Abstract
MicroRNA-223 plays an important role in the inflammatory response of macrophages. Recent studies have identified that miR-223 was highly expressed in H. pylori infection macrophages, the significance of the elevation, however, has not yet been investigated. In this study, we analyzed the impact of elevated miR-233 to macrophage inflammatory response and possible mechanisms. We found that miR-223 not only could inhibit the expression of inflammatory cytokines including IL-6, IL-8, IL-12 and TNF-α, but also was able to decrease the expression of CD40, CD68, CD80, and CD163. Furthermore, proteins relating to inflammatory signal pathways, such as IRAK-1, NF-κB and MAPK, in H. pylori infected macrophages were down-regulated. Taken together, these results indicated that miR-223 may act as an inflammatory inhibitory factor in H. pylori infected macrophages by IRAK-1, NF-κB or MAPK signal pathways. These findings contribute to the understanding of miR-223 in macrophages inflammatory responses induced by H. pylori.
Keywords: Macrophage, miR-223, Helicobacter pylori
Introduction
Helicobacter pylori (H. pylori) is a gram-negative microaerophilic bacterium causing a lifelong infection worldwide. It could inhabit in the stomach and cause gastritis, peptic ulcer, and gastric cancer [1]. In acute or chronic H. Pylori infection, inflammation is thought to be a major determinant of both peptic ulceration and gastric malignancy [2]. However, the regulatory mechanisms of H. pylori-induced inflammation are still not well understood.
In response to infection, the host launches a vigorous immune response, including the mucosal infiltration of neutrophils, lymphocytes, and macrophages. Host defense against pathogens requires the induction of appropriate innate immune responses, while excessive or inappropriate activation of the immune system can be deleterious to the host itself. Therefore, various immune regulators, including microRNAs (miRNA), also are involved in the immune responses [3].
MicroRNAs (miRNAs) are non-coding 20~22 nucleotide small RNAs that regulate gene expression by inducing translational repression or degradation of their mRNA targets [4]. Recent studies showed that a range of miRNAs, such as miR-155, miR-146a, miR-223 have critical functions in the regulation of inflammatory responses in macrophages or monocytes [5-8]. Changes in miRNA expression in response to bacterial infection have been reported, including H. pylori infection in the gastric mucosa, gastric epithelial cells and immune cells e.g. macrophages, dendritic cells (DCs) [9-12]. Among the reported miRNAs, miRNA-223 (miR-223) is a potent regulator of some inflammatory responses [13,14]. At present, studies have found that miR-223 over-expression inhibited the production of IL-1β from inflammasome [15]. Interestingly, miR-223 was critical for the control of tuberculosis and potentially other chronic inflammatory diseases by regulating leukocyte chemotaxis via chemoattractants [16]. Moreover, miR-223 can suppress the proinflammatory activation of macrophages [17], while the specific regulation mechanisms of miRNA-223 for cellular inflammation remain largely unknown.
Here, we investigated the functions of miR-223 in the inflammatory response of H. pylori infected macrophages, and its signal regulation pathways.
Materials and methods
Macrophage culture
The human acute monocytic leukemia cell line THP-1 was purchased from the Cell Resource Center of Shanghai Academy of Sciences, Chinese Academy of Sciences. Cells were cultured in wells or flasks at 37°C with 5% CO2, in RPMI 1640-GlutaMAX™ (HyClone Laboratories, GE Healthcare Lifesciences, Logan, UT, USA) containing 10% (v/v) fetal bovine serum (HyClone), 100 U/ml penicillin, 0.1 mg/ml streptomycin and 0.25 μg/ml amphotericin B. Differentiation of THP1 cells into macrophage-like cells was induced by stimulation with 0.1 mmol/L phorbol 12-myristate 13-acetate, PMA (Sigma, St Louis, MO, USA) for 24 h.
