Abstract
Background
Malignant pleural mesothelioma (MPM) is an aggressive cancer refractory to current therapies. Reduced expression of micro ribonucleic acid (miR)‐591 in a range of cancer types has suggested it is a potent tumor suppressor, and overexpression has been shown to inhibit tumor cell growth. The role of miR‐591 in MPM is largely unknown.
Methods
miR‐591 was over‐expressed in vitro using micro RNA mimics in three MPM cell lines (H513, H2052, H2373), and effects on tumor cell growth, proliferation, invasion, and target gene expression were assessed.
Results
miR‐591 mimic was introduced into MPM cell lines to overexpress this microRNA. The cellular growth, proliferation, and invasive capability was significantly inhibited after overexpression of miR‐591. Growth inhibition caused by miR‐591 correlated with upregulation of p21 and Bax. Reduced invasive capability correlated with downregulation of matrix metalloproteinase‐2 and transforming growth factor‐β1.
Conclusion
miR‐591 is a potent tumor suppressor in MPM. Overexpression of miR‐591 may represent a novel therapeutic approach for MPM.
Keywords: Malignant pleural mesothelioma, microRNA, miR‐591
Introduction
Malignant pleural mesothelioma (MPM) is an aggressive tumor arising from the pleura, highly associated with asbestos exposure.1 The number of MPM patients has increased worldwide, probably as a result of increased asbestos consumption in developing countries. The molecular mechanisms of MPM are poorly understood. Aside from exposure to asbestos, other cofactors, such as radiation, exposure to simian virus 40, and genetic predisposition have also been suggested.2 Unfortunately, MPM is resistant to all currently available treatments and patients usually die within a year. Thus, a more comprehensive understanding of the molecular biology of MPM and development of new therapeutic strategies for MPM are urgently required.
Micro ribonucleic acids (miRNAs) are a class of short (∼22 nucleotides) noncoding RNAs that regulate gene expression by binding to messenger (m)RNA, leading to mRNA degradation or the inhibition of translation.3 Increasing evidence indicates that miRNAs are key players in cancer initiation and progression, acting as either oncogenes or tumor suppressors. In common with other cancers, the miRNA expression in MPM displays characteristic alterations.4, 5, 6, 7, 8 In a previous study, we identified and listed altered miRNAs in MPM tissues and noted a significant downregulation of miR‐1 expression in tumor samples.8 Overexpression of miR‐1 could induce apoptosis, thus making it a candidate therapeutic target. We also found significant downregulation of miR‐591 expression in MPM tissues. Overexpression of miR‐591 significantly repressed tumor growth in orthotopic neuroblastoma xenograft.9 In ovarian cancer, miR‐591 resensitized paclitaxel‐resistant cancer cells to paclitaxel by enhancing apoptosis and inhibiting cell migration and proliferation.10, 11
In the present study, we overexpressed miR‐591 in MPM cells to investigate the impact of miR‐591 on the cellular biological effects of MPM cells and to elucidate the cancer mechanisms involved in MPM.
Methods
Cell lines
Malignant pleural mesothelioma cell lines H513 (epithelial), H2052 (sarcomatoid), and H2373 (sarcomatoid) were obtained from the National Cancer Institute (NCI, Bethesda, MD, USA) and used in subsequent experiments.
Chemicals and culture medium
RPMI‐1640, fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Life Technologies (Foster City, CA, USA). Cell lines were maintained in RPMI‐1640 medium supplemented with 10% fetal bovine serum and antibiotics. All cell culture wares were purchased from Corning Incorporated (Fremont, CA, USA). Unless otherwise specified, all other chemicals were obtained from Sigma‐Aldrich Corporation (St. Louis, MO, USA).
