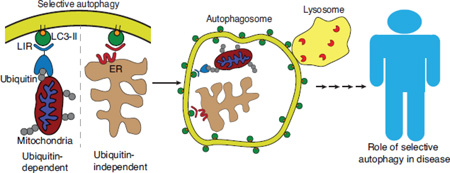
Abstract
Selective autophagy is critical for regulating cellular homeostasis by mediating lysosomal turnover of a wide variety of substrates including proteins, aggregates, organelles, and pathogens via a growing class of molecules termed selective autophagy receptors. The molecular mechanisms of selective autophagy receptor action and regulation are complex. Selective autophagy receptors link their bound cargo to the autophagosomal membrane by interacting with lipidated ATG8 proteins (LC3/GABARAP) that are intimately associated with the autophagosome membrane. The cargo signals that selective autophagy receptors recognize are diverse but their recognition can be broadly grouped into two classes, ubiquitin-dependent cargo recognition versus ubiquitin-independent. The roles of post-translational modification of selective autophagy receptors in regulating these pathways in response to stimuli are an active area of research. Here we will review recent advances in the identification of selective autophagy receptors and their regulatory mechanisms. Given its importance in maintaining cellular homeostasis, disruption of autophagy can lead to disease including neurodegeneration and cancer. The role of autophagy in cancer is complex as autophagy can mediate promotion or inhibition of tumorigenesis. Here we will also review the importance of autophagy in cancer with a specific focus on the role of selective autophagy receptors.
Keywords: Macroautophagy (autophagy), selective autophagy, ferritinophagy, mitophagy, cancer
Graphical abstract
INTRODUCTION
Macroautophagy (referred to hereafter as autophagy) is a conserved eukaryotic cellular catabolic pathway that degrades cellular organelles and other macromolecules via the lysosome as part of a recycling and protective process to maintain cellular fitness at a basal state as well as during stress[1,2]. The process involves the coordinated activity of a family of autophagy-related (Atg) proteins to mediate sequestration of cargo in a double membrane vesicle (autophagosome) that then fuses to a lysosome (autolysosome) filled with, among other components, lysosomal enzymes, allowing for degradation of cargo such as damaged organelles or toxic protein aggregates. This is followed by the recycling of breakdown products to be used in bioenergetic and anabolic pathways (Figure 1) [3,4]. While initially thought to be a bulk degradative pathway stimulated in response to cellular stressors, including starvation, research over the last decade has identified selectivity in the pathway for the identification of specific cargo for autophagic degradation[5,6]. Studies now suggest that multiple forms of selective autophagy are continuously active at some basal level in order to maintain cellular homeostasis whereas specific stimuli can activate selective autophagic pathways in order to address the particular stressor[7]. Coincident with its importance in maintaining cellular homeostasis, the disruption of selective autophagy pathways has been shown to play a role in diverse disease processes including neurodegeneration, atherosclerosis, and cancer[2,8–11]
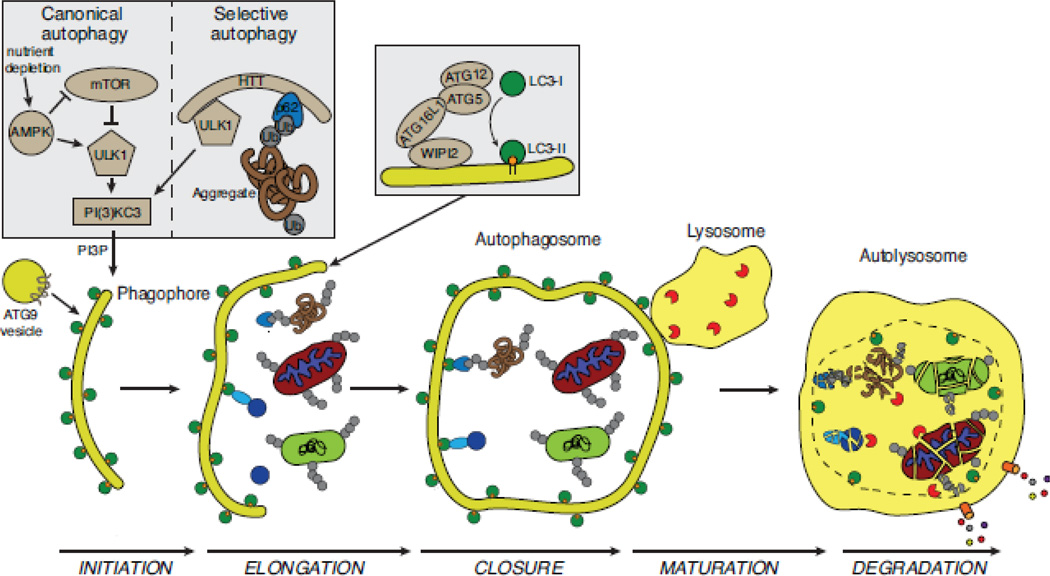
Figure 1.
The process and regulation of selective autophagy. The stages of autophagy (initiation, elongation, closure, maturation, and degradation) are shown. Cargo is sequestered in selective and bulk degradative manners via a double-membrane phagophore that fuses on itself to form the autophagosome. This subsequently fuses to the lysosome (autolysosome), where the cargo is degraded by lysosomal enzymes and degradation products are recycled to the cytosol by lysosomal transporters[198]. mTOR is a key regulator of bulk autophagy in response to changes in nutrient availability. During nutrient-replete conditions, mTOR is activated and autophagy is inhibited through repression of ULK1/2 (the mammalian homologs of ATG1). Upon nutrient depletion, the ULK1/2 complex is activated and can promote autophagy initiation. ULK1/2 is also activated at low energy states (an increased AMP/ATP ratio) by phosphorylation via AMPK as well as repression of mTORC1 activity. Activation of the selective autophagy pathway is via multiple specific stimuli and how and whether these stimuli engage the ULK complex in all circumstances is less well understood. Huntingtin (mammalian homology of yeast ATG11) was recently identified as a scaffold protein that can activate the ULK complex and bring a selective autophagy receptor into close proximity with activated ULK complex thereby linking activation of autophagosome formation with cargo destined for degradation. Also critical to autophagy initiation is the production of phosphatidylinositol-3-phosphate (PI3P) by the class III PI3K complex, here labeled ‘PI(3)KC3’ composed of Vps34, ATG14, ATG6/Beclin1 and p150 (Vps15). ATG9 containing vesicles contribute membrane to the growing autophagosome and likely participate in recruitment of other essential autophagy machinery (not shown)[29]. WIPI2 binds ATG16L1 to localize the ATG5–ATG12-ATG16L1 complex to autophagosomal membranes. This complex acts downstream of the ULK and PI(3)KC3 complexes in an E3-like manner to conjugate phosphatidylethanolamine (PE) to LC3-I to produce lipidated LC3-II that then associates with autophagosomal membranes and has roles in autophagosome membrane elongation. LC3-II is present on the outer and inner surfaces of the autophagosome (depicted as a green circle with orange PE moiety) and acts as the physical link between selective autophagy receptors and the autophagosome membrane. Selective autophagic cargos depicted here include ubiquitylated mitochondria, ubiquitylated bacteria (light green rounded rectangle), ubiquitylated protein aggregates recognized by a selective autophagy receptor (brown tangle), and nonubiquitylated cargo, such as ferritin (blue circle recognized by light blue oval depicting NCOA4).
Selective autophagy is characterized by the following principles, not necessarily stepwise or all required, as discussed below: 1. Presence of a degradation cue, 2. Cargo recognition via a so-called ‘selective autophagy receptor’ protein, 3. Recruitment of autophagosome machinery/membrane for eventual delivery to the lysosome for degradation [12]. Selective autophagic pathways are generally named for the cargo destined for degradation and include aggrephagy (protein aggregates), ferritinophagy (ferritin), mitophagy (mitochondria), xenophagy (pathogens, including bacteria), and ER-phagy (endoplasmic reticulum), among many (Tables 1–2) [5,13–18]. While not intended to be a comprehensive review of the rapidly expanding field of selective autophagy (we point the reader to several excellent reviews on the subject [5,6,19,20]), we will review the molecular mechanisms of the selective autophagic process and focus on recent progress in identification of new selective autophagic pathways. Lastly, we will further identify the current challenges and bottlenecks to future major advances in the field of selective autophagy and, most importantly, determine the relevance of selective autophagy pathways in the context of human physiology and pathophysiology.
Table 1.
Ubiquitin-dependent Selective Autophagy
| Pathway | Receptor(s) | Substrate | References |
|---|---|---|---|
| Mitophagy | OPTN, NDP52, TAX1BP1, p62 | Mitochondria | [60,61,67] |
| RNA granule disposal | NDP52, p62 | RNA granules | [68] |
| Pexophagy | NBR1, p62 | Peroxisome (PEX5) | [69,70] |
| Aggrephagy | p62, NBR1, OPTN TOLLIP, Cue5 | Protein Aggregates | [51,52,54,58,71] |
| Xenophagy | p62, OPTN, NDP52, TAX1BP1 | Bacteria | [55,57,72,73] |
| Proteaphagy | RPN10 | Proteasomes | [59] |
| Midbody disposal | p62, NBR1 | Midbody | [74,75] |
| Zymophagy | p62 | Zymogen | [76] |
Table 2.
