Abstract
The two-component signal transduction system CpxAR is especially widespread in Gram-negative bacteria. It has been reported that CpxAR contributes to the multidrug resistance (MDR) in Escherichia coli. CpxR is a response regulator in the two-component CpxAR system. The aim of this study was to explore the role of cpxR in the MDR of S. enterica serovar Typhimurium. The minimal inhibitory concentrations (MICs) of various antibiotics commonly used in veterinary medicine for strains JS (a multidrug-susceptible standard strain of S. enterica serovar Typhimurium), JSΔcpxR, JSΔcpxR/pcpxR, JSΔcpxR/pcpxR*, JSΔcpxRΔacrB, JSΔcpxRΔacrB/pcpxR, JSΔcpxRΔacrB/pcpxR*, 9 S. enterica serovar Typhimurium isolates (SH1–9), and SH1–9ΔcpxR were determined by the 2-fold broth microdilution method. The relative mRNA expression levels of ompF, ompC, ompW, ompD, tolC, acrB, acrD, acrF, mdtA, marA, and soxS in strains JS, JSΔcpxR, and JSΔcpxR/pcpxR were detected by real-time PCR. The results showed 2- to 4-fold decreases in the MICs of amikacin (AMK), gentamycin (GEN), apramycin (APR), neomycin (NEO), ceftriaxone (CRO), ceftiofur (CEF), and cefquinome (CEQ) for strain JSΔcpxR, as compared to those for the parental strain JS. Likewise, SH1–9ΔcpxR were found to have 2- to 8-fold reduction in resistance to the above antibiotics, except for NEO, as compared to their parental strains SH1–9. Furthermore, 2- to 4-fold further decreases in the MICs of AMK, GEN, APR, and CEF for strain JSΔcpxRΔacrB were observed, as compared to those for strain JSΔacrB. In addition, CpxR overproduction in strain JSΔcpxR led to significant decreases in the mRNA expression levels of ompF, ompC, ompW, ompD, tolC, acrB, marA, and soxS, and significant increases in those of stm3031 and stm1530. Notably, after all strains were induced simultaneously by GEN to the 15th passage at subinhibitory concentrations, strain JSΔcpxR/pcpxR showed significant increases in mRNA expression levels of the efflux pump acrD and mdtA genes, as compared to strain JSΔcpxR. Our results indicate that the two-component regulator CpxR contributes to resistance of S. enterica serovar Typhimurium to aminoglycosides and β-lactams by influencing the expression level of the MDR-related genes.
Keywords: S. enterica serovar Typhimurium, CpxR, aminoglycosides, β-lactams, resistance, AcrD
Introduction
Salmonella enterica serovar Typhimurium is a food-borne pathogen that causes gastroenteritis in humans (Scherer and Miller, 2001) and fowl typhoid in poultry (Barrow et al., 2004). The prevalence of multidrug-resistant (MDR) S. enterica species in many parts of the world has become a significant public health concern. Drug resistance in many cases is attributable to synergy between reduced drug intake (mainly due to low outer membrane permeability) (Pagès et al., 2008; Li and Nikaido, 2009) and active drug export (via efflux pumps) (Zgurskaya and Nikaido, 2000; Pagès et al., 2010). Resistance nodulation-cell division (RND)-family efflux systems (including AcrAB, AcrAD, AcrEF, MdtEF, and MdtABC) are especially effective in generating resistance in Gram negative bacteria (Nikaido, 1996) and often have a wide substrate specificity (Nikaido and Pagès, 2012). In Escherichia coli, all five RND-family drug exporters confer resistance to β-lactam antibiotics (Nishino et al., 2003), and acrD is also known to participate in the efflux of aminoglycosides (Rosenberg et al., 2000; Nishino and Yamaguchi, 2001a; Aires and Nikaido, 2005; Nishino et al., 2007). Some outer membrane proteins, especially OmpF, OmpC, and OmpW, have been shown to contribute to antibiotic resistance in E. coli and Salmonella typhimurium (Nikaido, 2003). In addition, OmpD, STM3031, and STM1530 are associated with ceftriaxone (CRO) resistance in S. enterica serovar Typhimurium (Hu et al., 2011), and OmpW participates in resistance to neomycin (NEO) and ampicillin (AMP) in E. coli (Wu et al., 2012).
Two-component signal transduction systems (TCSs) are key in the sensory response of bacteria (Parkinson and Kofoid, 1992). Studies have elucidated that the TCSs EvgA and BaeR contribute to MDR by modulating production of the multidrug transporter in E. coli (Nishino and Yamaguchi, 2001b; Baranova and Nikaido, 2002; Nishino and Yamaguchi, 2002). The Cpx envelope stress response is controlled by a TCS consisting of the membrane localized sensor kinase CpxA and the regulator CpxR. CpxR mediates the output response as a transcriptional regulator through phosphorylation of its receiver domain with an aspartate (D51) moiety (Stephenson and Hoch, 2002; MacRitchie et al., 2008). Phosphorylated CpxR (CpxR-P), which functions as a transcription factor, activates and, in a small number of cases, represses transcription of target genes by binding to the promoter of target genes at the consensus sequence 5′-GTAAAN5GTAAA-3′ (De Wulf et al., 2002; Price and Raivio, 2009). In addition, the response regulator CpxR is also activated by some signals without the involvement of CpxA. For example, some cytoplasmic or growth signals, as well as excess carbon (glucose or pyruvate) in growth medium both activate CpxR independently of CpxA (Cuny et al., 2007; Wolfe et al., 2008).
