs at Dpl 7, n = 4 controls and 3 mutant animals, p = 0.848, t = 0.202; for PDGFRα+ OPCs at Dpl 14, n = 3 controls and 4 mutant animals, p = 0.299, t = 1.158; Two-tailed unpaired Student’s t test).
(g) Immunostaining for CC1 in LPC lesions from control and Chd7cKO spinal cords at Dpl 14. Scale bar, 100 μm.
(h) Quantification of CC1+ OLs in LPC lesion sites at Dpl 14 (n = 4 controls and 4 mutant animals, ** p = 0.0015, t = 5.523; whiskers show minimum and maximum, box limits are first and third quartile and center lines are median; Two-tailed unpaired Student’s t test)
(i) EM images of LPC lesions from control and Chd7cKO spinal cords at Dpl 14. Scale bar, 2 μm.
(j) The percentage of remyelinated axons in LPC-induced lesions of control and Chd7cKO spinal cords at Dpl 14 (n = 4 controls and 4 mutant animals, * p = 0.0188, t = 3.191; whiskers show minimum and maximum, box limits are first and third quartile and center lines are median; Two-tailed unpaired Student’s t test).
(k) The myelin g-ratio in LPC-induced lesions of control and Chd7cKO mutants at Dpl 14 (for control, g-ratio = 0.8943 ± 0.005128, n = 128 remyelinated axons from 4 control mice; for Chd7cKO, g-ratio = 0.9330 ± 0.003088, n = 126 remyelinated axons from 4 mutant mice; p < 0.0001, t = 6.440; Two-tailed unpaired Student’s t test).
To determine the potential role of Chd7 in OL regeneration, we examined re-appearance of myelin genes and OLs in the lesion during remyelination in the control and Chd7cKO mice. At Dpl 7 and Dpl 14, Mbp and Plp1 expression and the number of Plp1+ differentiating OLs (Fig. 4d,e) were substantially reduced in Chd7cKO lesions compared to control. In addition, the number of OL lineage cells including CC1+ OLs, Sox10+ or Olig2+ cells in Chd7cKO mice during remyelination was significantly reduced (Fig. 4g,h; Supplementary Fig. 5). In contrast, loss of Chd7 did not appear to impair the recruitment of PDGFRα+ OPCs (Fig. 4d) and the number of OPCs in the lesion was comparable between control and Chd7cKO mice (Fig. 4d,f). Notably, far fewer myelinated axons were detected in the lesions of Chd7cKO mice than controls (Fig. 4i). The percentage of myelinating axons and the thickness of newly generated myelin sheaths around axons assessed by g-ratios at Dpl 14 were significantly reduced in Chd7cKO mutants (Fig. 4j,k). Similar results of reduction in OL regeneration were obtained for remyelination in the adult corpus callosum of Chd7cKO mice (Supplementary Fig. 6). Altogether, these observations indicate that Chd7 has a crucial role in OL remyelination in the context of white matter injury.
Chd7 regulates transcriptional programs for OL maturation
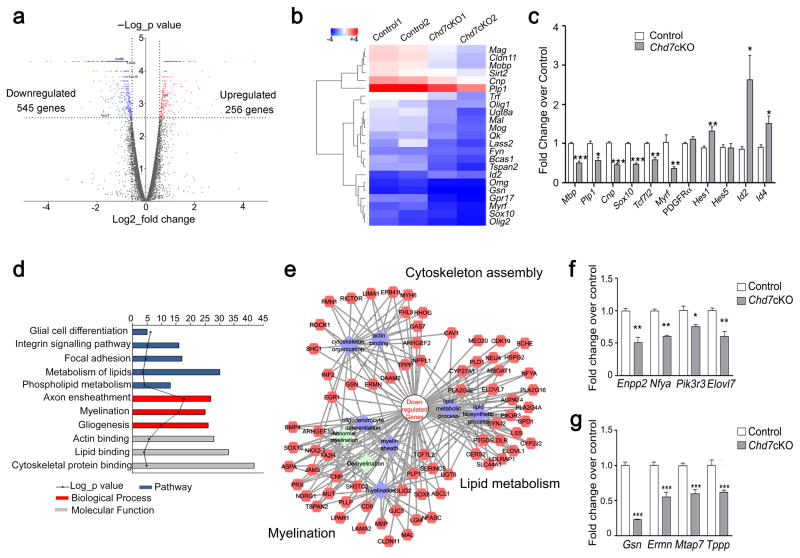
To investigate the underlying molecular mechanisms of dysmyelination in Chd7-deficient animals, we performed transcriptome profiling and analyzed global gene expression patterns in Chd7cKO and control spinal cords at P8. We identified genes altered in Chd7cKO pups that were either upregulated (n = 256) or downregulated (n = 545) relative to controls (fold change ≥ 1.5, p < 0.05; Fig. 5a). Among those, a cohort of myelin structural genes and critical myelination regulators including Myrf/Gm98 and Sox10 25, 26 were significantly reduced in the Chd7cKO spinal cord (Fig. 5b), consistent with deficient myelin formation phenotype. Similarly, quantitative real-time PCR analysis confirmed a dramatic reduction in myelination-promoting genes such as Myrf and Sox10, and an increase in differentiation inhibitors including Id2/4 and Hes1 (Fig. 5c). Gene ontology (GO) analysis further revealed that the genes downregulated in Chd7-deficient mice show an enrichment for those that function in cytoskeletal organization and lipid metabolism in addition to myelination (Fig. 5d). Congruent with physiological and morphological features of OLs in vivo, cytoskeleton and lipid metabolism pathways are highly active when OLs acquire complex morphology and assemble myelin sheaths around axons 27. Consistently, analysis of the potential gene regulatory network indicated dysregulation in the genes involved in actin assembly and lipid homeostasis, and in OL lineage-enriched genes in Chd7 mutants (Fig. 5e). Those alterations were further confirmed by qRT-PCR analysis of Chd7cKO spinal cords (Fig. 5f,g). Together, these data suggest that Chd7 regulates a gene regulatory network that controls OL differentiation and myelination.
Figure 5. Chd7 controls the core myelinogenic regulatory network.
(a) Volcano plots depict gene expression changes between control and Chd7cKO spinal cord at P8. Significantly differential transcripts were highlighted in color and totaled in each direction (FDR < 0.05).
(b) Heat map representing the expression of OL differentiation-related genes in control and Chd7cKO spinal cords from two independent animals.
(c) qRT-PCR analysis of myelination-associated genes in P8 control and Chd7cKO spinal cords (The data are presented as mean ± s.e.m. n = 3 controls and 3 mutant tissues; for Mbp, *** p = 0.0008, t = 9.11; for Plp1 * p = 0.028, t = 3.37; for Cnp, *** p = 0.0001, t = 14.63; for Sox10, *** p = 0.0005, t = 10.05; for Tcf7l2, ** p = 0.0049, t = 5.62; for Myrf, * p = 0.03, t = 3.29; for Pdgfrα, p = 0.168, t = 1.68; for Hes1, * p = 0.027, t = 3.38; for Hes5, p = 0.98, t = 0.02; for Id2, * p = 0.049, t = 2.81; for Id4, * p = 0.03, t = 3.29; Two-tailed unpaired Student’s t test).
(d) The gene ontology (GO) analysis of the significantly down-regulated genes between control and Chd7cKO.
(e) Protein-protein interaction networks among down-regulated genes in Chd7cKO are plotted using a ToppGene suite.