Micro ribonucleic acid (miR)‐591 mimics transfection
Pre‐miR miR‐591 mimic and scrambled oligonucleotides were purchased from Life Technologies. Cells from each MPM cell line were seeded into six‐well plates and allowed to attach overnight to achieve 60–70% confluency. miR‐591 mimic or scrambled control (75 pmol) in 200 μl of serum‐free antibiotic‐free medium was mixed with 7.5 μl of Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA), dissolved in 200 μl of the same medium, and allowed to stand at room temperature for 20 minutes. The resulting 400 μl transfection solutions were then added to each well containing 1.6 mL of RPMI‐1640 medium, supplemented with 10% FBS. Cells were grown and harvested 48 hours after transfection for additional analyses. RNA was harvested from the MPM cells at 48 hours post‐transfection to evaluate miR‐591 expression. Transfection efficiency was determined by quantitative real‐time‐polymerase chain reaction (qRT‐PCR).
Crystal violet staining assay
Micro RNA‐induced changes in cell growth were evaluated by crystal violet staining, as previously described.12 In brief, cells were fixed with 1% glutaraldehyde and stained with 0.1% crystal violet. Crystal violet bound to cells, including nuclei and cytosol elements. The unbound dye was removed with water. Bound crystal violet was photographed and solubilized with 0.2% Triton‐X‐100 (Sigma‐Aldrich, Steinheim, Germany) for quantitative analysis. Light extinction, which increases linearly with the cell number, was analyzed at 570 nm using an enzyme‐linked immunosorbent assay (ELISA)‐Reader (Dynex Technologies, Denkendorf, Germany).
XTT proliferation assay
Viability and proliferation of the transfected MPM cells were assessed by Cell Proliferation Kit II assay (XTT; Roche Diagnostics, Risch‐Rotkreuz, Switzerland). Cells were seeded into 96‐well dishes and allowed to attach overnight. At days one, three, and six post‐transfection, cells were labeled with the mixture of XTT labeling reagent and electron‐coupling reagent. Absorbance at 490 nm was quantitated by scanning spectrophotometer, and optical density readings from the cells of each group were recorded.
In vitro invasion assay
Transwell membranes coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used to quantify in vitro MPM cell invasion. Transfected cells were plated at 5 × 104 per well in the upper chamber in medium containing 0.5% FBS. Medium containing 15% FBS was added to the lower chamber. After 24 hours of incubation, non‐invasive cells were removed from the top well with a cotton swab, while the bottom cells were fixed with 3% paraformaldehyde, stained with 0.1% crystal violet, and photographed in three independent 10 × fields for each well. Cell counting was performed under light microscope. Data represents mean ± standard deviation (SD).
Quantitative real‐time‐polymerase chain reaction (qRT‐PCR)
Ribonucelic acid isolation and qT‐PCR were performed as previously described.8 Briefly, RNA and miRNA isolation were performed with the RNeasy Mini Kit and the miRNeasy Mini Kit, respectively (Qiagen Sciences, Valencia, CA, USA). Reverse transcription was performed using the TaqMan Reverse Transcription Kit for genes and the TaqMan miRNA Reverse Transcription Kit for miRNA (Life Technologies). qRT‐PCR was performed on the ViiA 7 Real‐Time PCR System with appropriate assay reagents for genes and miRNA (Life Technologies). PCR primer sequences for the genes examined in this study are shown in Table 1. Primers were tested to determine their optimal concentrations for PCR analysis, and the resulting products were run on 2% agarose gel to confirm the appropriate size and RNA integrity. Gene expression values were normalized to the endogenous control gene, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). Similarly, miRNA expression values were normalized to the endogenous control, RNU44.13 All PCR reactions were performed in triplicate. Multiple, independent experiments were conducted with high correlation (Pearson r > 0.70) among the results.
Table 1.
Primer sequences used in this study
| Gene Name | Accession Number | Primer Sequence (5′‐3′) |
|---|---|---|
| P21 | NM_ 078467 | Forward‐CAGGGGACAGCAGAGGAAGA |
| Reverse‐GGGCGGCCAGGGTATGTAC | ||
| BAX | NM_ 138763 | Forward‐GGCCGGGTTGTCGCCCTTTT |
| Reverse‐CCGCTCCCGGAGGAAGTCCA | ||
| GAPDH | NM_ 001256799 | Forward‐CCACCCATGGCAAATTCCATGGCA |
| Reverse‐AACAAAGCCTGGACAAAT |
GAPDH, glyceraldehyde 3‐phosphate dehydrogenase.
