Abstract
MicroRNAs-491-5p (miR-491-5p) has been found to involve in tumor initiation and development in several tumors. However, the biological function and underlying molecular mechanism of miR-491-5p in non-small lung cancer (NSCLC) remain unclear. This study was therefore to investigate biological role of and underlying molecular mechanisms of in NSCLC. It was found that miR-491-5p expression was significantly downregulated in NSCLC tissues when compared with corresponding adjacent normal tissues (P<0.01), and the value was negatively related to advanced and tumor-node-metastasis (TNM) stage and lymph node metastasis (both P<0.01). We also demonstrate that restoration of miR-491-5p suppressed NSCLC cell proliferation by arresting NSCLC cells in the G1/G0 phase and accelerating apoptosis. miR-491-5p also inhibited cell migration and invasion in NSCLC cells. Mechanically, IGF2BP1 was identified as direct targets of miR-491-5p. And IGF2BP1 expression was significantly upregulated, and correlated negative with miR-491-5p expression in NSCLC tissues. In vivo assay showed thatmiR-491-5p suppressed tumor growth in nude model by repressing IGF2BP1 expression. Collectively, miR-491-5p functioned as a tumor suppressor in NSCLC by targeting IGF2BP1. Restoration of miR-491-5p expression may represent a promising therapeutic approach for targeting malignant NSCLC.
Keywords: Non-small lung cancer, miR-491-5p, proliferation, invasion, IGF2BP1
Introduction
Non-small cell lung cancer (NSCLC), including adenocarcinoma (ADC), squamous cell carcinoma (SCC), adenosquamous cell carcinoma (ASC) and large cell carcinoma (LCC), represents the most frequent type of lung cancer, and accounts for approximately 80-85% of all lung cancer cases [1,2]. Despite radiotherapy, chemotherapy, and surgery have been recently used as standard treatment modalities for patients with NSCLC, the overall 5-year survival rate for NSCLC patients remains at approximately 15% since NSCLCs are often insensitive to chemotherapy and radiotherapy, and are fast-growing and highly invasive [3]. Therefore, it is urgent need to understand the molecular mechanisms underlying NSCLC development and progression for this disease diagnosis and treatment.
MicroRNAs (miRNAs) are small (18-25nt), single stranded, noncoding RNAs that regulate gene expression at post-transcriptional level by targeting the 3’-untranslated regions (3’UTR) of mRNA and leading to their translational inhibition or deregulation [4]. Growing evidence showed that miRNAs can regulate various biological processes, such as cell differentiation, proliferation, apoptosis, stress resistance, fat metabolism, and development [5,6]. It has been demonstrated that miRNAs involved in tumorigenesis and tumor progression, and function as oncogene or tumor suppressor by regulating tumor cell proliferation, cell cycle, apoptosis and metastasis [7,8]. miRNAs have been suggested to play crucial roles in NSCLC pathogenesis [9,10], which may potentially serve as novel NSCLC diagnostic markers and therapeutic agent.
miR-491, has been found to be lost in several cancers, such as hepato-cellular carcinoma (HCC) [11], glioblastoma [12], colorectal cancer [13] and breast cancer [14]. miR-491-5p, a mature form of miR-491, has been showed to suppress the growth and metastasis of ovarian cancer, pancreatic cancer, breast cancer and cervical cancer by targeting Bcl-XL, TP53, JMJD2B and hTERT genes, respectively [15-18]. These results suggesting that miR-491-5p functions as tumor suppressor in these type cancers. However, the biological role andprecise mechanisms of miR-491-5p in the progression of NSCLC have not been reported until now. Therefore, in the present study, we investigated the exact roles of the miR-491-5p and its underlying molecular mechanisms in NSCLC.
Materials and methods
Patients and tissue samples
36 paired NSCLC tissues and their corresponding adjacent normal lung tissues were obtained from patients who underwent NSCLC resection without preoperative treatment at Department of Thoracic Surgery, the First Hospital of Jilin University (Changchun, China), from April 2008 and June 2014, after receiving adequate informed consent. The corresponding adjacent normal tissues were obtained beyond 3 cm away from the boundary of NSCLC tissues. The tumor tissues and adjacent normal tissues were snap-frozen in liquid nitrogenimmediately after resection and stored at -80°C until use. The study was approved by the Medical Ethics Committee of Jilin University.
