Abstract
Revolutionary changes in sequencing technology and the desire to develop therapeutics for rare diseases have led to the generation of an enormous amount of genomic data in the last five years. Large-scale sequencing done in both research and diagnostic laboratories has linked many new genes to rare diseases, but has also generated a number of variants that we cannot interpret today. It is clear that we remain a long way from a complete understanding of the genomic variation in the human genome and its association with human health and disease. Recent studies identified susceptibility markers to infectious diseases and also the contribution of rare variants to complex diseases in different populations. The sequencing revolution has also led to the creation of a large number of databases that act as “keepers” of data, and in many cases give an interpretation of the effect of the variant. This interpretation is based on reports in the literature, prediction models, and in some cases is accompanied by functional evidence. As we move towards the practice of genomic medicine, and consider its place in “personalized medicine,” it is time to ask ourselves how we can aggregate this wealth of data into a single database for multiple users with different goals.
Keywords: Database, variant, mutation, inherited disease, data sharing
The Burden of Data from Large-Scale Sequencing Projects
We have come a long way since the first draft of the human genome was completed in 2001 (http://www.genome.gov; Venter 2000; Lander et al. 2001; Venter et al. 2001). Genomic data have revealed the complexity of the human genome, and the concept of one gene-one disease has changed. This has implications in all areas of medicine and is not limited to rare diseases (Macarthur 2012). Data from large-scale sequencing projects, such as the 1000 Genomes Project and projects focused on data aggregation, like the ExAC database, are now freely available for use in research and diagnostic settings (Watt et al. 2013; Lanthaler et al. 2014). These projects have made two things clear: 1. Certain regions of the human genome still cannot be sequenced with adequate depth or simply have not been assigned a sequence based on their content, strand orientation, or errors in assembly; and 2. There are thousands of rare variants in the general population that are difficult to classify. With respect to the latter, two influential papers published in 2015 are now widely used by research and diagnostic groups to classify variants. The Gunter et al publication (MacArthur et al. 2014) in Nature focuses on guidelines for investigating the causality of sequence variants in human disease by integrating gene and variant level evidence to support causality. The American College of Medical Genetics and Genomics (Richards et al, 2015) first published recommendations for sequence interpretation in 2005, and then again in 2008, with the most recent revision coming in 2015, which takes into account new gene discoveries and the significant advances in sequencing and introduces the five-term classification system (Maddalena et al. 2005; Richards et al. 2008; Richards et al. 2015). These guidelines are specifically directed towards inherited diseases testing in clinical laboratories, though they have also been used for somatic variant classification. Guidelines for classifying somatic variants are expected to be available in 2016 through a workgroup from the Association for Molecular Pathology. It is clear from both the Gunter et al and Richards et al recommendations that these guidelines must be used with caution and in reference to gene-disease association considering the still significant lack of variant data from many populations.
Variant Databases
Inherited Diseases
Inherited genetic disorders range from an incidence of 1:3000 to 1:200,000 or more. These are categorized as rare and ultra-rare disorders. Since the early discoveries of genes such as CFTR, DMD, FMR1, and PAH, there have been various efforts to create gene/disease-specific mutation databases. These efforts started with locus-specific databases (LSDBs) (Fokkema et al. 2005; Fokkema et al. 2011; Pan et al. 2011; Bean et al. 2013; International Alport Mutation et al. 2014; Lanthaler et al. 2014) made by individual research groups for genes such as TP53(Forbes et al. 2011). In the last decade as sequencing costs dropped, new genes were rapidly discovered. This made it necessary to create gene/disease-specific databases, and the Human Gene Mutation Database (HGMD) spearheaded much of this effort (Cooper et al. 1998; Krawczak et al. 2000; Cooper et al. 2006; George et al. 2008; Adzhubei et al. 2013; Peterson et al. 2013; Rivas et al. 2015); however, these efforts were not directed towards the collection of variants that are clinically curated. Databases like the HGMD act as a storehouse of genetic variation published in different databases. LSDBs have been more focused collecting curated variants from various genes in gene-specific databases. Overall, it is clear from available databases that only 25–30% of variants can be neatly classified as “pathogenic” or “benign,” with the remaining variants being classified as “variants of unknown clinical significance.”