Ubiquitin-independent Selective Autophagy
| Pathway | Receptor(s) | Substrate | References |
|---|---|---|---|
| ER-phagy | FAM134B, Atg40 | Endoplasmic Reticulum | [16,17] |
| Ferritinophagy | NCOA4 | Ferritin | [13,14] |
| Pexophagy | Atg30, Atg36 | Peroxisomes | [123,124] |
| Mitophagy | NIX, BNIP3, FUNDC1, Atg32 | Mitochondria | [64,125–128] |
| Aggrephagy | OPTN | Mutant HTT, SOD1 | [129] |
| Virophagy | TRIM5α, SMURF1, p62 | Viruses | [130–132] |
| Glycophagy | Stbd1 | Glycogen | [133] |
| Nucleophagy | Atg39 | Nuclear envelope | [17] |
| Lysophagy | Galectin-8/NDP52 | Lysosomes | [99] |
| Xenophagy | Galectin-8/NDP52 | Bacteria | [99] |
| Cvt targeting | Atg19, Atg34 | Ape1, Ams1 | [122,134] |
| Fatty acid synthase(FAS) disposal | FAS | FAS | [135] |
| Signalophagy | c-Cbl | Src | [136] |
| RHOA selective autophagy | SQSTM1 | RHOA | [137,138] |
| Nuclear lamina autophagy | Lamin B1 | Nuclear lamina/Nuclear | [139] |
| GATA4 selective autophagy | SQSTM1 | GATA4 | [140] |
Molecular mechanisms of autophagy initiation and autophagosome formation
Autophagy was initially characterized in mammalian cells as a cellular adaptive response to starvation stress[1,21]. However it is now clear that autophagy is active at some basal level in all cells and can be further activated by a variety of cellular stressors leading to a variety of selective autophagic events. Initial studies of the upstream molecular signaling apparatus for autophagy activation in response to starvation in yeast identified a complex set of over 30 different Atg genes, of which many are functionally conserved in higher eukaryotes [22,23]. Starvation-induced or canonical autophagy is controlled by multiple signaling components including those that interpret the cellular energy level (5’ AMP-activated protein kinase, AMPK) and nutrient/amino acid levels (Mammalian Target of Rapamycin, mTOR), as well as growth factors (Figure 1)[7,24]. These pathways converge on the unc-51 like autophagy activating kinase 1 (ULK1) (Atg1 ortholog) complex that mediates autophagy induction[25,26]. Following induction, the class III phophatidylinositol 3-kinase [PI(3)KC3)] complex composed of VPS34, p150, ATG14, and Beclin-1 (Atg6 ortholog) nucleates autophagosome formation, the ATG9 transmembrane protein mediates trafficking of source membrane for autophagosome elongation, and two ubiquitin-like conjugation systems described below participate in autophagosome maturation and recruitment of additional autophagy machinery including selective autophagy receptors[4,27–30]. How selective autophagy pathways engage the canonical autophagy activation machinery is less clear. Two recent studies have suggested a molecular link between engaged selective autophagy receptors and ULK1[31,32]. Notably, Atg11 in yeast and Huntingtin (HTT) in higher eukaryotes act as scaffolds to bind selective autophagy receptors and activate the ULK1/Atg1 kinase complexes, likely facilitating a physical link between substrate recognition and autophagosome initiation such that selective autophagy is localized to the recognized substrate. Finally, there are also non-canonical modes of autophagy activation not involving ULK1 or other core autophagy machinery that reflect the diverse mechanisms by which the autophagy program can be initiated[33,34].
The heart of the autophagosome maturation apparatus is the ubiquitin-like (UBL) protein lipidation system that catalyzes the conjugation of phosphatidylethanolamine to the C-terminus of Atg8 (there are seven mammalian homologs to the yeast Atg8), thereby facilitating attachment of ATG8 proteins to emerging autophagosomal membranes[35,36]. Briefly (for in-depth biochemical and structural reviews, readers are referred to the following references:[4,36]), ATG7 acts as an E1 enzyme with ATG10 as an E2 to conjugate the ubiquitin-like ATG12 to ATG5. This ATG12-ATG5 conjugate acts in a complex with ATG16L1 to facilitate ATG8 lipidation. Of note, the ATG16L1-ATG12-ATG5 complex is localized to autophagosomal membranes by WIPI2[37]. ATG8s are synthesized in a pro-ATG8 form that is cleaved by ATG4B to expose a C-terminal glycine residue. In the case of the mammalian ATG8 homolog, MAP1LC3B, this is dubbed the ‘LC3B-I form’. Subsequent action of ATG7 as an E1, ATG3 acting as an E2 and the ATG12-ATG5 conjugate in complex with ATG16L1 acting as an E3 then catalyzes the conjugation of phosphatidylethanolamine to the C-terminus of ATG8s. The lipidated ATG8 form of MAP1LC3B is termed ‘LC3B-II.’ This form is tightly associated with autophagosomal membranes.
Numerous studies indicate that ATG8 proteins function as adaptors for recruitment of selective autophagy receptors as well as regulatory proteins to autophagosomes[35,38]. Yeast contain a single ATG8 protein, but in mammals seven ATG8 proteins in two structurally related sub-families (MAP1LC3A,B/B2 & C, and GABARAP, GABARAPL1, & GABARAPL2 (also known as GATE-16)) are expressed, suggesting complex diversification of their functions[35]. Our understanding of how the seven mammalian ATG8 proteins control selective autophagy is limited, and the extent to which the two ATG8 subfamilies - LC3 and GABARAP - play unique or redundant roles in selective autophagy is not entirely clear. Many ATG8-associated proteins, including selective autophagy receptors, contain short linear peptide sequences that bind directly to ATG8s – the so-called LIR motif (LC3 interacting region)[39–41]. LIR motifs typically contain a W/F/Y-X-X-ψ sequence (ψ, hydrophobic residue: L/I/V, X is any residue), and often are preceded by acidic residues or by phosphorylation sites (as discussed in the section below), which modulate ATG8 binding. Non-canonical LIR sequences also exist suggesting further molecular determinants of ATG8 interacting proteins may yet be uncovered[42]. In-depth reviews of the importance of the LIR motif in selective autophagy and autophagosome machinery are presented elsewhere[39,41].
Upon autophagy activation, an incipient autophagosome is formed from pre-existing membrane sources. The source of autophagosomal membranes is an active area of research with multiple studies pointing to various sources (endoplasmic reticulum, Golgi complex, mitochondria, endosome, plasma membrane)[43,44]. The most likely scenario, and particularly relevant to selective autophagy, is that there are multiple membrane sources and that the source of membrane may be linked to the specific cargo being targeted due to spatial co-localization (e.g. in the case of ER-phagy or mitophagy). Regardless, the forming autophagosome engulfs cytoplasmic components, fuses to form a double-membrane autophagosome, and subsequently fuses with the lysosome leading to content degradation within an autolysosomal organelle. The products generated by lysosomal degradation are then released into the cytoplasm where they can be reused.
While macroautophagy is the most well-studied and robust pathway for selective autophagy, other lysosome-directed selective autophagic pathways exist including microautophagy and chaperone-mediated autophagy, reviewed elsewhere[45–47]. In addition, macroautophagy has been shown to be active in non-canonical pathways including LC3-associated phagocytosis, where additional research is necessary to understand the regulation and potential selectivity of these pathways[48,49].
Mechanisms of Selective autophagy
Selective autophagy is mediated by selective autophagy receptor proteins that physically link their cargo to the forming autophagosomal membrane for eventual delivery to the lysosome for degradation. There are multiple different types of selective autophagy receptors but they can be broadly grouped into two categories based on how they recognize cargo: ubiquitin-dependent versus non-ubiquitin mediated recognition[6]. As detailed above, the majority of autophagy receptors physically link to autophagosomal membranes via the phosphatidylethanolamine-conjugated ATG8 proteins, which are intimately associated with the initiation of autophagosomes as well as autophagosome and lysosome fusion steps. Selective autophagy receptors have been studied in detail in yeast and mammalian systems. While much of the core machinery for autophagosome initiation has been conserved and common pathways exist for specific cargo, there is significantly less conservation between yeast and mammalian cells with regards to the machinery for selective autophagy[50]. The study of selective autophagy pathways in mammalian cells has revealed important clues to their roles in disease-related processes[8,9]. Here we will focus on several aspects of mammalian selective autophagy receptors while also highlighting the parallel conserved pathways in yeast. Specifically, we will provide an overview of the two classes of selective autophagy receptors, how they recognize cargo, how they are regulated, and their importance in various disease processes. We will provide salient examples for each different class of selective autophagy receptor. At this point, given the expansion in number of identified selective autophagy receptors[6], no one review can fully encompass all known selective autophagy receptors but accompanying tables provide an ongoing resource of many of them (Table 1–2).
Selective autophagy receptors: ubiquitin-mediated cargo recognition
With the identification of the original mammalian selective autophagy receptors, it was clear that there was a central role for ubiquitin in selective autophagy. In the classic example, aggregates of either aberrantly folded or unused proteins are ubiquitylated and bound by ubiquitin-binding domain (UBD) containing receptors such as p62/SQSTM1 for subsequent delivery to the autophagosome for lysosomal degradation (Figure 2)[51,52]. It is clear that there is a cooperative function of the autophagy-lysosome system with the ubiquitin-proteasome system to manage the turnover of damaged proteins to maintain the proteome[53]. However in contrast to the ubiquitin-proteasome system that requires unfolding of substrates for degradation via the proteasome core, the autophagy-lysosome system is capable of handling much larger protein aggregates or tightly folded proteins without a requisite unfolding step[53]. Subsequent to the discovery of p62/SQSTM1 (hereafter referred to as p62), a slew of autophagy receptors that recognize ubiquitylated cargo were identified including NBR1, Optineurin (OPTN), TAX1BP1, NDP52/CALCOCO2, TOLLIP, and RPN10 (Table 1)[54–59]. Interestingly, there is some overlap in specificity for ubiquitylated cargo among selective autophagy receptors, including for ubiquitylated aggregates (p62, NBR1, OPTN, TOLLIP), bacteria (p62, OPTN, NDP52, TAX1BP1), mitochondria (OPTN, NDP52, TAX1BP1), and the midbody of cells (p62, NBR1). In some cases, as discussed below in the case of mitophagy, this overlap is cooperative to mediate delivery to autophagosomes[60,61]. In other cases, such as xenophagy, multiple different autophagy receptors appear capable of mediating the process individually[62]. This suggests that there is another layer of complexity and regulation with regards to recognition of cargo or activation of specific selective autophagy receptors in response to selective autophagy stimuli. Indeed, post-translational modifications of both the selective autophagy receptors as well as the cargo (and in some cases ubiquitin itself on the cargo) are integral to regulating autophagy receptor function[55,60]. There remain many questions with regards to the role of ubiquitin signals in selective autophagy, such as what the contribution of ubiquitin chain length and linkage are to cargo selectivity.
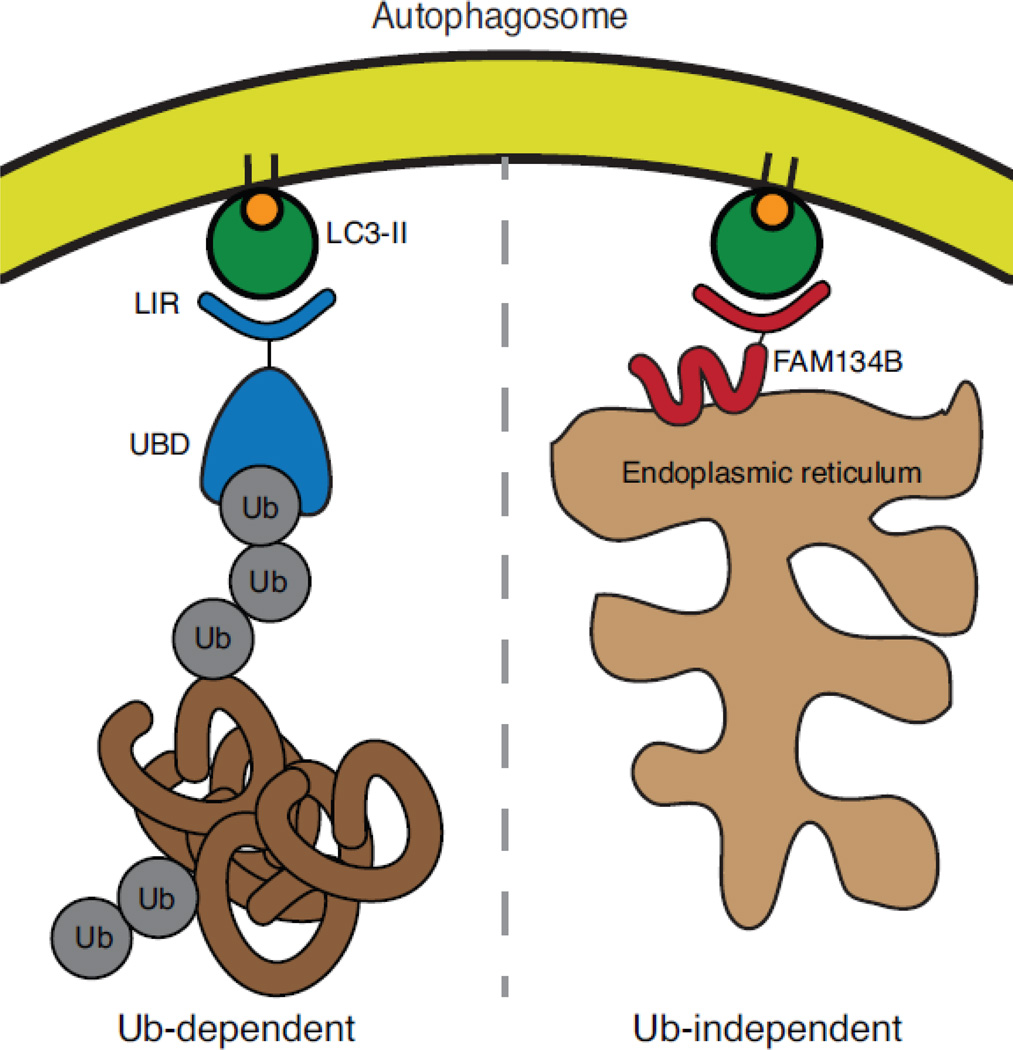
Figure 2.