Cell lines and culture
Four human lung cancer cell lines (A549, H1299, SPCA1 and H358) and normal lung cell (BEAS-2B) were brought from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and were routinely maintained in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Life Technologies, Inc., Grand Island, NY, USA), 100 U/ml penicillin sodium, and 100 mg/ml streptomycin sulfate at 37°C in a humidified air atmosphere containing 5% CO2. Cells were used when they were in the logarithmic growth phase.
RNA extraction and quantitative PCR
Total RNA containing miRNA and mRNA from tissue samples and cells was extracted by using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s instruction. For detection of miR-491-5p expression, complementary DNA (cDNA) was synthesized using the Taqman miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA), then were quantified using the miScript SYBR Green PCR kit (Qiagen, Hilden, Germany) under an ABI 7900 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). The primer of miR-491-5p and U6 were used as previously [18]. For detection of IGF2BP1 expression, cDNA synthesis was performed using PrimeScript RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions, then was quantified using Real-time PCR Mixture Reagent (Takara) under ABI 7900 Fast system. The primers for IGF2BP1 and GAPDH were used as previous described [19]. The relative expression levels of miR-491-5p and IGF2BP1 were calculated by the 2-ΔΔCt method. U6 and GAPDH were used as internal controls for miRNAs and mRNAs, respectively.
Cell transfection
miR-491-5pmimic (miR-491-5p) and corresponding negative control (miR-NC) (si-NC) were purchased from Ribobio (Guangzhou, Guangdong, China), and were transfected into A549 cells when cells were grown to 80-90% confluence, using the Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions at final concentration of 100 nM.
MTT assay
A total of 2×104 cells were plated onto 96-well plates overnight and transfected with 100 nM miR-491-5p or miR-NC. 10 μl of sterile MTT dye (5 mg/ml) was added into each well containing 100 ul medium after cells were cultured for 24 h-72 h. The cells were then incubated at 37°C for 4 h in a 5% CO2 incubator. Then MTT solution were removed and 150 μl dimethyl sulfoxide (DMSO, Sigma-Aldrich) were added to each well to dissolve the crystals for 10 min at 37°C. The absorbance in each well was measured using spectrophotometric analysis (BioTek, Grand Island, NY, USA) at 490 nm.
Cell cycle and cell apoptosis assay
The cells were seeded into 6-well plates and then transfected with the miR-491-5p mimic or miR-NC. After 48 h, the cells were harvested using trypsinization, washed in ice-cold PBS, and fixed in ice-cold ethanol in PBS. Then bovine pancreatic RNase (Sigma-Aldrich) was added to a final concentration of 2 mg/ml, and cultured at 37°C for 4 h in a 5% CO2 incubator for 30 min. For the cell cycle distribution, the cells were stained with 20 mg/ml propidium iodide (PI; Sigma-Aldrich) for 20 min at room temperature. For the cell apoptosis assays, the cells apoptosis was determined using the Annexin VApoptosis Detection Kit (Invitrogen) according to the manufacturer’s instructions. The cell cycle distribution and the cell apoptosis rates were quantified using an FACS Calibur flow cytometer (BD Biosciences, Mansfield, MA, USA).
Cell migration and invasion assay
To examine the migration ability of cells in vitro, a wound-healing assay was performed. In briefly, the 2×104 transfected cells were seeded into 24-well tissue culture plates for 24 h. Cells were then scratched using a sterile plastic micropipette tip to create an artificial wound. Migration of cells into the wound was observed and photographed under an inverted microscope at indicated time (0 h and 24 h).
Cell invasion was determined using 24-well Matrigel invasion chambers (Becton Dickinson) according to the manufacturer’s instructions. In briefly, transfected cells (5×104) were seeded per well in the upper well of thematrigel-coated invasion chamber in DMEM without serum. The lower chamber was filled with DMEM medium with 10% FBS to attract cells. After cells had been cultured at 37°C for 48 h, non-invading cells were removed from the top well with a cotton swab, while the bottom cells were fixed with 70% ethanol for 30 min and stained with 0.1% crystal violet for 10 min. The invaded cells were photographed and were counted in five randomly fields for each well under light microscope (Olympus, Tokyo, Japan).
