Abstract
The technology to reprogram human somatic cells to pluripotent state allows the generation of patient-specific induced pluripotent stem cells (iPSCs) and holds a great promise for regenerative medicine and autologous transplantation. Here we, for the first time, identified mesenchymal stem cells isolated from parotid gland (hPMSCs) as a suitable candidate for iPSC production. In the present study, hPMSCs were isolated from parotid gland specimens in patients with squamous cell carcinoma of the oral cavity. The mesenchymal stem cell properties of cultured hPMSCs were confirmed by expression of surface markers and induced differentiation into osteogenic, chondrogenic and adipogenic cell lineages. hPMSCs were then reprogrammed to pluripotent cells by episomal vector-mediated transduction of reprogramming factors (OCT3/4, SOX2, KLF4, c-MYC, LIN28 and TP53 shRNA). The resulting hPMSC-iPSCs showed similar characteristics as human embryonic stem cells (ESCs) with regard to morphology, pluripotent markers, global gene expression, and methylation status of pluripotent cell-specific genes OCT4 and NANOG. These hPMSC-iPSCs were able to differentiate into cells of all three germ layers both in vitro and in vivo. Our results indicate that hPMSCs could be an alternative cell source for generation of iPSCs and have the potential to be used in cell-based regenerative medicine.
Keywords: Induced pluripotent stem cell, mesenchymal stem cell, pluripotency, regenerative medicine, parotid gland
Introduction
Pioneering studies have demonstrated that mouse or human somatic cells can be reprogrammed into a pluripotent state without the use of embryonic material [1,2]. The reprogrammed cells, named induced pluripotent stem cells (iPSCs), are generated by ectopic expression of several defined transcription factors (OCT4 and SOX2, with KLF4 and c-MYC, or with NANOG and LIN28) [1-3]. Human iPSCs show similar characteristics as embryonic stem cells (ESCs) in terms of gene expression, pluripotency and epigenetic status, and thus provide valuable resources for derivation of patient- or disease-specific pluripotent stem cells devoid of ethical concerns, which could then be used in drug screening, drug development and aid understanding of specific disease mechanisms [4-7].
Human iPSCs can be generated from a wide range of somatic cell types, including neonatal or adult dermal fibroblasts [1,3,8-11], keratinocytes [12], neural stem cells [13], adipose stem cells [14], gut mesentery-derived cells [15], amnion-derived cells [16], and urine-derived cells [17]. The most common approach of iPSC reprogramming is introduction of key reprogramming factors into target cells mediated by lentiviral or retroviral vectors [1]. However, use of these integrating viruses has a number of disadvantages that may cause potential problems such as insertional mutagenesis, uncontrolled or undesirable gene silencing, residual expression, re-activation of transgenes, and immunogenicity. To cope with these problems, integration-free methods, for example the use of adenovirus [18], Sendai virus [19], piggy Bac systemical minicircle vector [20], episomal vectors [3], synthesized minicircle DNA [21] and direct protein delivery [22], have been developed.
Epigenetic memory is a concept first proposed by Kim et al. to describe the phenomenon that the iPSCs retain memory of the epigenetic signatures of their tissue of origin and thus favor the differentiation toward the donor-related lineages [23]. For example, the iPSCs derived from skin cells have a significantly higher potential to differentiate into keratinocytes than other cell types [24]. Further studies have demonstrated that in iPSCs, some imprinted genes that are involved in growth, metabolism and neurological development share the same epigenetical and transcriptional statuses with the initial somatic cells [25]. Such epigenetic memory could limit the full differentiation potential of iPSCs. Therefore, in regenerative cell medicine, it is critical to select the right donor cells to achieve optimal differentiation of target cell types.
Salivary glands are responsible for saliva production and crucial for food digestion and maintenance of oral health. Impaired salivary gland function caused by bacterial infection, Sjogren’s syndrome and cancer radiation therapy greatly decreases the patients’ quality of life [26]. Studies have been performed to develop tissue engineering techniques to repair the damaged saliva glands [27]. Parotid glands are the primary salivary glands in human. In this study, we highlighted the use of a combination of efficient episomal reprogramming vectors that consists of OCT3/4, SOX2, KLF4, L-MYC, LIN28, and shRNA targeting TP53 to generate iPSCs from human parotid gland mesenchymal stem cells (hPMSCs) [28].
Materials and methods
Isolation and culture of human parotid gland MSC cells (hPMSCs)
Samples of human parotid glands were obtained from patients with squamous cell carcinoma of the oral cavity during the neck dissection surgery. All medical procedures were approved by the Ethical Committee of Capital Medical University. Written informed consent was obtained from all patients. All patients were negative for the hepatitis B and C viruses, human immunodeficiency virus, and adult T-cell leukemia-associated antigen. None of the patients had received any other cancer treatment before the surgical procedure, or had a history of radiation to the neck.