(f,g) qRT-PCR validation of selected genes involved in lipid metabolism (f) and cytoskeleton organization in control vs. Chd7cKO spinal cords at P8 (g) (The data are presented as mean ± s.e.m. n = 3 controls and 3 mutant tissues; for Enpp2, ** p = 0.002, t = 6.67; for Nfya, ** p = 0.0018, t = 7.41, for Pik3r3, * p = 0.046, t = 2.86; for Elovl7, ** p = 0.0012, t = 8.22; for Gsn, *** p < 0.001, t = 14.809; for Ermn, *** p < 0.001, t = 8.659; for Mtap7, *** p < 0.001, t = 10.181; for Tppp, *** p < 0.001, t = 9.907; Two-tailed unpaired Student’s t test).
Chd7 selectively targets enhancers of myeliogenic genes
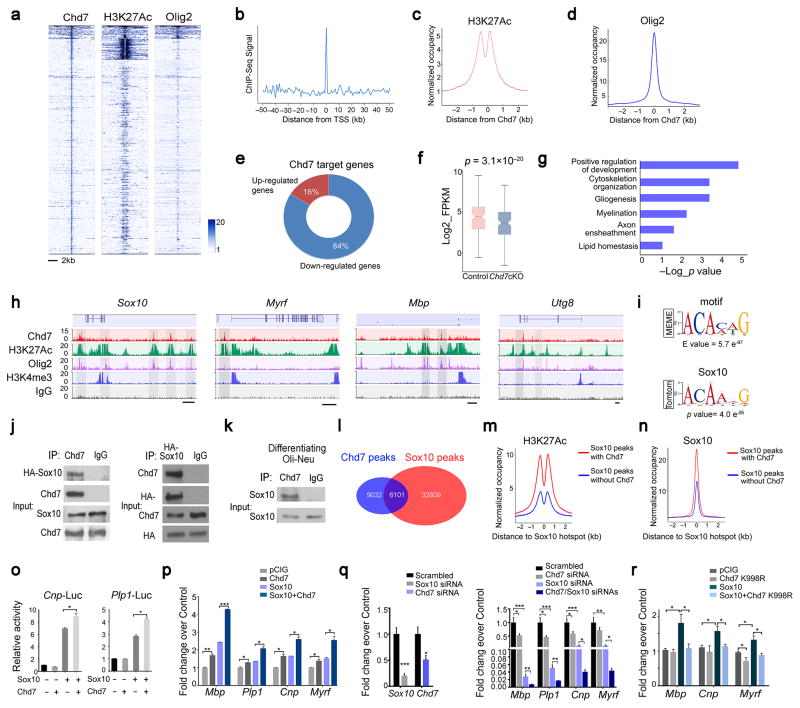
RNA-seq analysis does not establish whether a given regulated transcript is a direct Chd7 target. To identify Chd7 direct target genes, we utilized chromatin-immunoprecipitation and sequencing (ChIP-Seq) to assess Chd7 genome-wide occupancy in maturing OLs (mOL), which were differentiated from OPCs after three days of triiodothyronine (T3) exposure. Chd7 peak density was enriched within ± 5 kb elements proximal to transcriptional start sites (TSS; Fig. 6a,b). Chd7-occupied regions exhibit features of the active chromatin status, characterized by a bimodal distribution of H3K27Ac around Chd7 peaks and co-occupancy of Olig2 (Fig. 6a,c,d), an OL transcription factor previously shown to target to functional enhancers 22. Previous studies indicated that Brg1 targets to OL specific enhancers during the initiation of OPC differentiation, however, we note that the majority (~76%) of Chd7 binding regions are not targeted by Brg1 in mOLs (Supplementary Fig. 7), indicating that Chd7 harbors unique molecular functions that control OL maturation. Notably, among Chd7 target genes that were differentially expressed upon Chd7 deletion, the majority (~84%) was downregulated (Fig. 6e,f), suggesting that Chd7 predominantly functions as a transcriptional activator for promoting the OL differentiation program.
Figure 6. Chd7 interacts with Sox10 and selectively activates myelination-related genes during OL differentiation.
(a) ChIP-seq density heatmaps for Chd7, H3K27Ac and Olig2 within ± 5 kb around the Chd7 peak center.
(b) Histogram showing the distribution of ChIP-seq peaks relative to the TSS.
(c,d) Average H3K27Ac and Olig2 ChIP-seq enrichment profiles around the central position of Chd7 binding regions.
(e) Pie chart showing relative percentage of Chd7 target genes that are significantly upregulated or downregulated in Chd7cKO.
(f) Box plots for mRNA levels of Chd7 targeted genes that are differentially expressed in Chd7cKO mutants.
(g) Representative GO functional categories for Chd7 targeted genes.
(h) ChIP-seq showing Chd7 enrichment at selected gene loci (Sox10, Myrf, Mbp and Ugt8). Tracks represent sequence tag enrichments for Chd7, H3K27Ac, Olig2, Brg1, H3K4me3 and IgG. Genome scale bars: 5 kb.
(i) MEME analysis of the most highly represented de novo motif in the Chd7 cistrome in OLs. Sox10 binding motif was identified as the most significant matching binding site. Letter size indicates nucleotide frequency (presented as ‘bits’) at each position (horizontal axis).
(j,k) Co-immunoprecipitation of HA-Sox10 with Chd7 from transiently transfected 293T cells (j) or endogenous Sox10 with Chd7 in differentiating Oli-Neu cells (k). Full-length blots are presented in Supplemental Figure 13.
(l) Venn diagrams depicting overlap between Chd7 and Sox10 occupancy in OLs.
(m,n) ChIP reads density plots for levels of H3K27Ac and Sox10 at loci targeted by Sox10 only and co-targeted by Sox10/Chd7.
(o) Luciferase activity of Cnp (left) and Plp1 (right) promoter driven reporters in 293T cells co-transfected with Chd7, Sox10 or both. (The data are presented as mean ± s.e.m. For Cnp-Luc, n = 3 independent experiments, * p = 0.0138; for Plp1-Luc, n = 4 independent experiments, *p = 0.038; pairwise comparison for Sox10 compared with Sox10 + Chd7).
(p) qRT-PCR analyses of differentiating markers Mbp, Plp1, Cnp and Myrf in Oli-Neu cells transduced with Sox10, Chd7 or both under proliferation condition (The data are presented as mean ± s.e.m. n = 3 biological replicates; For Mbp, ANOVA F = 1661; multiple comparisons with t-test, ** pChd7 vs pCIG = 0.008, tChd7 vs pCIG = 6.397, *** pChd7+Sox10 vs Sox10 = 0.0002, tChd7+Sox10 vs Sox10 = 9.876. For Plp1, ANOVA F = 137.6; multiple comparisons with t-test, * pChd7 vs pCIG = 0.017, tChd7 vs pCIG = 4.792, * pChd7+Sox10 vs Sox10 = 0.01, tChd7+Sox10 vs Sox10 = 5.836. For Cnp, ANOVA F = 119.6; multiple comparisons with t-test, * pChd7 vs pCIG = 0.013, tChd7 vs pCIG = 5.344, * pChd7+Sox10 vs Sox10 = 0.01, tChd7+Sox10 vs Sox10 = 5.794. For Myrf, ANOVA F = 57.61; multiple comparisons with t-test, * pChd7 vs pCIG = 0.028, tChd7 vs pCIG = 4.042, * pChd7+Sox10 vs Sox10 = 0.02, tChd7+Sox10 vs Sox10 = 5.43).