Somatic Disease
Variant aggregation in somatic disease is focused mainly on variants in the COSMIC database, the cancer genome sequencing projects, and publications for developing targeted therapies for different cancers(Stoupel et al. 2005; Higgins et al. 2007; Forbes et al. 2010; Forbes et al. 2011; Medina et al. 2012; Sachdev et al. 2013; Watt et al. 2013). Although several variants have now been identified for their effects in different drug therapies, a great many variants still cannot be interpreted in a clinical context and fall into the category of “variants of unknown clinical significance.”
The Case for Data Submission and Variant Curation from Clinical Laboratories
Clinical laboratories develop new tests based on discoveries reported in the literature (Bean et al. 2013). These tests are largely “evidence based” for their disease causality. After the initial report of the gene discovery, most of the sequencing of a gene takes place in a clinical setting, yet these data do not find their way into the literature either due to lack of interest from the journals or the clinical laboratory in publishing the data. A vast amount of curated sequence data lies within the clinical laboratories. Many organizations, including the NIH, have taken steps to initiate the collection of this data through efforts like the ClinGen consortium (Peterson et al. 2013; Landrum et al. 2016)
US-Based Laboratories
Advances in genomic sequencing in the past decade have facilitated large-scale clinical laboratory sequencing of more than 2000 disease-associated genes, exomes, and genomes worldwide. These data have typically not been published or deposited into public databases and have therefore not been accessible to other clinical laboratories, researchers, and clinicians. Most clinical laboratories use ad hoc data-mining processes for variant interpretation and access large genomic data sets, such as dbSNP, 1000 Genomes, or the Broad ExAC database, published literature, locus-specific databases, data from experts performing research in these areas, and also internal laboratory data. Differences in interpretation can occur between laboratories and are caused not only by differences in the available data, but also the lack of standards surrounding variant classification. The Clinical Genome (ClinGen) Resource aims to improve our understanding of genomic variation through data sharing and collaboration, starting with aggregating sequence and structural variants in the National Center for Biotechnology Information (NCBI)'s publicly available ClinVar knowledge base(Peterson et al. 2013; Rehm et al. 2015; Landrum et al. 2016). More recent submissions to ClinVar have increased the number of variants to >90,000, providing a fast-growing resource for the community, as well as a substrate for catalyzing the much-needed harmonization of clinical variant classification standards. The ClinVar efforts have focused largely on submission from US-based laboratories from a grant supported by NHGRI. Clinical laboratories typically do not have dedicated efforts to make these submissions; so external funding is required to drive them.
The Human Variome Project
The Human Variome Project was the first large-scale effort to aggregate variants from clinical laboratories. The efforts from HVP are set up through country-specific nodes to collect variant data from various genes from different laboratories(Oetting 2011); Stanley et al. 2014; Oetting et al. 2016). This effort is extremely important since we still have so little knowledge of the rare variants versus disease-causing genomic variation in the different world populations. The ExAC database from the Broad Institute, which has aggregated variant data from over 66,000 exomes, has revealed many errors in the pathogenicity assignment of variants, but it is still not enough to classify rare variants in different populations.
Paucity of Submissions from Clinical Laboratories
As we move towards realizing the full potential of the human genome for personalized medicine, it is critical that we understand the impact of genomic variation in health and disease. The efforts made so far by groups in the US and Australasia and European countries have been tremendous in terms of aggregating, but are extremely fragmented. This is due mainly to a lack of funding to create a unified database, lack of engagement from key stakeholders and the use of different IT formats and frameworks for collecting data. According to the NIH Genetic Testing Registry, several hundred laboratories are performing genetic testing, but only 25 have submitted data a total of ~90,000 variants to ClinVar. Clinical laboratories generally receive very limited information about a patient’s clinical presentation when a test is ordered, making it impossible to deposit accurate phenotypic data into these databases. Therefore, caution must be exercised when trying to correlate phenotype with database genotype data since the database may contain serious errors in reporting phenotype. In addition, we must remember that our understanding of gene- disease causality is not static and interpretations may change with time.