H. pylori culture
The wild-type H. pylori strain 26695 obtained from the Cell Resource Center of Shanghai Academy of Sciences, Chinese Academy of Sciences was cultivated for 48 h at 37°C under microaerobic conditions (5% O2) on selective agar containing 21.5 g Wilkins Chalgren agar, 50 ml human blood, 10 μg/ml vancomycin, 10 μg/ml cefsulodin, 5 μg/ml trimethoprim, and 10 μg/ml amphotericin B.
Co-culture of macrophages and H. pylori
Immature macrophages were washed once in PBS and plated onto 24-well plastic plates at a density of 5×105 cells per well in 1 ml RPMI-1640. H. pylori were recovered from the agar plates using a swab and resuspended in RPMI-1640 at an optical density of 0.6 at 600 nm, which corresponds to 3×107 CFU/ml. The bacteria were added to macrophages at the indicated multiplicity of infection (MOI) 10:1 and the co-cultures were further incubated at 37°C in a 5% CO2 incubator for 24 h.
Quantitative reverse transcription PCR (qRT-PCR)
Small RNA and total RNA were purified by RNA purification kit according to the manufacturer’s instructions (RNAiso for Small RNA isolation, TaKaRa, China or TRIzol reagent for total RNA isolation, Invitrogen, USA). High quality RNA was used for first-strand cDNA synthesis by reverse transcription using M-MLV reverse transcriptase (TaKaRa, China). The quantitative real-time RT-PCR was performed in the Roche lightcycler (LightCycler 2.0) using TaKaRa SYBR Green I kit. The thermal cycling condition for PCR was 95°C for 30 s, 40 cycles of 95°C for 5 s, 60°C for 20 s and 72°C for 20 s, followed by 40°C for 20 min. The specificity of PCR was determined by sequencing. The following primers were used: miR-223 RT primers 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGACACTCACC-3’, Forward: 5’-GCCGAAACAUUCAACGCUGUC-3’, Reverse: 5’-CAGTGCAGGGTCCGAGGT-3’; U6, RT primers 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3’, Forward: 5’-TGCGGGTGCTCCGCTTCGG CAGC-3’, Reverse: 5’-CAGTGCAGGGTCCGAGGT-3’. All primers were synthetized by Invitrogen (New York, US).
Luciferase assay
Firefly luciferase reporter vectors with or without the intact putative miR-223 recognition sequence from the 3’-UTR of IRAK-1 were constructed using pMIR-REPORT miRNA Expression Reporter Vector (Applied Biosystems). All constructs were confirmed by sequencing. For the 3’UTR-luciferase assays, cells were co-transfected with 0.5 mg pMIR-REPORT or no-3’-UTR construct, 0.5 mg miR-223 mimics, miR-223 inhibitors, negative siRNA and 0.1 mg luciferase TransLipid Transfection Reagent. Twenty-four hours post transfection, luciferase assays were performed by the Dual Luciferase Reporter Assay system (TianGen, Beijing). Relative luciferase activity was normalized with Renilla luciferase activity.
siRNA transfection
THP-1 cells were first cultured in a 12-well plate for 24 h, and then were washed with PBS and resuspended in 500 μL OpitMEM (Naomi Biotech, WuXi, China). Then, 20 pmol miR-223 mimics, miR-223 inhibitors, negative siRNA, or a blank control were mixed with 5 μL IR in 100μL OpitMEM and heat shocked for 10 s at 37°C. Subsequently, the mixed solutions were added to the cells. After 6 h, the medium was replaced and the cells were continued to culture for 24 h. The transfected cells were observed by fluorescence microscopy. Differentiation of these cells into macrophage-like cells was induced with 0.1 mmol/L Phorbol 12-myristate 13-acetate, PMA (Sigma, St Louis, MO, USA) for 24 h.
Analysis of inflammatory cytokine genes
QPCR assays were performed with the Roche lightcycler (LightCycler 2.0). For qPCR, 2 μl cDNA, 0.5 μl forward primer (10 μmol/L), 0.5 μl reverse primer (10 μmol/L), 8 μl SYBR Premix ExTaq (TaKaRa, China) and 9 μl ddH2O were mixed and incubated for 35 cycles under the following cycling conditions: 95°C for 15 s, 52°C for 35 s, 72°C for 20 s. Dissociation curves were used to ensure specific amplification. Relative expression differences were calculated using the 2-ΔΔCt formula: ΔΔCt =ΔCtreference-ΔCtsample. Each condition was performed in duplicates.