In recent years, the CpxAR two-component system conferring resistance to antibacterial agent has received special attention. In E. coli, CpxR overproduction was found to confer resistance to β-lactams in an acrB-free background (Hirakawa et al., 2003a). CpxR-P also confers resistance to fosfomycin by directly repressing the expression of two genes, glpT and uhpT, in the enterohemorrhagic E. coli (EHEC) strain O157:H7 (Kurabayashi et al., 2014). Moreover, the CpxAR pathway contributes to E. coli resistance to antimicrobial peptides, such as ApoEdpl-W, polymyxin B, and melittin (Audrain et al., 2013) and protamine (Weatherspoon-Griffin et al., 2014). In Klebsiella pneumoniae, CpxR was able to directly bind to the promoter regions of ompCKPand kpnEF, which contribute to the K. Pneumoniae MDR phenotype (Srinivasan et al., 2012; Srinivasan and Rajamohan, 2013). In S. enterica serovar Typhimurium, studies about the effect of cpxR on the resistance are still very limited, only few reports showed CpxAR confers resistance to CRO (Hu et al., 2011) and the antimicrobial peptides protamine, magainin, and melittin (Weatherspoon-Griffin et al., 2011). However, whether CpxAR plays a role in resistance of S. enterica serovar Typhimurium especially clinical isolates to aminoglycosides and β-lactams and the molecular mechanisms underlying resistance to aminoglycosides and β-lactams remain unknown. In this study, we systematically investigated the role of cpxR in aminoglycoside and β-lactam resistance in both susceptible strains and clinical isolates of S. enterica serovar Typhimurium, and also explored the molecular mechanisms of CpxAR that confer resistance to aminoglycosides and β-lactams.
Materials and methods
Bacterial strains, plasmids, and bacteriophage
The bacterial strains, plasmids, and bacteriophage used in this study are listed in Table 1. Salmonella enterica serovar Typhimurium strain CVCC541, a clinical susceptible strain isolated from chicken in Changchun City, China, was supplied by the China Institute of Veterinary Drug Control (Beijing, China) and designated as JS in this report. Strains JSΔcpxR and JSΔacrB were generated from JS using the one-step inactivation of chromosomal genes method. Strain JSΔacrBΔcpxR was constructed by the phage P22-mediated transduction method using strain JSΔcpxR as the donor and JSΔacrB as the recipient. In this study, nine S. enterica serovar Typhimurium isolates were isolated from chickens collected from nine different regions of Henan province in China and named SH1–9.
Table 1.
Bacterial strains, plasmids, and phage used in this study.
| Strain, plasmid, or phage | Relevant characteristics | References or source |
|---|---|---|
| STRAINS | ||
| JS | S. enterica Serovar Typhimurium CVCC541 | Supplied by China Institute of Veterinary Drug Control |
| JSΔcpxR | Derivative of JS that lacks cpxR | Huang et al., 2015 |
| JSΔacrB | Derivative of JS that lacks acrB | Huang et al., 2016 |
| JSΔacrBΔcpxR | Derivative of JS that lacks both cpxR and acrB, ΔcpxR::kan | Huang et al., 2016 |
| SH(1–9) | Clinical isolates from chicken in Henan province in China | This study |
| SH(1–9) ΔcpxR | Derivative of SH(1–9) that lack cpxR, ΔcpxR::kan | This study |
| PLASMIDS | ||
| pKD4 | Gene knowout help vector: repR6Kγ ApR FRT KmR FRT | From E. coli Genetic Stock Center in Yale University |
| pKD46 | Gene knowout help vector: ApR ParaBAD γ β exo | |
| pBAD | Expression vector: reppBR322ApR araC PBAD | Invitrogen Corporation |
| pBAD-CpxR | cpxR gene cloned to pBAD; ApR | This study |
| pBAD-CpxR* | Mutation sequence CpxR* cloned to pBAD; ApR | This study |
| PHAGE | ||
| P22HT105/int | Transduction medium of Salmonella | Supplied by Microbial Genomics Research Center of Harbin Medical University |
Construction of the expression plasmids pBAD-CpxR and pBAD-CpxR*
The complete open reading frame of cpxR was amplified by PCR with primers XholI-cpxR-F/HindIII-cpxR-R (Table 2) from the genomic DNA of strain JS. The mutation sequence cpxR*, which encodes a CpxR variant with an alanine residue at position 51 in place of aspartate, was engineered by overlapping PCR (Urban et al., 1997; Huang et al., 1999). The mutation site was generated through the design of primers Fm and Rm (Table 2). Three PCR reactions were performed to obtain the mutation sequence cpxR*. Primers XholI-cpxR-F/Rm were used for amplification of the anterior segment of cpxR, primers Fm/HindIII-cpxR-R were used for amplification of the second part of cpxR, and the primers XholI-cpxR-F/HindIII-cpxR-R were used for splicing by overlap extension PCR. Finally, the expression plasmids pBAD-CpxR and pBAD-CpxR* were generated by inserting the target fragment to the multiple cloning site of vector pBAD. The expression level of target proteins were determined according to the concentration of the inducer L-arabinose (Guzman et al., 1995).
Table 2.
Sequences of primers used in this study.
| Function | Primer | Sequence (5′ → 3′) | References or source |
|---|---|---|---|
| Amplification of cpxR gene | XholI-cpxR-F HindIII-cpxR-R | CGCTCGAGATGAATAAAATCCTGTTAGT GCAAGCTTTCATGAAGCGGAAACCATCA | This study |
| Preparation of cpxR* | XholI-cpxR-F Rm | CGCTCGAGATGAATAAAATCCTGTTAGT ACTTTTGCTTGCCGTCATGATGCCGAAG | This study |
| Fm Hind III-cpxR-R | CTTCGGCATCATGACGGCAAGCAAAAGT GCAAGCTTTCATGAAGCGGAAACCATCA | This study | |
| REAL-TIME RELATIVE QUANTITATIVE PCR | |||
| ompF | ompF-F ompF-R | CCTGGCAGCGGTGATCC AAATTTCTGCTGCGTTTGCG | Tatavarthy and Cannons, 2010 |
| ompC | ompC-F ompC-R | TCGCAGCCTGCTGAACCAGAAC ACGGGTTGCGTTATAGGTCTGAG | Hu et al., 2011 |
| ompD | ompD-F ompD-R | GCAACCGTACTGAAAGCCAGGG GCCAAAGAAGTCAGTGTTACGGT | Hu et al., 2011 |
| ompW | ompW-F ompW-R | CAGCAGCAAAGTGCGTCCTTATGT AGACAGAGGCGCCAATTAACCAGT | Hu et al., 2011 |
| stm3031 | stm3031-F stm3031-R | TGCAAGCAGGGAGTAATAACGGGT TCACTTGGATACGCCCAGTCCCAT | Hu et al., 2011 |
| stm1530 | stm1530-F stm1530-R | CGTCTCGGTTTTGCTGGTTTGG GCCGTCATTTTTACCCTGATACTGC | Hu et al., 2011 |
| acrB | acrB-F acrB-R | CGTGAGCGTTGAGAAGTCCT GGCGTCAGTTGGTATTTGGT | Li et al., 2009 |
| acrD | acrD-F acrD-R | TCCGGCCAAATTGAATAGTT TCGGAACCGTCCTGATTAAC | Eaves et al., 2004 |
| acrF | acrF-F acrF-R | TATCTGGCTGGATGCGAATCTGCT ACTTTGCCGAACTCTTCCGGATCT | Eaves et al., 2004 |
| mdtA | mdtA-F mdtA-R | GAATGCGCGTCGTGATCTG TCCAGTTCCTGACGGGAAAC | Nishino et al., 2007 |
| marA | marA-F marA-R | ATACATCCGCAGCCGTAAAA GTGATTCGCCATGCATATTG | Li et al., 2009 |
| soxS | soxS-F soxS-R | TACGGTAACGCATCAAACA ACAGGCGGTGACGGTAAT | Li et al., 2009 |
| 16SrRNA | 16SrRNA-F 16SrRNA-R | TTAGATACCCTGGTAGTCCACGC TTGCGGGACTTAACCCAAC | Li et al., 2009 |
The underlined bases are restriction sites.