Parotid glands were isolated from patients and minced using a scalpel. After incubating in 30 mL ethylene glycol tetraacetic acid (EGTA) buffer at 37°C for 20 min, the samples were centrifuged at 100 x g for 5 min at room temperature. The obtained cell pellets were resuspended in 60 mL digestion buffer containing a 1:1 mixture of Dulbecco’s modified Eagles medium (DMEM) and Ham’s F12, 1 mg/mL collagenase type II, and 1 mg/mL hyaluronidase (all from Invitrogen, Carlsbad, CA, USA). After incubating for 60 min at 37°C with gentle agitation, the cell suspension was further incubated in dispersion buffer containing DMEM/F12 (1:1) and 1.5 mg/ml dispase (Invitrogen) for 30 min at 37°C. The resulting cell suspension was passed through a stainless filter with a 75 μm mesh and centrifuged at 100 x g for 5 min. The remaining cell pellet was resuspended and the resulting parotid gland cells were seeded at a density of 4 x 103 cells/cm2 in cell culture medium consisting of DMEM/F12 (1:1), 2 mM glutamine, penicillin (100 U/mL), streptomycin (100 U/mL) and fibroblast growth factors (FGF, 5 ng/mL) (Sigma-Aldrich, St. Louis, MO, USA). The cells were incubated under optimized growth conditions at 37°C with 5% CO2. To allow hPMSC to adhere, the medium remained changed within the first 48-72 h. Subsequently, the medium was replaced three times a week. When the hPMSCs reached 80-90% confluence, cells were detached using trypsin-EDTA solution (0.25%) (Thermo Fisher Scientific, Pittsburgh, PA, USA), and subcultured at a density of 2.5 x 103 cells/cm2.
Lineage differentiation of hPMSCs
To induce adipogenic differentiation, hPMSCs from passages 2 to 3 were seeded at a density of 1.5 x 104 cells/cm2 in 6- or 24-well plates in culture medium supplemented with 1 mM dexamethasone (DEX), 10 mg/L insulin and 0.5 mM isobutylmethylxanthine (IBMX) (all from Sigma-Aldrich) for 4 weeks. Culture medium was replaced every 3 days. To examine differentiation, lipid vesicles were visualized by Oil Red O staining. To induce osteogenic differentiation, hPMSCs from passages 2 to 3 were seeded at a density of 3 x 104 cells/cm2 in culture medium supplemented with 0.1 mM DEX, 10 mM β-glycerophosphate and 50 mg/mL vitamin C (all from Sigma-Aldrich) for 4 weeks. Culture medium was replaced every 3 days. Calcium deposition was illustrated by Alizarin Red staining to demonstrate osteogenic differentiation. To induce chondrogenic differentiation, 2 x 105 hPMSCs were centrifuged at 550 g in 15 mL polypropylene tubes (Falcon, Bedford, MA, USA) for 5 min. Supernatant was removed and DMEM medium supplemented with 10 ng/ml transforming growth factor-β1 (TGF-β1, BD Biosciences, Franklin Lakes, NY, USA), 50 mg/ml ITS Premix (BD Biosciences) and 37.5 mg/ml ascorbate 2-phosphate (Sigma-Aldrich) was added to the cell pellets. Medium was replaced every 3 days and the cell pellets were harvested after 21 days. Safranin-O/Fast Green staining was used to evaluate chondrogenesis.
Flow cytometry analysis
hPMSCs were washed with stain buffer (BD Pharmingen, San Jose, CA) and incubated with PE-conjugated mouse anti-human antibodies for CD34, CD44, CD45, CD90, CD105 and CD71 (all from BD Pharmingen). Stained cells were analyzed using a FACS Calibur Flow Cytometer and Cell Quest software (BD Biosciences).
Transfection of hPMSCs and generation of hPMSC-iPSCs
Ten micrograms of episomal plasmid mixtures (OCT3/4, SOX2, KLF4, c-MYC, LIN28, and TP53 shRNA, all purchased from Addgene, Cambridge, MA, USA) were electroporated into 5 x 106 hPMSCs using a Nucleofector 2b Device (Lonza, Basel, Switzerland) and Amaxa Nucleofector Kit for primary fibroblasts (Lonza) according to the manufacturer’s instructions. Transfected cells were seeded onto 12-well plates coated with vitronectin and grown in Reproeasy reprogramming medium (Cellapy, Beijing, China). When iPSCs were mature, individual colonies were mechanically isolated into a 6-well plate and cultured in PSCeasy human pluripotent stem cell medium (Cellapy) for colony expansion. iPSCs were passaged by non-enzymatic dissociation using 0.5 mM EDTA.
Alkaline phosphatase (AP) and immunofluorescence (IF) staining of iPSCs
AP staining was performed using a kit purchased from Roche Applied Science (Mannheim, Germany) according to the manufacturer’s instructions. For IF staining, cells were first fixed with 4% paraformaldehyde for 10 min at room temperature. After washing with PBS, cells were incubated with PBS containing 10% normal bovine serum albumin (Sigma-Aldrich) and 0.1% Triton X-100 for 30 min at room temperature. The cells were then incubated with primary antibodies at 4°C overnight. The primary antibodies were mouse anti-SSEA-4 (1:100, Chemicon, Temecula, CA, USA), mouse anti-TRA-1-60 (1:100, Chemicon), mouse anti-TRA-1-81 (1:100, Chemicon), rabbit anti-NANOG (1:100, Abcam, Cambridge, UK), rabbit anti-OCT4 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA), and rabbit anti-SOX2 (1:100, Chemicon). Normal mouse or rabbit serum was served as a negative control. Anti-rabbit or anti-mouse IgG secondary antibodies conjugated with fluorescein (Santa Cruz) were used for visualization of the interested antigens. Additionally, nuclei were counterstained with DAPI. Immunofluorescent images were examined under an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan).