Flow cytometry analysis
H. pylori infected macrophages (MOI of 10:1; circles) were first transfected with siRNAs in six well plates for 24 h, and then cells were harvested and washed twice with PBS containing 0.2% BSA. After washes, cells were stained with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-labeled monoclonal antibodies (BD, Franklin Lakes, NJ, USA)) to CD40, CD68 and CD80, CD163 respectively , or the appropriate isotype controls. Macrophages were washed and fixed in 10% (v/v) formaldehyde-PBS. Finally, cell sorting and analysis were performed on a flow cytometry platform (Beckman MOFLO XDP, USA). Median fluorescence intensities (MFI) and the percentages of positive expression cells were analyzed.
Western blotting analysis
Macrophages with or without H. pylori infection were first transfected with siRNAs and then cells were harvested and lysed for western blotting. Equal amount of proteins from cell lysates were loaded and separated by 10% SDS-PAGE gels, and transferred onto a polyvinylidene difluoride membrane (PVDF, Milipore, Bedford, MA). The membrane was blocked with 5% non-fat milk and incubated with antibodies against IRAK-1 (R&D Systems, Minneapolis, MN, USA, 1:400 dilution), NF-κB (R&D Systems, Minneapolis, MN, USA, 1:600 dilution) and MAPK (R&D Systems, Minneapolis, MN, USA, 1:800 dilution). After incubation, membrane was washed and incubated with HRP conjugated secondary antibodies. Immune-detected protein bands were quantified with Image J.
Statistical analysis
All data were expressed as mean ± SEM. Comparisons were performed with One-way ANOVA with SNK post hoc. P<0.05 was considered statistically significant. All analyses were performed using SPSS16.0 (SPSS).
Results
H. pylori influences the expression profile of miRNAs in macrophages
In order to investigate the potential function of miRNAs in response to H. pylori infection, the expression profile of six miRNAs known to be related to innate immunity in macrophages with H. pylori infection was determined by qRT-PCR (Table 1). After H. pylori infection, three miRNAs (miR-21, miR-155, miR-223) were significantly increased (P<0.05). Moreover, miR-223 expression was significantly elevated at all tested time point (P<0.05) (Table 1). These results implied that miRNAs might play an important regulatory role in macrophages against H. pylori infection.
Table 1.
MicroRNA expression profile in Helicobacter pylori infected macrophages
| miRNAs | Changes of miRNA expression over control | Control (non-infection) | |||
|---|---|---|---|---|---|
|
|
|||||
| 4 h | 8 h | 12 h | 24 h | 1 | |
| miR-21 | 3.21 ± 0.025* | 5.12 ± 0.031* | 7.55 ± 0.028* | 9.86 ± 0.24* | 1 |
| miR-155 | 5.12 ± 0.033* | 11.21 ± 0.030* | 16.5 ± 0.027* | 21.2 ± 0.029* | 1 |
| miR-223 | 4.44 ± 0.019* | 5.89 ± 0.032* | 10.8 ± 0.35* | 16.72 ± 0.27* | 1 |
| miR-26b | 0.08 ± 0.002* | 0.07 ± 0.001* | 0.12 ± 0.014* | 0.78 ± 0.002 | 1 |
| miR-202 | 0.60 ± 0.015* | 0.45 ± 0.023* | 0.56 ± 0.022* | 0.86 ± 0.025 | 1 |
| miR-205 | 0.24 ± 0.002* | 0.17 ± 0.021* | 0.009 ± 0.018* | 0.015 ± 0.013* | 1 |
P<0.05 compared with mock controls.
Results are from three independent triplicated experiments (N=8).
