Abstract
Chronic inflammation contributes to high risk of colorectal cancer (CRC) development. Thus, discovering inflammation biomarkers for monitoring of CRC progression is necessary. In this study, we performed isobaric tags for relative and absolute quantitation-based proteomic assay on CRC tissues and paired normal mucosal tissues to identify key components in CRC pathogenesis. A total of 115 altered protein expressions were found with over twofold difference as compared with normal controls, which were associated with various molecular functions and biological processes. Here, we found that peroxiredoxin 1 (PRDX1) expression was higher in CRC tissues than that of matched controls and was determined as a tumor biomarker by receiver operating characteristic curve. PRDX1 expression was significantly upregulated in NCM460 cells challenged by H2O2 in a dose-dependent manner. PRDX1 depletion in SW480 cells enhanced reactive oxygen species (ROS), NO, and ONOO- production and increased the mRNA and protein expressions of pro-inflammatory cytokines [tumor necrosis factor-α, interleukin (IL)-1β, and IL-6] and chemokines (IL-8 and CXCL1), and partly activated nuclear factor-κB p65. Overall, our findings provide data on global alteration in the proteome of CRC tissues and reveal the potential of PRDX1 as an inflammation marker in CRC development, suggesting a novel therapy against inflammation-associated CRC.
Keywords: Colorectal cancer, inflammation, iTRAQ, PRDX1, biomarker
Introduction
Colorectal cancer (CRC) is the third and second most commonly diagnosed cancer in males and females, respectively, with more than 1.2 million new cases and 600,000 deaths per year [1]. Despite significant advances in diagnosis and treatment, the mortality rate of CRC remains high [2]. CRC possesses a complicated pathologic course [3]. Chronic inflammation is known to play a key role in CRC development [4-6]. For instance, patients with inflammatory bowel disease, such as ulcerative colitis and Crohn’s disease, have higher risk of CRC development than the healthy population [7-9]. Thus, discovering specific and sensitive inflammation biomarkers for assessment of CRC progression is urgently needed. Protein biomarkers as indicators of specific physiological and pathological states can be used for diagnosis and prognosis of specific cancers and for predicting or monitoring response to treatments [10,11]. To date, only a few proteins are employed as inflammation biomarkers of CRC, including C-reactive protein, tumor necrosis factor-α (TNF-α), and serum amyloid A [12,13]. However, none of these proteins is recommended for clinical screening [14]. Over the past few years, several novel differentially expressed protein biomarkers, such as galectin (Gal)-1/4, phosphatase and tensin homolog, and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, have been reported by applying proteomics technologies coupled with mass spectrometry (MS) [15-17], but most of these biomarkers are insufficient in either specificity or sensitivity [18,19]. Thus, discovering reliable inflammation-related protein biomarkers that can be measured objectively and applied in CRC patients for diagnosis, prognosis, and predicting response to treatment remains a persevering task.
High-throughput and high-sensitivity proteomics technologies have enabled large-scale screening of novel cancer-specific protein biomarkers [20-22]. Isobaric tags for relative and absolute quantitation (iTRAQ) is a well-approved technique that is based on isotopic labeling of proteins or peptides with differential molecular weight tags. This technique is a great alternative to two dimensional (2-D) electrophoresis and possesses advantages in reproducibility, sensitivity, and quantitative precision [23]. In particular, differential tagging with iTRAQ coupled with multidimensional liquid chromatography (LC) and MS/MS analysis is an emerging highly efficient technological process in discovering an increasing panel of candidate protein biomarkers related to CRC development [24-26].