Construction of cpxR-deficient mutants of S. enterica serovar typhimurium isolates
The generation of strain JSΔcpxR::kan was described in our previous study (Huang et al., 2015). The deletions were then transferred to nine S. enterica serovar Typhimurium isolates (SH1–9) by P22HT105/int transductions as previously described (Davis et al., 1980; Mann and Slauch, 1997). Nine cpxR-deficient mutants were designated as SH1–9ΔcpxR in this study.
Antibiotic susceptibility testing
The minimal inhibitory concentrations (MICs) of selected antibiotics for all strains were determined by the 2-fold broth microdilution method according to the CLSI guidelines (Clinical and Laboratory Standards Institute, 2008, 2012). The antibiotics used for susceptibility determination were gentamycin (GEN), amikacin (AMK), apramycin (APR), NEO, CRO, ceftiofur (CEF), CEQ. E. coli ATCC 25922 was used for quality control in all susceptibility tests. All tests were performed independently at least three times.
GEN induction testing
A single colony of each tested strain (JS, JSΔcpxR, and JSΔcpxR/pcpxR) was cultured in Luria-Bertani (LB) medium containing a 50% MIC of GEN at 37°C for 18 h. After growth overnight at 37°C, the cultures was diluted 1:100 in LB medium and cultured at 37°C for 18 h, and simultaneously the inducer GEN was added at subinhibitory concentrations. GEN induction testing of the strains was performed for 15 generations in this way.
Expression levels of MDR-related genes
Total RNA was isolated from bacterial cultures using the MiniBEST Universal RNA Extraction Kit (TaKaRa Bio, Inc. Shiga, Japan) according to the manufacturer's instructions. OD260/OD280 values of total RNA were detected using a trace nucleic acid protein analyzer spectrophotometer (NanoDrop; Thermo Fisher Scientific, Waltham, MA, USA). Bulk cDNA samples were synthesized from total RNA using the PrimeScript™RT reagent Kit with gDNA Eraser (TaKaRa Bio, Inc.). The synthesized cDNA was confirmed by PCR and stored at –20°C until used. Real-time PCR was performed using the LightCycler®480 System (Roche Diagnostics, Indianapolis, IN, USA) with specific primer pairs (Table 2), cDNA template, and TaKaRa SYBR Premix Ex Taq II (TaKaRa Bio, Inc.). The 16S rRNA gene was chosen as a housekeeping gene. To precisely test the relative expression level of the genes of interest, standard curves of the amplification of all detected genes were individually established. CT values tested came within the linearity range for PCR amplification. Each sample was independently run at least twice. The 2−Δ(ΔCT) method was used to calculate altered folds of the gene tested in the mutants, as compared to that in JS. Three independent experiments were performed under the same conditions.
Statistical analysis
Statistical analysis was performed using SPSS version 17.0 software (IBM-SPSS, Inc., Chicago, IL, USA). Data were compared using the Student's t-test. A probability (p) value of > 0.05 was considered statistically significant.
Results
Deletion of cpxR increases susceptibility of JS to aminoglycosides and β-lactams
To examine whether the response regulator CpxR contributes to the drug resistance of S. enterica serovar Typhimurium, a cpxR deletion mutant, JSΔcpxR, was generated from strain JS, and the complementary strain JSΔcpxR/pcpxR was prepared through the introduction of the expression plasmid pBAD-CpxR into JSΔcpxR. The MICs of a number of antibiotics for strain JS and JSΔcpxR were then determined. As shown in Table 3, strain JSΔcpxR showed 2–4-fold decreases in the MICs of GEN, AMK, APR, NEO, CRO, CEF, and CEQ, as compared to the parental strain JS. The MICs of the above antibiotics increased by 4-fold for the complementary strain JSΔcpxR/pcpxR, as compared to those for JSΔcpxR. These results clearly suggest that cpxR plays an important role in conferring resistance of S. enterica serovar Typhimurium to aminoglycosides and β-lactams. In addition, JSΔcpxR/pcpxR*exhibited the same susceptibility as JSΔcpxR to the tested antibiotics except for CEQ, which demonstrates that the susceptibility changes of S. enterica serovar Typhimurium to the tested antibiotics was mediated by CpxR-P.
Table 3.