IRAK-1 mRNA is a target of miR-223
To validate whether IRAK-1 is a potential target of miR-223 in macrophage, a luciferase reporter vector containing a 3’UTR of IRAK-1 mRNA (pMIR-Report/IRAK-1 3’UTR), as well as a reporter containing a mutated 3’UTR (pMIR-Report/Mut-IRAK-1 3’UTR) were constructed (Figure 1A). Cells transfected with pMIRReport/IRAK-1 3’UTR were treated with either miR-223mimics or miR-223 inhibitor. Our results showed that miR-223 mimics reduced luciferase production by 1.8-fold reduced while miR-223 inhibitor increased the luciferase activity by 1.9-fold (Figure 1B). These data indicated that IRAK-1 might be a potential target for miR-223 in macrophage.
Figure 1.
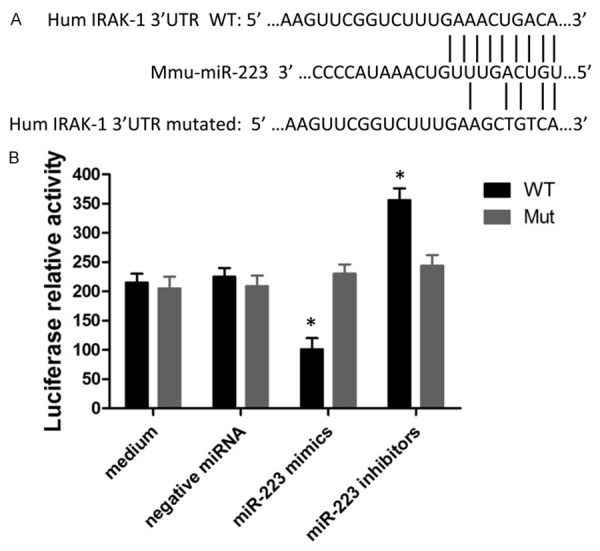
Validation of IRAK-1 mRNA as a target of miR-223. A: Sequence of potential binding site of miR-223 in the 3’UTR of IRAK-1 mRNA (top sequence), mutations were introduced into the binding site for generation of mutated IRAK-1 3’UTR (bottom sequence). B: Validation of miR-223 target using IRAK-1 3’UTR luciferase reporter. Data shown are mean ± SEM of 6 independent experiments (*P≤0.05).
The expression of miR-223 in macrophages with H. pylori infection and siRNA treatment
The expression levels of miR-223 were measured in H. pylori infected macrophages with treatment of miR-223 mimics, miR-223 inhibitors, or negative siRNA (Figure 2). Our results showed that miR-223 expression was significantly higher in H. pylori infected macrophages with miR-223 mimic treatment, while opposite results were observed in miR-223 inhibitors treated macrophages.
Figure 2.

The expression of miR-223 in macrophages. A: The expression of miR-223 in macrophages transfected with negative miRNA, miR-223 mimics or miR-223 inhibitors was determined by qRT-PCR assay. B: The fold changes of miR-223 expression relative to the non-transfected cells were used for analysis. Data shown are mean ± SEM of three independent experiments (*P≤0.05).
MicroiRNA-223 down-regulates expression of IL-6, IL-8, IL-12, TNF-a
To study the function of miR-223 in inflammatory regulation, the expression of inflammatory cytokines IL-6, IL-8, IL-12 and TNF-a were detected by qPCR (Figure 3). As expected, the expression levels of IL-6, IL-8, IL-12 and TNF-a were down-regulated in macrophages without H. pylori infection or H. pylori infected macrophages with miR-223 mimic treatment in comparison with control group (P<0.05). Contrarily, H. pylori infected macrophages with miR-223 inhibitor treatment increased the expression of IL-6, IL-8, IL-12 and TNF-a (P<0.05). These results suggested that miR-223 may play negative role in the inflammatory pathway in part by targeting some signal pathways, thus down-regulating the expression of its downstream pro-inflammatory mediators.
Figure 3.