Resolving Variants of Unknown Clinical Significance
Most clinical laboratories use ad hoc data mining processes for variant interpretation and use large genomic data sets discussed above as well as their internal laboratory data. Differences in interpretation can occur between laboratories due not only to differences in data available to a lab but also to a lack of standards for variant classification. The NHGRI funded Clinical Genome (ClinGen) Resource aims to improve understanding of genomic variation through data sharing and collaboration, starting with aggregating sequence and structural variants in the National Center for Biotechnology Information (NCBI)'s publicly available ClinVar knowledgebase. Emory Genetics Laboratory (EGL), ARUP, Laboratory for Molecular Medicine (LMM) and University of Chicago tested this process by submitting over 20,000 variants collectively in genes associated with various disorders (ACMG Annual meeting abstract, 2015). Of the 15,000 variants submitted by EGL, comparison to other laboratories' classifications revealed discrepancies for 82 variants (LMM), 15 variants (ARUP) and 28 (University of Chicago). Out of the 43 differences between EGL, ARUP and U of Chicago more than 95% were minor differences (likely benign/ benign) and 5% were more significant differences (VOUS/ benign or pathogenic). All minor differences were resolved based on additional data available in the laboratories (e.g. functional data, direct communication with a research investigator or in house testing of additional family members). Of the more significant differences, 97% were resolved. Those that could not be resolved were mainly based on differences in laboratory specific classification rules. An analysis of LMM/EGL discrepancies revealed three common reasons, including 1. threshold differences (e.g. the minor allele frequency above which a variant is classified as benign), 2. reporting differences influencing classification stringency (e.g. the inclusion of likely benign variants on clinical reports leading to a lower threshold for classifying them as such) and 3. the weight given to computational predictions. Of 82 variants compared, only one was deemed a discrepancy that could not be resolved without additional clinical expert input (an atypical Fabry disease variant). More recent submissions to ClinVar have increased the number of variants >90,000, providing a rapidly growing resource for the community and providing a substrate for catalyzing the much needed harmonization of clinical variant classification standards.
Big and Niche Pharma and Rare Disease
Rare diseases, though individually rare, are collectively common, and big pharma, which had until recently shied away from pursuing targeted therapeutic approaches for genetic disease, is now interested in developing drugs to treat these diseases. Many niche pharma companies have drugs in clinical trials, with a prominent example being the range of treatment options in different phases of clinical trails for Duchenne muscular dystrophy (DMD)(Falzarano et al. 2015). Also, the recent discovery of gene-editing approaches, such as the CRISPR/Cas system, has triggered further interest in developing approaches to treat rare diseases(Wojtal et al. 2016). This has clearly necessitated the creation of a unified global database for gene-disease association and their variants.
Patient Registries and Disease Advocacy Groups
Many patient advocacy groups have taken the lead in creating patient registries, driven mainly by patients who register themselves by submitting their gene test results and clinical details. These data are self reported and at times curated by physicians. Most of these databases are driven towards “clinical trial readiness” for new therapeutic strategies, which are coming up rapidly. It is clear from these registries that patients are driving the effort of “personalized medicine” and are no longer interested in keeping their results to themselves, but are willing to participate in such databases, which can give them access to new clinical trials and help them become part of a community that shares the goals and concerns of their particular disease (Peterson et al. 2013).
Privacy Issues in Databases
Most of the databases, which aggregate data from the literature or present gene-level data, do not contain any patient-level information or full clinical histories to avoid any identification issues. Clinical laboratories probably risk the most exposure and have policies in place for depositing data in various databases. Privacy issues naturally limit the amount of information that can be deposited into databases.