Luciferase activity assay
The wild-type 3’-UTR segment of the IGF2BP1 mRNA (not the full length of IGF2BP1 3’-UTR) containing a putative miR-491-5p-binding site, was amplified using PCR and subcloned intopGL3-control vector (Ambion, Austin, TX, USA) at the NheI and XhoI restriction sites, termed as: Wt-IGF2BP1-3’UTR. Amutant construct in miR-491-5p binding sites of IGF2BP1 3’UTR region also was generated using Quick Change Site-Directed Mutagenesis Kit (Agilent, Roseville City, CA), and subcloned intopGL3-control vector (Ambion, Austin, TX, USA) at the NheI and XhoI restriction sites, and named as Mut-IGF2BP1-3’UTR. For reporter assay, A549 cells were plated onto 24-well plate (2×104 cells/well) and transiently co-transfected with Wt/MutIGF2BP1 reported plasmid, along with miR-491-5p/miR-NC using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured 48 h after transfectionby the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase was used to normalize the Renilla luciferase.
Western blot
Cells or tissues were washed twice with ice-cold phosphate buffered saline, and lysed in RIPA buffer (Pierce, Waltham, MA, USA) on ice for 30 min, and then centrifuged at 13,000 g for 30 min. After centrifugation, proteins in the supernatants were quantified using a bicinchoninic acid (BCA) protein assay kit (Boster, China). Equal quantities (30 μg) of protein samples were loaded on 10% SDS-PAGE and transferred onto PVDF membrane (Milipore, Lake Placid, NY, USA). After blocked in 5% BSA, the membrane was incubated with antibody mouse anti-human IGF2BP1 (1:1000 dilution, Santa Cruz Biotechnology Inc., California, USA) and antibody mouse anti-human GAPDH overnight at 4°C, followed incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5000 dilution, Santa Cruz, USA) for 1 h at room temperature. Protein band were determined with chemiluminescence detection system (Pierce) and visualized on Bio-Rad ChemiDocXRS (Bio-Rad Laboratories, Hercules, CA, USA).
Xenografted tumor model
Ten of five-week-old BALB/C nude male mice (20-25 g) were brought from the Experimental Animal Center of Changchun Institute for Biological Sciences (Changchun, China). All mice were maintained in the pathogen-free (SPF) conditions. All animal experiments were approved by National Institutes of Health Animal Care and the Use Committee guidelines of the Jilin University (Changchun, China).
For the in vivo tumor assay, 2×106 A549 cells stably expressing the miR-491-5p or miR-NC were collected and suspended in 0.2 ml PBS for each mouse (five in each group), and the cells were injected into left side of the posterior flank of nude mouse. Tumors growth were measured with calipers to estimate volume from day 7 to day 35 after injection according to the formula Volume (mm3) =1/2 width2 × length. The animals were sacrificed after 35 days and the tumor tissue were removed for determination miR-491-5p and IGF2BP1 expression.
Statistical analysis
All data are presented as the means ± SD (standard deviation) from at least three independent experiments. Unpaired Student’s t test was used to determine the significance, using the GraphPadPrism version 6.0 software (GraphPad SoftwareInc., San Diego, CA, USA) and the SPSS 16.0 software (SPSS, Chicago, IL, USA). For all analyses, P<0.05 was considered statistically significant.
Results
MiR-491-5p expression was downregulated in NSCLC and associated with advanced clinical stage and NSCLC metastasis
To understand the potential biological significance of altered miR-491-5p expression in NSCLC progression, we evaluated miR-491-5p expression in 36 pairs of human NSCLC tissues and their corresponding normal tissues by quantitative RT–PCR (qRT-PCR). It was found that expression of miR-491-5p in NSCLC was significantly lower than that of corresponding adjacent normal tissue (Figure 1A). Then the relationship between the miR-491-5p expression levels and the clinicopathological characteristics of the NSCLC patients were investigated. As showed in Table 1, no significant correlations between miR-491-5p expression and age, gender, or tumor size were found, while we found that miR-491-5p expression was negatively associated with advanced TNM stage and lymph node metastasis. In addition, we also assessed the expression of miR-491-5p in four lung cancer line (A549, H1299, SPCA1 and H358) and normal lung cell (BEAS-2B), and found that the expression level of miR-491-5p was decreased in all four NSCLC cell lines compared with BEAS-2B (Figure 1B). The A549 cell line has lowest expression of miR-491-5p, thus it was selected for below study.
Figure 1.

miR-491-5p expression is downregulated in non-small lung cancer (NSCLC) tissues and cell lines. A. Detection of miR-491-5p expression in 36 paired NSCLC tissues corresponding by quantitative RT-PCR (qRT-PCR) **P<0.01 compared to normal tissue. B. Detection of miR-491-5p expression in four human NSCLC cell lines (A549, H1299, SPCA1 and H358) and normal lung cell (BEAS-2B) by qRT-PCR. *P<0.05; **P<0.01 compared to BEAS-2B.