RT 2 profiler PCR array for motility genes
The Human Target RT2 Profiler PCR Array from Qiagen (Cat# PAHS‐128Z; Hilden, Germany) was used to analyze the expression of motility‐related genes. This panel of 86 genes includes genes related with cell motility, such as matrix metalloproteinase‐2 (MMP2) and transforming growth factor‐β1 (TGF‐β1). Total RNA was isolated from miR‐591 transfected and control MPM cells. One microgram of total RNA was used to perform reverse transcription according to manufacturer instructions. cDNA was subsequently diluted and qRT‐PCR reactions were performed on 384‐well plates supplied by the manufacturer. Data were analyzed using the manufacturer's web‐based software. Data analysis was based on the ΔΔCT method with normalization of the raw data to multiple housekeeping genes in the RT2 Array from Qiagen.
Statistical analysis
Means and SDs were calculated from numerical data. In the figures, bar graphs depict the mean and SD. A student's t‐test was performed to calculate significance. P < 0.05 was considered to be significant.
Results
miR‐591 inhibits cell growth in malignant pleural mesothelioma (MPM) cells
We transfected three MPM cell lines (H513, H2052, and H2373) with miR‐591 mimic and a scramble control and confirmed that miR‐591 expression increased in all of the miR‐591‐transfected cells, compared with the scramble control: there was a 9.3‐fold increase for H513, 8.2‐fold for H2052 and 7.9‐fold for H2373. To observe the impact of miR‐591 overexpression on the mesothelioma phenotype, we performed crystal violet staining assay. We found that cell growth was markedly repressed in MPM cell lines with induced miR‐591 overexpression at day three (Fig 1a). We dissolved and quantitated the crystal violet dye and found a significant decrease in the crystal violet dye reading in all three miR‐591 transfected MPM cell lines (P < 0.05; Fig 1b).
Figure 1.
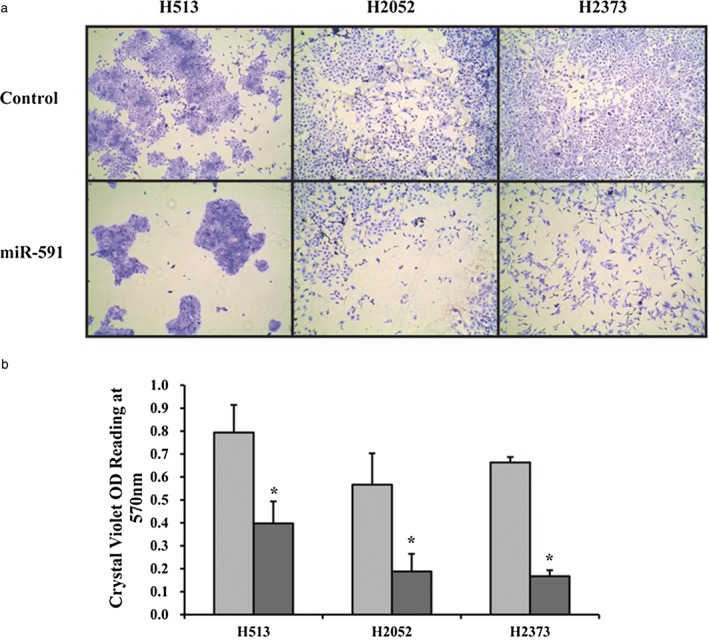
Growth effects of micro ribonucleic acid (miR)‐591 overexpression in mesothelioma cell lines. (a) Crystal violet staining of cells on day three post‐transfection of miR‐591. Decreased cell growth was observed in miR‐591‐transfected cells (miR‐591) compared with a scrambled control. Magnification, 40x. (b) Quantitative analysis of the staining assay indicated significant changes (P < 0.05). *Indicates significant change (P < 0.05).