The Need for an Aggregated Global Genomic Variant Database
The multiple fragmented efforts to deposit, collect, store, and interpret data have created conflict in the community and resulted in the use of resources that could otherwise be directed towards a single streamlined effort with a uniform format for data collection and reporting. Many new efforts, such as the Global Alliance for Genomics and Health, are focused on developing unified strategies for responsible, voluntary, and secure sharing of genomic and clinical data. Despite a tremendous amount of work, there are still multiple parallel approaches being taken in the community at times leading to collecting duplicate data, which creates errors in our estimation of gene-variant burden in the population.
Given global web accessibility, it is impossible to limit the creation of databases. Today, only 5200 out of the 22,000 genes in the human genome are known to be disease associated. Many more discoveries are expected to come at a rapid pace in the next decade. It is also clear that some genes previously shown to be disease associated no longer cause disease and are either redundant functionally or may be implicated in an as yet unknown disorder.
High-quality, accurate classification of the clinical significance of an identified sequence variant is key in personalized genomic medicine. The increasing availability of population data on genetic variation covered above has greatly improved our ability to interpret normal sequence variation; however, sequence data from most patients with genetic disease resides with the clinical laboratories. It is critical that clinical laboratories recognize the importance of the data they hold and share these data with the medical community.
What Will a Global Genomic Variant Database Look Like?
We must design a global approach to gather genetic information in different databases and registries into a common global database, which would not only act as a repository, but also give appropriate recognition to efforts already in place (Figure 1). This can be done through micro attribution, which needs to be recognized by funding agencies as a positive effort made by the investigator. Without support, investigators may not submit the data to databases due to efforts focused on publications, which is needed to secure grants for research purposes.
Figure 1.
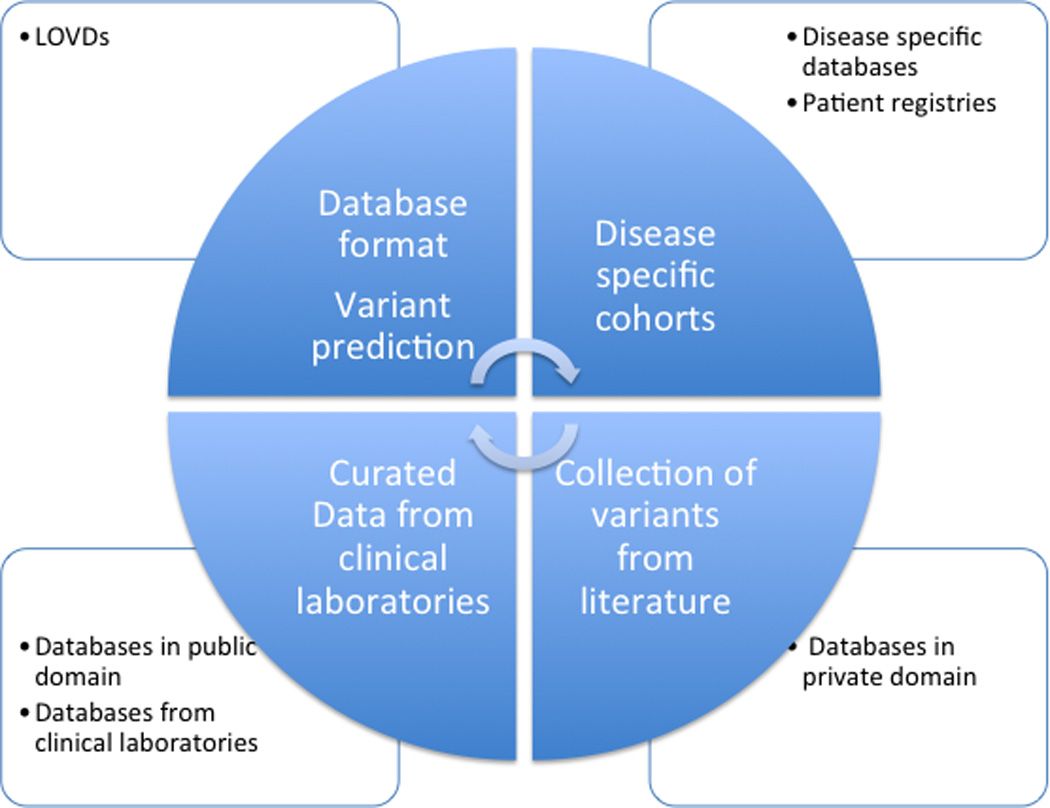
Amalgamation of databases to create a global database (blue circle). Data can assimilated from the four main domains where current variant data resides (open squares). These four domains include the LSDBs, disease specific cohorts and patient registries, databases in the public domain, such as ClinVar, and clinical laboratories databases, such as ClinVitae, EmVClass and ARUP, as well as databases in private domain, such as HGMD. Each of these arms brings a different strength to the global database and creates uniformity in data accessibility and presentation for usage for clinical trials and understanding the role genomic variants in personalized medicine.