(q) Left: qPCR validation of knockdown efficiency of Sox10 and Chd7 in primary rat OPCs under differentiation conditions (The data are presented as mean ± s.e.m. n = 5 biological replicates; for Sox10 *** p = 0.0003, t = 6.017; for Chd7 * p = 0.016, t = 3.041; Two-tailed unpaired Student’s t test). Right: qRT-PCR analyses of OL-differentiation associated genes following treatments with scrambled (n = 3), Sox10 (n = 5), Chd7 (n = 5), and both Sox10 and Chd7 siRNAs (n = 5), respectively. (mean ± s.e.m. n, numbers of biological replicates; for Mbp, ANOVA F (3, 14) = 36.26; multiple comparisons with t-test, * pChd7 vs scrambled = 0.03, tChd7 vs scrambled = 2.82, *** pSox10 vs scrambled = 0.0004, tSox10 vs scrambled = 6.84, ** pSox10+Chd7 vs Sox10 = 0.006, tSox10+Chd7 vs Sox10 = 3.7. For Plp1, ANOVA F (3, 14) = 38.12; multiple comparisons with t-test, * pChd7 vs scrambled = 0.015, tChd7 vs scrambled = 3.34, *** pSox10 vs scrambled = 0.0005, tSox10 vs scrambled = 6.68, ** pSox10+Chd7 vs Sox10 = 0.004, tSox10+Chd7 vs Sox10 = 3.86. For Cnp, ANOVA F (3, 14) = 37.99; multiple comparisons with t-test, * pChd7 vs scrambled = 0.033, tChd7 vs scrambled = 2.74, *** pSox10 vs scrambled = 0.0002, tSox10 vs scrambled = 7.55, * pSox10+Chd7 vs Sox10 = 0.01, tSox10+Chd7 vs Sox10 = 3.15. For Myrf, ANOVA F (3, 14) = 21.45; multiple comparisons with t-test, ** pSox10 vs scrambled = 0.002, tSox10 vs scrambled = 5.74, * pSox10+Chd7 vs Sox10 = 0.03, tSox10+Chd7 vs Sox10 = 2.69).
(r) qRT-PCR analyses of differentiated OL markers Mbp, Cnp and Myrf in Oli-Neu cells transduced with Sox10, Chd7 K998R mutant form or both under proliferation condition (The data are presented as mean ± s.e.m. n = 4 independent experiments; for Mbp, ANOVA F (3, 12) = 8.803; multiple comparisons with t-test, * pSox10 vs pCIG = 0.022, tSox10 vs pCIG = 3.062, * pSox10+Chd7 K998R vs Sox10 =0.025, tSox10+Chd7 K998R vs Sox10 =2.97. For Cnp, ANOVA F (3, 12) = 4.802; multiple comparisons with t-test, * pSox10 vs pCIG = 0.035, tSox10 vs pCIG = 2.706, * pSox10+Chd7 K998R vs Sox10 =0.035, tSox10+Chd7 K998R vs Sox10 =2.71. For Myrf, ANOVA F (3, 12) = 12.76; multiple comparisons with t-test, * pChd7 K998R vs pCIG = 0.031, tChd7 K998R vs pCIG = 2.79, * pSox10 vs pCIG = 0.014, tSox10 vs pCIG = 3.447, * pSox10+Chd7 K998R vs Sox10 =0.015, tSox10+Chd7 K998R vs Sox10 =3.41).
Gene ontology analyses of Chd7-targeted regions revealed significant association with several regulatory arms of pathways and modules related to OL maturation (Fig. 6g). Among Chd7-targeted genes were those encoding molecules involved in transcriptional control of myelinogenesis (Sox10, Myrf, Olig1, Zfp191 and Sip1/Zeb2), lipid metabolism and cytoskeleton assembly, as well as key myelin components (Mbp, Utg8, Plp1 and Cnp; Fig. 6h; Supplementary Fig. 8a,b). When comparing Chd7 binding with those in embryonic stem cells (ESCs) 10 or neural stem cells (NSCs)8, the binding sites of Chd7 in OLs minimally overlapped with those in ESCs or in NSCs (Supplementary Fig. 8c), suggesting a unique role of Chd7 in OL differentiation. Accordingly, Chd7 target genes were generally expressed at relatively high levels in the OL lineage compared with other cell types (Supplementary Fig. 8d). The significant correlation between Chd7 occupancy and elevated myelination-associated gene expression further suggests that Chd7 selectively activates specific gene sets for OL differentiation.
To investigate whether any specific DNA sequence motifs were enriched at Chd7 binding sites, we utilized the de novo DNA motif–discovery program MEME 28 to identify consensus-sequence motifs associated with Chd7 targeted sites. This analysis revealed that Chd7 binding sites with the consensus binding motif [C(T/A)TTG(T/A)(T/A)] for Sox10 (MEME motif score E = 5.7 e−97; Fig. 6i), a transcription factor required for terminal differentiation of the OL lineage 29, were most significantly overrepresented (Supplementary Fig. 9a).
Chd7 interacts with Sox10 to activate myelination program
To determine whether Chd7 and Sox10 colocalization result from a direct interaction between these two proteins or unrelated occupancy, we performed co-immunoprecipitation by expressing Sox10 and Chd7 in 293T cells or by pulling-down endogenous Sox10 using a Chd7 antibody in an oligodendroglial cell line, Oli-neu, under differentiating conditions 30. We found that Chd7 formed a complex with Sox10 in the 293T cells transfected with Sox10 and Chd7 and differentiating Oli-neu cells (Fig. 6j,k), suggesting that Chd7 and Sox10 targeting to these sites occurred through a physical complex of these proteins. To determine whether Sox10 and Chd7 co-occupied the same regulatory elements in mOLs, we then charted genome-wide occupancy of Sox10 in mOL using ChIP-seq. We found that approximately 40% of Chd7 peaks overlapped with those of Sox10 in mOL (Fig. 6l; Supplementary Fig. 9b,c), which match with Sox10 peaks in developing spinal cord 29. Sox10 and Chd7 targeted to a common set of OL regulatory genes including myelination-promoting genes Myrf, Nkx2.2, Sip1, Olig1 and Mbp (Supplementary Fig. 10).
Chd7/Sox10 co-occupied sites were highly enriched with the activating histone mark H3K27ac as compared to the Sox10 loci without Chd7 co-localization (Fig. 6m). Importantly, Sox10 binding exhibited a preference for Chd7-occupied sites (Fig. 6n). This observation indicates Chd7 occupancy defines enhancer elements favoring Sox10 recruitment and a more accessible chromatin state associated with gene activation. Consistently, Chd7 and Sox10 overexpression in combination enhances the activity of Cnp and Plp1-promoter reporters (Fig. 6o) and promotes expression of OL differentiation genes greater than when expressed individually in Oli-neu cells (Fig. 6p). Reduction of both Chd7 and Sox10 by siRNA-mediated knockdown caused much stronger inhibition in expression of myelination-related genes than individual Chd7 or Sox10 downregulation during OPC differentiation (Fig. 6q), raising the possibility of genetic interaction and cooperativity between Chd7 and Sox10 in control of OL differentiation. Notably, Sox10-mediated activation of myelin-associated genes was inhibited by the presence of the ATPase-defective mutant variant of Chd7 (Chd7K998R) (Fig. 6r) 9, suggesting an inhibitory effect of the Chd7 K998R mutant on Sox10 function. Together, these findings indicate cooperative functions of Chd7 and Sox10 in activating the transcriptional program for OL differentiation.