RNA isolation, reverse transcription and real-time (RT) PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Approximately 1 µg of total RNA from each sample was used for Oligo(dT)20-primed reverse transcription (Invitrogen). The resulting complementary DNA (cDNA) was used in RT-PCR with the conditions including an initial denaturation step of 5 min at 95°C, 35 cycles of 30 s denaturation at 95°C, 30 s annealing and 1 min extension at 72°C, and a final elongation step at 72°C for 10 min. Amplified PCR products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. Images were obtained using a Bio-Rad Gel document system (Hercules, CA, USA). Primer sequences are shown in Supplementary Table 1.
Bisulfite sequencing
Genomic DNA (1 μg) was treated using the CpGenome DNA modification kit (Chemicon) according to the manufacturer’s recommendations. The treated DNA was purified using a QIAquick column (Qiagen, Shanghai, China). The promoter regions of the human OCT4 and NANOG genes were amplified by PCR, and the PCR products were subcloned into the pCR2.1-TOPO vector. Five clones of each sample were verified by sequencing using the M13 universal primer.
Karyotyping
PMSC-iPSCs were prepared for karyotype analysis by incubation in medium containing 0.1 mg/ml colcemid for 5 hours. Cells were trypsinized, resuspended in 0.075 M KCl incubated at 37°C for 30 min, and fixed in 3:1 methanol:acetic acid at room temperature for 5 min. Centrifugation and fixing steps were repeated three times. Harvested cells were stained using a standard G-banding technique.
Microarray analysis
The gene expression profiles of H1 human ESCs, hPMSCs and hPMSC-iPSCs were obtained using GeneChip Human Genome U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA). All microarray experiments were performed and analyzed by Capital Bio Corporation (Beijing, China). Briefly, total RNA from each sample was subjected to probe preparation and cRNA was hybridized to the GeneChip. Arrays were scanned using an Affymetrix GCS3000 device and images were analyzed by the GCOS software (Affymetrix). The raw gene expression data were extracted using the GeneChip Operating Software, and the signal intensities from the mRNA channel in all arrays were normalized using the Robust Multiple-chip Analysis (RMA) algorithm and the DNA-Chip Analyzer (dChip) software (www.dchip.org) [29]. The raw mRNA expression data are available from the NCBI Gene Expression Omnibus (GEO), accession number GSE72984.
In vitro and in vivo differentiation
For in vitro differentiation, hPMSC-iPSCs were harvested using 0.5 mM EDTA and cultured in human ESC media without basic FGF (bFGF) in non-tissue culture treated plates. After 8 days in suspension culture, embryoid bodies (EB) were transferred to gelatin-coated plates and cultured in differentiation medium (high glucose DMEM supplemented with 15% FBS) for additional 8 days. The cells that underwent spontaneous differentiation were harvested and the specific gene expression was measured by RT-PCR.
To evaluate in vivo pluripotency, hPMSC-iPSCs were harvested, suspended in PBS, and 3 x 106 cells were injected subcutaneously into the hind limbs of SCID mice (Vital River Laboratories, Beijng, China). Approximately eight weeks after injection, the tumors that formed were dissected, fixed in 4% paraformaldehyde, embedded in paraffin blocks, and examined histologically by hematoxylin and eosin (H&E) staining. All animal experiments were approved by the Ethical Committee of Capital Medical University.
Results
Characterization of hPMSCs
hPMSCs were isolated from human parotid glands and cultured in optimized growth media. As shown in Figure 1A, the adherent cells exhibited mesenchymal cell morphology. Expression of mesenchymal stem cell (MSC) markers CD44, CD71, CD90 and CD105 were confirmed by flow cytometry analysis, while expression of non-MSC markers CD34 and CD45 was not detected in those cells (Figure 1B). To evaluate the potential of hPMSCs to differentiate into several mesenchymal lineages, hPMSCs after 3-5 passages were cultured in tissue-specific condition media to induce differentiation. Calcium deposition in cell cultures was detected with Alizarin Red staining, confirming the osteogenic differentiation of cultured hPMSCs (Figure 1C). Chondrogenesis of induced hPMSCs was demonstrated by Safranin-O/Fast Green staining in a 21 day pellet culture (Figure 1D). After 14-day induction of adipogenic differentiation, cultured cells exhibited evident accumulation of lipid droplets as illustrated by Oil Red O staining (Figure 1E). Taken together, these results indicated that the hPMSCs could be isolated from human parotid glands and preserved the stem cell-like properties.
Figure 1.