OD, optical density, measured in this assay as a correlate of a number of viable cells.  , Control;
, Control;  , miR‐591.
, miR‐591.
The XTT viability and proliferation experiments showed that cell proliferation in transfected cells decreased compared with the scrambled control at days one, three and six, respectively. Significant (P < 0.05) changes in XTT dye incorporation were quantitated at days three and six (Fig 2).
Figure 2.
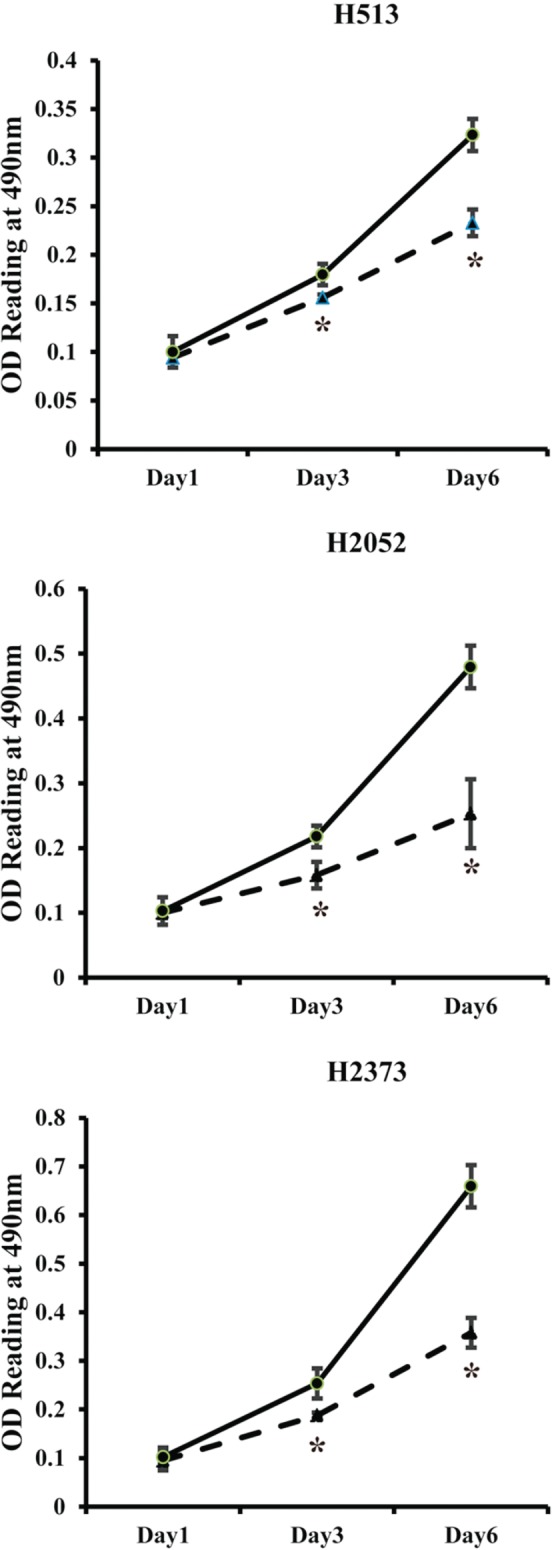
XTT growth assay. Significantly decreased cell proliferation was observed in micro ribonucleic acid (miR)‐591‐transfected cells (miR‐591) compared with a scrambled control by days three and six in all three MPM cell lines. *Indicates significant change (P < 0.05). OD, optical density, measured in this assay as a correlate of a number of viable cells.  , miR‐591;
, miR‐591;  , Control.
, Control.
miR‐591 inhibits cell invasion in MPM cells
To estimate the effect of miR‐591 on invasion potential in MPM, cell invasion was examined by transwell assay. Microscopic images of the transwell assay are shown in Figure 3a. In all 3 MPM cell lines, the number of invasive cells were significantly suppressed in miR‐591 transfected cells, compared with the scrambled control (232 ± 18 vs. 658 ± 58 for H513, P < 0.01; 17 ± 7 vs. 180 ± 20 for H2052, P < 0.01; 165 ± 75 vs. 452 ± 112 for H2373, P < 0.01; Fig 3b). These data demonstrated that tumor cell motility changed with the overexpression of miR‐591.