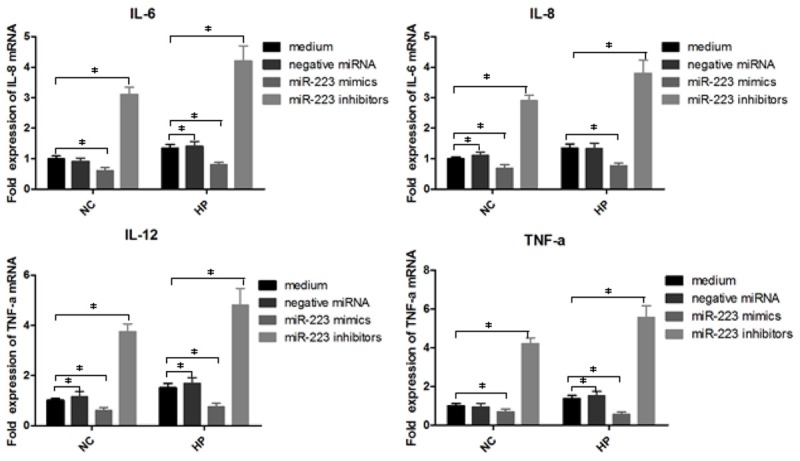
The impact of miR-223 on the expression of IL-6, IL-8, IL-12 and TNF-a. H. pylori infected macrophages were transfected with miR-223 mimics or miR-223 inhibitors. The total RNA was harvested at 12 h post infection. The expression of IL-6, IL-8, IL-12 and TNF-a were determined by qRT-PCR. Data shown are mean ± SEM of 6 independent experiments (*P≤0.05).
Expression of CD40, CD68, CD80 ,CD163 in H. pylori infection macrophages interferenced by siRNA (flow cytometry)
In order to confirm the expression of CD40, CD68, CD80, CD163 proteins in H. pylori infection macrophages interferenced by miR-223 mimics, miR-223 inhibitors and negative miRNA to study the role of miR-223 on molecular signaling proteins in cell surface, we quantified CD40, CD68, CD80, CD163 levels by flow cytometry. H. pylori non-infection or infection macrophages were interferenced by miR-223 mimics, miR-223 inhibitors and negative miRNA for 24 h, then subjected to flow cytometry for the detection of CD40, CD68, CD80, CD163 (Figure 4). The results show that CD40, CD68, CD80, CD163 levels decreased in H. pylori infection macrophages transfected with miR-223mimics (*P<0.05), but increased in miR-223 inhibitors transfected macrophages (*P<0.05). Meanwhile, the expression of CD40, CD68, CD80 decreased in macrophages transfected with miR-223mimics (*P<0.05), but only CD40, CD80 was enhanced in macrophages interfered by miR-223inhibitors (*P<0.05).
Figure 4.
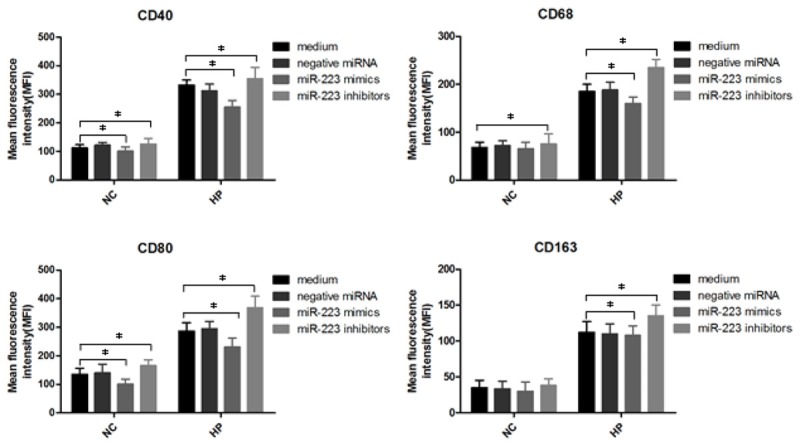
Analysis of surface CD40, CD68, CD80 and CD163 on H. pylori infected macrophages with siRNA treatment. H. pylori infected macrophages were first treated with negative miRNA, miR-223 mimics or miR-223 inhibitors, and then 106 cells were analyzed by flow cytometry. Bar graphs show the mean fluorescence intensities (MFI) of CD40, CD68, CD80 and CD163. Data shown are mean ± SEM of 3 independent experiments (*P≤0.05).