Who Can Fund a Global Genomic Variant Database?
Funding for the various databases comes either from the organization creating the databases or from large funding agencies. Pharmaceutical companies, which need access to genomic data and patients, harness the information via collaborative efforts with patient advocacy groups or joint efforts with different entities. The total amount of funds that goes into these efforts is large, and it is clear that if a common fund were created through a non-profit entity, it could probably achieve the goals of a global database for the community.
Should the Database Be Freely Accessible and How Can We Make It Sustainable?
In considering the creation of a global database, it is important to put mechanisms in place to make it sustainable. This can be achieved in multiple ways by creating different levels of accessibility to the depth of data that needs to be accessed. It is important that the data are freely available to patient advocacy groups and patients, but entities using the data for clinical interpretation or developing therapeutic strategies could gain fee-based access to the database. This approach will make the database sustainable and support the huge effort required to create and maintain the database.
Summary
Technological advances have made it necessary for the community to consider approaches to the large-scale collection of data from genome sequencing and clinical outcomes. It is time to investigate how we can collect data from individual data sets and the current pattern, where data exist in silos and are kept in a non-standardized format. Large community efforts to aggregate data should be supported to create a global database, which would have multiple levels of data and grant different levels of accessibility to users to make it sustainable. Above all, it is critical that the key stakeholders come together now to drive the implementation of a global genomic variation database.
References
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Chapter 7: Unit7 20. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean LJ, Tinker SW, da Silva C, Hegde MR. Free the data: one laboratory's approach to knowledge-based genomic variant classification and preparation for EMR integration of genomic data. Hum Mutat. 2013;34:1183–1188. doi: 10.1002/humu.22364. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Ball EV, Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Stenson PD, Chuzhanova NA. The Human Gene Mutation Database (HGMD) and its exploitation in the study of mutational mechanisms. Chapter 1: Unit 1 13. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0113s12. [DOI] [PubMed] [Google Scholar]
- Falzarano MS, Scotton C, Passarelli C, Ferlini A. Duchenne Muscular Dystrophy: From Diagnosis to Therapy. Molecules. 2015;20:18168–18184. doi: 10.3390/molecules201018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema IF, den Dunnen JT, Taschner PE. LOVD: easy creation of a locus-specific sequence variation database using an "LSDB-in-a-box" approach. Hum Mutat. 2005;26:63–68. doi: 10.1002/humu.20201. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA, Smith TD, Callaghan S, Hardman L, Pierides C, Horaitis O, Wouters MA, Cotton RG. General mutation databases: analysis and review. J Med Genet. 2008;45:65–70. doi: 10.1136/jmg.2007.052639. [DOI] [PubMed] [Google Scholar]
- Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Alport Mutation C. Savige J, Ars E, Cotton RG, Crockett D, Dagher H, Deltas C, Ding J, Flinter F, Pont-Kingdon G, et al. DNA variant databases improve test accuracy and phenotype prediction in Alport syndrome. Pediatr Nephrol. 2014;29:971–977. doi: 10.1007/s00467-013-2486-8. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Fenton I, Stenson PD, Abeysinghe S, Thomas N, Cooper DN. Human gene mutation database-a biomedical information and research resource. Hum Mutat. 