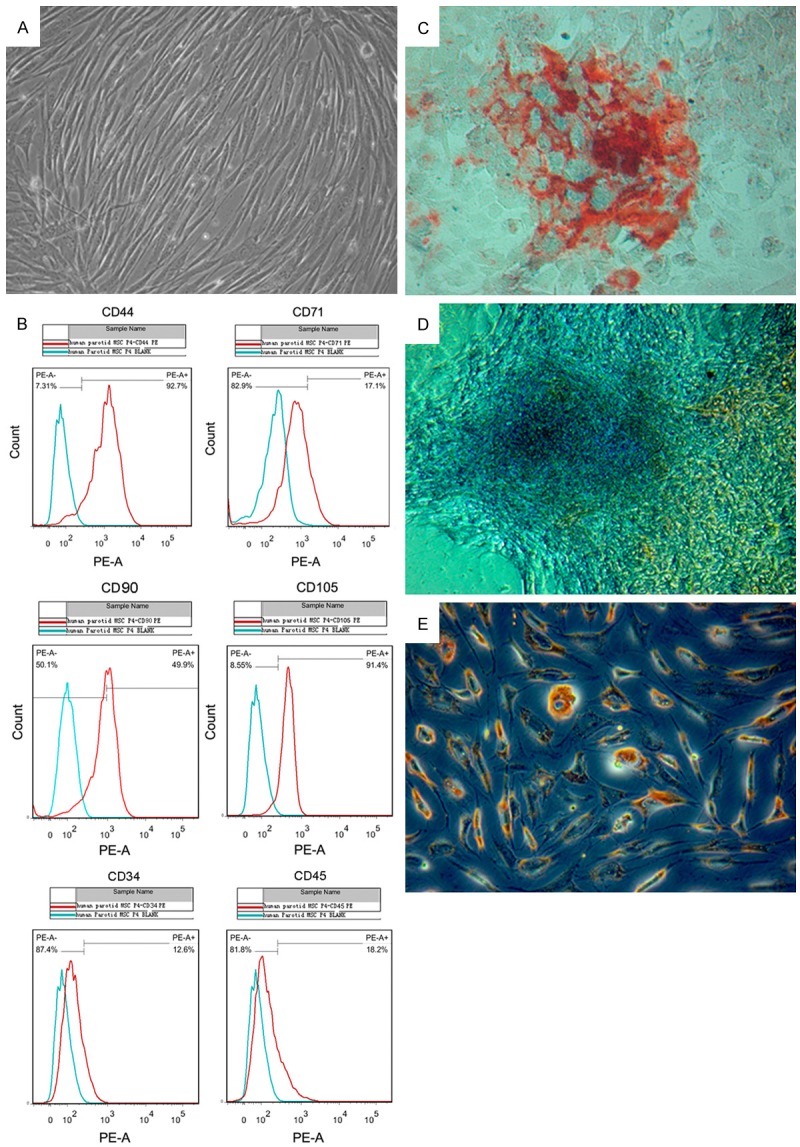
Characterization and lineage differentiation of cultured hPMSCs. (A) Phase-contrast microscopic images of hPMSCs in primary passage. (B) Flow cytometry analysis of mesenchymal stem cell surface markers (CD44, CD71, CD90 and CD105) and negative markers (CD34 and CD45) in cultured hPMSCs. (C) After induction, osteogenic differentiation of hPMSCs was demonstrated by formation of calcium deposits (red bone nodules outside the cells) detected with Alizarin Red staining (D) Safranin-O/Fast Green stain showed chondrogenic differentiation of hPMSCs in a 21 day pellet culture. (E) Adipogenic differentiation of hPMSCs was characterized by formation of intracytoplasmic lipid droplets stained with Oil Red O. All microscopic images were acquired at 200X magnification.
Generation of hPMSC-iPSCs
To generate iPSCs using an integration-free system, cultured hPMSCs were electroporated with episomal plasmid vectors for expression of OCT3/4, SOX2, KLF4, L-MYC, LIN28 and shRNA targeting TP53. The cell viability and transfection efficiency in our laboratory were 70-80% and 40-60%, respectively. Two days post transfection, cells were detached and plated onto vitronectin-coated plates. On day 20, colonies appeared and exhibited typical human ESC-like morphology (Figure 2A). The resulting cells were designated as hPMSC-iPSCs, and the ESC-like morphology was maintained during passaging on vitronectin-coated plates in chemical defined medium as described in the Materials and methods (Figure 2B). Of the original 5X104 PMSCs transduced per experiment, we routinely observed a yield of about 50 human ES-like colonies, with an efficiency of about 0.1%. Previous research has shown that iPSCs have increased AP activity compared to parental cells [1]. As shown in Figure 2C, the hPMSC-iPSC colonies were positive for AP staining (Figure 2C) compared to hPMSCs (Figure 2D). Besides ESC-like morphology, hPMSC-iPSCs also possessed the similar proliferation properties with ESCs (Figure 2E). The doubling time of hPMSC-iPSCs (29 hr) was equivalent to that of ESCs (28 hr).
Figure 2.