MicroiRNA-223 down-regulates IRAK-1 in macrophages in response to H. pylori infection
To explore whether miR-223 is capable of regulating IRAK-1 expression in macrophages with H. pylori infection, macrophages with or without H. pylori infection were treated by miR-223 mimics, miR-223 inhibitors or negative miRNAs and IRAK-1, NF-κB and MAPK expression was determined by Western blotting (Figure 5). Interestingly, miR-223 could down-regulate the expression of IRAK-1, NF-κB and MAPK in macrophages with and without H. pylori infection (P<0.05). Unlike miR-223 mimics, miR-223 inhibitors increased the expression of IRAK-1, NF-κB and MAPK in cells with both treatments (P<0.05). These data indicated that miR-223 was able to regulate the immune response in macrophages against H. pylori infection possibly through down-regulation of IRAK-1, NF-κB and MAPK expression.
Figure 5.
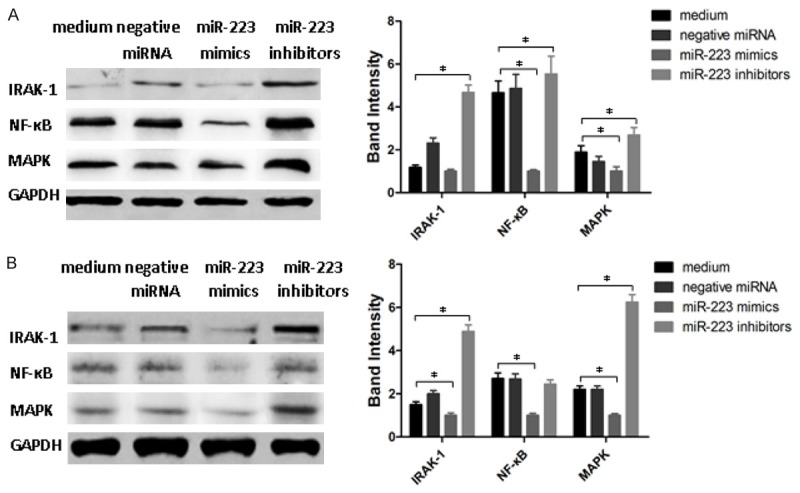
Analysis of the expression of IRAK-1, NF-κB and MAPK proteins by Western blotting. A: Analysis of IRAK-1, NF-κB and MAPK proteins in H. pylori free macrophages with miR-223 mimics, miR-223 inhibitors or negative miRNA treatment. B: Analysis of IRAK-1, NF-κB and MAPK proteins in H. pylori infected macrophages with miR-223 mimics, miR-223 inhibitors or negative miRNA treatment. Data shown are mean ± SEM of 3 independent experiments (*P<0.045).
Discussion
In this study, an alteration of several immunity-related miRNAs was observed in macrophages infected with H. pylori, with decreased expression of miR-205 and increased expression of miR-26b, miR-202, miR-21, miR-155 and miR-223. More importantly, miR-223 demonstrated a capacity to play a negative role in the regulation of immune response to H. pylori infection, which was in part through a mechanism by targeting IRAK-1 and post-transcriptionally downregulating IRAK-1 expression and sequentially inhibiting the production of inflammatory mediators, such as IL-6, IL-8, IL-12, and TNF-a. These results strongly suggest that miR-223 plays a critical role in macrophage-mediated immune response against H. pylori infection.
It has been recognized that miRNAs are involved in host immunity, not only by governing the maturation, proliferation, differentiation and activation of immune cells, but also by regulating the immune responses through many pathways [18]. Increasingly evidence have suggested that miR-223 plays multiple roles in the organogenesis, as well as pathogenesis of a variety of diseases, including tumors, immune diseases, and infectious diseases [16]. When challenged by endotoxin, miR-223 deficient mice exhibited increased inflammatory lung lesions [19], During monocytes differentiation into macrophages, miR-223 is down-regulated [14]. However, the role of miR-223 in regulating processes such as macrophage activation, subsequent adipose tissue inflammation and systemic insulin resistance is still unknown. The present study provides evidence to support a novel role of miR-223 in modulating macrophage polarization.