Table 1.
Correlation between miR-491-5p status and clinical characteristics in patients with NSCLC
| Feature | N | miR-491-5p expression | P value |
|---|---|---|---|
| Age(years) | P>0.05 | ||
| <60 | 16 | 0.35±0.08 | |
| ≥60 | 20 | 0.37±0.09 | |
| Gender | P>0.05 | ||
| Female | 17 | 0.38±0.11 | |
| Man | 19 | 0.34±0.07 | |
| Tumor size | P>0.05 | ||
| T1/T2 | 23 | 0.41±0.11 | |
| T3/T4 | 13 | 0.27±0.05 | |
| Lymph node metastasis | P<0.01 | ||
| No | 26 | 0.46±0.13 | |
| Yes | 10 | 0.11±0.02 | |
| TNM stage | P<0.01 | ||
| I-II | 22 | 0.49±0.13 | |
| III-IV | 14 | 0.15±0.03 |
miR-491-5p inhibited cell proliferation in NSCLC cells
To elucidate potential effects of miR-491-5p in the progression of NSCLC, miR-491-5p was over-expressed in A549 cells (Figure 2A) by transfection of miR-491-5p. Alterations in cell proliferation, cell cycle distribution and apoptosis induced by miR-491-5p were then examined. MTT assays revealed that restoration of miR-491-5p significantly decreased cell proliferation of A549 cells at the second and third days (Figure 2B, P<0.05). To explore the possible mechanisms of miR-491-5p’s function in cell proliferation, we determined the cell cycle distribution of A549 cells transfected with miR-491-5p using flow cytometry, and found that overexpression of miR-491-5p in A549 cells induced a significant increase in the percentage of cells inthe G1/G0 peak and a decrease in the percentage of cells inthe S peaks (Figure 2C). In addition, we also measured cell apoptosis ratio in A549 cells after transfected with miR-491-5p. It was found that restoration of miR-491-5p in A549 cells obviously increased the rate of apoptosis (Figure 2D). These results suggested that miR-491-5p could inhibit cell proliferation by arresting the tumor cells at the G1/G0 phase and increasing tumor cell apoptosis.
Figure 2.

miR-491-5p inhibited cell growth in NSCLC cells. A. The miR-491-5p expression level in A549 cells after transfected with miR-491-5p or miR-NC were detected by qRT-PCR. B-D. The effect on cell proliferation, cell cycle distribution and apoptosis of miR-491-5p overexpression in A549 cells were determined. *P<0.05, **P<0.01 compared with miR-NC.
miR-491-5p inhibited cell migration and invasion in NSCLC cells
We next assessed the effect of miR-491-5p on the migration and invasion of NSCLC cells by wound healing and invasion chamber assay, respectively. The results showed that overexpression of miR-491-5p in A549 cells markedly decreased the migratory and invasive capabilities (P<0.05, Figure 3A and 3B).
Figure 3.

miR-491-5p inhibited cell migration and invasion in NSCLC cells. A. The effect on cell migration of miR-491-5p overexpression in A549 cells were determined by wound healing assay. B. The effect on cell invasion of miR-491-5p overexpression in A549 cells were determined bychamber invasion analyses. **P<0.01 compared with miR-NC.