Ubiquitin-dependent and ubiquitin–independent selective autophagy. On the left, a prototypical selective autophagy receptor with an ubiquitin-binding domain (UBD) recognizes an ubiquitylated substrate (protein aggregate) and physically links the aggregate to the autophagosome via a LIR motif that binds to lipidated and autophagosome membrane associated ATG8 (LC3-II). On the right, FAM134B-mediated endoplasmic reticulum-phagy (ER-phagy), a ubiquitin-independent selective autophagy pathway is depicted. Here, FAM134B is an ER-membrane protein with a LIR motif that is recognized by autophagosome associated LC3-II in order to deliver predominantly cisternal ER to autophagosomes for degradation.
Clues to the regulation of activation of selective autophagy receptors by post-translational modifications came from a study on OPTN[55]. Here, a conserved TANK binding kinase 1 (TBK1) phosphorylation site just upstream of the LIR motif (serine 177) was identified on OPTN as important for enhancing LC3 binding affinity thereby promoting increased xenophagy (discussed below). Notably, TBK1 is activated by microbe-derived lipopolysaccharide (LPS) thereby providing a link between sensing of bacterial burden with activation of selective autophagy for clearance[63]. Subsequent work has demonstrated a similar importance of phosphorylation adjacent to a LIR motif for enhancing LC3 interaction by the mitophagy receptor BNIP3. Conversely, dephosphorylation adjacent to the FUNDC1 LIR motif enhances LC3 interaction and regulates hypoxia-induced mitophagy[64,65]. While not a conserved mechanism for all selective autophagy receptors, integration of a degradation signal with phosphorylation/dephosphorylation adjacent to a LIR motif leading to enhanced selective autophagy receptor activity is nonetheless one example of how selective autophagy receptors can be regulated.
Further work is needed to precisely define how selective autophagy receptors encode specificity within cells, as well as potentially different selectivity in distinct cell types. In addition, there is further complexity in these pathways given many of the selective autophagy receptors have non-autophagy functions[66]. Below we highlight the role of p62 in selective autophagy as well as the process of PINK1/PARKIN mediated mitophagy to highlight the above-discussed principles of selective autophagy receptors that recognize ubiquitylated cargo.
p62: multi-functional selective autophagy receptor with roles in aggrephagy and xenophagy
p62 is a multi-functional protein with roles in selective autophagy as well as many signaling pathways such as the NF-κB pathway[66,77]. Here we will consider its role in multiple ubiquitin-mediated selective autophagy processes, including aggrephagy, xenophagy, and later mitophagy (considered in the next section). While review of p62 involvement in multiple signaling pathways is beyond the scope of this review, we will also briefly consider the role of p62 in the Keap1-Nrf signaling pathway given the intersection with the autophagy pathway.
p62 is a multi-domain protein that includes an UBA (ubiquitin-associated domain) domain, a PB1 oligomerization domain important for the aggregation of ubiquitylated aggregates, and a LIR motif for ATG8 interaction[52,78–80]. p62 was initially shown to be incorporated in ubiquitylated protein aggregates, including neurodegenerative inclusion bodies associated with Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease[81–83]. The importance of this association became clear with the identification of a conserved LIR motif on p62 that is recognized by autophagosome-associated ATG8 proteins in order to link ubiquitylated protein aggregates bound by p62 to autophagosomes for eventual degradation[52]. This represented the first description of a LIR motif and thereby how a selective autophagy receptor physically links itself to the autophagosomal membrane for delivery of cargo. Interestingly, while delivery of ubiquitylated cargo to the autophagosome via p62 requires interaction with ATG8s, p62 is able to localize to autophagosome sites without an active LIR motif[84]. This speaks to a further level of regulation of cargo recruitment and autophagosome initiation that may be in part explained by the discovery of scaffold proteins such as HTT, as described above and discussed in more detail below[32]. Knockout studies in mice and Drosophila confirmed p62 is important for the aggregation of ubiquitylated proteins and plays a role in their autophagic degradation in vivo[85,86]. These findings in part explained the neurodegenerative phenotypes associated with accumulation of ubiquitylated protein aggregates seen in mice with central nervous system (CNS) loss of autophagy (ATG5/7 knockout)[87,88]. With the discovery of p62 as a selective autophagy receptor, it became clear that selective autophagy of protein aggregates may play an important role in neurodegenerative diseases[83]. Given the importance of this degradation pathway, it is not surprising that there is a significant amount of redundancy with multiple selective autophagy receptors also mediating aggrephagy including NBR1 and OPTN[54,71].
Further study is required in order to understand the regulation of aggrephagy and how specific selective autophagy receptors are activated for this process. Research on p62 has revealed some of the regulation of aggrephagy via p62 intrinsic and extrinsic mechanisms and suggests potentially conserved mechanisms for selective autophagy receptor regulation in general. The affinity of p62 for ubiquitin is low but is enhanced by phosphorylation at residue 403 within the p62 UBA domain via kinases, including Casein kinase 2 (CK2)[89]. While CK2 phosphorylation is not stimulated in response to aggregation, one model suggests that a pool of phosphorylated p62 surveys the cell for poly-ubiquitylated aggregates and binds them for autophagic targeting. As binding of poly-ubiquitin to the UBA domain inhibits dephosphorylation, an increase in poly-ubiquitylated aggregates in the cell such as in the situation of proteasome overload or inhibition then shifts the balance of p62 to the phosphorylated form thereby enhancing aggrephagy[89]. Another intrinsic feature of p62 that contributes to aggrephagy is its ability to oligomerize via its PB1 domain thereby increasing the avidity of p62 for membrane surface clustered LC3B and ubiquitylated aggregates[79,90,91]. Oligomerization therefore links selection of phagophore and ubiquitylated cargo. Furthermore, p62 oligomer complexes facilitate membrane bending and in combination with the tight membrane association of these complexes promote completion of capture of p62-bound aggregates in the autophagosome for delivery to the lysosome. These mechanisms may in part be conserved as other selective autophagy receptors including OPTN are phosphorylated near their UBDs to modulate poly-ubiquitin-binding[60] and both NBR1 and OPTN have the potential to oligomerize [54,92].
Several recent studies highlight the complexity of extrinsic regulation of p62 function in aggrephagy. Similar to the above-described role of HTT in coordinating the activation of selective autophagy by binding ULK1 and p62 simultaneously, there are additional auxiliary autophagy machinery that can regulate and promote selective autophagic clearance of aggregates. For example, the ubiquitin-binding deacetylase HDAC6 can bind poly-ubiquitylated aggregates recognized by p62 and subsequently recruit components of the actin cytoskeleton important in mediating autophagosome-lysosome fusion[93]. Notably, HDAC6 functions selectively during basal autophagy as opposed to bulk-starvation induced autophagy, thereby adding another layer of specificity to the selective autophagic process. Another additional regulator of selective autophagy is ALFY, a large multi-domain protein that interacts with PI3P containing membranes and has been shown to be recruited to ubiquitylated inclusion bodies[94]. Furthermore, ALFY acts as a scaffold to mediate a complex between p62-poly-ubiquitylated cargo complexes and autophagic machinery (including the ATG5-12-16L1 complex and LC3) and promotes selective autophagic degradation of protein aggregates[95]. Clearly, selective autophagy receptor regulation is a complex process, with multiple layers of regulation yet to be discovered. It is not surprising that selective autophagy is a tightly regulated process given the energetic and mechanistic costs of inappropriately degrading functional protein complexes.
Of note, p62 has no yeast homolog and until recently it was unclear whether ubiquitin-mediated selective autophagy existed in yeast. A recent study with the express purpose of identifying yeast proteins that had both ubiquitin-binding and ATG8-interacting regions identified Cue5 and the corresponding mammalian protein, TOLLIP, as ubiquitin-mediated selective autophagy receptors[58]. Further work showed that these may be important in selective autophagic degradation of ubiquitylated protein aggregates including those found in Huntington’s disease. This further highlights the importance of this selective autophagic pathway for cellular homeostasis with conservation from yeast to mammals. While the ubiquitin E3 ligases (Rsp5, Ubc4, and Ubc5) responsible for ubiquitylating a portion of Cue5 autophagic cargos were identified, other E3 ligases likely exist for this process[58]. Indeed, while the machinery for ubiquitylation of autophagic cargo substrates is shared with that for the proteasome and some of the specific E3 ligases and substrates for ubiquitylation within aggregates have been identified, this is an area for further research[6].
p62 is also involved in the selective autophagy of ubiquitylated cytosolic bacteria in a process called xenophagy (selective autophagy of pathogens including bacteria and viruses)[18,62]. Indeed, autophagy had been implicated in the degradation of cytosolic bacteria as part of the innate immune system for several decades but the molecular details for targeting have only recently become clear. While initially demonstrated for Rickettsia conorri bacteria and subsequently shown to be a conserved mechanism across many bacterial types, much of the work demonstrating selective autophagy receptor dependence in xenophagy has centered on the Salmonella enterica, serovar Typhimurium (S. typhimurium)[96]. S. typhimurium invades cells, proliferates and evades the immune system in a specialized Salmonella containing vacuole (SCV)[97]. S. typhimurium will occasionally damage the SCV leading to their extrusion into the cytosol where they are rapidly ubiquitylated by unknown E3 ligases on unknown substrate proteins[98]. Cytosolic poly-ubiquitylated bacteria are subsequently recognized by selective autophagic receptors, including p62, for selective autophagic degradation[72]. Xenophagy limits intracellular bacterial burden and loss of autophagy leads to increased bacterial replication. Multiple subsequent studies showed that, NDP52, OPTN, and TAX1BP1 also act as selective autophagy receptors for xenophagy[55,57,73]. Of note, there are additional non-ubiquitin signals that are recognized in xenophagy. In one example, damaged SCVs expose a β-galactoside sugar which is recognized by cytosolic galectin 8 which is in turn recognized by NDP52 for degradation of the SCV in autophagosomes[99]. While it is clear that each of the studied selective autophagy receptors contribute to xenophagy, further work is required to clarify the relative contributions of each. Much work has been performed examining the role of autophagy and selective autophagy and how it is integrated with the host immune response in a wide variety of bacterial and viral infections and is reviewed elsewhere[18,62,100,101]. Understanding the role of selective autophagy here may prove relevant for future therapeutic intervention.
