Abstract
Background: Emerging evidence indicated that dysregulated long non-coding RNAs (lncRNAs) was implicated in the tumorigenesis and progression. LncRNA TUSC7 was involved in various malignancies. However, the role of TUSC7 in human non-small-cell lung cancer (NSCLC) remains unclear. Methods: Expression of TUSC7 was analyzed in 112 cases of NSCLC tissues and six lung cancer cell lines by quantitative real-time PCR (qRT-PCR). Then the correlation of TUSC7 expression with clinicopathological features and prognosis was aslo studied. Furthermore, overexpression of TUSC7 was performed and its role in tumor progression was explored. Results: The expression level of TUSC7 was lower in NSCLC tissues and lung cancer cells compared with their normal counterparts. Lower expression of TUSC7 in NSCLC tissues was associated with larger tumor size and higher TNM stage. Patients with lower TUSC7 expression had worse overall survival compared with the high expression cases. Univariate and multivariate analyses suggested that low expression of TUSC7 was an independent poor prognostic indicator for NSCLC patients. Moreover, upregulation of TUSC7 expression could inhibit proliferation of lung cancer cell in vitro. Conclusions: Our results suggested that TUSC7 was a potential biomarker for NSCLC prognosis, and the dysregulation of TUSC7 may play an important role in the NSCLC progression.
Keywords: Long noncoding RNA, TUSC7, non-small-cell lung cancer, prognosis, function
Introduction
Non-small-cell lung cancer (NSCLC) represents the major type of primary lung cancer and is often diagnosed at moderate or later stage. The migration and invasion of neoplasms is the leading cause of cancer mortality [1]. Recently, various long non-coding RNAs (lncRNAs, RNA gene products consisting of 200 to 100, 000 nt), including HNF1A-AS1 [2], MALAT1 [3] and HOTAIR [4], have been identified as new modulators in the origination and progression of NSCLC. These findings indicated that lncRNAs could be employed as novel therapeutic targets. Therefore, to explore the molecular mechanisms involved in the development of NSCLC and to investigate for the effective therapeutic targets as well, more efforts are needed to illustrate the expression profile and role of lncRNA in NSCLC.
Tumor suppressor candidate 7 (TUSC7), also known as LOC285194 (LSAMP-AS3), was a lncRNA originally found as a down-regulated gene in in osteosarcoma. Its gene is located at chromosome 3q13.31 [5], acting as a p53-regulated tumor suppressor gene in human colorectal and gastric cancer, in part by repressing miRNAs [6,7]. Exogenous expression of TUSC7 in cancer cells significantly inhibited the cellular growth, while low expression of TUSC7 indicated poor prognosis of patients [5,8,9]. However, little is known about the expression and the role of TUSC7 in NSCLC.
In this study, we detected the expression of lncRNA TUSC7 in NSCLC tissues and cell lines, and analysed its association with clinicopathological features. Kaplan-Meier curves compared by the log rank test was also conducted in order to determine whether lncRNA TUSC7 predictor for the prediction of clinical outcomes in NSCLC patients. Finally, in vitro assays were explored to demonstrate the biological functions of lncRNA TUSC7 on NSCLC progression.
Materials and methods
Patients and specimens
A total of 112 primary NSCLC cases who underwent surgery in the Second Affiliated Hospital of Xi’an Jiaotong University between 2008 and 2010 were enrolled. Tissue collecting include paired cancer tissues and adjacent non-tumor (ANT) lung tissues. All tissues were immediately frozen and stored in liquid nitrogen until use. None of the patients received any preoperative chemotherapy or radiotherapy. The clinical stages were determined according to the classification of malignant tumors by the International Association for the Study of Lung Cancer (IASLC) Tumor-Node-Metastasis (TNM) classification, 7th edition [10]. This study was approved by the Research Ethics Committee of the Second Affiliated Hospital, School of Medicine, Xi’an Jiaotong University. Informed consent were obtained from all of the patients. The clinicopathological features of patients, including age, gender, histopathological type, TNM stage, were summarized in Table 1.
Table 1.