Figure 3.
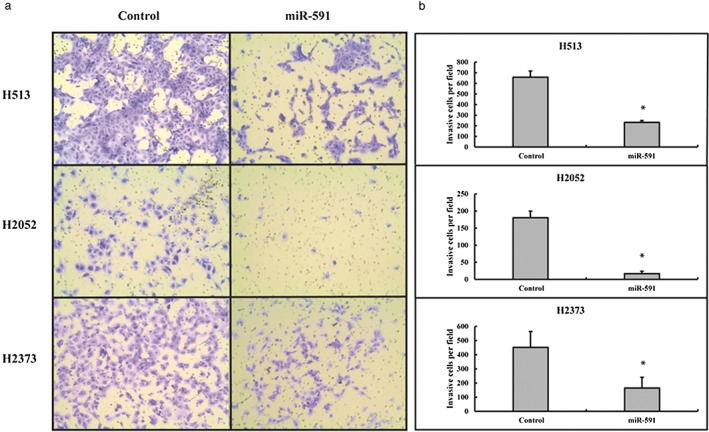
Transwell assay. Micro ribonucleic acid (miR)‐591 suppressed tumor cell invasive capability in vitro. (a) Representative images of the transwell assay of H513, H2052, and H2373 after transfection with miR‐591 or a scrambled control. Magnification, 100x. (b) Quantitative analysis of the transwell assay indicated significant changes (P < 0.05). *Indicates significant change (P < 0.05).
Phenotype changes in miR‐591 transfected cells is associated with alterations of apoptotic and cell cycle genes
To confirm the phenotypic changes in growth arrest, we examined genes associated with cell cycle homeostasis and pro‐apoptosis using qRT‐PCR. Total RNA was isolated from transfected MPM cells at 48 hours post‐transfection. Gene expressions of Bax (2.5‐fold increase for H513, P < 0.01; 3.6‐fold increase for H2052, P < 0.01; 1.8‐fold increase for H2373, P < 0.01) and p21 (4‐fold increase for H513, P < 0.01; 5.8‐fold increase for H2052, P < 0.01; 2.4‐fold increase for H2373, P < 0.01) were both significantly increased in miR‐591 overexpressed cells (Fig 4).
Figure 4.
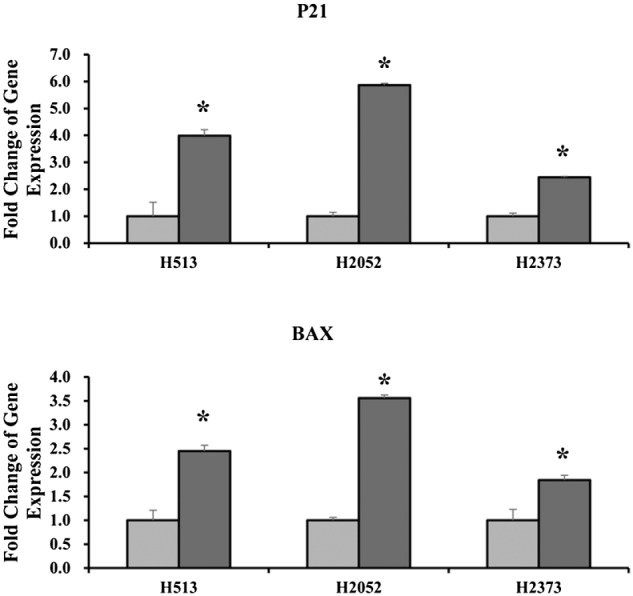
Gene expression changes associated with the growth arrest phenotype in micro ribonucleic acid (miR)‐591 transfected mesothelioma cells. Significant increases in P21 and BAX were observed in miR‐591‐transfected cells compared with a scrambled control. Fold changes were calculated by the ΔΔCt method. *Indicates significant change (P < 0.05).  , Control;
, Control;  , miR‐591.