Remarkably, p62 is also involved in selection of autophagic cargo via non-ubiquitin signals. One example involves Nrf2, a transcription factor with critical roles in the anti-oxidant response[102]. p62 mediates selective autophagic degradation of the E3 ligase Keap1 that constitutively targets Nrf2 for proteasomal turnover thereby regulating the Keap1-Nrf2 oxidative stress response pathway[103]. p62 has a Keap1 interacting region (KIR) that competes for Nrf2 binding on Keap1 thereby leading to stabilization of Nrf2[104]. Therefore alterations in p62 levels via autophagy activation or inhibition can lead to alterations in Nrf2 activity, comprising a selective autophagic pathway for regulation of Nrf2. Furthermore, p62 is a target of the Nrf2 transcriptional response thereby leading to a p62-mediated positive feedback loop after Nrf2 activation[105]. The above examples highlight the complexity of just one of the selective autophagy receptors. Indeed, p62 has further roles in selective autophagy including participating in mitochondrial autophagy, namely mitophagy (described below).
PINK1/PARKIN mediated mitophagy
The most well studied selective autophagic pathway to date involves maintenance of mitochondrial fitness, known as mitophagy. There are multiple selective pathways for mitophagy that are cue and context specific including FUNDC1 mediated hypoxia-induced mitophagy, NIX mediated mitophagy during erythroid differentiation, and PINK1/PARKIN mitophagy implicated during non-hypoxic mitochondrial damage and in neurodegenerative diseases including Parkinson’s disease and amyotrophic lateral sclerosis (ALS)[15,60,64,106–108] (Table 1 and 2). Through a series of elegant biochemical and cell biological studies, the molecular mechanisms of PINK1/PARKIN mediated mitophagy have recently become clear[60,61]. Here we will discuss this pathway as an example of the complex nature of selective autophagy regulation. The upstream events of selective autophagy receptor recruitment are critical to mitophagy and involve a signal transduction pathway including the protein kinase PINK1 and ubiquitin E3 ligase PARKIN[109]. PINK1 is stabilized on the mitochondrial outer membrane in response to mitochondrial damage and PINK1 phosphorylates both cytoplasmic PARKIN as well as ubiquitin present on mitochondria[110–114]. PARKIN phosphorylation as well as binding to phosphorylated ubiquitin on the mitochondria leads to PARKIN activation with the generation of ubiquitin chains of varying linkages on multiple mitochondrial outer membrane (MOM) proteins[114–116]. Multiple studies have identified the selective autophagy receptors, OPTN, p62, TAX1BP1, and NDP52 as critical for the recognition of these ubiquitylated mitochondrial proteins for mitophagy[60,61,67]. This recognition occurs through UBAN and ZNF domains in OPTN, a UBA domain in p62 as above, and a UBZ domain in NDP52[60].
A recent study unraveled a further upstream step in the regulation of this pathway. After being activated by an unknown kinase in response to mitochondrial damage, TBK1 promotes phosphorylation of OPTN at the LIR site to promote ATG8 interaction. TBK1 also phosphorylates near the OPTN ubiquitin-binding UBAN domain to enhance ubiquitin binding, leading to retention of OPTN on ubiquitylated mitochondria thereby promoting mitophagy. This cascade further activates TBK1 in a self-reinforcing loop ([60] Figure 7). The relevance of mitophagy to neurodegenerative diseases is clear given work in murine models and the existence of mutated forms of PINK1 and PARKIN in forms of Parkinson’s Disease as well as ALS patient-derived mutations in p62, OPTN, and TBK1[67,109,117–120]. Beyond its role in neurodegenerative diseases, further work is required to understand the importance of mitophagy in other tissues and pathologic conditions for which mouse models to monitor mitophagy have been recently developed[121]. The molecular mechanisms of PINK1/PARKIN mediated mitophagy described above illustrate how complex systems have evolved in order to carefully regulate selective autophagy pathways as inappropriate activation or deactivation could lead to pathologic states. Further work is required in order to unravel the likely equally complex regulatory systems for other selective autophagic pathways.
Here we have highlighted a few salient examples of the known ubiquitin-dependent selective autophagy pathways highlighting the principles of selective autophagy including: 1. Presence of a degradation cue (aggregated proteins, damaged mitochondria, intracellular bacteria), 2. Cargo recognition via a so-called ‘selective autophagy receptor’ protein (recognition of ubiquitylated cargo via UBDs in selective autophagy receptors), and 3. Recruitment of autophagosome machinery and membrane for eventual delivery to the lysosome for degradation (LC3 interacting motifs on selective autophagy receptors). Additional modes of ubiquitin-dependent selective autophagy exist as detailed in Table 1 and more are likely to be discovered.
Ubiquitin-independent selective autophagy
Recently there has been a significant increase[6] in the number of recognized ubiquitinindependent selective autophagy pathways (Table 2). Ubiquitin-independent selective autophagy receptors identify a wide variety of protein, lipid or sugar-based signals (Table 2). While initially identified in yeast where it was thought ubiquitin-dependent pathways did not exist, more recent studies have identified a large number of ubiquitin-independent processes in higher eukaryotes[6,122]. Important work has allowed the field to progress beyond mechanistic questions to further understand the in vivo relevance of these pathways. Here we highlight two recently identified ubiquitin-independent selective autophagy receptors as models of this process: NCOA4 for ferritinophagy (ferritin autophagy) and the FAM134 protein family for ER-phagy (endoplasmic reticulum autophagy)[13,14,16].
NCOA4-mediated ferritinophagy
In work from our laboratory, we reasoned that proteins intimately involved in cargo selection and recruitment to autophagosomes would be tightly associated with autophagosomal membranes. Therefore, in an effort to identify new selective autophagy receptor-cargo pairs, we devised a quantitative autophagosomal proteomics approach that would allow us to identify proteins selectively enriched in autophagosomes[13]. While previously identified as a nuclear receptor coactivating protein[141], NCOA4 was highly enriched in autophagosomes and interaction proteomics revealed ferritin heavy and light chains (FTH1, FTL), the intracellular iron storage complex, as high confidence interacting proteins[13]. Iron has an essential role in multiple cellular processes, therefore regulating iron levels in the cell is critical[142]. Prior research showed that when iron levels in the cell are low, ferritin is degraded via autophagy allowing the release of iron for use by the cell[143]. We found that NCOA4 was important for ferritin delivery to autophagosomes, and as NCOA4-deficient cells were impaired in their ability to degrade ferritin and release their iron intracellularly, this led to decreased bioavailable intracellular iron. This work as well as subsequent work by Dowdle et al. identified NCOA4 as a selective autophagy receptor for autophagic degradation of ferritin (ferritinophagy), which is critical for cellular iron homeostasis (Figure 3)[13,14].
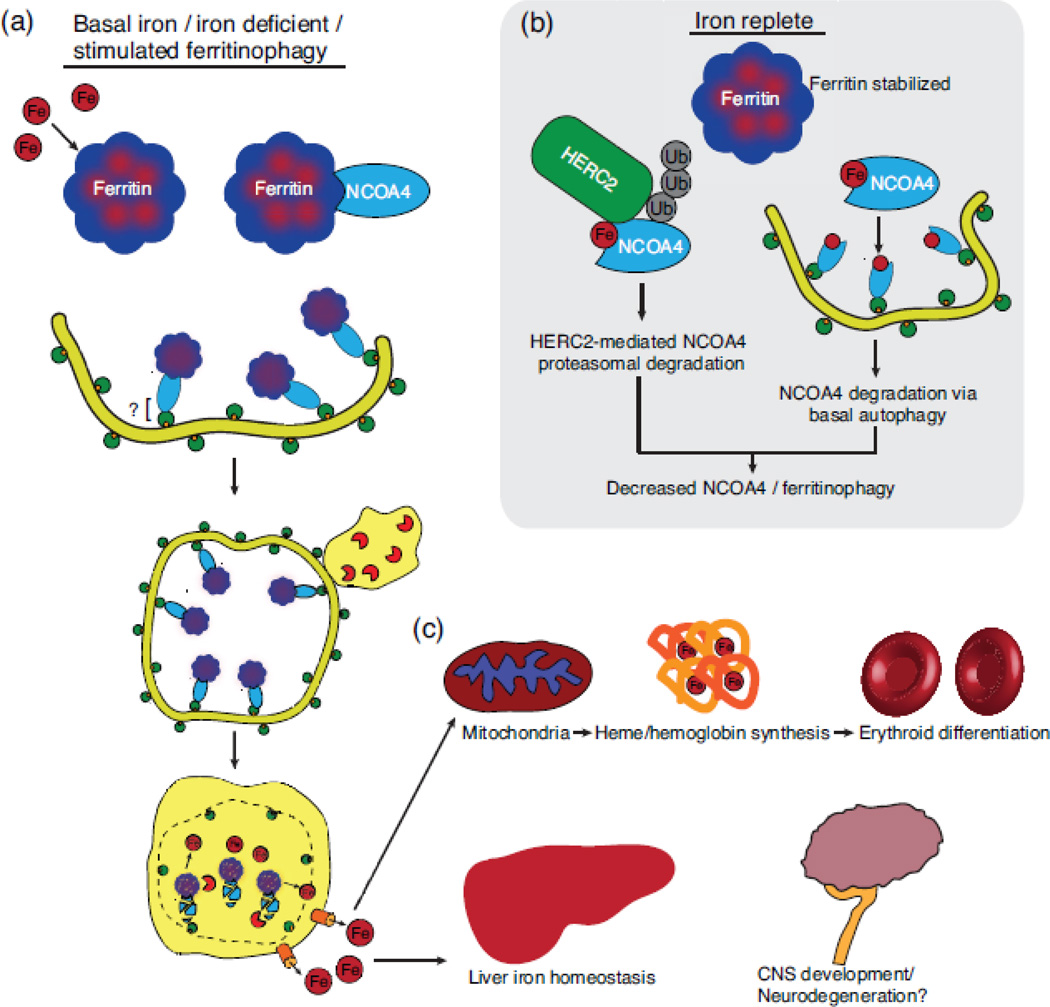
Figure 3.