The relationship between TUSC7 expression and clinicopathologic parameters of NSCLC patients
| Characteristics | No. of patients | Low expression | High expression | P value |
|---|---|---|---|---|
| Age (years) | 0.122 | |||
| ≤60 | 80 (71.4%) | 43 (53.8%) | 37 (46.2%) | |
| >60 | 32 (28.6%) | 13 (40.6%) | 19 (59.4%) | |
| Gender | 0.171 | |||
| Male | 57 (50.9%) | 25 (43.8%) | 32 (56.2%) | |
| Female | 55 (49.1%) | 31(56.4%) | 24 (43.6%) | |
| Histology | 0.376 | |||
| SCC | 66 (58.9%) | 32 (48.5%) | 44 (51.5%) | |
| AC | 46 (41.1%) | 24 (52.2%) | 12 (47.8%) | |
| Differentiation | 0.225 | |||
| Moderate-well | 37 (33.0%) | 25 (67.6%) | 12 (32.4%) | |
| Poorly | 75 (67.0%) | 31 (41.3%) | 44 (58.7%) | |
| Tumor size | 0.007* | |||
| ≤3 cm | 63 (56.2%) | 39 (61.9%) | 24 (38.1%) | |
| >3 cm | 49 (43.8%) | 17 (36.7%) | 32 (63.3%) | |
| Lymphatic metastasis | 0.162 | |||
| Negative | 47 (42.0%) | 26 (55.3%) | 21 (44.7%) | |
| Positive | 65 (58.0%) | 30 (46.2%) | 35 (53.8%) | |
| TNM stage | 0.012* | |||
| I+II | 63 (66.1%) | 41 (65.1%) | 22 (34.9%) | |
| III | 49 (33.9%) | 15 (30.6%) | 34 (69.4%) |
TNM, tumour-node-metastasis staging system.
P<0.05.
Cell lines and culture conditions
Six NSCLC cancer cell lines (A549, H1299, H1650, H1975, H23, and H358) and a normal human bronchial epithelial cell line (16HBE) were obtained from the Cell Engineering Research Center of The Fourth Military Medical University. Cells were maintained in RPMI 1640 (GIBCO-BRL; Invitrogen, Carlsbad, CA) or Dulbecco’s modified Eagle’s medium (DMEM; GIBCO-BRL; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA), 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA) and cultured at 37°C in humidified air with 5% CO2.
RNA preparation, reverse transcription, and quantitative real-time PCR
Total RNA was extracted from frozen tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For qRT-PCR, the isolated RNA was reverse transcribed to cDNA using a Reverse Transcription Kit (Takara, Dalian, China) following the manufacturer’s protocol. Real-time PCR analyses were conducted with SYBR® Premix Ex TaqTM (Takara, Dalian, China). The results were normalized to the expression of β-actin. The primer sequences can be found in the Supplementary Table S1. qRT-PCR and data collection were performed using an ABI 7500 instrument (Applied Biosystems, Foster City, CA). All qRT-PCR reactions were performed in duplicate. Relative expression of TUSC7 was calculated using the comparative cycle threshold (CT) (2-ΔΔCT) method.
Cell transfection
Human TUSC7 gene (NR_015391.1) was ligated into pLenti6.3-MCS/V5 DEST vector (Invitrogen, Shanghai, China). The following primers were used to construct the TUSC7 vectors: forward: 5’-CGA TCT TAA TTA AGG GGT ACC AAA GTC CAC TCT G-3’ and reverse: 5’-TCA GTG GCG CGC CTT TTT CGT GAG TAC ACA ATA GTC ATC-3’. The empty vector was used as a negative control (NC). Lung cancer cells at approximately 60%-70% confluence were transfected using Opti-MEM and Lipofectamine 2000 reagents (Invitrogen, CA, USA) according to the manufacturer’s instructions after 24 h of culture.
Cell proliferation assay
The in vitro cell proliferation of lung cancer cells was measured using the MTT (Sigma, St. Louis, MO) method according to the manufacturer’s instruction. In brief, cells were seeded into 96-well plates and transfected with 0.2 ug pLenti-TUSC7 or pLenti-NC for 24 h. In the indicated time periods, 0.1 ml of spent medium was replaced with an equal volume of fresh medium containing MTT 0.5 mg/ml. Plates were incubated at 37°C for 4 hours, and then the medium was replaced with 0.1 ml of DMSO (Sigma) and plates were agitated at room temperature for 10 min. The absorbance was measured at 490 nm using an enzyme-labeled analyzer.