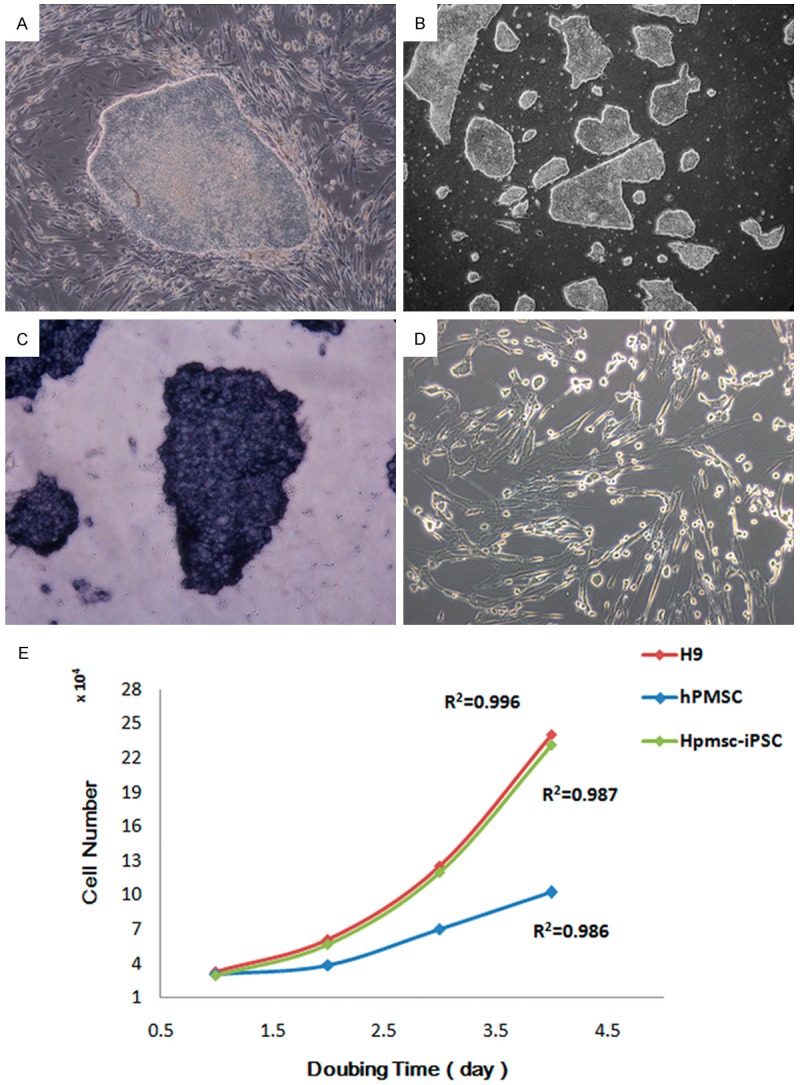
Generation of iPSCs from hPMSCs. A. hPMSCs were transfected with episomal plasmid vectors for pluripotent stem cell induction. After 20 days, the typical ES-like colonies emerged. B. The hPMSC-iPSC colonies at passage 30 were maintained on vitronetion in chemical defined medium. C. The hPMSC-iPSC colonies were stained positive for AP. D. The hPMSCs were stained negative for AP. Images were acquired at 200X magnification. Representative images are shown. E. Growth curves of ESCs, hPMSC-iPSCs and hPMSCs.
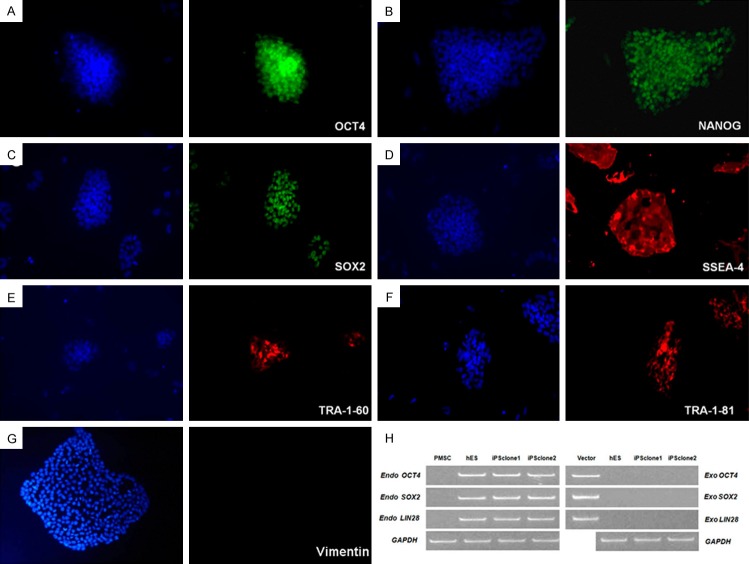
To further confirm the pluripotency of these colonies, immunofluorescence staining was used to assess the expression of stem cell-specific genes including OCT4, NANOG, SOX2, SSEA-4, TRA-1-60 and TRA-1-81 (Figure 3A-F).The maker of mesenchymal cells, Vimentin, was used for negative control (Figure 3G). We examined the expression of endogenous and exogenous OCT4, SOX2, and LIN28 by RT-PCR, and showed that the OCT4, SOX2, and LIN28 transgenes were efficiently silenced in PMSC-iPSCs, while the expression of endogenous OCT4, SOX2 and LIN28 was activated to levels comparable with human ES cells (Figure 3H). These results indicated that the hPMSCs could be reprogrammed to become pluripotent stem cells.
Figure 3.
Immunofluorescence staining of hPMSC-iPSC colonies. Expression of pluripotent markers OCT4 (A), NANOG (B), SOX2 (C), SSEA-4 (D), TRA-1-60 (E), TRA-1-81 (F) in hPMSC-iPSC colonies was detected by immunofluorescence staining using specific antibodies. hPMSC-Ipsc colonies were negative for Vimentin (G). Nuclei were counterstained with DAPI and shown in blue. Images were acquired at 200X magnification. (H) Expression of exogenous and endogenous OCT4, SOX2 and LIN28 were assessed by RT-PCR. Human ES cells and hPMSCs were used as controls.