Susceptibility of S. enterica serovar Typhimurium to several antibiotics.
| Strain | MICs (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | APR | NEO | CRO | CEF | CEQ | ENR | CIP | |
| JS | 0.5 | 0.25 | 2 | 0.4 | 0.02 | 0.32 | 0.08 | 0.032 | 0.016 |
| JSΔcpxR | 0.125 | 0.0625 | 1 | 0.1 | 0.01 | 0.08 | 0.04 | 0.032 | 0.016 |
| JSΔcpxR/pcpxR | 0.5 | 0.25 | 4 | 0.4 | 0.04 | 0.32 | 0.16 | 0.032 | 0.016 |
| JSΔcpxR/pcpxR* | 0.125 | 0.0625 | 1 | 0.1 | 0.01 | 0.08 | 0.16 | 0.032 | 0.016 |
| JSΔacrB | 0.25 | 0.25 | 2 | 0.2 | 0.01 | 0.0025 | 0.02 | 0.001 | 0.001 |
| JSΔacrBΔcpxR | 0.0625 | 0.0625 | 0.5 | 6.4▴ | 0.01 | 0.00125 | 0.02 | 0.001 | 0.001 |
| JSΔacrBΔcpxR/pcpxR | 0.5 | 0.25 | 2 | 25.6▴ | 0.02 | 0.005 | 0.08 | 0.001 | 0.001 |
| JSΔacrBΔcpxR/pcpxR▴ | 0.0625 | 0.0625 | 0.5 | 6.4▴ | 0.01 | 0.00125 | 0.04 | 0.001 | 0.001 |
AMK, Amikacin; GEN, Gentamycin; APR, Apramycin; NEO, Neomycin; CRO, Ceftriaxone; CEF, Ceftiofur; CEQ, Cefquinome; ENR, Enrofloxacin; CIP, Ciprofloxacin.
High NEO resistance is present in strains because of the replacement of cpxR gene by kanamycin-resistant gene (ΔcpxR::kan).
Effects of deletion of acrB on cpxR-mediated multidrug resistance
In susceptible S. enterica serovar Typhimurium, the AcrAB efflux pump is constitutively expressed and plays a predominant role in intrinsic and acquired resistance (Mazzariol et al., 2000; Nishino et al., 2006). It has wild substrate spectrum and can capture substrates from the periplasm or the outer leaflet of the cytoplasmic membrane (Yu et al., 2003). Therefore, AcrAB may mask partial function of some efflux pumps located in the cytoplasmic membrane (Hirakawa et al., 2003a,b; Eaves et al., 2004; Nishino et al., 2007). To clarify the role of CpxR in resistance conferred by other efflux pumps, an acrB deletion mutant (JSΔacrB) and a double deletion mutant (JSΔacrBΔcpxR) were generated from strain JS. The cpxR complementary strain JSΔacrBΔcpxR/pcpxR was prepared as described above. The MICs of various antibiotics for strains JSΔacrB, JSΔacrBΔcpxR, and JSΔacrBΔcpxR/pcpxR were then determined. As shown in Table 3, strain JSΔacrBΔcpxR showed 2–4-fold decreases in the MICs of GEN, AMK, APR, NEO, and CEF, as compared to strain JSΔacrB, while the complementary strain JSΔacrBΔcpxR/pcpxR exhibited 2–8-fold increases in the MICs of GEN, AMK, APR, NEO, CRO, CEF, and CEQ, as compared to strain JSΔacrBΔcpxR. These results revealed that CpxR can modulate resistance of S. enterica serovar Typhimurium to aminoglycosides and β-lactams in both acrB and ΔacrB backgrounds.
Role of cpxR in drug resistance of S. enterica serovar typhimurium isolates
To determinate the role of cpxR in regulating drug resistance of S. enterica serovar Typhimurium isolates, nine cpxR-deficient mutants (SH1–9ΔcpxR) derived from nine S. enterica serovar Typhimurium isolates (SH1–9) were constructed. The MICs for SH1–9 and SH1–9ΔcpxR to the above antibiotics were then determined. As shown in Table 4, among the nine cpxR deletion strains, all showed 2–4-fold decreases in the MICs of GEN, AMK, APR, and CEF, six revealed 2–4-fold decreases in the MIC of CRO, and six revealed 2–4-fold decreases in the MIC of CEQ, compared with their parental strains. These results indicate that cpxR also plays an important role in resistance of S. enterica serovar Typhimurium isolates to aminoglycosides and β-lactams.
Table 4.
Susceptibilities of S. enterica serovar Typhimurium isolates to antibiotics after cpxR were deleted.
| Strains | MICs (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| AMK | GEN | APR | NEO | CRO | CEF | CEQ | |
| SH1 | 1.25 | 1 | 2 | 0.8 | 0.5 | 4 | 1 |
| SH1ΔcpxR | 0.625 | 0.125 | 0.5 | 12.8▴ | 0.5 | 2 | 1 |
| SH2 | 5 | 32 | 4 | 0.4 | 0.5 | 4 | 0.5 |
| SH2ΔcpxR | 1.25 | 16 | 2 | 6.4▴ | 0.25 | 2 | 0.25 |
| SH3 | 2 | 0.2 | 4 | 0.4 | 0.1 | 0.8 | 0.05 |
| SH3ΔcpxR | 0.5 | 0.1 | 1 | 6.4▴ | 0.05 | 0.4 | 0.05 |
| SH4 | 1 | 0.4 | 2 | 0.4 | 0.05 | 0.8 | 0.1 |
| SH4ΔcpxR | 0.5 | 0.1 | 1 | 6.4▴ | 0.05 | 0.4 | 0.05 |
| SH5 | 1 | 0.4 | 4 | 0.4 | 0.05 | 0.8 | 0.1 |
| SH5ΔcpxR | 0.5 | 0.2 | 2 | 3.2▴ | 0.05 | 0.4 | 0.05 |
| SH6 | 0.4 | 0.2 | 8 | 0.8 | 0.1 | 0.2 | 0.1 |
| SH6ΔcpxR | 0.1 | 0.05 | 2 | 12.8▴ | 0.05 | 0.2 | 0.1 |
| SH7 | 1.6 | 12.8 | 1024 | 1.6 | 0.1 | 1.6 | 0.2 |
| SH7ΔcpxR | 0.4 | 3.2 | 256 | 12.8▴ | 0.05 | 0.8 | 0.1 |
| SH8 | 0.4 | 0.2 | 4 | 0.8 | 0.2 | 0.4 | 0.1 |
| SH8ΔcpxR | 0.2 | 0.1 | 2 | 12.8▴ | 0.05 | 0.1 | 0.05 |
| SH9 | 0.8 | 0.4 | 4 | 0.8 | 0.1 | 0.8 | 0.05 |
| SH9ΔcpxR | 0.1 | 0.05 | 1 | 12.8▴ | 0.05 | 0.4 | 0.025 |
AMK, Amikacin; GEN, Gentamycin; APR, Apramycin; NEO, Neomycin; CRO, Ceftriaxone; CEF, Ceftiofur; CEQ, Cefquinome.