Cell cycle analysis
Cells were seeded at a density of 1 × 106 cells/well in six-well plates. After 24 h, cells were washed with PBS and fixed in ice-cold 70% ethanol for 1 h and then treated with 100 uL of 50 mg/L propidium iodide for 30 min at 4°C in the dark. The cell-cycle profiles were assayed using the Elite ESP flow cytometer at 488 nm, and data were analyzed with the CELL Quest software (BD Biosciences,San Jose, CA, USA).
Statistical analysis
All statistical analyses were performed using the SPSS 20.0 software (IBM, SPSS, Chicago, IL, USA). Differences between groups were evaluated by the Student’s t test for continuous variables and χ2 test for categorical variables. Disease free survival (DFS) was calculated from registration to detection of tumor recurrence or until last follow-up date. Overall survival (OS) was defined as the time from registration to death or until last follow-up date. The Kaplan-Meier curves was used to calculate the DFS and OS, compared by the log rank test. Variables with a value of p < 0.05 in the univariate analysis were included in the subsequent multivariate analysis (Cox proportional hazards regression model). All statistical analysis were two sided and p values less than 0.05 were considered significant.
Results
TUSC7 is downregulated in human NSCLC tissues
To analyze the expression profile of TUSC7 in NSCLC, we firstly assessed the expression of TUSC7 in 112 cases of paired cancer tissues and adjacent normal tissues. As shown in Figure 1A, TUSC7 expression was significantly decreased in NSCLC tissues compared with the ANT (p < 0.001) and downregulation of TUSC7 was observed in 88 (78.6%) cases (Figure 1B). To assess the correlation of TUSC7 expression with clinicopathologic data, all cases were categorized as low or high in relation to the median value. Chi-Square tests showed that TUSC7 expression in NSCLC was significantly correlated with tumor size (p = 0.007) and TNM stage (p = 0.012). However, TUSC7 expression in NSCLC was not associated with other parameters such as age (p = 0.122), gender (p = 0.171), histology (p = 0.376), differentiation (p = 0.225), or lymph node metastasis (p = 0.162) (Table 1).
Figure 1.
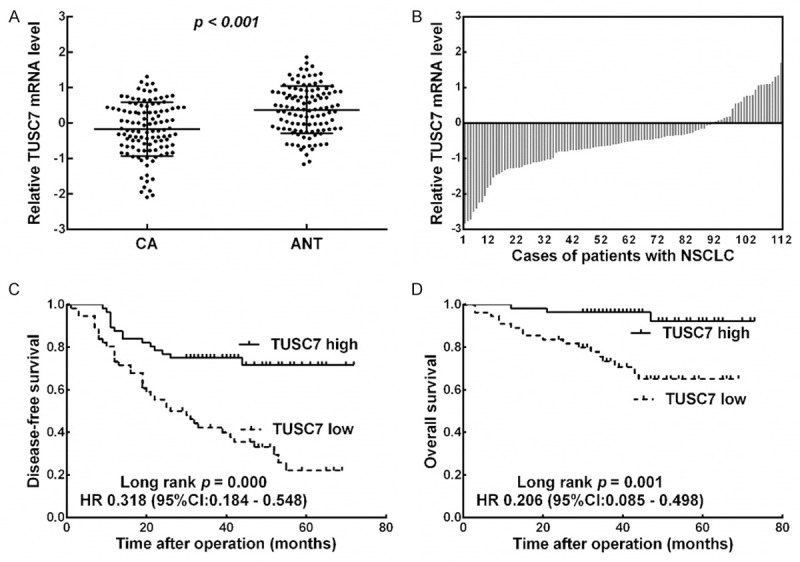
Downexpression of TUSC7 in NSCLC confers poor prognosis. A. qRT-PCR was performed to detect the TUSC7 expression in 112 pairs of NSCLC (CA) and corresponding noncancerous lung tissues (ANT). The mean expression level of TUSC7 in NSCLC tissues was significantly lower than that in ANT (P < 0.001). B. Low TUSC7 expression levels were observed in NSCLC tissues in 88 (78.6%) cases. β-actin was used as an internal control. C. Kaplan-Meier DFS analysis showed that the DFS rates in the TUSC7 low-expression group were significantly lower than those of patients in the TUSC7 high-expression group. D. Kaplan-Meier OS analysis showed that the OS rates in the TUSC7 low-expression group were significantly lower than the TUSC7 high-expression group.