Osterix/Sp7 and Creb3l2 are Chd7 targets for OL maturation
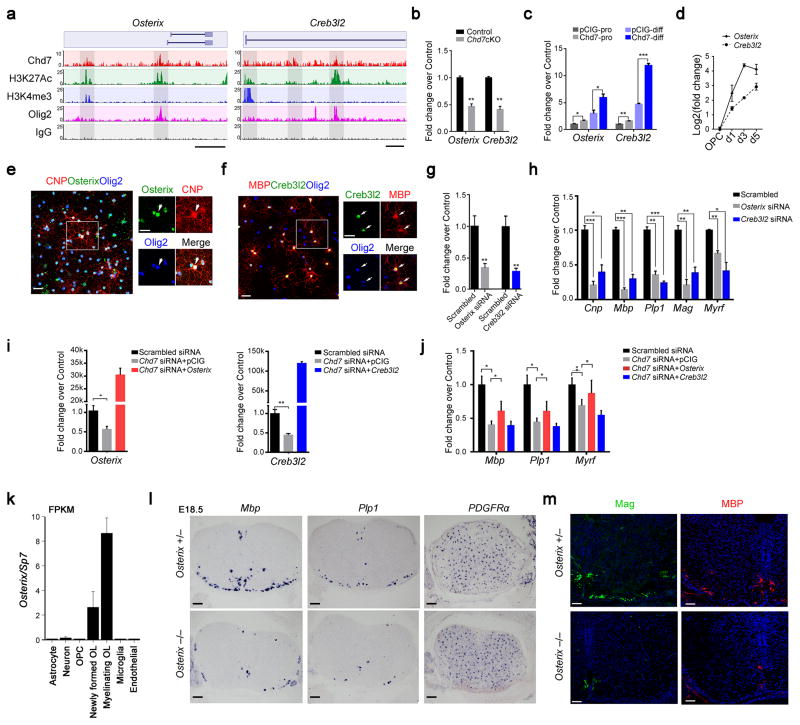
The observation that Chd7 transcriptionally activates an OL-specific gene network prompted us to search for Chd7-driven candidate genes that contribute to OL differentiation. By integrating Chd7 genome occupancy and transcriptome profiling, and considering genes downregulated in the Chd7cKO and occupied by Chd7 as candidate targets, we identified a cohort of potential transcriptional regulators that have yet to be characterized in OL development including Osterix/Sp7, Creb3l2, Zfp365, Zfp167, and Sorbs3. Intriguingly, Osterix encodes a zinc finger-containing transcription factor essential for osteoblast differentiation and bone formation31, and Creb3l2 encodes cAMP-responsive element binding protein 3-like 2, a basic leucine zipper (bZIP) transcription factor, which is critical for chondrogenesis32. We chose Osterix and Creb3l2 for further characterization because malformations of the temporal bone are also a common feature in CHARGE syndrome 33, 34. Chd7 targets the proximal elements of both gene loci around transcription start sites marked by H3K4me3 as well as distal enhancer elements marked by H3K27Ac (Fig. 7a). Expression of Osterix and Creb3l2 was significantly diminished in the spinal cord of Chd7cKO animals (Fig. 7b) while Chd7 overexpression significantly induced Osterix and Creb3l2 expression in OPCs under both proliferation- and differentiation-promoting conditions (Fig. 7c). Collectively, these data suggest that Chd7 directly activates Osterix and Creb3l2 in the OL lineage.
Figure 7. Osterix and Creb3l2 are direct Chd7 transcriptional targets and required for proper OL differentiation.
(a) ChIP-seq tracks for Chd7, H3K27Ac, Olig2, H3K4me3 and IgG at Osterix (left) and Creb3l2 (right) loci. Genome scale bars: 5 kb.
(b) qRT-PCR analyses of Osterix and Creb3l2 in P8 control and Chd7cKO spinal cords (The data are presented as mean ± s.e.m. n = 3 controls and 3 mutant tissues; for Osterix ** p = 0.005, t = 5.64; for Creb3l2 ** p = 0.0019, t = 7.26; Two-tailed unpaired Student’s t test).
(c) qRT-PCR analyses of Osterix and Creb3l2 in control and Chd7-overexpressing Oli-Neu cells under proliferation (pro) or differentiation (diff) conditions spinal cords (The data are presented as mean ± s.e.m. n = 3 biological replicates; for Osterix under proliferation condition, * p = 0.024, t = 3.551; for Osterix under differentiation condition, * p = 0.031, t = 3.269; for Creb3l2 under proliferation condition, * * p = 0.005, t = 5.565; for Creb3l2 under differentiation condition, *** p < 0.0001, t = 17.158; Two-tailed unpaired Student’s t test).
(d) qRT-PCR analyses of Osterix and Creb3l2 expression during OL differentiation (The data are presented as mean ± s.e.m. n = 3 biological replicates).
(e,f) Immunolabeling of Osterix or Creb3l2, CNP or MBP and Olig2 in primary mouse OLs. Scale bars, 50 μm. Boxed area (scale bar: 25 μm) was shown at a high magnification at right panels. Arrows indicate the co-labeling cells.
(g) qPCR validation of knockdown efficiency of Osterix and Creb3l2 in primary rat OLs (The data are presented as mean ± s.e.m. For Osterix, n = 3 independent experiments, ** p = 0.0067, t = 5.17; for Creb3l2, n = 3 independent experiments, ** p = 0.0022, t = 6.97; Two-tailed unpaired Student’s t test).
(h) qRT-PCR analyses of OL differentiation-associated genes following treatments with scrambled, Osterix or Creb3l2 siRNAs (The data are presented as mean ± s.e.m. n = 3 independent experiments; for Osterix siRNA vs scrambled siRNA, *** pCnp = 0.0008, tCnp = 9.013, *** pMbp = 0.00081, tMbp = 16.48, ** pPlp1 = 0.0011, tPlp = 8.49, ** pMag = 0.002, tMag = 7.43, ** pMyrf = 0.0014, tMag = 7.91; for Creb3l2 siRNA vs scrambled siRNA, * pCnp = 0.01, tCnp = 4.60, ** pMbp = 0.001, tMbp = 8.73, *** pPlp1 = 0.0002, tPlp = 12.97, ** pMag = 0.005, tMag = 5.65, * pMyrf = 0.014, tMag = 4.11; Two-tailed unpaired Student’s t test).
(i) qRT–PCR analysis of Osterix (left) and Creb3l2 (right) expression in OPCs transfected with siRNAs against Chd7 with control vector or vectors overexpressing Osterix and Creb3l2, respectively (The data are presented as mean ± s.e.m. For Osterix, n = 4 independent experiments, * p = 0.013, t = 3.468; for Creb3l2, n = 5 independent experiments, ** p = 0.001, t = 5.07; Two-tailed unpaired Student’s t test).
(j) qRT–PCR analysis of differentiation-associated genes (Mbp, Plp1, and Myrf) in OPCs transfected with siRNAs against Chd7 with control vector or vectors overexpressing Osterix and Creb3l2, respectively (The data are presented as mean ± s.e.m. n = 3 (scrambled siRNA), 3 (Chd7 siRNA + pCIG), 4 (Chd7 siRNA + Osterix), 4 (Chd7 siRNA + Creb3l2); n, numbers of biological replicates. For Mbp, ANOVO F (3, 11) = 10.77; multiple comparisons with t-test, * pscrambled siRNA vs Chd7 siRNA = 0.012, tscrambled siRNA vs Chd7 siRNA = 4.30, * ppCIG vs Osterix = 0.036, tscrambled siRNA vs Chd7 siRNA = 2.83. For Plp1, ANOVO F (3, 11) = 10.99; multiple comparisons with t-test, * pscrambled siRNA vs Chd7 siRNA = 0.022, tscrambled siRNA vs Chd7 siRNA = 3.63, * ppCIG vs Osterix = 0.045, tscrambled siRNA vs Chd7 siRNA = 2.65. For Myrf, ANOVO F (3, 11) = 7.42; multiple comparisons with t-test, * pscrambled siRNA vs Chd7 siRNA = 0.047, tscrambled siRNA vs Chd7 siRNA = 2.84, * ppCIG vs Osterix = 0.049, tscrambled siRNA vs Chd7 siRNA = 2.62).
k) Graph depicts RNA abundance represented as FPKM in indicated neural cell types.
l) RNA in situ hybridization for Mbp, Plp1 and PDGFRα on transverse spinal cord sections from Osterix +/− and Osterix −/− mice at E18.5. Scale bar, 100 μm.
m) Immunolabeling for Mag and MBP on transverse spinal cord sections from Osterix+/− and Osterix −/− mice at E18.5. Scale bars, 50 μm.