High NEO resistance is present in strains because of the replacement of cpxR gene by the kanamycin-resistant gene (ΔcpxR::kan).
Effects of cpxR on the expression levels of a series of MDR-related genes
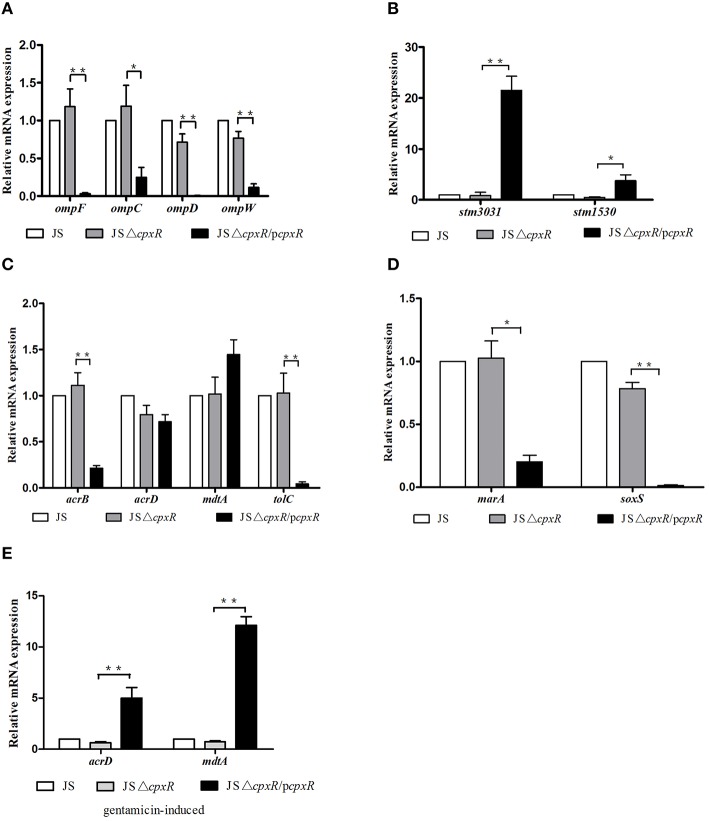
In E. coli, it has been confirmed that cpxR can modulate the expression of the outer membrane proteins OmpF and OmpC (Batchelor et al., 2005), and the transporter MdtABC (Hirakawa et al., 2005). In S. enterica serovar Typhimurium, the expression of OmpD, STM3031, and STM1530 plays important roles in cpxR-mediated CRO resistance (Hu et al., 2011). In order to determine whether the drug resistance mediated by cpxR is due to altered expression levels of MDR-related genes, we detected the relative mRNA expression of a series of MDR-related genes. As shown in Figure 1, JSΔcpxR showed no significant differences in the mRNA expression levels of all tested genes, as compared to strain JS, while the mRNA expression levels of ompF, ompC, ompW, ompD, acrB, tolC, marA, and soxS genes in strain JSΔcpxR/pcpxR were significantly decreased (p < 0.01 or p < 0.05) relative to strain JSΔcpxR (Figures 1A,C,D) and the mRNA levels of stm3031 and stm1530 in strain JSΔcpxR/pcpxR were significantly increased (p < 0.01 or p < 0.05) relative to strain JSΔcpxR (Figure 1B). There were no significant differences in mRNA expression levels of acrD and mdtA among strains JS, JSΔcpxR, and JSΔcpxR/pcpxR (Figure 1C). The expression levels of AcrF in these three strains were all very low and almost undetectable (data not shown). However, after all strains were induced by GEN at subinhibitory concentrations to the 15th passage simultaneously, strain JSΔcpxR/pcpxR showed significant (p < 0.01) increases in the mRNA expression levels of mdtA and acrD, as compared to JSΔcpxR (Figure 1E). These results suggest that the overexpression of cpxR can downregulate the expression levels of OmpF, OmpC, OmpW, OmpD, AcrB, TolC, MarA, and SoxS and upregulate those of STM3031 and STM1530 in susceptible S. enterica serovar Typhimurium strains, and it also upregulated the expression levels of the efflux pumps AcrD and MdtA under the pressure of GEN.
Figure 1.
Relative mRNA expression levels (n-fold) determined by real-time PCR. The expression level of each mRNA in strain JS represents 1-fold. The expression of the 16S rRNA gene was used as an internal control. Each bar represents the average value of three independent experiments. (A) Relative mRNA expression levels of the outer membrane proteins genes ompF, ompC, ompD, and ompW; (B) Relative mRNA expression levels of the outer membrane protein genes stm3031 and stm1530; (C) Relative mRNA expression levels of the efflux pumps genes acrB, acrD, mdtA, and tolC; (D) Relative mRNA expression levels of the transcription factor genes marA and soxS. (E) Relative mRNA expression levels of the efflux pump genes acrD and mdtA in all tested strains induced by GEN to the 15th passage at subinhibitory concentrations. *p < 0.05, **p < 0.01.
Discussion
In this study, we analyzed the effect of CpxR on the drug resistance of a susceptible strain and nine clinical isolates of S. enterica serovar Typhimurium and found 2- to 4-fold decreases in resistance to aminoglycosides and β-lactams by deletion of cpxR (Tables 3, 4). These results are similar to those of previous studies reporting that the overexpression of cpxR in E. coli caused 2-fold increases in resistance to β-lactams (Hirakawa et al., 2003a), but different from the findings that the cpxA-cpxR deleted mutant R200(ΔcpxAR) showed more remarkable decreases (>2048-fold) than strain JSΔcpxR in the MIC of CRO, as compared to their parental strain (Hu et al., 2011). Obviously, R200, generated by a multistep resistance selection method, is a CRO-resistant strain. Therefore, we concluded that the influence of cpxR on the drug resistance of resistant strains is greater than that of susceptible strains.