Macrophages are the first line of host immune cells to encounter H. pylori in gastric tissue. MicroRNAs have been demonstrated to play a regulatory role in the macrophage immune response towards eradication of H. pylori infection while avoiding excessive damage [13]. For example, miR-155 was shown to be a negative regulator of TLR signaling SOCS1 and SHIP1 during the macrophage inflammatory response, while repression of miR-223 expression seemed to affect downstream of TLR signaling molecules such as IRAK-1 and IRAK-2 or TRAF-6, and lead to an elevated inflammatory response [20]. Consistent with these findings, miR-223 in the current study showed an ability to negatively regulate the production of pro-inflammatory mediators IL-6, IL-8, IL-12 and TNF-a in macrophage by targeting IRAK-1 and post-transcriptionally down-regulating IRAK-1, IRAK-1, NF-κB and MAPK proteins expression.
Upon H. pylori infection, microphages could be activated and produce IL-6, IL-10 and IL-23 and express CD40 and CD80 [21]. CD68 and CD163 are associated with a large range of inflammatory diseases including macrophage activation syndrome, HIV infection, rheumatoid arthritis [22]. The expression of CD40, CD68, CD80 and CD163 was enhanced in H. pylori infected macrophages with miR-223 inhibitors treatment, but decreased in miR-223 mimics treated macrophages.
Taken together, our findings provide a new explanation characterizing the molecular mechanism underlying the regulation of miR-223 in H. pylori infected macrophage. this may provide useful information for further understanding of molecular mechanisms by which the miR-223/NF-κB pathway controls the inflammatory response and promotes the pathogenesis of inflammatory diseases.
Acknowledgements
This work was supported by Science and Technology Project of Jiangsu Province in China (NO: KS1425) and by Medical science and technology development fund of Jiangsu University in China (JLY20140047). We thank XiangYa Hospital and Xiangya Medical College for technical assistance.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237–259. doi: 10.1016/j.bpg.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 6.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 10.Fehri LF, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One. 2010;5:e9500–e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belair C, Baud J, Chabas S, Sharma CM, Vogel J, Staedel C, Darfeuille F. Helicobacter pylori interferes with an embryonic stem cell micro RNA cluster to block cell cycle progression. Silence. 2011;2:7. doi: 10.1186/1758-907X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 13.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-[kappa] B pathway by regulating expression of the kinase IKK [alpha] during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 15.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O’Neill LA, Masters SL. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 16.Dorhoi A, Iannaccone M, Farinacci M, Faé KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ, Oberbeck-Müller D, Jörg S, Heinemann E, Hahnke K, Löwe D, Del Nonno F, Goletti D, Capparelli R, Kaufmann SH. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest. 2013;123:4836. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S. A novel regulator of macrophage activation: miR-223 in obesity associated adipose tissue inflammation. Circulation. 2012;125:2892–903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 18.Olivieri F, Rippo MR, Prattichizzo F, Babini L, Graciotti L, Recchioni R, Procopio AD. Toll like receptor signaling in “inflammaging”: microRNA as new players. Immun Ageing. 2013;10:11. doi: 10.1186/1742-4933-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trinchieri G. Cancer and Inflammation: An Old Intuition with Rapidly Evolving New Concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 20.Yao Y, Li G, Wu J, Zhang X, Wang J. Inflammatory response of macrophages cultured with Helicobacter pylori strains was regulated by miR-155. Int J Clin Exp Pathol. 2015;8:4545–54. [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 22.Fehlings M, Drobbe L, Moos V, Viveros PR, Hagen J, Beigier-Bompadre M, Pang E, Belogolova E, Churin Y, Schneider T. Comparative analysis of the interaction of Helicobacter pylori with human dendritic cells, macrophages, and monocytes. Infect Immun. 2012;80:2724–2734. doi: 10.1128/IAI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