IGF2BP1 is a direct target of miR-491-5p
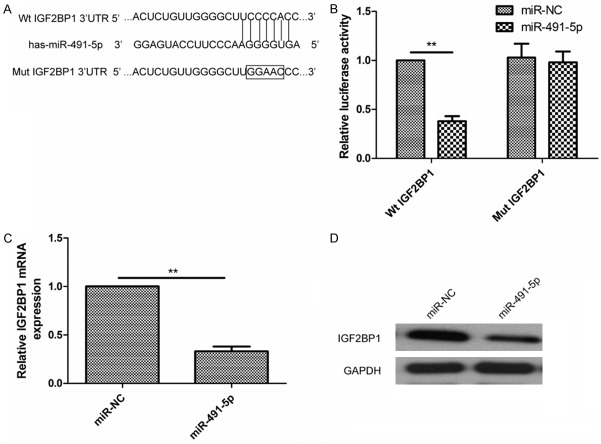
To investigate the underlying molecular mechanism of growth inhibition by miR-491-5p in NSCLC cells, we identified the potential targets of miR-491-5p using two publicly available algorithms (Targetscan6.2 and miRanda). IGF2BP1, one of the predicted potential targets of miR-491-5p, was selected as the candidate for further study due to its with a putative conserved binding site for miR-491-5p at position 763-769 (Figure 4A). To further determine whether miR-491-5p could directly associate with the 3’UTR of IGF2BP1mRNA, we constructed a reporter plasmid harboring the wild-type or mutant binding site sequence in IGF2BP13’-UTR, which located downstream of luciferase reporter gene, then luciferase reporter assay were performed in A549 cells co-transfected with wild type 3’-UTR IGF2BP1 reporter plasmid or mutant type 3’UTR IGF2BP1, along with miR-491-5p mimic or miR-NC. It was found that restoration of miR-491-5p significantly decreased the luciferase activity of the IGF2BP1 3’-UTR-Wt in A549 cells (Figure 4B), while had no inhibition effect on the mutant IGF2BP1-3’UTR reporter activity in A549 cells (Figure 4B). In addition, overexpression of miR-491-5p in A549 cells markedly inhibited the expression of IGF2BP1 at both mRNA and protein level (Figure 4C and 4D). These results suggest that IGF2BP1 is a direct target of miR-491-5p.
Figure 4.
IGF2BP1 was a direct target of miR-491-5p. A. Schematic construction of wild-type (WT) and mutant (Mut) 3’-UTR of IGF2BP1 were shown. B. The effects of miR-491-5p overexpression on the activity of the 3’UTRs of IGF2BP1 in A549 cells were analyzed by the dual luciferase reporter assay. C, D. The effects of miR-491-5p overexpression on expression of IGF2BP1 at mRNA level and protein level was detected by qRT-PCR or western blot. GAPDH was used as an internal control. *P<0.05, **P<0.01 compared with miR-NC.
IGF2BP1 expression was upregulated and inversely correlated with miR-491-5p expression inNSCLC tissues
Since the above results showed that the miR-491-5p could regulate IGF2BP1 expression through directly targeting its 3’UTR in NSCLC cells, we investigated the IGF2BP1 expression in NSCLC tissues and corresponding adjacent normal tissues. It was found that the expression of IGF2BP1 on both mRNA and protein level was upregulated in NSCLC tissues compared with corresponding adjacent tissues (Figure 5A and 5B). Through spearman’s correlation analysis, we found that IGF2BP1 mRNA expression was inversely correlated with miR-491-5p expression in NSCLC tissues (Figure 5C; r=-0.522, P<0.05).
Figure 5.

IGF2BP1 was up-regulated and inversely correlated with miR-491-5p expressionin NSCLC tissues. A, B. IGF2BP1 mRNA expression and protein expression in human NSCLC tissues and their corresponding normal tissues were determined by qRT-PCR and western blot, respectively. GAPDH was used as an internal control. **P<0.01 versus normal tissue. C. The reverse relationship between IGF2BP1 mRNA expression and miR-491-5p expression was analyzed in NSCLC tissues by Spearman’s correlation.
miR-491-5p suppressed tumor growth in a mouse xenograft model by repressing IGF2BP1
Finally, anin vivo model was applied to evaluate the effect of miR-491-5p restoration ontumorigenicity. A549 cells stable expression miR-491-5p or miR-NC were subcutaneously inoculated in nude mice (n=5 for each group), respectively. It was found that the tumor growth was slower in A549/miR-491-5p group than that of A549/miR-NC group (Figure 6A). When mice were killed 35 days after injection, the tumor tissues were stripped and weighted. We found that the weight of tumor tissues in A549/miR-491-5p group was markedly reduced compared with A549/miR-NC group (Figure 6B). In addition, we also investigate the expression of miR-491-5p and IGF2BP1 expression. It was found that miR-491-5p expression remarkably upregulated, while IGF2BP1 expression obviously downregulated in A549/miR-491-5p group compared to A549/miR-NC group (Figure 6C and 6D). These results suggested that miR-491-5p suppresses tumor growth in a mouse xenograft model by repressing IGF2BP1.
Figure 6.

miR-491-5p suppressed tumor growth in a xenograft model by repressing IGF2BP1. A. Tumor growth curves for tumor volumes in xenografts of nude mice were established based on the tumor volume measured every week until five weeks. B. Tumor tissues weight was measured. C. Detection of miR-491-5p expression in tumor tissue by qRT-PCR. D. Detection of IGF2BP1 protein expression in tumor tissues by Western blot. GAPDH was used as internal control. *P<0.05, **P<0.01 versus the miR-NC group.