It is known that OmpF and OmpC are the most abundant outer membrane proteins of S. enterica serovar Typhimurium. Many antimicrobial agents have been found to alter the expression of these proteins. Moreover, it has been confirmed that decreased level of OmpD, and increased levels of STM3031 and STM1530 are associated with S. enterica serovar Typhimurium CRO resistance (Hu et al., 2009, 2011). In this study, we found significant reductions in the levels of OmpF, OmpC, OmpD, and OmpW, and significant increases in levels of STM3031 and STM1530 when cpxR was reverted to strain JSΔcpxR. Thus, the altered levels of the above-mentioned outer membrane proteins influenced by CpxR may be closely associated with the CpxR-mediated resistance of S. enterica serovar Typhimurium to β-lactams.
In Gram-negative bacteria, transporters belonging to the RND family are particularly effective in generating resistance, and MDR often results from the overexpression of multidrug efflux transporters (Grkovic et al., 2002). In this study, before the strains were induced with GEN, CpxR overexpression led to significant reductions in levels of AcrB, TolC, MarA, and SoxS. MarA and SoxS are global regulatory factors (Wall et al., 2009). Once overexpressed, MarA further activates AcrAB/TolC efflux and alters the expression of some membrane proteins (Sulavik et al., 1997). To our knowledge, the influence of CpxR on the mRNA levels of marA and soxS genes has not been demonstrated. Our results can give two suggestions. One is that the decrease in AcrAB–TolC, mediated by the complementation of cpxR, is associated with the decrease of the regulatory factors MarA and SoxS. The other is the expression levels of AcrB and TolC do not play a decisive role in CpxR-mediated resistance of S. enterica serovar Typhimurium to aminoglycoside and β-lactams. Our finding that CpxR can influence the susceptibility of S. enterica serovar Typhimurium to aminoglycosides and β-lactams in both acrB and ΔacrB backgrounds also supports the second suggestion. Nevertheless, more studies should be carried out to elucidate the reciprocal relationship among CpxR, outer membrane protein genes, efflux genes, and regulative genes.
In this study, the up-regulatory effect of CpxR on the expression levels of AcrD and MdtA were observed in the GEN-induced strains. Aminoglycoside uptake in Gram-negative bacteria includes three consecutive steps. The first step is an electrostatic interaction between aminoglycosides and the bacteria cell envelope through displacement of Mg2+ and Ca2+ ions that link adjacent lipopolysaccharide molecules, which damages the bacteria outer membrane and enhances its permeability. The second step is energy-dependent phase I of uptake, which leads to a small quantity of antibiotic molecules transversing the cytoplasmic membrane. The third step is energy-dependent phase II of uptake, in which misfolded proteins are produced due to the binding of incoming antibiotics to the ribosome. Some of these proteins are incorporated in the cytoplasmic membrane leading to the loss of membrane integrity. Therefore, additional quantities of aminoglycosides are transported across the damaged cytoplasmic membrane (Taber et al., 1987). Thus, CpxR may be activated by GEN in the inducing experiment in vitro. It has been reported that the promoter regions of acrD and mdtABC harbor binding sites for the response regulator BaeR (Nishino et al., 2007). CpxR also can bind to the cpxR box located in the promoter region of target genes. In common, the consensus cpxR box includes a tandem repeated GTAAA sequence that is separated by a 5-bp space (Batchelor et al., 2005). The DNA binding feature of CpxR encouraged us to analyze the promoter region of acrD and mdtA in the chromosome of S. enterica serovar Typhimurium LT2 (accession number: AE006468) for the presence of putative CpxR binding sites. Interestingly, our analysis revealed the presence of two similar sequences located 173 bp (site 1: GTAAA-gaacg-GCAAA) and 106 bp (site2: GTAAA-agcgc-ATGAT) upstream of the acrD translational start site, respectively. Among them, site 1 was also found 328 bp upstream of the kpnEF translational start site. Furthermore, it has been confirmed that purified CpxR from a strain of K. Pneumoniae can directly bind to site 1 (Srinivasan and Rajamohan, 2013). Because CpxR of K. Pneumoniae exhibits the highest level of homology to CpxR of S. enterica serovar Typhimurium (96%), CpxR of S. enterica serovar Typhimurium may directly bind to the promoter region of acrD. As we know, AcrD participates in the efflux of aminoglycosides, thus our analysis indicates that CpxR contributes to AcrD-mediated resistance of S. enterica serovar Typhimurium to GEN, which belongs to aminoglycosides.
Moreover, in this study, there were no significant differences in the mRNA expression levels of all tested genes in strain JSΔcpxR, as compared to strain JS. It has been demonstrated that histidine kinase (HK) also possess response regulator phosphatase activity, which may ensure that the response regulator remains inactive in the absence of activating signals (Raivio and Silhavy, 1997). Therefore, we think that the response regulator CpxR is always in a pre-stimulated resting state and does not modulate mRNA levels at physiological levels. Biochemical data suggest that CpxR can become phosphorylated by the low-molecular-weight phospho-donor acetyl phosphate and, further, that this form (CpxR-P) has a greater affinity for binding to the promoters of target genes (Pogliano et al., 1997). Similarly, it has been demonstrated that, in the absence of CpxA, CpxR can transcriptionally activate downstream target genes, suggesting that CpxR-P is responsible for transcriptional activation of target genes (Danese et al., 1995; De Wulf and Lin, 2000; Batchelor et al., 2005). In this scenario, we conclude that CpxR overproduction in JSΔcpxR encourages the emergence of CpxR-P, which acts as a modulator of gene expression.