Oxidative stress plays an important role in the pathogenesis of various cancers [27,28]. The occurrence of tumor by oxidant stress caused mainly by excessive generation of reactive oxygen species (ROS) leading to structural damages of DNA, such as insertions, deletions, base-pair mutations, or rearrangements [29]. In addition, ROS can directly activate nuclear and cytoplasmic signal transduction pathways which contact with malignant transformation [30]. Therefore, antioxidases have been suggested to play a functional role in tumorgenesis. Peroxiredoxins (PRDX) may have a similar function owing to their peroxidase enzyme activity. PRDX are a protein family with complex oligomeric structures whose members a class of thiol peroxidases that degrade hydroperoxides to water [31-33]. Many species have PRDX from prokaryotes to eukaryotes, and mammals have six different PRDX [34]. PRDX contain essential catalytic cysteine residues and are reduced mainly by thioredoxin (Trxs) [35]. Various types of PRDX have diverse and even opposing functions [36], and reports link them to both the prevention and promotion of cancer; the exact roles of PRDX are not clearly elucidated [37-39]. PRDX1, which belongs to the PRDX family, is composed of thiol-specific antioxidant enzymes that reduce H2O2 and peroxynitrite [40,41] and is associated with mitigation of oxidative damage [42]. PRDX1 has been proved to be overexpressed in breast cancer [43], malignant mesothelioma [44] and lung cancer [45]. PRDX1 has two conserved cysteines and belongs to 2-Cys subgroup accordingly. The N-terminal Cys-SH group in PRDX1 has been shown to be the primary site of oxidation and when oxidized, it rapidly reacts with another conserved cysteine in C-terminus to form an intermolecular disulfide bond [46]. Moreover, PRDX1 interacts with the cellular oncogene products c-Abl and c-Myc and thereby inhibits c-Abl kinase activity [47] and c-Myc-mediated transformation [48] independent of its antioxidant activity. Therefore, PRDX1 acts as a tumor suppressor. However, the biological function of PRDX1 in inflammation-associated CRC remains unclear.
In this study, we obtained comprehensive differential protein profiles of CRC tissues and normal controls by comparative proteomics approach (iTRAQ). Protein expression alterations were further identified by MALDI-TOF-MS. In particular, mRNA and protein levels of PRDX1 were markedly increased in CRC tissues as compared with controls, and receiver operating characteristic (ROC) curve suggests its potential as a tumor biomarker. The upregulation of PRDX1 and its biological function were also elucidated in the oxidative stress-stimulated normal colon cell line NCM460. We found the positive effect of PRDX1 knockdown on the expression changes of pro-inflammatory cytokines and chemokines as well as nuclear factor-κB (NF-κB) p65 activation in the colon cancer cell line SW480. Collectively, our data indicated PRDX1 as an inflammation biomarker and therapeutic target for CRC.
Materials and methods
Patients and specimens
CRC tissues and paired normal mucosal tissues (located at least 10 cm away from the tumor margin) were harvested from four patients for iTRAQ and western blot assays. These four patients underwent CRC surgery at the 150th Central Hospital of Chinese People’s Liberation Army (PLA). Sample tissues were excised, flash frozen in liquid nitrogen, and stored at -80°C until use. The overall clinical pathological data of the four CRC cases are listed in Table 1. Another 80 cases of CRC patients (mean age 58 years old, ranging from 22 to 86) who underwent CRC resection in our hospital between January 2010 and June 2012 were enrolled for immunohistochemical (IHC) studies. The clinical characteristics of these 80 cases are summarized in Table 2. Clinical staging was classified according to the “UICC TNM Classification of Malignant Tumors” [49]. Two independent experienced pathologists performed histological diagnosis for each sample. Freely tendered informed consents were obtained from all patients. The study was approved by the local ethics committee of the 150th Central Hospital of PLA.
Table 1.
Clinicopathological characteristics of fresh frozen tissues of four CRC cases for iTRAQ and western blot analysis
| Sample No. | Gender | Age (year) | Location | Pathological grade | Size (cm) | Lymphatic invasion | TNM Stage |
|---|---|---|---|---|---|---|---|
| 1 | Female | 59 | Colon | Moderately differentiated | 3 | Negative | IIa |
| 2 | Male | 68 | Colon | Moderately differentiated | 4 | Negative | IIb |
| 3 | Female | 64 | Colon | Well differentiated | 3 | Negative | IIb |
| 4 | Male | 56 | Colon | Poorly differentiated | 5 | Negative | IIa |
Table 2.