, miR‐591.
miR‐591 affects multiple genes related with cell motility
We assessed the likely gene targets of miR‐591 to gain further insight into potential mechanisms to explain why miR‐591 inhibits invasion in MPM cells. RT2 Profiler PCR Array analysis revealed downregulation of multiple genes, shown in Table 2. Notably, both MMP2 and TGF‐β1 were consistently downregulated in the three miR‐591 overexpressed MPM cell lines.
Table 2.
Genes regulated by miR‐591 examined by RT 2 array
| H513 | |
|---|---|
| Gene Symbol | Fold Regulation |
| ITGB2 | −3.155 |
| MMP2 | −2.0732 |
| MYL9 | −2.3631 |
| RHO | −2.0472 |
| TGFB1 | −2.043 |
| H2052 | |
|---|---|
| Gene Symbol | Fold Regulation |
| CAPN1 | −2.3109 |
| DPP4 | −2.0272 |
| MMP2 | −3.1987 |
| TGFB1 | −3.1394 |
| H2373 | |
|---|---|
| Gene Symbol | Fold Regulation |
| CAV1 | −3.8933 |
| ENAH | −2.0907 |
| IGF1R | −2.3054 |
| ITGB1 | −2.0777 |
| MET | −3.625 |
| MMP2 | −5.148 |
| MYH10 | −2.7019 |
| MYL9 | −2.5385 |
| MYLK | −2.1435 |
| PAK1 | −2.0307 |
| PAK4 | −2.7721 |
| PIK3CA | −2.1615 |
| PRKCA | −2.6592 |
| PTPN1 | −2.3214 |
| RASA1 | −2.3022 |
| RHOC | −2.5315 |
| RND3 | −2.181 |
| TGFB1 | −2.6574 |
| VIM | −2.528 |
| WASF1 | −2.6354 |
| WIPF1 | −2.9221 |
Gene symbol, the full names of gene symbols are available from URL: (http://www.sabiosciences.com/genetable.php?pcatn=PAHS‐128Z). (−) value indicates fold regulation of downregulated genes. Fold regulation is the negative inverse of the fold change. Mir, micro ribonucleic acid.
Discussion
Malignant pleural mesothelioma was once an uncommon malignancy, but its incidence has increased worldwide, probably resulting from widespread exposure to asbestos. MPM is resistant to all currently available treatments and patients usually die within a year; therefore, new therapeutic strategies to treat patients with MPM are needed.
Numerous miRNAs are dysregulated in cancers, including MPM, and a single miRNA can have multiple targets involved in different oncogenic pathways. Therefore, targeting aberrant miRNA expression to simultaneously control multiple genes within the same and/or related pathways may be more effective than any single‐target agents.14, 15 To date, a limited number of studies have evaluated the differential expression of miRNAs of MPM using different sample sources.4, 5, 6, 8 Most recently, we compared miRNA expression in MPM tissues with miRNAs in normal parietal pleura and found significant downregulation of miR‐591 expression in MPM tissues.8 Several studies have investigated the effects of restoring the expression of downregulated miRNAs in MPM.7, 16, 17, 18, 19 The expression of miR‐16 was consistently downregulated in MPM tissues and cell lines, and re‐expression led to growth inhibition both in vitro and in vivo.17 The expression of miR‐34b and miR‐34c (miR‐34b/c) was frequently downregulated by methylation in MPM cell lines and primary tumors and the forced overexpression of miR‐34b/c had a significant antitumor effect that induced apoptosis in MPM cells in vitro.18 miR‐34b/c could enhance the radiosensitivity of MPM cells by promoting radiation‐induced apoptosis.19
In previous studies, the overexpression of miR‐591 resulted in significantly decreased tumor growth and migration in neuroblastoma and ovarian cancer.9, 10 Consistent with these results, our study demonstrated that the overexpression of miR‐591 inhibited proliferation and invasion in MPM cell lines.