In summary, we have reported the first systematical and extensive study about the role of CpxR in aminoglycoside and β-lactams resistance in both susceptible strains and clinical isolates of S. enterica serovar Typhimurium. Our results not only clearly confirmed that CpxR contributes to resistance of S. enterica serovar Typhimurium to aminoglycoside and β-lactams but also indicated that the effect of CpxR on the expression levels of MDR-related genes is closely associated with CpxR-mediated resistance of S. enterica serovar Typhimurium to aminoglycoside and β-lactams. This is the first time that the effect of CpxR on the expression levels of marA and soxS genes have been investigated in S. enterica serovar Typhimurium. Further studies are obviously required to investigate the reciprocal relationship among CpxR, MDR-related outer membrane protein genes, efflux pump genes and regulative genes including marA and soxS.
Author contributions
HH, YS, and GH conceived of the study, and participated in its design and coordination. YG, CM isolated the S. enterica Serovar Typhimurium isolates. HH carried out the antibiotics susceptibility testing and molecular biology studies, including gene deletion, construction of expression vector and RT-PCR. HH and YP performed the statistical analysis. HH drafted the manuscript. YS, LY, and GH revised the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31372481) and the Key Research Program of Higher Education of Henan Province (15A230006).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Chenzhang Wang for assistance with the real-time PCR.
References
- Aires J. R., Nikaido H. (2005). Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187, 1923–1929. 10.1128/JB.187.6.1923-1929.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audrain B., Ferrières L., Zairi A., Soubigou G., Dobson C., Coppée J. Y., et al. (2013). Induction of the Cpx envelope stress pathway contributes to Escherichia coli tolerance to antimicrobial peptides. Appl. Environ. Microbiol. 79, 7770–7779. 10.1128/AEM.02593-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova N., Nikaido H. (2002). The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184, 4168–4176. 10.1128/JB.184.15.4168-4176.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow P. A., Bumstead N., Marston K., Lovell M. A., Wigley P. (2004). Faecal shedding and intestinal colonization of Salmonella enterica in in-bred chicked: the effect of host genetic background. Epidemiol. Infect. 132, 117–126. 10.1017/S0950268803001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E., Walthers D., Kenney L. J., Goulian M. (2005). The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins OmpF and OmpC. J. Bacteriol. 187, 5723–5731. 10.1128/JB.187.16.5723-5731.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Laboratory Standards Institute (2008). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard, 3rd Edn. Wayne, PA: CLSI; (Informational Supplement, M31–A3). [Google Scholar]
- Clinical Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-second Informational Supplement. Wayne, PA: CLSI (Twenty-Two Informational Supplement M100–S22). [Google Scholar]
- Cuny C., Lesbats M., Dukan S. (2007). Induction of a global stress response during the first step of Escherichia coli plate growth. Appl. Environ. Microbiol. 73, 885–889. 10.1128/AEM.01874-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese P. N., Snyder W. B., Cosma C. L., Davis L. J., Silhavy T. J. (1995). The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9, 387–398. 10.1101/gad.9.4.387 [DOI] [PubMed] [Google Scholar]
- Davis R. W., Bolstein D., Rorh J. R. (1980). Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- De Wulf P., Lin E. C. (2000). Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P Levels underlie CpxA* phenotypes. J. Bacteriol. 182, 1423–1426. 10.1128/JB.182.5.1423-1426.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McGuire A. M., Liu X., Lin E. C. (2002). Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277, 26652–26661. 10.1074/jbc.M203487200 [DOI] [PubMed] [Google Scholar]
- Eaves D. J., Ricci V., Piddock L. J. (2004). Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica Serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48, 1145–1150. 10.1128/AAC.48.4.1145-1150.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S., Brown M. H., Skurray R. A. (2002). Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66, 671–701. 10.1128/MMBR.66.4.671-701.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Inazumi Y., Masaki T., Hirata T., Yamaguchi A. (2005). Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55, 1113–1126. 10.1111/j.1365-2958.2004.04449.x [DOI] [PubMed] [Google Scholar]
- Hirakawa H., Nishino K., Hirata T., Yamaguchi A. (2003b). Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185, 1851–1856. 10.1128/JB.185.6.1851-1856.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H., Nishino K., Yamada J., Hirata T., Yamaguchi A. (2003a). β-Lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52, 576–582. 10.1093/jac/dkg406 [DOI] [PubMed] [Google Scholar]
- Hu W. S., Chen H. W., Zhang R. Y., Huang C. Y., Shen C. F. (2011). The expression levels of outer membrane proteins STM1530 and OmpD, which are influenced by the CpxAR and BaeSR two-component systems, play important roles in the ceftriaxone resistance of Salmonella enterica Serovar Typhimurium. Antimicrob. Agents Chemother. 55, 3829–3837. 10.1128/AAC.00216-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Lin J. F., Lin Y. H., Chang H. Y. (2009). Outer membrane protein STM3031 (Ail/OmpX-like) plays a key role in the ceftriaxone resistance of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53, 3248–3255. 10.1128/AAC.00079-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Liu B. G., Sun Y. W., Ma C. H., Cheng L. P., Yuan L., et al. (2016). Construction of cpxR and acrB double gene deletion strain of Salmonella enterica Serovar Typhimurium and analysis of its susceptibility to antibacterial agents. Acta Vet. Zootech. Sin. 47, 595–602. 10.11843/j.issn.0366-6964.2016.03.00 [DOI] [Google Scholar]
- Huang H., Tian Y. K., Sun Y. W., Cheng L. P., Yuan L., Liu J. H., et al. (2015). Construction of cpxR gene deleted Salmonella typhimurium and analysis of its susceptibility to antibiotics. Chin. J. Prev. Vet. Med. 37, 756–760. 10.3969/j.issn.1008-0589.2015.10.06 [DOI] [Google Scholar]
- Huang P. T., Yu W. Y., Chen T. M. (1999). PCR Lab Manual (Translate). Beijing: Science Press. [Google Scholar]
- Kurabayashi K., Hirakawa Y., Tanimoto K., Tomita H., Hirakawa H. (2014). Role of the CpxAR two-component signal transduction system in control of fosfomycin resistance and carbon substrate uptake. J. Bacteriol. 196, 248–256. 10.1128/JB.01151-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang Y. P., Wu Y. N., Wu C. M., Shen J. Z. (2009). Association of marR, soxR and acrR mutations with multiple antibiotic resistance in ciprofloxacin-selected Salmonella mutants. Food Saf. Qual. Detect. Technol. 1, 1–9. [Google Scholar]