NCOA4-mediated ferritinophagy pathway. (a) Iron (Fe) is sequestered in 24-mer ferritin complexes containing ferritin heavy and light chains. NCOA4 recognizes ferritin and delivers it to an incipient autophagosome. The molecular nature of the NCOA4-autophagosome interaction is unclear but likely involves interaction with LC3-II potentially via a non-canonical LIR motif. Degradation of ferritin in an autolysosome releases iron, which is exported to the cytosol. (b) NCOA4 levels and thereby ferritinophagy are regulated by iron levels in the cell. Under high iron/iron replete conditions, NCOA4 is recognized by HERC2, an E3 ubiquitin ligase, in an iron-dependent manner and targeted for proteasomal degradation. In tandem, NCOA4 is also targeted for autophagic degradation. A lower level of NCOA4 therefore decreases flux through the ferritinophagy pathway. (c) Liberated iron can be used in many iron-dependent processes including heme synthesis, which is required for hemoglobin synthesis during erythroid differentiation. Furthermore, NCOA4-mediated ferritinophagy is important for other physiological processes such as maintenance of liver iron homeostasis. Further research will be required to understand the role of NCOA4 in CNS development and pathophysiology of iron-associated neurodegenerative disorders.
While NCOA4 associated with multiple Atg8 proteins in an in vitro binding assay, we were unable to identify a canonical LC3-interacting region (LIR) motif in NCOA4, although the existence of non-canonical ATG8-binding motifs may suggest that NCOA4 uses such an alternative motif for interaction (Figure 3a)[42]. Of note, Mizushima and colleagues subsequently published an ultrastructural analysis study of MEFs knocked out for various ATG proteins and showed that despite lacking ATG8s localized to the autophagosome membrane, ferritin clusters accumulated at the autophagosome formation site, similar to p62 not requiring ATG8s for autophagosome localization as discussed above[144]. Therefore, additional research is required to understand the recruitment of NCOA4 and thereby ferritin to autophagosomes.
In terms of cargo recognition, NCOA4 appears to be specific for ferritin as other autophagic substrates have yet to be identified in interaction proteomics experiments. We showed that NCOA4 interacts directly with the FTH1 subunit of ferritin via a conserved NCOA4 C-terminal domain and a key conserved residue on FTH1[145]. Mutation at these binding sites abrogates binding in vivo and abolishes ferritinophagy. Further structural work would be required to elucidate the molecular mechanisms of this interaction.
The ferritinophagy pathway is regulated by cellular iron levels via a NCOA4-interacting protein we identified, the multifunctional HERC2 ubiquitin E3 ligase[13]. HERC2 uses its ‘CUL7-homology domain’ to recognize a C-terminal domain in NCOA4 under high iron conditions to mediate NCOA4 turnover via the ubiquitin-proteasome system, thereby reducing the steady-state NCOA4 levels and increasing ferritin for iron storage (Figure 3b). Surprisingly, we found that this C-terminal domain within NCOA4 binds iron and the iron-bound state of NCOA4 determines HERC2 binding, suggesting an iron-dependent switch for NCOA4 turnover (Figure 3b)[145]. Further work is required in order to understand whether other levels of regulation exist for the pathway including a potential link to cellular oxidative stress given the intimate link between iron levels and reactive oxygen species[146,147].
Having established the importance and regulation of NCOA4 for intracellular iron homeostasis, we sought to understand the role of NCOA4 in iron-dependent processes on an organismal level. Initial links between NCOA4 and processes with a requirement for high iron availability came from expression studies where ncoa4 mRNA is high at sites of erythropoiesis during zebrafish development[148]. Using a zebrafish model and a hemin-induced erythroid-like K562 cell line, we showed that NCOA4-mediated ferritinophagy is important for erythropoiesis in vivo given its role in mobilizing iron from ferritin for use in heme synthesis (Figure 3c)[145]. This study established the importance of NCOA4 as a critical regulator of cellular and organismal iron metabolism and revealed the mechanism of its iron-dependent regulation.
A recent study by Carlomagno further examined the role of NCOA4 in regulating systemic iron homeostasis in a genetically engineered mouse model of NCOA4 deficiency[149]. Consistent with the role of NCOA4 in degrading ferritin, NCOA4-null mice inappropriately accumulate iron-laden ferritin in all tissues evaluated. As NCOA4-null mice are unable to mobilize iron from ferritin, they develop a hypochromic microcytic anemia that is significantly worsened when they were fed a low-iron diet. Interestingly, when these mice were fed a high iron diet, they developed significant increases in ferritin deposits as well as increases in serum concentrations of alanine and aspartate liver transferases, fatty liver degeneration, and premature death compared to a wild-type cohort. The authors hypothesize that enhanced basal levels of tissue iron in NCOA4-null mice saturate anti-oxidant pathways leading to liver damage. As such, scavenger proteins implicated in the anti-oxidative stress response such as superoxide dismutase (SOD) and glutathione peroxidase (GPx) were expressed at significantly higher levels in NCOA4-null mice. As part of its role in maintaining iron homeostasis, NCOA4 may also play an important role in modulating organismal oxidative stress levels[149].
It remains to be determined what the physiologic importance of NCOA4-ferritinophagy is in non-erythroid tissues, including in CNS development and neurodegenerative diseases given the recent discovery of a group of patients with neurodegeneration and brain iron accumulation with mutations in autophagic proteins (Figure 3c)[142,150,151].
ER-phagy
As with mitochondria, maintaining organellar quality and quantity is important for cellular homeostasis. Recent work from Khaminets et al. and Mochida et al. identified selective autophagy receptors for the endoplasmic reticulum (ER) in mammalian cells (FAM134B) and yeast cells (Atg40) respectively[16,17]. Khaminets and colleagues used results from an LC3 interactome study to identify potential selective autophagy receptors, initially identifying NBR1 as a selective autophagy receptor[54]. Also identified in this screen and confirmed as ATG8 interactors containing a LIR motif were members of the reticulon-homology-domain-containing FAM134 protein family (FAM134A, B, and C). They focused on this hit given mutations (truncations Q145X and S309X) resulted in a sensory neuropathy including impaired pain reception in affected patients[152]. Significantly, these truncations end prior to the identified LIR motif already suggesting a potential molecular pathogenic explanation related to a selective autophagy function. In their study, FAM134B co-localized predominantly with cisternal endoplasmic reticulum markers CLIMP-62 and SEC61B and was found to be ER-membrane anchored with the ability to remodel membranes via its reticulon homology domain. The authors went on to show that FAM134B co-localized with LC3-positive autophagosomes and over-expression led to increased accumulation of ER-structures in lysosomes. FAM134B knockdown or LIR-deficient mutant expressing cells conversely showed less ER localization with LC3-positive autophagosomes and decreased ER fragmentation and degradation. The author’s work identified FAM134B as a selective autophagy receptor for the delivery of ER into lysosomes (ER-phagy) that is critical for overall ER homeostasis. In cell culture studies, FAM134B was also critical for cell survival in the setting of stress conditions as FAM134B-deficient cell lines showed decreased cell viability upon stress. They extended their work to the in vivo setting with Fam134b −/− mice, where phenotypic analysis of aged mice recapitulated findings from the associated human diseases, including increased sensitivity towards noxious heat and decreased sensory axon numbers, suggesting affects on neuronal health.
The importance of this pathway was highlighted by a parallel report showing an evolutionarily conserved pathway in yeast involving Atg40 as a mediator of ER-phagy that was critical for ER turnover and cellular fitness in the setting of various stress conditions[17]. Much remains to be discovered with regards to ER-phagy including the regulation of the pathway and whether this involves any post-translational modifications.
NCOA4-mediated ferritinophagy and ER-phagy are representative of a growing class of selective autophagy receptors that recognize cargo via ubiquitin-independent signals and in many instances appear specialized for degradation of one substrate cargo. These selective autophagy receptors are involved in a diverse set of disease pathways thereby further implicating selective autophagy in human health and disease.
Autophagy in cancer
In addition to the role of selective autophagy in inherited diseases (e.g. ER-phagy) or many neurodegenerative diseases (where loss of autophagy activity associated with aging can lead to disease), there is a growing recognition that selective autophagy is involved in regulating cancer. The role of autophagy in cancer is extremely complex, as demonstrated by a growing literature describing situations where autophagy can either promote or inhibit tumorigenesis[2,10,11,153]. The most likely explanation is that the role of autophagy in cancer is dynamic with both tumor suppressive and pro-tumorigenic roles, which depend on multiple factors including tumor stage, cellular context and tissue of origin. Recent work has brought to light tumor suppressive selective autophagy pathways that can mitigate oncogenic signals and conversely selective autophagic pathways that support tumor maintenance and progression. Here we will briefly review the role of autophagy in cancer (for more in-depth reviews, please see [2,10,11,153]) and highlight recent advances in our understanding of the importance of selective autophagy in cancer first as a tumor suppressive and second as pro-tumorigenic mechanism.
Autophagy in tumor suppression
Autophagy was initially considered a mechanism by which to suppress tumor initiation. Indirect evidence of this tumor suppressive role was based on oncogene and tumor suppressor gene alterations such as AKT amplification, PI3K mutation or Pten loss that were associated with a loss of autophagy activation[2]. This suggested that active autophagy prevented the oncogenic transformation of cells. More direct evidence of autophagy as a tumor suppressor came from mouse genetic studies of core autophagy machinery including ATG5, ATG7, and BECN1 (Beclin-1) showing that when autophagy is impaired, there is an increase in tumor initiation[154–157]. In most instances, the tumors that develop in models where autophagy is completely ablated are benign lesions[156]. One notable exception is the BECN1 study, where heterozygous mice were used. In this case, tumors were able to progress in some instances to malignant lesions. This may reflect the fact that these mice, while autophagy impaired, still were autophagy competent[154]. Together the data suggests that while autophagy loss may predispose to tumor initiation, it also supports the transformation to invasive cancers. Of note, while initial studies on BECN1 showed that many breast and ovarian cancers have a single copy loss of this gene, more recent large-scale sequencing studies suggest that this may be a passenger alteration given the proximity of BRCA1 and the lack of any BECN1-only alterations in cancers[158]. It remains to be seen whether loss of function of other autophagy machinery, including selective autophagy receptors may play a role in tumor initiation.
From a mechanistic standpoint, inhibition of autophagy leads to an accumulation of reactive oxygen species, increased DNA damage, and mitochondrial defects, all implicated in tumorigenesis[10]. Interestingly, studies on p62 and tumorigenesis suggest a potential mechanistic link to selective autophagy. In mouse models with defective autophagy, subsequent p62 ablation reduces tumorigenesis suggesting that p62 accumulation upon autophagy loss can contribute to tumorigenesis[157]. Conversely, p62 over-expression promotes oxidative stress and tumor growth[159]. Indeed, amplification of chromosome 5q and thereby p62 expression is implicated in clear cell renal cell carcinoma[160]. As p62 has roles in selective autophagy as well as many signaling pathways, more research is required to understand the precise roles and importance of p62 in tumor progression.