Gene expression analysis and promoter methylation status of ESC-specific genes in hPMSC-iPSCs
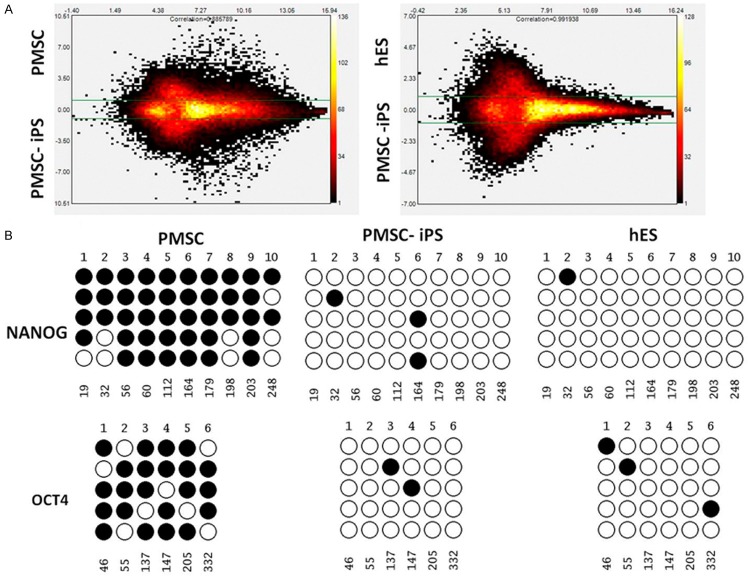
Global gene expression analyses were performed using microarrays to compare the gene expression profiles among hPMSCs, hPMSC-iPSCs and H1 human ESCs. The results were shown in scatter plots, which further demonstrated that the hPMSC-iPSCs were highly similar to human ESCs but completely different from the parental hPMSCs at the global transcriptional level (Figure 4A).
Figure 4.
Gene expression profiling and promoter methylation analysis in hPMSC-iPSCs. A. MA plots are shown to compare the global gene expression profiles between hPMSCs and hPMSC-iPSCs, and between human ESCs and hPMSC-iPSCs. B. Results from bisulfite genomic sequencing are presented to show the CpG methylation status in the promoter regions of OCT4 and NANOG in hPMSCs, hPMSC-iPSCs and human ESCs. Open and closed circles indicate unmethylated and methylated CpGs, respectively.
To confirm the epigenetic remodeling in reprogrammed cells, we evaluated the methylation status of CpG dinucleotides in promoters of ESC-specific genes OCT4 and NANOG. Results from bisulfite sequencing analysis showed that the OCT4 and NANOG promoter regions were demethylated in hPMSC-iPSCs relative to the parental hPMSCs and were thus similar to those in human ESCs (Figure 4B). These results further suggested that the hPMSC-iPSCs generated by gene reprogramming possessed ESC-like characteristics.
Pluripotency and karyotype analysis of hPMSC-iPSCs
To examine the in vitro differentiation potential of hPMSC-iPSCs, we evaluated the spontaneous formation of EBs from hPMSC-iPSCs in a suspension culture. EBs were clearly visible after 8 days in suspension (Figure 5A). After transferring to gelatin-coated plates and culturing for additional 8 days in differentiation medium, RNA was isolated from these differentiated cells, and RT-PCR was performed to examine the expression of genes specific for the three germ layers, including AFP (endoderm), PDX1 (endoderm), GATA4 (mesoderm), BRACHYURY (mesoderm), MAP2 (ectoderm), and PAX6 (ectoderm). As shown in Figure 5B, all these lineage-specific genes were upregulated, while the ESC-specific genes OCT4 and NANOG were downregulated in the differentiated cells. Immunofluorescence staining showed that the detected cells were positive for β III-tubulin (ectoderm), α-SMA (mesoderm) and AFP (endoderm) (Figure 5C-E). These results suggested that the hPMSC-iPSCs were capable of differentiating into various cell types in vitro.
Figure 5.
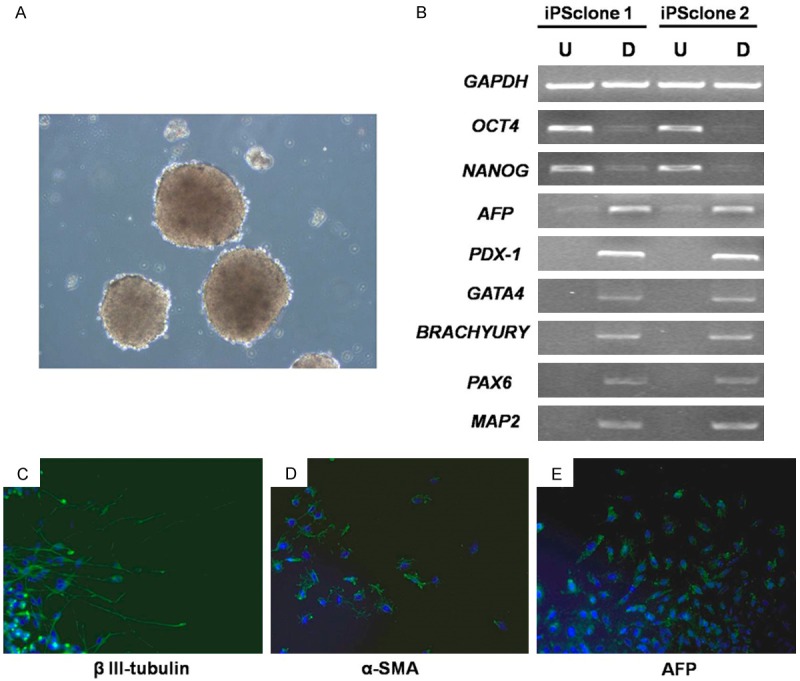
In vitro differentiation of hPMSC-iPSCs. A. hPMSC-iPSCs were grown in suspension and EB formation (three-dimensional aggregates of cells) was detected after 8 days. B. RT-PCR analysis of various differentiation markers for the three germ layers in two iPS clones. U: Undifferentiated; D: Differentiated. Expression of GAPDH was used as internal control. C-E. Immunofluorescence staining showed differentiation of hPMSC-iPSCs into cells expressing markers of the three germ layers.