Discussion
Accumulating evidence showed that the aberrant expression of miRNAs play crucial roles in the occurrence and development of non-small lung cancer by regulating target genes [9,10,20]. For example, miR-30b/c could inhibit NSCLC cell proliferation by targeting Rab18 [21]; miR-98 suppress proliferation, migration, and invasion of lung cancer cells by directly binding to the 3’-UTR of ITGB3 Mrna [22]. miR-92a could promotes growth, metastasis, and chemoesistance in non-small cell lung cancer cells at least partially by targeting PTEN [23], and so on. In this study, we found that miR-491-5p expression was decreased in NSCLC tissues and cell lines, and that miR-491-5p overexpression inhibited proliferation, migration, and invasion in NSCLC cells by targeting IGF2BP1. These results might provide a new insight into the pathophysiological mechanism of NSCLC and a novel therapy target for NSCLC treatment.
miR-491-5p, a mature form of miR-491, has been found to be downregulated in several cancer, such as ovarian cancer, pancreatic cancer, breast cancer and cervical cancer [15-18], suggesting that miR-491-5p functions as tumor suppressor in these type cancers. However, a report showed that the level of miR-495-5p was increased in colon cancer, especially in patients aged 70 years and older, and high miR-491-5p expression correlated with poor overall survival of patients with colon cancer [24], suggesting that miR-491-5p functions as oncogene in colon cancer. These controversial findings suggest that the role miR-491-5p has in tumor progression depending on theorgan-specific actions and different cellular contexts. However, the biological function and underlying mechanism of miR-491-5p in NSCLC remains largely unclear. Here, we found that the level of miR-491-5p expression is significantly downregulated in NSCLC tissue and cell lines, and its expression level correlated with key pathological characteristics including TNM stage, and lymph node metastasis. Further, function studies revealed thatoverexpression of miR-491-5p inhibited cell proliferation, migration, and invasion and induced cell cycle arrest at G0/G1 stage and apoptosis in vitro, and suppressed tumor growth in a nude mice model. These results suggested that miR-491-5p play a suppressor role in NSCLC procession.
To investigate the possible molecular mechanism of miR-491-5p suppressive NSCLC growth, thus we used two bioinformatics algorithms (TargetScan and miRanda algorithm) to predict gene targets for miR-491-5p. IGF2BP1 were selected as the potential target of miR-491-5p for further validation since IGF2BP1 had been reported to involve in NSCLC procession and development [25]. IGF2BP1, as an RNA binding protein, negatively regulates IGF2 mRNA [26], has been showed to act as oncogene in various cancer, including NSCLC [27]. In addition, IGF2BP1 was identified as a target of several miRNAs, including miR-494 [27], miR-150 [19] miR-625 [28], miR-196b [29] and miR-873 [30]. Here we further confirmed that IGF2BP1 was a target of miR-491-5p by luciferase reporter assays, qRT-PCR and western blotting assay. In addition, we also confirmed that IGF2BP1 expression was upregulated in NSCLC tissue, is negatively correlated with miR-491-5p in NSCLC tissues. Collectively, these results suggested that miR-491-5p exerts suppressive function partially by targeting IGF2BP1.