Clinicopathological characteristics and Pearson chi square test of 80 CRC cases for expression of PRDX1 by IHC analysis
| Variable | Cases | PRDX1 | ||
|---|---|---|---|---|
|
| ||||
| Up-regulated | Down- or un-regulated | P Valuea | ||
| Gender | ||||
| Male | 44 | 35 | 9 | 0.707 |
| Female | 36 | 24 | 12 | |
| Age | ||||
| > 60 | 47 | 34 | 13 | 0.842 |
| ≤ 60 | 33 | 22 | 11 | |
| Location | ||||
| Colon | 22 | 12 | 10 | 0.130 |
| Sigmoid | 23 | 19 | 4 | |
| Rectum | 35 | 29 | 6 | |
| Pathological grade (differentiated) | ||||
| Well | 4 | 3 | 1 | 0.274b |
| Moderately | 37 | 31 | 6 | |
| Poor | 39 | 24 | 15 | |
| Lymph nodes invasion | ||||
| Positive | 27 | 25 | 2 | 0.014* |
| Negative | 53 | 32 | 19 | |
| TNM stage | ||||
| I | 11 | 8 | 3 | 0.032* |
| IIa | 26 | 23 | 3 | |
| IIb | 20 | 10 | 10 | |
| III | 23 | 16 | 7 | |
Pearson chi square.
Correctional χ2 tests by combining cells having “Expected Count” less than 5.
P < 0.05.
Sample preparation
For iTRAQ and western blot analysis, tissue samples were washed three times with 1 mL of PBS containing a mixture of protease inhibitors [1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 10 μM leupeptin, 1 μg/ml aprotinin, and 1 μM pepstatin] as previously described [50]. The samples were then homogenized in 0.5 ml of PBS with protease inhibitors using a handheld homogenizer. Homogenates were flash frozen in liquid nitrogen and stored at -80°C until use. For IHC assay, CRC tissues and paired normal mucosal tissues were obtained, fixed in 10% formalin, embedded in paraffin, and stored at 4°C until use.
Protein digestion, iTRAQ labeling, and strong cation-exchange (SCX) fractionation
Extracted protein samples from the four pairs of CRC tissues and matched normal mucosal tissues were pooled separately and purified by centrifugation. Protein concentrations were determined by Bradford assay (Bio-Rad, Richmond, CA, USA). iTRAQ labeling was performed as previously described [51,52]. In brief, 100 μg of tissue proteins was denatured, alkylated, and then digested. The proteins were labeled with iTRAQ tags as follows: CRC tissues-113 isobaric tag and paired normal mucosal tissues-115 isobaric tag. The labeled samples were combined, desalted with Sep-Pak Vac C18 cartridge 1 cm3/50 mg (Waters, USA), and then fractionated on a Shimadzu HPLC system (Shimadzu, Japan) with an SCX column (Phenomenex Luna SCX, 4.6 mm × 250 mm, 5 μm, 100 Å, USA). SCX separation was performed using an integrated gradient of 0% to 100% buffer B [2 M KCl, 10 mM KH2PO4 in 25% acetonitrile (CAN), pH 3.0] in buffer A (10 mM KH2PO4 in 25% ACN, pH 3.0) and 5% to 30% linear binary gradient at a flow rate of 1 ml/min for 60 min. A total of 30 fractions were collected every 1 min (collection was paused when no peaks were detected) and incorporated to 10 final fractions for HPLC-ESI-MS/MS analysis.
HPLC-ESI-MS/MS analysis
Each fraction was dried and redissolved in buffer C (5% CAN). Fractions were desalted with Strata-X C-18 Column (Phenomenex, USA). All SCX fractions were analyzed using QSTAR XL LC MS/MS system (Applied Biosystems, USA) and HPLC column (Domestic C18 column, 5 μm, 300 Å, 0.75 mm × 100 mm, Bonna-Agela Technologies, China). The HPLC integrated gradient consisted of 5% to 80% (linear binary gradient 5% to 35%) buffer D (95% ACN, 0.1% formic acid) in buffer C at a flow rate of 400 nl/min for 90 min.
A Q-TOF instrument was operated in positive-ion mode with ion-spray voltage maintained at 2.5 kV. MS-based iTRAQ-labeled samples were acquired in an information-dependent acquisition mode. The scan type was TOF-MS (TOF masses of 400 Da to 1800 Da) with an accumulation time of 0.25 s. Precursor ion selection was based on ion intensity ( > 25 counts of peptide signal intensity) and charge state (2+ to 5+). Polarity pattern was positive with ion-spray voltage floating at 2.5 kV. Parameters for rolling collision energy (automatically set according to the precursor m/z and charge state) were manually optimized according to the iTRAQ tags. iTRAQ-labeled peptides were fragmented under collision-induced dissociation to produce reporter ions at 113.1 and 115.1. Fragment ions of peptides were simultaneously produced, resulting in sequencing of the labeled peptides and identification of the corresponding proteins. The ratios of the peak areas of the iTRAQ reporter ions reflected the relative abundances of peptides and proteins in the samples. The mass spectrometer was calibrated using BSA tryptic peptides.