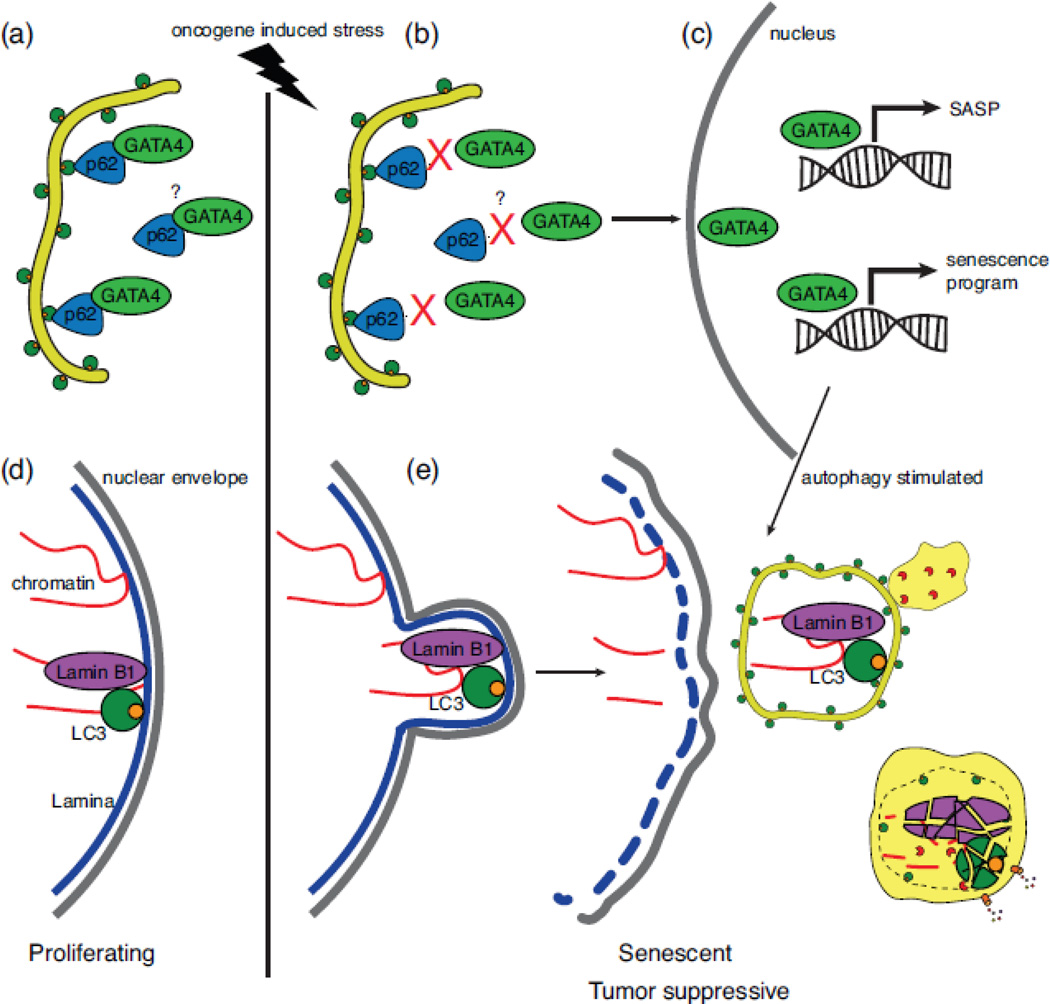
Another potential mechanism for tumor suppression by autophagy is via its role in cellular senescence. Cellular senescence is a program of permanent arrest of the cell division cycle that can be induced by cells in response to oncogenes in order to prevent malignant transformation[161]. Notably, many studies have shown autophagy is upregulated during the cellular senescence program. However, there are conflicting reports that suggest autophagy inhibition promotes cellular senescence[162]. A recent study unraveled this conflicting evidence regarding the role of autophagy as either supportive or inhibitory of cellular senescence and identified a selective autophagic process important in regulating cellular senescence[140]. The authors identify the transcription factor GATA4 as a master regulator of the cellular senescence program and the senescence associated secretory phenotype (SASP)[163]. Furthermore, GATA4 levels are regulated by p62-mediated selective autophagy. They first show that GATA4 is turned over basally via p62 selective autophagy. Upon senescence activating stimuli such as oncogene-induced stress or DNA damage, GATA4 selective autophagy is disrupted thereby stabilizing GATA4 and allowing for translocation of GATA4 to the nucleus where it activates the cellular senescence program and SASP (Figure 4).
Figure 4.
Selective autophagy in cancer. (a) Cellular senescence is a barrier to tumorigenesis and selective autophagy explains the conflicting reports about the role of autophagy in senescence. Normal proliferating cells basally degrade the pro-senescence transcription factor GATA4 via p62 selective autophagy. The molecular details of the GATA4-p62 interaction and its regulation are currently unclear. (b) Upon oncogene-induced stress, the GATA4-p62 interaction is abrogated by an unknown mechanism thereby stabilizing GATA4. (c) GATA4 translocates to the nucleus where it activates a senescence transcriptional program as well as a senescence associated secretory phenotype (SASP) transcriptional program. General autophagy is also stimulated at this time and is required to drive senescence. This pathway clarifies the apparent paradox in the literature that autophagy can suppress senescence (basal selective autophagy of GATA4) and promote senescence (autophagy is required to support senescence phenotype). (d) Basally, Lamin B1 binds nuclear LC3 and chromatin but is not targeted for autophagic degradation. (e) Upon oncogene-induced stress, Lamin B1 is autophagocytosed with bound chromatin to mediate nuclear lamina selective autophagy contributing to a senescence phenotype and thereby tumor suppression.
With the identification of GATA4 as a regulator of senescence and a substrate for selective autophagy, the authors were able to experimentally untangle the role of autophagy in senescence. Transiently inhibiting autophagy led to the accumulation of GATA4, thereby inducing the cellular senescence program. Autophagy activity was subsequently restored allowing general autophagy to support a senescence program. The cellular response to transient autophagy inhibition was induction of the cellular senescence program. Conversely with longterm inhibition of autophagy, cellular senescence was not induced. While GATA4 levels were high with long-term autophagy inhibition, senescence initiation was inhibited as general autophagy was not available to support the senescence phenotype. p62 depletion leads to a robust activation of the senescence response due to accumulation of GATA4 further supporting this model. The authors conclude that p62-directed selective autophagy of GATA4 is an antisenescence mechanism while general autophagy acts as a pro-senescence factor.
Examples of selective autophagy as a tumor suppressor
Selective autophagy has been directly implicated as tumor suppressive in multiple studies, several of which are highlighted here. Dou et al. identified Lamin B1, a nuclear lamina protein, as a selective autophagy receptor for nuclear lamina that may be important for initiating cellular senescence and thereby suppressing tumorigenesis[139]. The same group had previously identified a lysosome-mediated processing of chromatin during senescence. However, it was unclear whether this was autophagy-related[164]. Furthermore, prior work had shown pools of autophagy proteins present in the nucleus, including ATG8/LC3. Whether they were involved in degrading nuclear components was unclear[165,166]. The authors identified an LC3-Lamin B1 interaction in the nucleus basally in the absence of any stimuli, nonetheless Lamin B1 was not found in autophagosomes basally. As prior work showed that Lamin B1 is lost during oncogenic insults and that most primary cells and tissues induce senescence (and autophagy) in response to oncogenic Ras expression, the authors reasoned that oncogenic stress may lead to Lamin B1 mediated nuclear lamina autophagy[167]. Ras-oncogene expression led to Lamin B1 colocalization within autophagosomes, induction of autophagy, reduction of perinuclear heterochromatin, nuclear membrane blebbing, and presence of cytoplasmic chromatin fragments (Figure 4). Lamin B1-mediated selective autophagy of nuclear lamina led to impaired cell proliferation and global alterations of the epigenome and transcriptome all supporting a program of cellular senescence and therefore tumor suppression[139,168]. Of note, Lamin B1 associates with transcriptionally inactive heterochromatin domains raising the question as to why already silenced chromatin would be degraded via this pathway to mediate senescence. In addition, it would be interesting to correlate the specific chromatin regions identified as degraded in this study with large-scale genomic analyses in KRas mutant tumors.
There are a number of other studies supporting the role of selective autophagy in maintaining genomic stability. For instance, p62 and NDP52 were recently identified as selective autophagy receptors for RNA retrotransposon containing granules, thereby implicating selective autophagy in the control of retrotransposon insertion into the genome[68]. Loss of autophagy leads to an increase in retrotransposon genomic insertions and a loss of selective autophagy could therefore be implicated in tumorigenesis. Another link between selective autophagy and maintenance of cellular homeostasis to prevent tumorigenesis is that of the selective degradation of activated RHOA via p62 to maintain appropriate RHOA levels and localization. Loss of autophagy leads to an inappropriate accumulation of active RHOA at the midbody leading to increased cell size, cytokinesis failure, increased DNA content, aneuploidy, and escaped apoptosis[137,138]. The authors show a positive correlation between autophagy defects and the higher expression of RHOA in human lung carcinomas.
While additional examples of selective autophagy acting in a tumor suppressive manner exist[10,169], further work is required to broaden our understanding of the complex role of autophagy in tumor suppression, what other selective autophagy pathways are involved and how they are regulated by cells.
Pro-tumorigenic functions of autophagy
A substantial amount of evidence suggests autophagy also functions in multiple cancer types including pancreatic ductal adenocarcinoma (PDAC), Kras and B-raf driven lung cancers[170,171], melanoma, and CNS malignancies to sustain growth of fully formed tumors[10,172,173]. Although autophagy is present in all tissues at low levels for homeostatic functions, many groups have identified that autophagy is elevated in RAS-driven cancers and is crucial for tumor growth[174–177]. Inhibition of autophagy pharmacologically with chloroquine (CQ) or hydroxychloroquine (HCQ), as well as genetically, demonstrated significant responses in cancers using both in vitro and in vivo systems. These systems include Kras-driven autochthonous pancreatic and lung cancer models[174,178–181]. Of note, a recent study called into question the reliance of tumor cell lines on autophagy[182]. This discrepancy with a large number of prior studies may be due to multiple experimental differences, and many studies continue to support the idea that autophagy inhibition has anti-tumor effects in multiple cancer types through both cell autonomous and non-autonomous mechanisms[2,10,11,153,171].