Downexpression of TUSC7 was associated with poor prognosis of NSCLC
Kaplan-Meier analysis indicated that patients with low expression of TUSC7 had significantly shorter DFS and OS than the high expression group (p = 0.000 and p = 0.001; log rank test, Figure 1C and 1D). Univariate Cox proportional hazards regression analysis of DFS and OS also revealed that patients with low expression of TUSC7 had significantly worse prognosis than the high expression group (Tables 2 and 3). Moreover, multivariate analysis demonstrated that decreased expression of TUSC7 was an independent indicator for both DFS (Table 2) and OS (Table 3) ofin NSCLC patients.
Table 2.
Univariate and multivariate analysis of different prognostic factors for disease-free survival in 112 patients with NSCLC
| Prognostic factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (≤60/>60) | 0.720 | 0.420-1.236 | 0.234 | |||
| Gender (Male/Female) | 0.753 | 0.409-1.385 | 0.361 | |||
| Tumor size (≤3 cm/>3 cm) | 2.183 | 1.214-3.926 | 0.009* | |||
| Lymphatic metastasis (Negative/Positive) | 3.338 | 1.903-5.855 | 0.000* | |||
| TNM stage (I+II/III) | 2.800 | 1.638-4.787 | 0.000* | 4.326 | 1.310-3.903 | 0.003* |
| TUSC7 (High/Low) | 0.820 | 0.394-0.976 | 0.000* | 0.695 | 0.294-0.822 | 0.000* |
HR, hazard ratio; CI, confidence interval.
P<0.05.
Table 3.
Univariate and multivariate analysis of different prognostic factors for overall survival in 112 patients with NSCLC
| Prognostic factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (≤60/>60) | 0.701 | 0.254-1.934 | 0.492 | |||
| Gender (Male/Female) | 0.736 | 0.301-1.801 | 0.502 | |||
| Tumor size (≤3 cm/>3 cm) | 5.053 | 1.476-17.298 | 0.010* | 4.545 | 1.327-15.570 | 0.016* |
| Lymphatic metastasis (Negative/Positive) | 4.672 | 1.693-12.892 | 0.003* | |||
| TNM stage (I+II/III) | 2.174 | 0.904-5.226 | 0.083 | |||
| TUSC7 (High/Low) | 0.307 | 0.113-0.682 | 0.002* | 0.259 | 0.068-0.443 | 0.003* |
HR, hazard ratio; CI, confidence interval.
P<0.05.
Overexpression of TUSC7 decreases the proliferation of NSCLC cancer cells
To investigate the roles of TUSC7 in NSCLC, we performed qRT-PCR to evaluate the levels of TUSC7 in six NSCLC cell lines and one normal human bronchial epithelial cell line (16HBE). The expression of TUSC7 was lower in all six cancer cell lines compared with the level observed in 16HBE cell, with the lowest in H23 and H358 cells (Figure 2A). Thus H23 and H358 cells were selected and transfected with pLenti-TUSC7 or pLenti-NC. The RT-qPCR results confirmed that, compared with the control transfections, TUSC7 expression was effective increased in the presence of pLenti-TUSC7 (Figure 2B). We investigated the impact of TUSC7 on the growth of lung cancer cells, MTT assay showed that compared with the control, TUSC7 overexpression significantly inhibited the proliferation of H23 (Figure 2C) and H358 (Figure 2D) cells (both P < 0.05).
Figure 2.