Database searching, criteria, and bioinformatics
Protein identification and quantification for iTRAQ experiments were performed with Mascot version 2.3.01 (Matrix Science Inc, USA). The Scaffold version 3.0 (Proteome Software Inc, Portland, USA) in the Mascot software was used for peptide identification and isoform-specific quantification. A strict cut-off value of unused ProtScore ≥ 2, which corresponds to a confidence limit of 95%, was applied for protein identification to minimize false-positive results. At least two peptides with 95% confidence were considered for protein quantification. The resulting data set was auto bias-corrected to preclude any variations that may be imparted because of unequal mixing during combination of different labeled samples. For iTRAQ quantitation, the peptides were automatically selected by “Pro Group algorithm” (at least one peptide with 99% confidence) to calculate the reporter peak area, error factor (EF), and P value [53,54]. An EF < 2 was set to satisfy the quantification quality. P < 0.05 was considered significant for protein quantification. Fold changes over 2 (log2 value > 0.6 or > -0.6) was set as cut-off values in determining significant changes in protein expression [55].
The subcellular location and function of the identified proteins were elucidated using the UniProt Knowledgebase (Swiss-Prot/TrEMBL, www.expasy.org) and Gene Ontology (GEO) database (http://www.geneontology.org/).
Cell culture
The normal colon cell line NCM460 and the colon cancer cell line SW480 were purchased from American Type Culture Collection. Both cell lines were cultured in Roswell Park Memorial Institute 1640 medium (Gibco, BRL, UK) supplemented with 10% fetal bovine serum (Gibco), 100 U/ml penicillin (Invitrogen, Carlsbad, CA, USA), and 100 µg/ml streptomycin (Invitrogen) under humidified air with 5% CO2 at 37°C.
Western blot analysis
Cytosolic and nuclear extracts were prepared with a nuclear extract kit (Active Motif, Carlsbad, CA, USA). NF-κB p65 levels were quantified in nuclear fractions, whereas all other protein levels were quantified in cytosolic fractions. The final cytosolic and nuclear protein extracts were boiled, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred onto nitrocellulose membranes, and then immunoblotted with primary antibodies. An equivalent amount of sample was loaded and probed with mouse anti-β-actin monoclonal antibody (mAb) and rabbit anti-laminin B mAb (Sigma, CA, USA). The bands were detected using an enhanced chemiluminescence assay kit (Pierce, Rockford, IL, USA) and quantified with Quantity One software (Bio-Rad).
Immunohistochemistry and scoring
CRC and paired control tissues were collected to evaluate the pathological changes. All specimens were fixed with 10% formalin overnight, embedded with paraffin, non-serially sectioned (4 µm), and mounted on polylysine-covered slides. The sections were deparaffinized in xylene and rehydrated in a graded series of ethanol solutions. Then, the sections were submerged in citrate buffer (pH 6.0) and boiled in an autoclave at 121°C for 3 min to retrieve their antigenicity. The slides were then cooled at room temperature. Endogenous peroxidase was quenched with 0.3% H2O2 in methanol for 15 min. Nonspecific adsorption was minimized by incubating the sections in 10% normal goat serum (Gibco) in PBS for 20 min. The sections were incubated overnight with 1:200 dilution of primary anti-PRDX1 polyclonal antibody (Abcam, Cambridge, UK) or with control solutions of buffer or nonspecific purified rabbit immunoglobulin G (Sigma-Aldrich, Saint Louis, USA). Subsequently, sections were incubated with a biotinylated secondary antibody using ChemMate EnVision Kit (Dako, Hamburg, Germany) for 15 min. The reaction products were visualized with diaminobenzidine (DAB) substrate (Maixin Biotech., Fuzhou, China) as chromogen. The sections were counterstained with commercial hematoxylin (Maixin Biotech.), dehydrated, and mounted under light microscope (Leica, Wetzlar, Germany).