To test for pluripotency in vivo, hPMSC-iPSCs were transplanted into the hind limbs of SCID mice. Teratomas were found eight weeks following inoculation. Histological examination revealed that the teratomas contained the derivatives of all three germ layers, including neural epithelium, cartilage and glandular epithelium (Figure 6A-C). Immunohistochemistry staining confirmed the presence of derivatives of all three germ layers within the tumor, including neural epithelium, muscle, and glandular epithelium (Figure 6D-F). This result demonstrated the in vivo differentiation ability of hPMSC-iPSCs. Karyotyping was conducted on iPSCs at passage 5 and 60, and revealed a normal karyotype of 46XY (Figure 6G, 6H).
Figure 6.
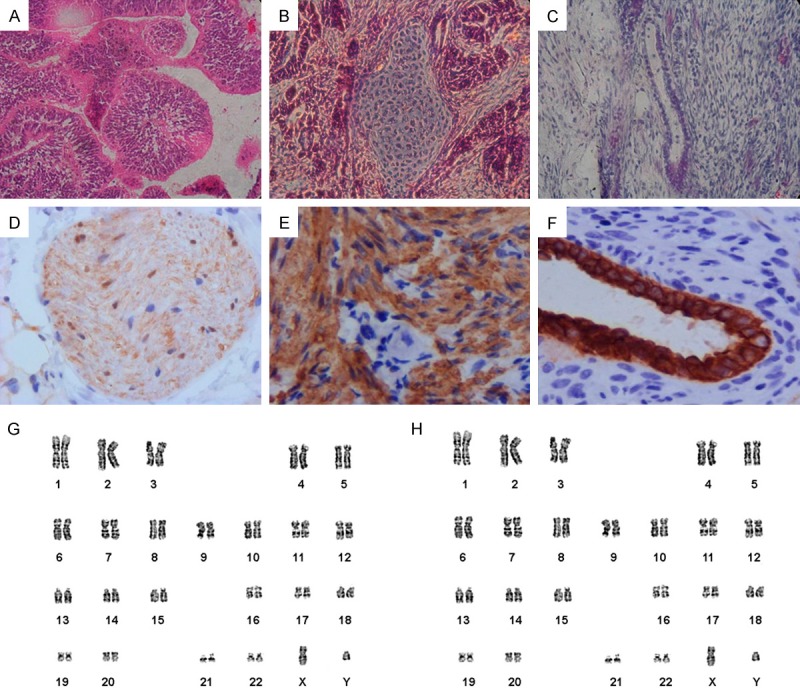
In vivo differentiation and karyotype analysis of hPMSC-iPSCs. (A-C) H&E staining of teratomas derived from hPMSC-iPSCs revealed the presence of differentiated tissues from all three germ layers: neural epithelium (C), cartilage (D), and glandular epithelium (E). (D-F) Immunohistchemistry staining of teratomas indicated positive for anti-β-III-tubulin (D), smooth muscle actin (E), pan-cytokeratin (F). (G, H) Karyotyping of hPMSC-iPSCs at passage 5(G) and passage 60 (H).
Discussion
Since its discovery, iPSC technology holds a great promise for regenerative medicine. Genetic reprogramming through ectopic expression of key transcription factors restores pluripotency in mature somatic cells, which gives them potential to differentiate into any desired cell types. To date, a variety of human cell types have been tested to be reprogrammed into iPSCs. Here, a novel human cell source for iPSCs production has been reported. In this study, we showed that the hPMSCs isolated from the parotid glands in patients with squamous cell carcinoma of the oral cavity could be reprogrammed into iPSCs using an integration-free system.
The first step towards efficient reprogramming of somatic cells and generation of iPSCs is the choice of ideal donor cells. Fibroblast is the first cell source used as a proof of concept that revolutionized the field of stem cell research [1]. Fibroblasts are easy to transfect and maintain in culture, but the reprogramming efficiency is indicated at 0.01-0.5%, which is relatively low. Great efforts have then been made to find out the reprogramming capacity of different cell types. Recently, multipotent stem cells and tissue-specific progenitor cells have been tested for iPSC generation and the results shown superior reprogramming capacity of those cells [30-32]. The increased reprogramming efficiency is likely due to the self-renewal capacity and existing transcriptional and epigenetic changes that contribute to the stemness of these cells.