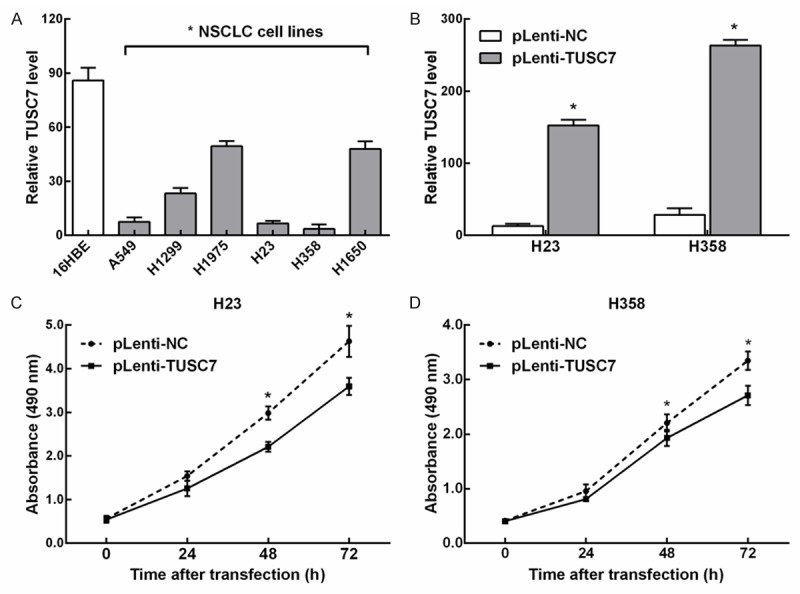
Overexpression of TUSC7 affects NSCLC cell proliferation. A. TUSC7 expression in 6 NSCLC cell lines were all significant downregulated compared with 16HBE cell line. B. TUSC7 expression levels were determined using RT-qPCR in H23 and H358 cells transfected with pLenti-TUSC7 or pLenti-NC. C. Compared with the control, knockdown of TUSC7 expression significantly inhibited H23 cell proliferation as measured using a MTT assay. D. Compared with the control, knockdown of TUSC7 expression significantly inhibited H358 cell proliferation as measured using a MTT assay. *Significant difference compared with the control (mean ± SD; *p < 0.05, n = 3).
Cell cycle analysis showed a decrease in the G1 population of TUSC7-overexpression NSCLC cells (Figure 3A). We then examined changes induced by overexpression of TUSC7 on transcription of a panel of proliferation-associated genes. Consistent with the results of cell cycle analysis, Bcl2, BimEL, cyclin A2, and cyclin B1 were all upregulated by TUSC7-overexpression, while Cyclin D1, p21, and p27 were all downregulated (Figure 3B). Taken together, these data implicate TUSC7 as growth suppressor in NSCLC through regulation of cell cycle transcripts and p21/p27.
Figure 3.
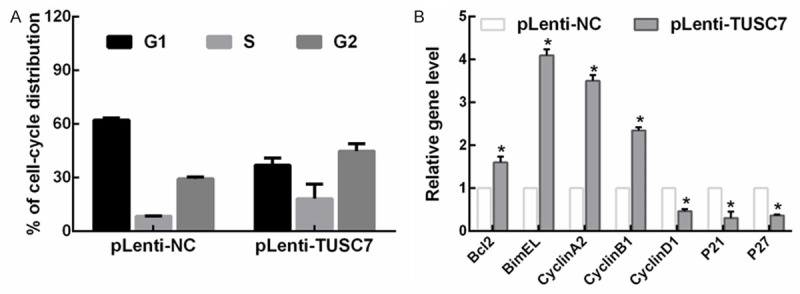
Overexpression of TUSC7 affects NSCLC cell cycle. A. Cell cycle analysis showed a mild decrease in the G1 population of TUSC7-overexpression NSCLC cells. Cell cycle distribution was analyzed by flow cytometry. B. TUSC7 regulated NSCLC cell growth by modulating a panel of proliferation-associated genes, as measured by RT-qPCR (mean ± SD; *p< 0.05, n = 3).
Discussion
Lung cancer is one of the most prevalent malignancies worldwide. As the most frequent type of lung cancer, non-small-cell lung cancer (NSCLC) accounts for 80-85% of all lung cancer cases, with the 5-year survival rate about 20-30% after surgery [11]. Although great improvements have been brought in adjuvant treatments including pre-/post-operative radiotherapy, chemotherapy and sequential/concurrent chemoradiation therapy, recurrences and metastases are still common in patients with advanced disease, and the overall survival for NSCLC patients remains dismal. Therefore, more efforts are needed to illustrate the underlying mechanism of tumorigensis and progression, in order to select the optimum therapy, minimize the drug resistance and improve the therapeutic effect.