The antiapoptotic potential of MPM makes it highly resistant to chemotherapy and radiotherapy.20 Our results show that miR‐591 probably induced cell cycle arrest and apoptosis by upregulating p21 and Bax. P21 is a potent negative regulator of the G1‐S checkpoint, while Bax is a pro‐apoptotic gene that belongs to the B cell lymphoma 2 family. Mutations and deletions of the TP53 gene are rare in MPM.21 The antiapoptotic potential of MPM has been considered to arise downstream of p53.22 Both p21 and Bax are the downstream targets of p53. Whether miR‐591 directly or indirectly regulates these genes remains to be determined, as none of the predicted gene targets of miR‐591 are the canonical mediators of apoptosis. miR‐591 may trigger a regulatory signaling cascade and exert an indirect effect on cell cycle and apoptotic pathways.
In our study, the suppressive effects of miR‐591 on invasion were confirmed in MPM cell lines by transwell assay. We found significant downregulation of MMP2 and TGF‐β1 in all three miR‐591 overexpressed MPM cell lines. MMP2 plays a key role in MPM invasion and metastasis.23, 24 TGF‐β promotes tumor progression by enhancing epithelial–mesenchymal transition, metastasis, angiogenesis, etc.25 TGF‐β could increase the expression, secretion, and activity of MMP‐2 and downregulate the expression of the protease tissue inhibitors of metalloproteinases.26, 27 Targeting TGF‐β signaling is emerging as an anti‐cancer therapeutic strategy. PF‐03446962 is an anti‐TGF‐β type I receptor monoclonal antibody. A Phase II clinical trial testing PF‐03446962 in patients with advanced MPM already treated with cytotoxic chemotherapy is currently ongoing.28, 29
In conclusion, our results show that overexpressing miR‐591 in vitro results in significantly decreased proliferation and invasion in MPM cell lines. Our study provides new insights into the molecular pathogenesis of MPM and suggests that miR‐591 could be a potential therapeutic target for MPM.
Disclosure
No authors report any conflict of interest.
Acknowledgments
We thank Xiaodong Tian, Shai Ben‐Shoshan and Sawa Keymeulen for assistance with data management. We thank Janice Zhang for careful assistance in manuscript preparation.
References
- 1. Guo G, Chmielecki J, Goparaju C et al Whole‐exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res 2015; 75: 264–269. [DOI] [PubMed] [Google Scholar]
- 2. Hashim D, Boffetta P. Occupational and environmental exposures and cancers in developing countries. Ann Glob Health 2014; 80: 393–411. [DOI] [PubMed] [Google Scholar]
- 3. Schirle NT, Sheu‐Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science 2014; 346: 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balatti V, Maniero S, Ferracin M et al MicroRNAs dysregulation in human malignant pleural mesothelioma. J Thorac Oncol 2011; 6: 844–851. [DOI] [PubMed] [Google Scholar]
- 5. Busacca S, Germano S, De Cecco L et al MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol 2010; 42: 312–319. [DOI] [PubMed] [Google Scholar]
- 6. Guled M, Lahti L, Lindholm PM et al CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma ‐A miRNA microarray analysis. Genes Chromosomes Cancer 2009; 48: 615–623. [DOI] [PubMed] [Google Scholar]
- 7. Ueno T, Toyooka S, Fukazawa T et al Preclinical evaluation of microRNA‐34b/c delivery for malignant pleural mesothelioma. Acta Med Okayama 2014; 68: 23–26. [DOI] [PubMed] [Google Scholar]
- 8. Xu Y, Zheng M, Merritt RE et al miR‐1 induces growth arrest and apoptosis in malignant mesothelioma. Chest 2013; 144: 1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shohet JM, Ghosh R, Coarfa C et al A genome‐wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res 2011; 71: 3841–3851. [DOI] [PubMed] [Google Scholar]