As CQ and HCQ have been used in patients for many years for a variety of indications, testing this as a therapeutic approach in cancer patients is feasible and currently ongoing[2,183]. As these agents act on the lysosome by inhibiting lysosomal acidification they inhibit the late stages of autophagy at the level of autophagosome maturation and cargo degradation[183]. In addition, these drugs may have other non-autophagy specific therapeutic effects as they block other processes that depend on lysosomal activity. Given the poor pharmacokinetics and unclear pharmacodynamic properties of HCQ at the doses currently used in the clinic, multiple other potentially targetable proteins critical to autophagy (kinases such as ULK1 and VPS34, E1-like proteins such as ATG7, and proteases such as ATG4b) are being pursued as targets for autophagy inhibition[184,185]. As described below, there is the potential for identification of selective autophagic cargo that are important for sustaining tumor proliferation. Identification of these selective autophagy pathways may provide an even more targeted approach for autophagy inhibition in cancer.
Examples where selective autophagy supports tumor growth
From a mechanistic standpoint, the role of autophagy in supporting tumor proliferation is complex but likely centers around an increased metabolic and biosynthetic need in rapidly dividing cells in an austere tumor microenvironment[172]. The question of how a tumor cell maintains high basal autophagic flux at all times suggests that unlike bulk autophagy or starvation-induced autophagy there is some selectivity built into the process. Selective autophagy has been shown to be involved in promoting cancer cell survival both at a basal level as well as in response to stressors such as chemotherapy or targeted therapies[169]. Selective autophagy has been shown to be important in tumor cells undergoing hypoxia, which is commonly found in the tumor microenvironment[64,65]. Here, tumors may rely on hypoxia-inducible mitophagy pathways to support cellular fitness. Genomic instability can lead to a high degree of protein misfolding and while the proteasome is equipped to degrade many misfolded proteins, at times the ubiquitin proteasome system can be overwhelmed or unable to deal with large protein aggregates[53]. In these cases where the proteasome is overwhelmed, selective autophagy via various autophagy receptors can play a role in aggrephagy in order to maintain tumor cell fitness. Given this dual reliance on the proteasome and autophagy, one therapeutic strategy being tested currently is dual inhibition of the proteasome and the autophagy lysosome system[186]. Here we present several recent examples of selective autophagy supporting tumor cell fitness that may suggest targets for therapy with potential therapeutic windows.
Multiple groups have studied the importance of autophagy in tumors driven by oncogenic Ras. One study looked at the selective remodeling of the proteome in response to autophagy inhibition[187]. Prior work had shown that autophagy ablation in non-small cell lung carcinomas causes elevated inflammatory responses and suppresses tumor growth[188]. The relation of selective autophagy to this process was unclear, therefore the authors compared the proteomes of Ras-driven cancer cells with or without autophagy loss. They found a selective remodeling of the proteome in the autophagy competent tumors whereby proteins essential for survival were retained but pro-inflammatory proteins were eliminated. While the details of this selective targeting are unclear at this time, the authors noted that this discovery may translate to effective combinations of autophagy inhibition and immunotherapy.
Another potential role of selective autophagy in promoting or maintaining cancer cell survival would be in maintaining appropriate levels of signaling complexes or degrading proapoptotic proteins. In one example of tumor cells co-opting selective autophagy to maintain appropriate levels of oncogenic stimuli, Sandilands and colleagues defined a selective autophagy pathway important for regulating active Src levels in tumor cells to promote survival following loss of focal adhesion kinase (FAK) signaling[136]. The Src family of non-receptor tyrosine kinases is involved in signaling to promote cell adhesion, invasion, proliferation and survival[189]. Indeed Src proteins are frequently over-expressed and activated in solid cancers[190]. FAK is a critical binding partner that helps maintain Src at focal adhesions where its activity is regulated and directed[191]. In cancer cells where FAK expression is lost (due to cell detachment or FAK inhibition) or signaling is impaired, inappropriate Src activity can lead to reduced cancer cell viability. Sandilands examined the consequences of impaired FAK expression and activity on Src in a skin cancer model (squamous cell carcinoma) as well as pancreatic cancer model and noted that active Src is targeted for autophagic degradation upon loss of FAK activity[136]. They identified c-Cbl, an E3 ubiquitin ligase as a LIR-containing selective autophagy receptor essential for mediating selective autophagy of Src. Abrogation of autophagy generally or by specifically depleting c-Cbl in the setting of FAK loss led to a decrease in cell survival. The authors noted that FAK deletion likely leads to a transition of active Src from an oncogenic driver to an overactive kinase that is toxic and that cancer cells have adapted a selective autophagic pathway for degradation of overactive Src in this situation in order to survive. In subsequent work the authors also characterized a similar autophagic pathway for Ret, a receptor tyrosine kinase normally important in development but identified in multiple cancers as an oncogenic driver[192]. Here active Ret was degraded via autophagy upon altered or reduced FAK signaling. In this case the c-Cbl ligase was not required suggesting the existence of a Ret specific selective autophagy receptor yet to be discovered. The authors note that this work could impact the clinical use of FAK inhibitors and that dual inhibition of FAK and autophagy may be an efficacious combination.
Understanding the reliance of tumor cells on specific selective autophagic pathways may suggest vulnerabilities that can be exploited for therapeutic gain.
CONCLUSIONS AND PERSPECTIVES
Autophagic degradation of cellular contents is employed by eukaryotes from yeast to man in order to maintain cellular homeostasis and protect against disease. It is clear that selectivity is required for this system to function effectively at a basal level and in response to specific stimuli given the prospect of a routine bulk autophagic degradative response can be both an energetically and mechanistically unfavorable proposition. Selectivity is built into the autophagy system via a complex series of molecular machines with selective autophagy receptors central to this process, given they interact directly with the cargo as well as the autophagosomal targeting machinery (ATG8s). Selective autophagy receptors recognize their cargo by ubiquitin-dependent and - independent mechanisms. What is not yet clear is how the ubiquitin-dependent selective autophagy receptors determine specificity for their specific ubiquitylated substrates and what other molecules may play a role in determining this. While the overlap of selective autophagy receptors in mediating processes such as aggrephagy, mitophagy, and xenophagy suggests the importance of these pathways and some requirement for redundancy, we do not yet understand the relative contribution of each selective autophagy receptor for a pathway and how that may be encoded. We are now beginning to elucidate some of the regulation of these selective autophagy pathways, including how specific stimuli or stressors are sensed by the cell and then integrated by selective autophagy receptors and associated machinery. It is clear that post-translational modifications including phosphorylation and dephosphorylation play an important role. Identifying the kinases and phosphatases involved in this regulation should be a key goal of future studies. It remains to be determined what other types of post-translational modifications may play a role in regulating the network. The recent increase in selective autophagy receptors identified is impressive, but not unexpected given the likely requirement for a multitude of selective autophagic processes in order to maintain cellular homeostasis and that multicellular organisms may even rely on distinct processes in different tissues. Indeed, selective autophagy receptors for multiple pathways including lipophagy[193], granulophagy[194], myelinophagy[195], and ribophagy[196] have yet to be identified. Additionally, unidentified selective autophagic pathways likely exist including ones potentially similar to ferritinophagy in that they may be specific for regulating intracellular levels of critical metals or ions.
Understanding the role of selective autophagy receptors in vivo is of paramount importance given the phenotype of dysregulation of the pathway may only become apparent once tested in vivo. Recent work by Dikic and colleagues on ER-phagy illustrate the importance of studying these pathways in vivo as it more clearly defined how disrupted ER-phagy is relevant to an inherited human disease[16]. Further work understanding the importance of NCOA4-mediated ferritinophagy in vivo will identify the tissue compartments that rely on this process and how dysregulation of ferritinophagy may lead to disease. Likewise, the field will continue to learn from studying selective autophagy pathways using in vivo models[121,197]. These same in vivo systems will be of use when attempting to understand the role of selective autophagy in cancer. Studying the role of selective autophagy pathways in cancer has in part unraveled the complex role of autophagy in cancer as tumor suppressive and pro-tumorigenic dependent on context. Importantly, understanding the precise roles of various forms of selective autophagy in maintaining tumor growth provides the opportunity to target these processes more selectively.
HIGHLIGHTS.