In summary, the present study demonstrated that miR-491-5p was downregulated in NSCLC tissues and cell lines, and its expression level was significantly negative corrected with TNM stage and lymph node metastasis, and that restoration of miR-491-5p in NSCLC cells drastically decreased cell proliferation, migration, invasion, increased cell apoptosis and cell cycle arrest at G0/G1 stage in vitro, as well as suppressed tumor growth in nude mice model by targeting IGF2BP1. These findings suggested that miR-491-5p functions as a tumor suppressor in NSCLC by repressing IGF2BP1 expression, and that miR-491-5p might serve as a promising therapeutic target in NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Carrizosa DR, Gold KA. New strategies in immunotherapy for non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:553–559. doi: 10.3978/j.issn.2218-6751.2015.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNAMediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skrzypski M, Dziadziuszko R, Jassem J. MicroRNA in lung cancer diagnostics and treatment. Mutat Res. 2011;717:25–31. doi: 10.1016/j.mrfmmm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Guan P, Yin Z, Li X, Wu W, Zhou B. Metaanalysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res. 2012;31:54. doi: 10.1186/1756-9966-31-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Qiu Y. Ginsenoside Rh2 Targets EGFR by Up-Regulation of miR-491 to Enhance Anti-tumor Activity in Hepatitis B Virus-Related Hepatocellular Carcinoma. Cell Biochem Biophys. 2015 doi: 10.1007/s12013-014-0456-9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Li X, Liu Y, Granberg KJ, Wang Q, Moore LM, Ji P, Gumin J, Sulman EP, Calin GA, Haapasalo H, Nykter M, Shmulevich I, Fuller GN, Lang FF, Zhang W. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2015;34:1619–1628. doi: 10.1038/onc.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano H, Miyazawa T, Kinoshita K, Yamada Y, Yoshida T. Functional screening identifies a microRNA, miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal cancer cells. Int J Cancer. 2010;127:1072–1080. doi: 10.1002/ijc.25143. [DOI] [PubMed] [Google Scholar]
- 14.Leivonen SK, Sahlberg KK, Makela R, Due EU, Kallioniemi O, Borresen-Dale AL, Perala M. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104. doi: 10.1016/j.molonc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denoyelle C, Lambert B, Meryet-Figuiere M, Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH, Gauduchon P, Juin P, Poulain L. miR-491-5p-induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL-XL and EGFR leading to BIM activation. Cell Death Dis. 2014;5:e1445. doi: 10.1038/cddis.2014.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo R, Wang Y, Shi WY, Liu B, Hou SQ, Liu L. MicroRNA miR-491-5p targeting both TP53 and Bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules. 2012;17:14733–14747. doi: 10.3390/molecules171214733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui Z, Yiling C, Wenting Y, XuQun H, ChuanYi Z, Hui L. miR-491-5p functions as a tumor suppressor by targeting JMJD2B in ERalphapositive breast cancer. FEBS Lett. 2015;589:812–821. doi: 10.1016/j.febslet.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, Zhai YX, Liu HQ, Shi YA, Li XB. MicroRNA-491-5p suppresses cervical cancer cell growth by targeting hTERT. Oncol Rep. 2015;34:979–986. doi: 10.3892/or.2015.4013. [DOI] [PubMed] [Google Scholar]
- 19.Qu Y, Pan S, Kang M, Dong R, Zhao J. MicroRNA-150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biol. 2015 doi: 10.1007/s13277-015-4389-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Wu J, Li Y, Li X, Yang T, Yang Q, Jiang Y. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int. 2014;2014:364316. doi: 10.1155/2014/364316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong K, Chen K, Han L, Li B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. doi: 10.1186/1471-2407-14-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni R, Huang Y, Wang J. miR-98 targets ITGB3 to inhibit proliferation, migration, and invasion of non-small-cell lung cancer. Onco Targets Ther. 2015;8:2689–2697. doi: 10.2147/OTT.S90998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren P, Gong F, Zhang Y, Jiang J, Zhang H. MicroRNA-92a promotes growth, metastasis, and chemoresistance in non-small cell lung cancer cells by targeting PTEN. Tumour Biol. 2015 doi: 10.1007/s13277-015-4150-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 25.Lukanova A, Toniolo P, Akhmedkhanov A, Biessy C, Haley NJ, Shore RE, Riboli E, Rinaldi S, Kaaks R. A prospective study of insulinlike growth factor-I, IGF-binding proteins-1, -2 and -3 and lung cancer risk in women. Int J Cancer. 2001;92:888–892. doi: 10.1002/ijc.1265. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohdaira H, Sekiguchi M, Miyata K, Yoshida K. MicroRNA-494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 2012;45:32–38. doi: 10.1111/j.1365-2184.2011.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ, Liu LL, Yi C, Fu J, Hu W, Wen JM, Yun JP. miR-625 suppresses tumour migration and invasion by targeting IGF2BP1 in hepatocellular carcinoma. Oncogene. 2015;34:965–977. doi: 10.1038/onc.2014.35. [DOI] [PubMed] [Google Scholar]
- 29.Rebucci M, Sermeus A, Leonard E, Delaive E, Dieu M, Fransolet M, Arnould T, Michiels C. miRNA-196b inhibits cell proliferation and induces apoptosis in HepG2 cells by targeting IGF2BP1. Mol Cancer. 2015;14:79. doi: 10.1186/s12943-015-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL, Zhang MH, Liang HQ. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]