LncRNAs emerged as a new kind of regulators involved in the malignancies by acting on multiple crucial aspects including genetics, epigenetics, and transcriptomics, which have dramatically altered our understanding of the biology of human tumor entities. A large number of studies have shown that dysregulation of lncRNAs participates in the carcinogenesis of NSCLC [12-16]. Here, we focused on lncRNA-TUSC7, which was previously reported to be downregulated and function as a tumor suppressor in a variety of malignant tumors, such as osteosarcoma [5], colorectal cancer [6], and gastric cancer [7]. In addition, downregulation of TUSC7 in tumors was found to be associated with poor prognosis [5,8,9]. In the present study, by assessing the expression level of TUSC7 in 112 NSCLC patients using qRT-PCR, we firstly reported that TUSC7 was significantly downregulated in NSCLC tissues compared with adjacent noncancerous tissues. Meanwhile, its remarkable lower expression also presented in six lung cancer cell lines compared with that of normal human bronchial epithelial cells.
Then, we analysed the relationship between TUSC7 expression and clinicopathologic features of NSCLC patients, by categorizing the 112 patients into TUSC7-low group and the TUSC7-high group using the median expression level of TUSC7 as the cutoff value. Results showed that the expression of TUSC7 was closely associated with tumor size and TNM stage of NSCLC. As is known, TUSC7 maybe an indicator of the intrinsic characteristics of cancer progression, and we found that patients with low TUSC7 expression had a shorter DFS and OS rate than the high TUSC7 group. By further univariate and multivariate analyses, the expression of TUSC7 was demonstrated to be an independent factor for predicting the prognosis of NSCLC patients. Taken together, these results suggested that TUSC7 might play a key role in the pathogenesis and progression of NSCLC.
Furthermore, we studied the effect of lncRNA TUSC7 on the biological function of cancer cells, and found that overexpression of TUSC7 significantly reduced lung cancer cells proliferation in vitro. Mechanically, TUSC7 function as a growth suppressor in NSCLC through regulation of cell cycle transcripts and p21/p27. Previous studies have shown that several miRNAs, such as miR-211 and miR-23b [6,7], could exhibit reciprocal repression with TUSC7, which may play a role in the TUSC7-mediated inhibition of cancer cell growth. However, the molecular mechanisms underlying how downregulation of TUSC7 influenced the development and progression of NSCLC haven’t been fully stated. We speculated that TUSC7 might regulate NSCLC cell proliferation through different potential miRNAs. Nevertheless, as the target genes of one lncRNA may differ between specific tissues and cell types, genes controlled by TUSC7 in NSCLC remain unknown, further studies are required to solve this problem. In future work, high-throughput techniques should be applied to obtain a global view of the changes in genomics and proteomics and to elucidate the mechanisms.
Conclusions
Our study demonstrated that TUSC7 expression was significantly downregulated in NSCLC tissues and cell lines compared with their normal counterparts. In addition, TUSC7 expression was closely associated with tumor size and TNM stage. Kaplan-Meier analysis indicated that NSCLC patients with lower TUSC7 expression levels had worse DFS and OS rates. Moreover, a multivariate analysis revealed that TUSC7 is an independent predictor of DFS and OS in NSCLC patients. Overexpression of TUSC7 in lung cancer cell lines caused a decrease in cellular proliferation. Together, these results suggested that TUSC7 might represent a novel prognostic indicator and a target for gene therapy in NSCLC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40:558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelialmesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 4.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan C, Weng WW, Sheng WQ, Zhou XY, Du X. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;37:1269–78. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 8.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX, Lv J, Yang JS, Zhu B, Cao XF. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 11.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–550. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 12.Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, Lu BB, Yin R, Zhang EB, Xu L, De W, Wang ZX. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Xu Q, Liu F, Ye X, Wang J, Meng X. Identification and validation of long noncoding RNA biomarkers in human non-small-cell lung carcinomas. J Thorac Oncol. 2015;10:645–654. doi: 10.1097/JTO.0000000000000470. [DOI] [PubMed] [Google Scholar]
- 14.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt LH, Gorlich D, Spieker T, Rohde C, Schuler M, Mohr M, Humberg J, Sauer T, Thoenissen NH, Huge A, Voss R, Marra A, Faldum A, Muller-Tidow C, Berdel WE, Wiewrodt R. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9:1294–1304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 16.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