- Li X. Z., Nikaido H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623. 10.2165/11317030-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRitchie D. M., Buelow D. R., Price N. L., Raivio T. L. (2008). Two-component signaling and gram negative envelope stress response systems. Adv. Exp. Med. Biol. 631, 80–110. 10.1007/978-0-387-78885-2_6 [DOI] [PubMed] [Google Scholar]
- Mann B. A., Slauch J. M. (1997). Transduction of low-copy number plasmids by bacteriophage P22. Genetics 146, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzariol A., Cornaglia G., Nikaido H. (2000). Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coli K-12 to β-lactams. Antimicrob. Agents Chemother. 44, 1387–1390. 10.1128/AAC.44.5.1387-1390.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (1996). Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Pagès J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363. 10.1111/j.1574-6976.2011.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Latifi T., Groisman E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 1, 126–141. 10.1111/j.1365-2958.2005.04940.x [DOI] [PubMed] [Google Scholar]
- Nishino K., Nikaido E., Yamaguchi A. (2007). Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189, 9066–9075. 10.1128/JB.01045-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamada J., Hirakawa H., Hirata T., Yamaguchi A. (2003). Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to β-Lactams[J]. Antimicrob. Agents Chemother. 47, 3030–3033. 10.1128/AAC.47.9.3030-3033.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2001a). Analysis of the complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. 10.1128/JB.183.20.5803-5812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2001b). Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183, 1455–1458. 10.1128/JB.183.4.1455-1458.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A. (2002). EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184, 2319–2323. 10.1128/JB.184.8.2319-2323.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès J. M., Alibert-Franco S., Mahamoud A., Bolla J. M., Davin-Regli A., Chevalier J., et al. (2010). Efflux pumps of gram-negative bacteria, a new target for new molecules. Curr. Top. Med. Chem. 8, 1848–1857. 10.2174/156802610793176620 [DOI] [PubMed] [Google Scholar]
- Pagès J. M., James C. E., Winterhalter M. (2008). The porin and the permeating antibiotic: a selective diffusion barrier in Gram-megative bacteria. Nat. Rev. Microbiol. 6, 893–903. 10.1038/nrmicro1994 [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. (1992). Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26, 71–112. 10.1146/annurev.ge.26.120192.000443 [DOI] [PubMed] [Google Scholar]
- Pogliano J., Lynch A. S., Belin D., Lin E. C. C., Beckwith J. (1997). Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182. 10.1101/gad.11.9.1169 [DOI] [PubMed] [Google Scholar]
- Price N. L., Raivio T. L. (2009). Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191, 1798–1815. 10.1128/JB.00798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio T. L., Silhavy T. J. (1997). Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179, 7724–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E. Y., Ma D., Nikaido H. (2000). AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182, 1754–1756. 10.1128/JB.182.6.1754-1756.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer C. A., Miller S. I. (2001). Molecular pathogenesis of Salmonella, in Principles of Bacterial Pathogenesis, ed Groisman E. A. (New York, NY: Academic Press, Inc.), 266–333. [Google Scholar]
- Srinivasan V. B., Rajamohan G. (2013). KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-Type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 57, 4449–4462. 10.1128/AAC.02284-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V. B., Vaidyanathan V., Mondal A., Rajamohan G. (2012). Role of the two-component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 7:e33777. 10.1371/journal.pone.0033777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K., Hoch J. A. (2002). Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr. Opin. Pharmacol. 2, 507–512. 10.1016/S1471-4892(02)00194-7 [DOI] [PubMed] [Google Scholar]
- Sulavik M. C., Dazer M., Miller P. F. (1997). The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179, 1857–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Mueller J. P., Miller P. F., Arrow A. S. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51, 439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavarthy A., Cannons A. (2010). Real-time PCR detection of Salmonella species using a novel target: the outer membrane porin F gene (ompF). Lett. Appl. Microbiol. 50, 645–652. 10.1111/j.1472-765X.2010.02848.x [DOI] [PubMed] [Google Scholar]
- Urban A., Neukirchen S., Jaeger K. E. (1997). A rapid and efficient method for site-directed mutagenesis using one-step over-lap extension PCR. Nucleic. Acids Res. 25, 2227–2228. 10.1093/nar/25.11.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M. E., Markowitz D. A., Rosner J. L., Martin R. G. (2009). Model of transcriptional activation by MarA in Escherichia coli. PLoS Comput. Biol. 5:e1000614. 10.1371/journal.pcbi.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherspoon-Griffin N., Yang D., Kong W., Hua Z., Shi Y. (2014). The CpxA/CpxR two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 289, 32571–32582. 10.1074/jbc.M114.565762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherspoon-Griffin N., Zhao G., Kong W., Kong Y., Morigen Andrews-Polymenis, H., et al. (2011). The CpxR/CpxA two-component system up-regulates two tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J. Biol. Chem. 286, 5529–5539. 10.1074/jbc.M110.200352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Parikhm N., Lima B. P., Zemaitaitis B. (2008). Signal integration by the two-component signal transduction response regulator CpxR. J. Bacteriol. 190, 2314–2322. 10.1128/JB.01906-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. B., Zou H. J., Tian L. H., Pan J. Y., Zhao F. K. (2012). Construction of ompW knock-out mutants of Escherichia coli to increase sensitivity to neomycinsulphate and ampicillin. Acta Microbiol. Sin. 52, 1021–1026. 10.13343/j.cnki.wsxb.2012.08.012 [DOI] [PubMed] [Google Scholar]
- Yu E. W., McDermott G., Zgurskaya H. I., Nikaido H., Koshland D. E., Jr. (2003). Structural basis of multiple drug-binding capacity of the AcrB multi drug efflux pump. Science 300, 976–980. 10.1126/science.1083137 [DOI] [PubMed] [Google Scholar]
- Zgurskaya H. I., Nikaido H. (2000). Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37, 219–225. 10.1046/j.1365-2958.2000.01926.x [DOI] [PubMed] [Google Scholar]