In our study, we adopted a previously published plastic adhesion protocol to isolate hPMSCs from parotid glands without any selection [33]. The resulting hPMSCs showed homogenous mesenchymal morphology and were positive for expression of CD44, CD71 and CD105, which are generally considered as MSC markers [34]. The multipotency of cultured hPMSCs was confirmed by induced lineage differentiation in conditioned media. The therapeutic potential of MSCs in cell-based regenerative medicine has been highlighted in many studies [35]. Although hPMSCs are multipotent and can be induced to differentiate into several lineages including osteoblasts, chondrocytes and adipocytes as shown in the present study, the number of hPMSCs that can be isolated from parotid glands is limited and it remains difficult to expand these cells and maintain their multipotency in culture [36]. In addition, a growing body of evidence from recent investigation has challenged the view that organ-specific stem cells can give rise to the entire organ or all the cell types within that organ [37]. Consequently, the pluripotent and ES-like iPSCs appears to be a better alternative for tissue engineering, regenerative medicine and cell-based therapy. The hPMSC-iPSCs generated in our study maintained their ES-like morphology and expression of stem cell-specific markers including OCT4, NANOG, SOX2, SSEA-4, TRA-1-60 and TRA-1-81 after 30 passages. Global gene expression profiling analysis showed that the transcriptome of hPMSC-iPSCs closely resembled the human ESCs but significantly different from the parental hPMSCs. The promoter regions of OCT4 and NANOG were similarly hypomethylated in hPMSC-iPSCs and human ESCs, while hypermethylated in the parental hPMSCs. The pluripotency and differentiation capacity of hPMSC-iPSCs after long-term culture was demonstrated by in vitro and in vivo assays. With growing knowledge of stem cell biology, we think iPSCs possess many advantages over MSCs in terms of proliferative capacity, sustainability, multi-lineage differentiation potential, and acquisition of other ESC-like properties.
Fibroblast is the most commonly used cell type for iPSC generation. In recent years, hematopoietic stem cells, squamous cells, keratinocytes and hepatocytes have been studied for the reprogramming capacity and differentiation potential [38]. However, if iPSCs indeed have memory of the donor tissue [23], when it comes to parotid gland regeneration, hPMSC-iPSCs should be the best candidate for producing parotid gland cells including epithelial, ductal and acinar cells to treat parotid gland-related problems. In the next investigation, we will assess the efficiency of hPMSC-iPSCs in regenerating damaged parotid glands and restoration of impaired parotid gland function. Future studies will focus on the use of hPMSC-iPSCs in generating other tissues or organs. Such study will be of great interest to development of novel therapy in regenerative medicine.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant 81070844, 31571517), Training program of High levels of health technical personnel Beijing (No. 2013-3-065).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratajczak MZ, Zuba-Surma EK, Wysoczynski M, Wan W, Ratajczak J, Wojakowski W, Kucia M. Hunt for pluripotent stem cell -- regenerative medicine search for almighty cell. J Autoimmun. 2008;30:151–162. doi: 10.1016/j.jaut.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci U S A. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengerke C, Daley GQ. Autologous blood cell therapies from pluripotent stem cells. Blood Rev. 2010;24:27–37. doi: 10.1016/j.blre.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 10.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 12.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 13.Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–643. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 14.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT, Wu JC. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zhao H, Lan F, Lee A, Chen L, Lin C, Yao Y, Li L. Generation of human-induced pluripotent stem cells from gut mesentery-derived cells by ectopic expression of OCT4/SOX2/NANOG. Cell Reprogram. 2010;12:237–247. doi: 10.1089/cell.2009.0103. [DOI] [PubMed] [Google Scholar]
- 16.Zhao HX, Li Y, Jin HF, Xie L, Liu C, Jiang F, Luo YN, Yin GW, Li Y, Wang J, Li LS, Yao YQ, Wang XH. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation. 2010;80:123–129. doi: 10.1016/j.diff.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, Hao L, Chan YC, Ng KM, Ho JC, Wieser M, Wu J, Redl H, Tse HF, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 19.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgenefree human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Almeida Pdel V, Gregio AM, Machado MA, de Lima AA, Azevedo LR. Saliva composition and functions: a comprehensive review. J Contemp Dent Pract. 2008;9:72–80. [PubMed] [Google Scholar]
- 27.Carpenter GH, Cotroneo E. Salivary gland regeneration. Front Oral Biol. 2010;14:107–128. doi: 10.1159/000313710. [DOI] [PubMed] [Google Scholar]
- 28.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 31.Yan X, Qin H, Qu C, Tuan RS, Shi S, Huang GT. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells Dev. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebau S, Mahaddalkar PU, Kestler HA, Illing A, Seufferlein T, Kleger A. A hierarchy in reprogramming capacity in different tissue microenvironments: what we know and what we need to know. Stem Cells Dev. 2013;22:695–706. doi: 10.1089/scd.2012.0461. [DOI] [PubMed] [Google Scholar]
- 33.Rotter N, Oder J, Schlenke P, Lindner U, Bohrnsen F, Kramer J, Rohwedel J, Huss R, Brandau S, Wollenberg B, Lang S. Isolation and characterization of adult stem cells from human salivary glands. Stem Cells Dev. 2008;17:509–518. doi: 10.1089/scd.2007.0180. [DOI] [PubMed] [Google Scholar]
- 34.Chagraoui J, Lepage-Noll A, Anjo A, Uzan G, Charbord P. Fetal liver stroma consists of cells in epithelial-to-mesenchymal transition. Blood. 2003;101:2973–2982. doi: 10.1182/blood-2002-05-1341. [DOI] [PubMed] [Google Scholar]
- 35.Wei X, Yang X, Han ZP, Qu FF, Shao L, Shi YF. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol Sin. 2013;34:747–754. doi: 10.1038/aps.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung S, Panchalingam KM, Rosenberg L, Behie LA. Ex vivo expansion of human mesenchymal stem cells in defined serum-free media. Stem Cells Int. 2012;2012:123030. doi: 10.1155/2012/123030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGFbeta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raab S, Klingenstein M, Liebau S, Linta L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014;2014:768391. doi: 10.1155/2014/768391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.