Consistent with the Chd7 expression pattern, Osterix and Creb3l2 expression levels increased significantly as OPCs differentiated into mature OLs (Fig. 7d). Similarly, we detected intense Osterix and Creb3l2 protein expression in cultured differentiating OLs bearing complex myelin processes (Fig. 7e,f). To evaluate the role of these OL-enriched genes in OL differentiation, we knocked down endogenous Osterix and Creb3l2 expression in OPCs using RNA interference (Fig. 7g). Downregulation of Osterix and Creb3l2 significantly inhibited expression of myelination genes including Cnp, Mbp, Plp1, Mag and Myrf (Fig. 7h). Intriguingly, exogenous overexpression of Osterix, but not Creb3l2, restored, at least partially, the defects in expression of myelination-associated genes caused by Chd7 knockdown in OPCs (Fig. 7i,j), pointing to the possibility that Osterix may mediate Chd7 function in OL differentiation.
Osterix is uniquely expressed in the OL lineage in the CNS, particularly in newly formed OL according to neural cell-type transcriptome database (Fig. 7k)35. To further determine the role of Osterix in OL development in vivo, we examined expression of markers for mature OLs and OPCs in Osterix-null animals, which die at birth. At E18.5, the latest collectable stage for Osterix-null embryos, expression of Mbp and Plp1 was markedly diminished in the spinal cord while the number of PDGFRα-expressing OPCs is comparable to that of control littermates (Fig. 7l). Consistently, expression of MAG and MBP assayed by immunostaining was profoundly reduced as compared to the control (Fig. 7m). These observations indicate that the Chd7 downstream regulator Osterix, a new OL-enriched transcriptional regulator, is required for the onset of OL differentiation.
DISCUSSION
The mechanisms underlying neurological deficiency, particularly the white matter defects, in CHARGE syndrome are poorly defined. Chd7 function in OL development has not been explored in any tissues or animal models. In the present study, strikingly, we find that Chd7 is highly enriched in differentiating/maturing OLs in the CNS. By generating mice with Chd7 mutated in the OL lineage, we demonstrate for the first time that Chd7 is required for proper onset of OL differentiation and myelination, as well as remyelination after demyelinating injury. Using a combination of RNA-seq and stage-specific ChIP-seq analyses, we present an unbiased whole genome occupancy map for Chd7 direct transcriptional targets for myelinogenesis and reveal Sox10 as a co-regulator of Chd7. We further identify new Chd7 targets such as an osteoblast-differentiation factor, Osterix, as a critical OL differentiation regulator. Thus, our studies identify hitherto previously uncharacterized Chd7 functions in white matter pathogenesis, and reveal that Chd7 acts as a molecular nexus of the critical regulatory networks that contribute to defects in distinct cells lineages affected in CHARGE syndrome.
Chd7 controls the onset of myelination and remyelination
Epigenetics and chromatin remodeling are increasingly appreciated to play critical roles in development and human diseases. We demonstrate here that loss-of-function in Chd7 leads to defects in CNS myelination and remyelination. The dysmyelinating phenotype in our mouse model resembles some of the features observed in CHARGE syndrome with proven CHD7 mutations, including delayed myelination and white matter defects (Fig. 1m,n) 13, 14.
Our data suggest that Chd7 regulates OPC differentiation in a cell-autonomous manner. Deletion of Chd7 does not appear to affect the developmental transition from Olig+ progenitors to PDGFRα+ OPCs, but blocks proper transitioning from OPC to OL. Consistently, temporal specific Chd7 deletion in PDGFRα+ OPCs leads to a failure of OPCs to differentiate. In addition, our studies using a LPC-induced demyelinating animal model further demonstrate that Chd7 is critical for remyelination, and is therefore an important determinant of effective myelin repair.
The genome-wide, base-resolution mapping of the Chd7 targeted cistrome here reveals that Chd7 selectively targets the enhancers of myelinogenic factors encompassing Myrf, Nkx2.2, Sip1 and Olig1, and activates a transcriptional program for OL differentiation, consistent with the Chd7 function as a transcriptional rheostat to regulate, or fine-tune gene expression levels 10. Notably, we find that the majority of targeting sites by Chd7 are distinct from those by Brg1, another ATP-dependent chromatin-remodeling factor in OLs (Supplementary Fig. 7), suggesting that distinct ATP-dependent chromatin remodeling factors such as Chd7 and Brg1 exert unique functions to regulate the OL maturation process. We further identify a cohort of previously uncharacterized Chd7-targets in OL such as the bone formation regulators Osterix and Creb3l2, and demonstrate that these transcriptional factors are also critical for oligodendroglial maturation. Despite its critical role in osteoblast lineage differentiation 31, Osterix is expressed predominantly in maturing OLs in the CNS. We find that Osterix positively regulates OL differentiation in vitro and in vivo. It is worth noting that Osterix regulates expression of osteopontin, which is associated with multiple sclerosis pathogenesis36, 37. Since malformations of the temporal bone are a common feature in CHARGE syndrome 33, 34, it is possible that regulation of Osterix by Chd7 may also contribute to temporal bone defects in CHARGE patients. These observations suggest that Chd7-controlled gene regulatory networks for OL differentiation may serve as an important intracellular cue to control the myelination process in the CNS and other seemingly unrelated malformations of other organs in CHARGE patients.
Chd7 interacts with Sox10 to regulate myelinogenic program
As an ATP-dependent nucleosome-remodeling factor, Chd7 requires additional transcriptional cofactors to target and activate gene expression. Our studies identify Sox10 as an important co-activator of Chd7 for target gene transcription during OL differentiation. Interestingly, mutations in Sox10 and Chd7 are both reported in Kallmann syndrome, a congenital disorder that exhibits hypogonadotropic hypogonadism and anosmia, features also frequently present in CHARGE syndrome 38. The clinical relevance between Sox10 and Chd7 mutations in these human diseases indicate a potential convergent role in developmental and pathogenic processes. Although the interaction of Chd7 with Sox10 provided the mechanistic basis for the role of Chd7 in myelinogenesis, the Chd7 mutant phenotype in mice seems less severe than that caused by deletion of Sox1025. These data suggest that Chd7 is an important accessory factor that quantitatively enhances the OL differentiation and myelination program that can proceed in its absence, but at a lower level. Alternative possible explanation could be redundancy with other CHD family proteins, such as CHD8, which is also highly expressed in oligodendrocytes (data not shown) and has been implicated in CHARGE syndrome pathogenesis through binding with Chd7 39.