Macroautophagy is a protective cellular catabolic pathway for both bulk and selective degradation of cellular components
Selective autophagy receptors mediate delivery of specific substrates to autophagosomes for degradation
Selective autophagy pathways are activated by specific stimuli but the molecular details of activation are not yet clear
Selective autophagy pathways are important in multiple disease processes including neurodegeneration and cancer
Distinct selective autophagy pathways can mediate either tumor suppression or promotion in cancer
Acknowledgments
The authors apologize for the omission of any primary references. This work was supported by NIH grant GM095567, National Cancer Institute Grants R01CA15749 and R01CA188048, ACS Research Scholar Grant RSG-13-298-01-TBG, and the Lustgarten Foundation, to A.C.K. J.D.M. is supported by a KL2/Catalyst Medical Research Investigator Training award (TR001100), and a Burroughs Wellcome Fund Career Award for Medical Scientists. A.C.K. is a consultant for Forma Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature Cell Biology. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & Development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 4.Noda NN, Inagaki F. Mechanisms of Autophagy. Annu. Rev. Biophys. 2015;44:101–122. doi: 10.1146/annurev-biophys-060414-034248. [DOI] [PubMed] [Google Scholar]
- 5.Svenning S, Johansen T. Selective autophagy. Essays Biochem. 2013;55:79–92. doi: 10.1042/bse0550079. [DOI] [PubMed] [Google Scholar]
- 6.Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends in Cell Biology. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic Control of Autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Choi AMK, Ryter SW, Levine B. Autophagy in Human Health and Disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 10.White E. The role for autophagy in cancer. J. Clin. Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends in Cell Biology. 2015;25:37–45. doi: 10.1016/j.tcb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto K. Quality control: Organellophagy: Eliminating cellular building blocks via selective autophagy. The Journal of Cell Biology. 2014;205:435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nature Cell Biology. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 15.Melser S, Lavie J, Bénard G. Mitochondrial degradation and energy metabolism. BBA - Molecular Cell Research. 2015;1853:2812–2821. doi: 10.1016/j.bbamcr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 17.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 18.Sorbara MT, Girardin SE. Emerging themes in bacterial autophagy. Current Opinion in Microbiology. 2015;23:163–170. doi: 10.1016/j.mib.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nature Cell Biology. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 20.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Molecular Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 21.De Duve C, Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 23.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO Journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung CH, Ro S-H, Cao J, Otto NM, Kim D-H. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zavodszky E, Vicinanza M, Rubinsztein DC. Biology and trafficking of ATG9 and ATG16L1, two proteins that regulate autophagosome formation. FEBS Lett. 2013;587:1988–1996. doi: 10.1016/j.febslet.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 29.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, et al. Early Steps in Autophagy Depend on Direct Phosphorylation of Atg9 by the Atg1 Kinase. Molecular Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao Y, Perna MG, Hofmann B, Beier V, Wollert T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nature Communications. 2015;7:1–13. doi: 10.1038/ncomms10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamber RA, Shoemaker CJ, Denic V. Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Molecular Cell. 2015;59:372–381. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rui Y-N, Xu Z, Patel B, Chen Z, Chen D, Tito A, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nature Cell Biology. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. U.S.a. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 35.Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nature Structural Molecular Biology. 2014;21:336–345. doi: 10.1038/nsmb.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Molecular Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. Journal of Cell Science. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 40.Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP, et al. iLIR. Autophagy. 2014;10:913–925. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Molecular Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Research. 2013;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 45.Li W-W, Li J, Bao J-K. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X-M, Sun L-L, Hu W, Ding Y-H, Dong M-Q, Du L-L. ESCRTs Cooperate with a Selective Autophagy Receptor to Mediate Vacuolar Targeting of Soluble Cargos. Molecular Cell. 2015;59:1035–1042. doi: 10.1016/j.molcel.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 47.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Research. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pimentel-Muiños FX, Boada-Romero E. Selective autophagy against membranous compartments. Autophagy. 2014;10:397–407. doi: 10.4161/auto.27244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nature Cell Biology. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Research. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. The Journal of Cell Biology. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 53.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 54.Kirkin V, Lamark T, Sou Y-S, Bjørkøy G, Nunn JL, Bruun J-A, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Molecular Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 55.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman AC, Scholefield CL, Kemp AJ, Newman M, McIver EG, Kamal A, et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS ONE. 2012;7:e50672. doi: 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 58.Lu K, Psakhye I, Jentsch S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 59.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Molecular Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heo J-M, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Molecular Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castrejón-Jiménez NS, Leyva-Paredes K, Hernández-González JC, Luna-Herrera J, García-Pérez BE. The role of autophagy in bacterial infections. Biosci Trends. 2015;9:149–159. doi: 10.5582/bst.2015.01035. [DOI] [PubMed] [Google Scholar]
- 63.Radtke AL, Delbridge LM, Balachandran S, Barber GN, O'Riordan MXD. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nature Cell Biology. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, et al. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. Febs J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 67.Wong YC, Holzbaur ELF. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U.S.a. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nature Communications. 2014;5:1–11. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]
- 69.Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, et al. NBR1 acts as an autophagy receptor for peroxisomes. Journal of Cell Science. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 70.Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nature Cell Biology. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen W-C, Li H-Y, Chen G-C, Chern Y, Tu P-H. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy. 2015;11:685–700. doi: 10.4161/auto.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 73.Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, et al. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015;11:e1005174–e1005126. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nature Cell Biology. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 75.Kuo T-C, Chen C-T, Baron D, Onder TT, Loewer S, Almeida S, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nature Cell Biology. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grasso D, Ropolo A, Lo Ré A, Boggio V, Molejón MI, Iovanna JL, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J. Biol. Chem. 2011;286:8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 78.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget's disease of bone. J. Biol. Chem. 2003;278:37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 79.Lamark T, Perander M, Outzen H, Kristiansen K, Øvervatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 80.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Molecular Cell. 2006;23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, et al. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J. Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 83.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 84.Itakura E, Mizushima N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. The Journal of Cell Biology. 2011;192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. The Journal of Cell Biology. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom A-L, Kemppainen R, Cox N, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J. Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 87.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 88.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J-I, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 89.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Molecular Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 90.Wurzer B, Zaffagnini G, Fracchiolla D, Turco E, Abert C, Romanov J, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJH, et al. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015;11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 92.Gao J, Ohtsubo M, Hotta Y, Minoshima S. Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: possible relationship with glaucoma pathology. PLoS ONE. 2014;9:e101206. doi: 10.1371/journal.pone.0101206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J-Y, Koga H, Kawaguchi Y, Tang W, Wong E, Gao Y-S, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO Journal. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simonsen A, Birkeland HCG, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. Journal of Cell Science. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 95.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat. Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- 97.LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nat. Rev. Microbiol. 2015;13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collins CA, Brown EJ. Cytosol as battleground: ubiquitin as a weapon for both host and pathogen. Trends in Cell Biology. 2010;20:205–213. doi: 10.1016/j.tcb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 99.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein Á, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jackson WT. Viruses and the autophagy pathway. Virology. 2015;479–480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 2015;16:1014–1024. doi: 10.1038/ni.3273. [DOI] [PubMed] [Google Scholar]
- 102.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes & Development. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, et al. Sestrins Activate Nrf2 by Promoting p62-Dependent Autophagic Degradation of Keap1 and Prevent Oxidative Liver Damage. Cell Metabolism. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 104.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 105.Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2015:1–21. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pickrell AM, Huang C-H, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, et al. Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron. 2015;87:371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. The Journal of Cell Biology. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080–120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 114.Ordureau A, Sarraf SA, Duda DM, Heo J-M, Jedrychowski MP, Sviderskiy VO, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Molecular Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nature Cell Biology. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- 117.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 118.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 119.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 121.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, et al. Measuring In Vivo Mitophagy. Molecular Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Molecular Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Farré J-C, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Developmental Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. The EMBO Journal. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Developmental Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 128.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. Journal of Cell Science. 2013;126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Orvedahl A, Sumpter R, Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, et al. TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Developmental Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Orvedahl A, MacPherson S, Sumpter R, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shpilka T, Welter E, Borovsky N, Amar N, Shimron F, Peleg Y, et al. Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc. Natl. Acad. Sci. U.S.a. 2015;112:1434–1439. doi: 10.1073/pnas.1409476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nature Cell Biology. 2011;14:51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- 137.Belaid A, Cerezo M, Chargui A, Corcelle-Termeau E, Pedeutour F, Giuliano S, et al. Autophagy Plays a Critical Role in the Degradation of Active RHOA, the Control of Cell Cytokinesis, and Genomic Stability. Cancer Research. 2013;73:4311–4322. doi: 10.1158/0008-5472.CAN-12-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Belaid A, Ndiaye PD, Cerezo M, Cailleteau L, Brest P, Klionsky DJ, et al. Autophagy and SQSTM1 on the RHOA(d) again. Autophagy. 2013;10:201–208. doi: 10.4161/auto.27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dou Z, Xu C, Donahue G, Shimi T, Pan J-A, Zhu J, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612–aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proceedings of the National Academy of Sciences. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. 2012;51:5705–5724. doi: 10.1021/bi300752r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Asano T, Komatsu M, Yamaguchi-Iwai Y, Ishikawa F, Mizushima N, Iwai K. Distinct Mechanisms of Ferritin Delivery to Lysosomes in Iron-Depleted and Iron-Replete Cells. Molecular and Cellular Biology. 2011;31:2040–2052. doi: 10.1128/MCB.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kishi-Itakura C, Koyama-Honda I, Itakura E, Mizushima N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. Journal of Cell Science. 2014;127:4089–4102. doi: 10.1242/jcs.156034. [DOI] [PubMed] [Google Scholar]
- 145.Mancias JD, Pontano Vaites L, Nissim S, Biancur DE, Kim AJ, Wang X, et al. Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife. 2015;4:e10308. doi: 10.7554/eLife.10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Terman A, Kurz T. Lysosomal iron, iron chelation, and cell death. Antioxid. Redox Signal. 2013;18:888–898. doi: 10.1089/ars.2012.4885. [DOI] [PubMed] [Google Scholar]
- 147.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 148.Weber GJ, Choe SE, Dooley KA, Paffett-Lugassy NN, Zhou Y, Zon LI. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–530. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bellelli R, Federico G, Matte A, Colecchia D, Iolascon A, Chiariello M, et al. NCOA4 Deficiency Impairs Systemic Iron Homeostasis. Cell Rep. 2016;14:411–421. doi: 10.1016/j.celrep.2015.12.065. [DOI] [PubMed] [Google Scholar]
- 150.Biasiotto G, Di Lorenzo D, Archetti S, Zanella I. Iron and Neurodegeneration: Is Ferritinophagy the Link? Mol. Neurobiol. 2015:1–33. doi: 10.1007/s12035-015-9473-y. [DOI] [PubMed] [Google Scholar]
- 151.Ebrahimi-Fakhari D, Saffari A, Wahlster L, Lu J, Byrne S, Hoffmann GF, et al. Congenital disorders of autophagy: an emerging novel class of inborn errors of neuro-metabolism. Brain. 2015;139:317–337. doi: 10.1093/brain/awv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet. 2009;41:1179–1181. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- 153.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. The EMBO Journal. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes & Development. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. The Journal of Cell Biology. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol. Cancer Res. 2014;12:485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen H-Y, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R, et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell. 2013;24:738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pérez-Mancera PA, Young ARJ, Narita M. Inside and out: the activities of senescence in cancer. Nat. Rev. Cancer. 2014;14:547–558. doi: 10.1038/nrc3773. [DOI] [PubMed] [Google Scholar]
- 162.Gewirtz DA. Autophagy and senescence: a partnership in search of definition. Autophagy. 2013;9:808–812. doi: 10.4161/auto.23922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, et al. Lysosome-mediated processing of chromatin in senescence. The Journal of Cell Biology. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Drake KR, Kang M, Kenworthy AK. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS ONE. 2010;5:e9806. doi: 10.1371/journal.pone.0009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Molecular Cell. 2015;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 167.Freund A, Laberge R-M, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Leidal AM, Debnath J. Autophagy Devours the Nuclear Lamina to Thwart Oncogenic Stress. Developmental Cell. 2015;35:529–530. doi: 10.1016/j.devcel.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 169.Dikic I, Johansen T, Kirkin V. Selective Autophagy in Cancer Development and Therapy. Cancer Research. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 170.Levy JMM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thorburn A. Autophagy and its effects: making sense of double-edged swords. PLoS Biol. 2014;12:e1001967. doi: 10.1371/journal.pbio.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kimmelman AC. Metabolic Dependencies in RAS-Driven Cancers. Clinical Cancer Research. 2015;21:1828–1834. doi: 10.1158/1078-0432.CCR-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes & Development. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & Development. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim M-J, Woo S-J, Yoon C-H, Lee J-S, An S, Choi Y-H, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J. Biol. Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & Development. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 182.Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc. Natl. Acad. Sci. U.S.a. 2016;113:182–187. doi: 10.1073/pnas.1515617113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Piao S, Amaravadi RK. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2015 doi: 10.1111/nyas.12953. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–1306. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2016;35:1–11. doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vogl DT, Stadtmauer EA, Tan K-S, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Molecular Cell. 2014;55:916–930. doi: 10.1016/j.molcel.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mathew R, White E. Eat this, not that! How selective autophagy helps cancer cells survive. Molecular & Cellular Oncology. 2014;2:e975638–4. doi: 10.4161/23723556.2014.975638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 190.Irby RB, Mao W, Coppola D, Kang J, Loubeau JM, Trudeau W, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat. Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 191.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sandilands E, Serrels B, Wilkinson S, Frame MC. Src-dependent autophagic degradation of Ret in FAK-signalling-defective cancer cells. EMBO Rep. 2012;13:733–740. doi: 10.1038/embor.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, Palomo-Irigoyen M, Varela-Rey M, Griffith M, et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. The Journal of Cell Biology. 2015;210:153–168. doi: 10.1083/jcb.201503019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature Cell Biology. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 197.Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends in Cell Biology. 2015;25:376–387. doi: 10.1016/j.tcb.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mizushima N. Autophagy: process and function. Genes & Development. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]