- 10. Huh JH, Kim TH, Kim K et al Dysregulation of miR‐106a and miR‐591 confers paclitaxel resistance to ovarian cancer. Br J Cancer 2013; 109: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gadducci A, Sergiampietri C, Lanfredini N, Guiggi I. Micro‐RNAs and ovarian cancer: The state of art and perspectives of clinical research. Gynecol Endocrinol 2014; 30: 266–271. [DOI] [PubMed] [Google Scholar]
- 12. Gillies RJ, Didier N, Denton M. Determination of cell number in monolayer cultures. Anal Biochem 1986; 159: 109–113. [DOI] [PubMed] [Google Scholar]
- 13. Gee HE, Buffa FM, Camps C et al The small‐nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer 2011; 104: 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garofalo M, Leva GD, Croce CM. MicroRNAs as anti‐cancer therapy. Curr Pharm Des 2014; 20: 5328–5335. [DOI] [PubMed] [Google Scholar]
- 15. Sethi S, Ali S, Sethi S, Sarkar FH. MicroRNAs in personalized cancer therapy. Clin Genet 2014; 86: 68–73. [DOI] [PubMed] [Google Scholar]
- 16. Ivanov SV, Goparaju CM, Lopez P et al Pro‐tumorigenic effects of miR‐31 loss in mesothelioma. J Biol Chem 2010; 285: 22809–22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reid G, Pel ME, Kirschner MB et al Restoring expression of miR‐16: A novel approach to therapy for malignant pleural mesothelioma. Ann Oncol 2013; 24: 3128–3135. [DOI] [PubMed] [Google Scholar]
- 18. Kubo T, Toyooka S, Tsukuda K et al Epigenetic silencing of microRNA‐34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res 2011; 17: 4965–4974. [DOI] [PubMed] [Google Scholar]
- 19. Maki Y, Asano H, Toyooka S et al MicroRNA miR‐34b/c enhances cellular radiosensitivity of malignant pleural mesothelioma cells. Anticancer Res 2012; 32: 4871–4875. [PubMed] [Google Scholar]
- 20. Krug LM, Wozniak AJ, Kindler HL et al Randomized phase II trial of pemetrexed/cisplatin with or without CBP501 in patients with advanced malignant pleural mesothelioma. Lung Cancer 2014; 85: 429–434. [DOI] [PubMed] [Google Scholar]
- 21. Zucali PA, Ceresoli GL, De Vincenzo F et al Advances in the biology of malignant pleural mesothelioma. Cancer Treat Rev 2011; 37: 543–558. [DOI] [PubMed] [Google Scholar]
- 22. de Assis LV, Isoldi MC. The function, mechanisms, and role of the genes PTEN and TP53 and the effects of asbestos in the development of malignant mesothelioma: A review focused on the genes' molecular mechanisms. Tumour Biol 2014; 35: 889–901. [DOI] [PubMed] [Google Scholar]
- 23. Roomi MW, Kalinovsky T, Niedzwiecki A, Rath M. Modulation of u‐PA, MMPs and their inhibitors by a novel nutrient mixture in human lung cancer and mesothelioma cell lines. Int J Oncol 2013; 42: 1883–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edwards JG, McLaren J, Jones JL, Waller DA, O'Byrne KJ. Matrix metalloproteinases 2 and 9 (gelatinases A and B) expression in malignant mesothelioma and benign pleura. Br J Cancer 2003; 88: 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Principe DR, Doll JA, Bauer J et al TGF‐beta: Duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst 2014; 106: djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF‐beta1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer 2012; 12: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiercinska E, Naber HP, Pardali E, van der Pluijm G, van Dam H, ten Dijke P. The TGF‐beta/Smad pathway induces breast cancer cell invasion through the up‐regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Res Treat 2011; 128: 657–666. [DOI] [PubMed] [Google Scholar]
- 28. Remon J, Reguart N, Corral J, Lianes P. Malignant pleural mesothelioma: New hope in the horizon with novel therapeutic strategies. Cancer Treat Rev 2015; 41: 27–34. [DOI] [PubMed] [Google Scholar]
- 29. Kotova S, Wong RM, Cameron RB et al New and emerging therapeutic options for malignant pleural mesothelioma: Review of early clinical trials. Cancer Manag Res 2015; 7: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]