How Chd7 is temporally and spatially targeted to specific sites in chromatin remains to be determined. Recruitment of pioneer transcription factors by DNA consensus motifs clustered on regulatory elements could prime chromatin, which, in turn, recruits remodeling factors for activating downstream target gene expression 40, 41. In line with this, Olig2 has been shown to act as a prepatterning factor that directs the recruitment of Brg1 to OL lineage-specific regulatory elements during the transition from OPC to immature OL 22. Our analysis of the Chd7 and Sox10 binding cistrome indicates that Sox10 preferentially occupies Chd7-targeted elements, while the frequency of Chd7 occupancy is comparable between elements with or without Sox10 targeting (Supplementary Fig. 11a). Since Sox10 occupancy in genomic loci increases gradually over the course of OPC maturation42, it seems likely that Chd7 is targeted to the chromatin first and then recruits Sox10. Intriguingly, Olig2 and Chd7 genome-wide binding cistromes indicate that Olig2 might possibly be prerequisite in the recruitment of Chd7 to regulatory elements (Supplementary Fig. 11b). Therefore, a potential sequential recruitment of priming factors such as Olig2, chromatin remodeling machinery such as Brg1 and Chd7, following by late phase regulators such as Sox10 may occur during OL lineage progression.
Implications for human diseases
Our study reports dysmorphic white matter in a significant portion of CHARGE patients with proven CHD7 mutations. The critical role for Chd7 in CNS myelination discovered here through OL lineage-specific mutagenesis implies that mutations in CHD7 may contribute to myelination and white matter defects seen in patients with CHARGE syndrome. A significant proportion of CHARGE patients develop growth retardation or delay, ranging from mild to severe. Given that white matter abnormalities caused by CHD7 mutations appear more severe in some of CHARGE patients compared with those in Chd7cKO mice; it is possible that altered expression or activity of Chd7 in other neural cell types such as neurons or neural stem cells 43 may also contribute to the human phenotype. Nonetheless, our studies show that Chd7 is highly enriched in differentiating OLs and critical for CNS myelination and remyelination. These observations suggest that dysmyelination caused by CHD7 mutations might contribute, at least partially, to developmental white matter defects seen in CHARGE patients 14. Whilst heterozygous Chd7 knockout mutants in the OL lineage are phenotypically normal, depletion of Chd7 expression in mice causes a myelination deficiency, indicating that a minimum threshold level of Chd7 expression is necessary for myelinogenesis. The phenotypic variation between the CHD7 haploinsufficient human mutants 5 and heterozygous knockout mice could be due to a differential sensitivity to gene dosage across species or dominant negative effects of human mutant proteins.
Our studies further provide multiple lines of functional genomics data that shed light on the molecular basis whereby Chd7 directly regulates not only a network of genes essential for OL myelination, but also genes associated with development and morphogenesis of multiple organs including craniofacial, eye and ear structures that are affected in CHARGE patients 5. Examples include genes involved in the developmental processes of craniofacial and bone development such as Osterix, Creb3l2, Col2a, Fgfr1/2 and Rarg 31, 44, eye development such as Pax6, Mitf and Rarg 45, 46, and ear development such as Ddr1, Prkra, Foxg1, Fgfr1/2 and Rarg 47–49 (Supplementary Fig. 12). Notably, retinoic acid receptor gamma (Rarg) acts as a direct target of Chd7 in OLs (Supplementary Fig. 12), consistent with recent reports that Chd7 regulates the expression of Rarg 43. Since Rarg signaling serves as a positive regulator of OL myelination and remyelination 50, its downregulation in Chd7 mutants may also contribute to CNS myelination defects in CHARGE syndrome. Together, our findings indicate that Chd7 serves as a nodal point of the regulatory network for the development of a diverse array of lineages spanning OLs and other cell types including osteoblasts, encompassing the phenotypes in CHARGE patients. Our unbiased genome-wide target gene analysis could provide a molecular framework to identify signaling pathways and molecules as therapeutic targets for promoting myelin regeneration in patients with demyelinating diseases and CHARGE syndrome.
Methods
Animals/Subjects
Mice homozygous for floxed alleles of Chd7 (Chd7f/f) 51 were crossed with mice carrying Cre recombinase driven by the Olig1 promoter (Olig1-Cre+/−) 52 to generate Chd7cKO (Chd7f/f;Olig1-Cre+/−) and heterozygous control (Chd7f/+;Olig1-Cre+/−) mice. Generation of Brg1cKO mice has been described previously22. Briefly, Brg1f/f mice 53 were crossed with Olig1-Cre mice to generate Brg1cKO (Brg1f/f;Olig1-Cre+/−) and heterozygous control (Brg1f/+;Olig1-Cre+/−) mice. Osterix/Sp7 mutant mice have been described previously 31, in which an IRES-lacZ-polyA/loxP-flanked PGK neo-bpA cassette is inserted into the second coding exon of Osterix. PDGFRα-CreERT BAC mice 23 and Rosa26tdTomato reporter mice (Ai14, Jax laboratory) where crossed with Chd7f/f mice to generate the OPC-specific iKO. Animals of either sex were used in the study and littermates were used as controls unless otherwise indicated. The mouse strains used in this study were generated and maintained on a mixed C57Bl/6;129Sv background and housed (three or less animals per cage) in a vivarium with a 12-hour light/dark cycle. All animal studies were approved by the Institutional Animal Care and Use Committees of the Cincinnati Children’s Hospital Medical Center, USA, and Sorbonne Universités, UPMC University Paris 06, France. MRI images of CHARGE patients including males and females from age 2 to age 15 were evaluated by neuroradiologists and compared with age-matched controls. All human patient images were obtained with informed consent as outlined by the institutional review board at the Cincinnati Children’s Hospital Medical Center.
Tissue processing and in situ hybridization
Mice at various developmental stages were anesthetized with ketamine/xylazine and perfused with PBS followed by 4% paraformaldehyde (PFA). Spinal cords or brains were dissected, fixed in 4% PFA overnight, dehydrated in 20% sucrose at 4°C, embedded in OCT and cryosectioned at 16 μm. For tamoxifen treatment of PDGFRα-CreERT-Chd7f/f mice, tamoxifen (T5648) was dissolved in corn oil (Sigma, C-8267) and injected intraperitoneally at 3 mg/40 gram (body weight) per day. In situ hybridization was performed as previously described 54. Digoxigenin-labeled riboprobes used in the study were: murine Mbp, Plp/Dm-20, and Pdgfra.
Electron microscopy
Tissue processing was performed essentially as described previously 22. Briefly, mice were deeply anesthetized with ketamine/xylazine, perfused with 0.1M cacodylate, followed by 2.5% paraformaldehyde/2.5% glutaraldehyde in 0.1 M cacodylate (pH 7.2). Spinal cord and optic nerves were dissected and postfixed in 1% OsO4. Ultrathin sections were stained with lead citrate for electron microscopy imaging.
Immunohistochemistry and immunoblotting
Cryosections (12-μm thick) or vibratome sections (50-μm thick) were permeabilized and blocked in blocking buffer (0.3% Triton X-100 and 5% normal donkey serum in PBS) for 1 h at room temperature and overlaid with primary antibodies overnight at 4 °C. Antibodies used in the study were: rabbit anti-Olig2 (Millipore, AB9610, 1:5000), rat anti-PDGFRα (BD Bioscience, 558774, 1:600), mouse anti-APC (CC1, Oncogene Research, OP80, 1:150), goat anti-MBP (Santa Cruz Biotechnology, sc-13914, 1:400), mouse anti-MOG (8-18C5, DSHB, 1:500), rabbit anti-Chd7 (Cell signaling, 6505, 1:300), rabbit anti-Chd7 (Abcam, AB38124, 1:600), goat anti-Osterix/Sp7 (Santa Cruz Biotechnology, sc-22538, 1:300), rabbit anti-Creb3l2 (Santa Cruz Biotechnology, sc-366044, 1:300). After washing with 0.4% Triton X-100 in PBS, cells or sections were incubated with secondary antibodies conjugated to Cy2, Cy3 or Cy5 (Jackson ImmunoResearch Laboratories, 1:1000) for 2 h at room temperature, stained in DAPI for 5 min, washed in PBS and mounted with Fluoromount-G (SouthernBiotech). Cell images were quantified in a double-blinded manner. For immunoblotting, whole cell lysates were prepared 48 hr after transfection by using 1× Passive Lysis Buffer (Promega, Madison) supplemented with a protease inhibitor cocktail (1:200, Sigma, St. Louis, MO). After western blotting, proteins were detected with appreciate secondary antibodies by using chemiluminescence with the ECL kit (Pierce) according to the instructions of the manufacturer.