2000;15:45–51. doi: 10.1002/(SICI)1098-1004(200001)15:1<45::AID-HUMU10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanthaler B, Wieser S, Deutschmann A, Schossig A, Fauth C, Zschocke J, Witsch-Baumgartner M. Genotype-based databases for variants causing rare diseases. Gene. 2014;550:136–140. doi: 10.1016/j.gene.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Macarthur DG. Challenges in clinical genomics. Genome Med. 2012;4:43. doi: 10.1186/gm342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalena A, Bale S, Das S, Grody W, Richards S Committee ALQA. Technical standards and guidelines: molecular genetic testing for ultra-rare disorders. Genet Med. 2005;7:571–583. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- Medina I, De Maria A, Bleda M, Salavert F, Alonso R, Gonzalez CY, Dopazo J. VARIANT: Command Line, Web service and Web interface for fast and accurate functional characterization of variants found by Next-Generation Sequencing. Nucleic Acids Res. 2012;40:W54–W58. doi: 10.1093/nar/gks572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting WS. Exploring the functional consequences of genomic variation: the 2010 Human Genome Variation Society Scientific Meeting. Hum Mutat. 2011;32:486–490. doi: 10.1002/humu.21443. [DOI] [PubMed] [Google Scholar]
- Oetting WS, Brenner SE, Brookes AJ, Greenblatt MS, Hart RK, Karchin R, Sunyaev SR, Taschner PE. Pathogenicity Interpretation in the Age of Precision Medicine: The 2015 Annual Scientific Meeting of the Human Genome Variation Society. Hum Mutat. 2016 doi: 10.1002/humu.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Cong P, Wang Y, Lin C, Yuan Y, Dong J, Banerjee S, Zhang T, Chen Y, Zhang T, et al. Novel LOVD databases for hereditary breast cancer and colorectal cancer genes in the Chinese population. Hum Mutat. 2011;32:1335–1340. doi: 10.1002/humu.21588. [DOI] [PubMed] [Google Scholar]
- Peterson TA, Doughty E, Kann MG. Towards precision medicine: advances in computational approaches for the analysis of human variants. J Mol Biol. 2013;425:4047–4063. doi: 10.1016/j.jmb.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE Molecular Subcommittee of the ALQAC. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev PS, Lipnicki DM, Kochan NA, Crawford JD, Rockwood K, Xiao S, Li J, Li X, Brayne C, Matthews FE, et al. COSMIC (Cohort Studies of Memory in an International Consortium): an international consortium to identify risk and protective factors and biomarkers of cognitive ageing and dementia in diverse ethnic and sociocultural groups. BMC Neurol. 2013;13:165. doi: 10.1186/1471-2377-13-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CM, Sunyaev SR, Greenblatt MS, Oetting WS. Clinically relevant variants - identifying, collecting, interpreting, and disseminating: the 2013 annual scientific meeting of the Human Genome Variation Society. Hum Mutat. 2014;35:505–510. doi: 10.1002/humu.22516. [DOI] [PubMed] [Google Scholar]
- Stoupel E, Kalediene R, Petrauskiene J, Starkuviene S, Abramson E, Israelevich P, Sulkes J. Suicide-homicide temporal interrelationship, links with other fatalities, and environmental physical activity. Crisis. 2005;26:85–89. doi: 10.1027/0227-5910.26.2.89. [DOI] [PubMed] [Google Scholar]
- Venter JC. Remarks at the human genome announcement. Funct Integr Genomics. 2000;1:154–155. doi: 10.1007/s101420000026. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Watt S, Jiao W, Brown AM, Petrocelli T, Tran B, Zhang T, McPherson JD, Kamel-Reid S, Bedard PL, Onetto N, et al. Clinical genomics information management software linking cancer genome sequence and clinical decisions. Genomics. 2013;102:140–147. doi: 10.1016/j.ygeno.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Wojtal D, Kemaladewi DU, Malam Z, Abdullah S, Wong TW, Hyatt E, Baghestani Z, Pereira S, Stavropoulos J, Mouly V, et al. Spell Checking Nature: Versatility of CRISPR/Cas9 for Developing Treatments for Inherited Disorders. Am J Hum Genet. 2016;98:90–101. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