Stereological estimation of white matter volume
White matter area was determined by measuring the MBP immunoreactive region as previously described 55. The volumetric analysis of MBP-positive tissue was performed according to the method described previously 56 using the following formula: V=SA · p · T, where V is the total volume, SA is the sum of the area measured, p is the inverse of the section sampling fraction, and T is the section thickness.
OL culture and transduction
Primary rat OPCs were isolated as described 57 with slight modifications. Briefly, mixed glial cells were initially cultured in DMEM-F12 medium supplied with 15% FBS, then switched to B104 conditioned medium for 2 days before isolating OPCs by mechanical detachment in an orbital shaker. Isolated rat OPCs were grown in Sato growth medium supplemented with mitogens 10 ng/ml PDGF-AA and 20 ng/ml bFGF, and differentiated in OL Differentiation Medium (Sato medium supplemented with 15 nM T3 and 10 ng/ml ciliary neurotrophic factor). Mouse OPCs were isolated from P5-P6 cortices by immunopanning with antibodies against Ran-2, GalC and O4 sequentially as previously described 58. OPCs were transduced with adenoviral vectors carrying GFP control or Cre-GFP and analyzed by immunofluorescence.
Lysolecithin-induced demyelinating injury
Lysolecithin-induced demyelination was carried out in the ventrolateral spinal white matter of approximately 8-week-old mice. Anesthesia was induced and maintained by peritoneal injection of a mixture of ketamine (90 mg/kg) and xylazine (10 mg/kg). After exposing the spinal vertebrae at the level of T9-T12, meningeal tissue in the intervertebral space was cleared, and the dura was pierced with a dental needle. 0.5 μl of 1% lysolecithin (L-a-lysophosphatidylcholine, Sigma L4129) via a Hamilton syringe attached a glass micropipette was injected into the ventrolateral white matter via a stereotactic apparatus. Spinal cord tissues carrying the lesions were collected at different time points. For demyelination in the brain, focal demyelinating lesions were induced by stereotaxic injection of 1.5 μl lysolecithin solution (1% in 0.9% NaCl) into the corpus callosum at coordinates: 1 mm lateral, 1.3 mm rostral to bregma, 1.7 mm deep to brain surface) using a glass-capillary connected to a 10 μl Hamilton syringe. Animals were left to recover in a warm chamber before being returned into their housing cages. LPC-induced injuries were conducted in a genotype-blinded manner.
RNA extraction and qRT-PCR
Analyses were conducted with RNA extracts from spinal cord tissue of mutant mice and their littermate controls. Total RNA was extracted per the Trizol (Life Technologies) protocol. cDNA was generated with iScript™ cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed using the ABI Prism 7700 Sequence Detector System (Perkin-Elmer Applied Biosystems). qRT-PCR primers for mouse gene sequences were: Plp-f, tgctcggctgtacctgtgtacatt, Plp-r, tacattctggcatcagcgcagaga; mouse Mbp-f, tcacagaagagaccctcaca; Mbp-r, gccgtagtgggtagttcttg; Cnp1-f, tccacgagtgcaagacgctattca, Cnp1-r, tgtaagcatcagcggacaccatct; Chd7-f, gcctctcatcacgtacagca, Chd7-r, ggatgggggatttgtcctac; Elovl1-f, gaaagggctggacacttactt, Elovl1-r, cctcttcagtgtgaggagaaag; Creb3l2-f, aagaatacatggacagcctgg; Creb3l2-r, ttccccatcaccaaagtctg; Sorbs3-f, agatacactggactccgtacc; Sorbs3-r, caaatttctgagttcgccgg; Creb3l2-f, catcaccagcacctctcatc; Creb3l2-r, ccatttctcactctccacctc; Gapdh-f, tgccaaatatgatgacatcaagaa, Gapdh-r, ggagtgggtgtcgctgttg.
Chromatin-immunoprecipitation sequencing (ChIP-Seq) and data analysis
Rabbit anti-Chd7 (Cell signaling, 6505) and anti-Sox10 (Abcam, ab155279) antibodies were used for immunoprecipitation experiments. ChIP-seq assays were performed essentially as previously described 22. Briefly, nuclear suspensions of OLs were sonicated with a Covaris S220 sonicator (total time 8 min). Antibody (chromatin: antibody = 4:1) was added to chromatin and incubated at 4°C overnight. Chromatin-protein complex was immunoprecipitated with protein A/G plus agarose beads and washed sequentially. The ChIP-seq libraries were prepared using NEBNext ChIP-seq Library Prep Master Mix Set for Illumina (NEB cat # E6240L), and then run on the Illumina sequencer HS2000.
FASTQ files of ChIP-seq data were aligned to March 2012 rat genome assembly (RGSC Rnor_5.0/rm5) using Bowtie with the following options: -p 8, -m 1. Resulting SAM files were converted to BAM format using SAM tools. Peak calling was performed using MACS (Model-based Analysis of ChIP-Seq) (http://liulab.dfci.harvard.edu/MACS) with a p value cut off of 1x e9.
RNA-seq and data analysis
RNA-seq libraries from control and Chd7cKO spinal cord were prepared using Illumina RNA-Seq Preparation Kit and sequenced by a HiSeq 2000 sequencer. All RNA-Seq data were aligned to mm9 using TopHat 59 with default settings. We used Cuff-diff 60 to (1) estimate FPKM values for known transcripts and (2) analyze differentially expressed transcripts. In all differential expression tests, a difference was considered significant if the q value was less than 0.05 (Cuff-diff default). Heatmap of gene expression was generated using R language (http://www.r-project.org).
Gene ontology
GO-analysis of genes repressed and increased in Chd7cKO mutants was performed using Gene Set Enrichment (GSEA, http://www.broadinstitute.org/gsea/index.jsp). We used ToppCluster (https://toppcluster.cchmc.org/) to construct the network of induced and suppressed genes belonging to over-represented GO-term categories. ToppCluster (https://toppcluster.cchmc.org/) was used to plot functional protein association networks in a certain gene sub-network or module. Enriched motifs of Chd7 binding peaks in OLs were identified by MEME (http://meme-suite.org).
Statistical analysis
All analyses were done using GraphPad Prism 6.00 (San Diego California, www.graphpad.com). Data are shown as mean ± s.e.m. or as a Box-and-whisker plot. Data distribution was assumed to be normal, but this was not formally tested. Statistical significance was determined using two-tailed Student’s t tests or Wilcoxon rank-sum and signed-rank tests as indicated. One-way ANOVA test was performed by multiple comparisons or pairwise comparisons following Turkey’s ranking tests when comparing multiple groups. Significance was set as * for p < 0.05, ** for p < 0.01, and *** p < 0.001, unless otherwise indicated. In MEME motif analysis, E value shows the significant enrichment of de novo motifs while p value is shown the significant similarity between de novo motif and known motif. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. Quantifications were performed from at least three independent experiments. No randomization was used to collect all the data, but they were quantified blindly.
A methods checklist is available with the supplementary materials.
Supplementary Material
Acknowledgments
Authors would like to thank Z. Ma, Y. Yang and Z. Tao for technical support. We thank J. Wysocka for Chd7-expressing vectors and comments, and E. Hurlock for suggestions, and P. Wight for the Oli-neu cell line. This study was funded in part by grants from the US National Institutes of Health R01NS072427 and R01NS075243 to Q.R.L. and the National Multiple Sclerosis Society (NMSS-4727) to Q.R.L and the Foundation for “l’Aide à la Recherche sur la Sclérose en Plaques” (ARSEP), the National Multiple Sclerosis Society grants RG-4318A1/1 & RG-1501-02851 (to C.P. and Q.R.L.) and the program “Investissements d’avenir” ANR-10-IAIHU-06 to C.P.
Footnotes
ACCESSION CODES:
All the RNA-seq and ChIP-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE72727.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
D.H., Q.R.L and C.P. conceived the studies, designed experiments and analyzed data. C.M., M.F., and C.P. performed the studies of Chd7-iKO mice; C.Z. assisted with ChIP-seq experiments; B.K. assisted with histological analysis. J.W. assisted with co-immunoprecipitation experiments; Y.Q. and M.F., assisted with LPC-induced demyelination injury; B.V.J. and D.I.C. provided MRI data; D.W. provided human brain samples; X.Z. provided Osterix mutant animals; A.C., H.W., X.H., H.H., and B.Z. provided technical support and intellectual inputs on the manuscript; D.M.M. provided Chd7 floxed mice and advice on the study. D.H. and Q.R.L wrote the manuscript with inputs from the co-authors.
References
- 1.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luna-Zurita L, Bruneau BG. Chromatin modulators as facilitating factors in cellular reprogramming. Curr Opin Genet Dev. 2013;23:556–561. doi: 10.1016/j.gde.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brookes E, Shi Y. Diverse epigenetic mechanisms of human disease. Annu Rev Genet. 2014;48:237–268. doi: 10.1146/annurev-genet-120213-092518. [DOI] [PubMed] [Google Scholar]
- 5.Martin DM. Chromatin remodeling in development and disease: focus on CHD7. PLoS Genet. 2010;6:e1001010. doi: 10.1371/journal.pgen.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman JE, et al. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J Med Genet. 2014;48:334–342. doi: 10.1136/jmg.2010.087106. [DOI] [PubMed] [Google Scholar]
- 7.Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 9.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouazoune K, Kingston RE. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci U S A. 2012;109:19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu T, et al. Deregulated FGF and homeotic gene expression underlies cerebellar vermis hypoplasia in CHARGE syndrome. Elife. 2013;2:e01305. doi: 10.7554/eLife.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory LC, et al. Structural pituitary abnormalities associated with CHARGE syndrome. J Clin Endocrinol Metab. 2013;98:E737–743. doi: 10.1210/jc.2012-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Yu T, Wang L, Mo X, Yu Y. A novel CHD7 mutation in a Chinese patient with CHARGE syndrome. Meta Gene. 2014;2:469–478. doi: 10.1016/j.mgene.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenzie IA, et al. Motor skill learning requires active central myelination. Science. 2014;346:318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mar S, Noetzel M. Axonal damage in leukodystrophies. Pediatr Neurol. 2010;42:239–242. doi: 10.1016/j.pediatrneurol.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Trapp BD, et al. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 18.Gallo V, Deneen B. Glial development: the crossroads of regeneration and repair in the CNS. Neuron. 2014;83:283–308. doi: 10.1016/j.neuron.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuchero JB, Barres BA. Intrinsic and extrinsic control of oligodendrocyte development. Curr Opin Neurobiol. 2013;23:914–920. doi: 10.1016/j.conb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bercury KK, Macklin WB. Dynamics and Mechanisms of CNS Myelination. Dev Cell. 2015;32:447–458. doi: 10.1016/j.devcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen JA, Hudson LD, Armstrong RC. Nuclear organization in differentiating oligodendrocytes. J Cell Sci. 2002;115:4071–4079. doi: 10.1242/jcs.00103. [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franklin RJ. Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 25.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes & development. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emery B, et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swiss VA, et al. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6:e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic acids research. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Anido C, et al. Differential Sox10 genomic occupancy in myelinating glia. Glia. 2015 doi: 10.1002/glia.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster N, et al. TGF-beta induces cell death in the oligodendroglial cell line OLI-neu. Glia. 2002;40:95–108. doi: 10.1002/glia.10110. [DOI] [PubMed] [Google Scholar]
- 31.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 32.Saito A, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nature cell biology. 2009;11:1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 33.Amiel J, et al. Temporal bone anomaly proposed as a major criteria for diagnosis of CHARGE syndrome. Am J Med Genet. 2001;99:124–127. doi: 10.1002/1096-8628(20010301)99:2<124::aid-ajmg1114>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Friedmann DR, et al. Venous malformations of the temporal bone are a common feature in CHARGE syndrome. Laryngoscope. 2012;122:895–900. doi: 10.1002/lary.23205. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braitch M, Constantinescu CS. The role of osteopontin in experimental autoimmune encephalomyelitis (EAE) and multiple sclerosis (MS) Inflamm Allergy Drug Targets. 2010;9:249–256. doi: 10.2174/187152810793358778. [DOI] [PubMed] [Google Scholar]
- 37.Comi C, et al. The impact of osteopontin gene variations on multiple sclerosis development and progression. Clin Dev Immunol. 2012;2012:212893. doi: 10.1155/2012/212893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jongmans MC, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome--the clinical overlap with CHARGE syndrome. Clin Genet. 2009;75:65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batsukh T, et al. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum Mol Genet. 2010;19:2858–2866. doi: 10.1093/hmg/ddq189. [DOI] [PubMed] [Google Scholar]
- 40.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao C, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. 2016 doi: 10.1038/ncomms10883. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Micucci JA, et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014;23:434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahlman J, et al. Premature vertebral endplate ossification and mild disc degeneration in mice after inactivation of one allele belonging to the Col2a1 gene for Type II collagen. Spine (Phila Pa 1976) 2001;26:2558–2565. doi: 10.1097/00007632-200112010-00008. [DOI] [PubMed] [Google Scholar]
- 45.Graw J. Eye development. Curr Top Dev Biol. 2010;90:343–386. doi: 10.1016/S0070-2153(10)90010-0. [DOI] [PubMed] [Google Scholar]
- 46.Bharti K, et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 2012;8:e1002757. doi: 10.1371/journal.pgen.1002757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schimmang T. Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming. Hear Res. 2013;297:84–90. doi: 10.1016/j.heares.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer zum Gottesberge AM, Gross O, Becker-Lendzian U, Massing T, Vogel WF. Inner ear defects and hearing loss in mice lacking the collagen receptor DDR1. Lab Invest. 2008;88:27–37. doi: 10.1038/labinvest.3700692. [DOI] [PubMed] [Google Scholar]
- 50.Huang JK, et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat Neurosci. 2011;14:45–53. doi: 10.1038/nn.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin M, et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 55.Hedtjarn M, Mallard C, Arvidsson P, Hagberg H. White matter injury in the immature brain: role of interleukin-18. Neurosci Lett. 2005;373:16–20. doi: 10.1016/j.neulet.2004.09.062. [DOI] [PubMed] [Google Scholar]
- 56.Roughton K, Bostrom M, Kalm M, Blomgren K. Irradiation to the young mouse brain impaired white matter growth more in females than in males. Cell Death Dis. 2013;4:e897. doi: 10.1038/cddis.2013.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, et al. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nature protocols. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- 58.Chan JR, et al. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron. 2004;43:183–191. doi: 10.1016/j.neuron.2004.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.