Two experienced pathologists observed and evaluated all stained sections in a blinded manner for DAB-positive staining. Five views were examined per slide, and 200 cells were observed per view at × 200 magnification. Cytoplasmic and nuclear immunostaining in cells was considered positive staining. In case of discrepancy, the sections were re-evaluated until a consensus was obtained. The scoring approach for the assessment of IHC staining was relatively simple and highly reproducible based on percentage prevalence and intensity of positive cells within the tumor [56]. The scores representing the extent of positively stained tumor cells were as follows: 0, 0%; 1, 1%-30%; 2, 31%-60%; and 3, > 60%. Intensity was estimated and expressed as 0, 1, 2, and 3 for negative, weak, moderate, and strong staining, respectively. The combination of the extent (E) and intensity (I) was obtained by the product of E × I called EI, which varied from 0 to 9 for each spot and was employed as the final staining score. Based on the final scores, tumor tissues were divided into two types, namely, low-level PRDX1 group (with a score ≤ 3) and high-level PRDX1 group (with a score > 3).
Immunocytochemical staining
After different treatments, the cells were fixed with 4% formaldehyde diluted in PBS for 5 min and then washed three times with PBS. Subsequently, the cells were mixed with anti-PRDX1 antibody, applied to the sections, and incubated overnight at 4°C. The cells were washed three times again with PBS, incubated with Alexa Fluor 488-labeled secondary antibodies at room temperature for 1 h, and observed under a fluorescent microscope (BX51, Olympus, Tokyo, Japan). Positive cells in six fields of each culture were counted.
Transit transfection with siRNA
Small interfering RNA (siRNA) duplexes corresponding to PRDX1 and scrambled siRNA were obtained from Invitrogen. Transient transfection of siRNAs was carried out using lipofectamine® 2000 transfection reagent (Invitrogen). siRNA (100 nM) was formulated with lipofectamine® 2000 transfection reagent according to the manufacturer›s instructions.
Measurement of intracellular ROS
Intracellular accumulation of ROS was determined by measuring the oxidative conversion of 2’,7’-dichlorofluorescin diacetate (DCFH-DA) to the fluorescent compound dichlorofluorescin (DCF). In brief, cells that were seeded in 96-well plates and underwent various treatments for the indicated time intervals were incubated with DCFH-DA solution (15 µM, final concentration) for 0.5 h at 37°C. DCF fluorescence was determined at 485 nm excitation and 520 nm emission using a fluorescence microplate reader (Safire2, Tecan, Switzerland). All measurements were performed in triplicate.
Measurement of NO production
At the indicated time points, cells subjected to various treatments were harvested and NO production was assayed. The chemical determination of NO is based on the diazotization of sulfanilamide with NO at acidic pH and the subsequent oxidation of scopoletin, which can be detected by fluorophotometry as described previously [57].
Analysis of ONOO- release ratio
ONOO- was synthesized, titrated, and stored as previously described [58,59]. ONOO- was measured by luminol-amplified chemiluminescence. All emitted photons were measured using a Berthold AutoLumat LB953 Luminometer (Dr Berthold GmbH & Co. KG, Wildbad, Germany). Cells (106 cells per 0.5 ml of PBS) subjected to various treatments were placed in a vial. Light emission was recorded by a computer interface and reported as the integrated light emission for a total period of 0.05 s-1.00 s. The results were calculated in counts per second. Chemiluminescence responses were converted to picomoles of ONOO- using a standard curve constructed with various concentrations of pure ONOO-.
Measurement of cytokine and chemokine production
Cytokine and chemokine levels of TNF-α, interleukin (IL)-1β, IL-6, IL-8, and CXCL1 in the cell culture medium were measured using commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN, USA). Optical density was measured on an ELISA plate scanner (CA94089, Molecular Devices, Sunnyvale, Canada). All experiments were performed according to the manufacturers’ instructions [60].
RNA extraction and quantitative real-time PCR
Total RNA was extracted from 100 mg of tissue samples or 1 × 106 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The harvested RNA was diluted to a concentration of 1 µg/µl, packaged, and preserved at -80°C. cDNA was generated using a Reverse Transcription Kit (Promega, Madison, WI, USA). Quantitative real-time PCR (qPCR) was performed using a standard IQTM SYBR Green Supermix kit (Bio-Rad), and PCR-specific amplification was assessed by Mastercycler® ep realplex (Eppendorf, Hamburg, Germany). β-actin was used as endogenous control. The relative level of PRDX1 was calculated via the comparative 2-ΔΔCt method [61]. Primer sequences used are listed in Table 3.
Table 3.
Details of primers for quantitative RT-PCR
| Gene | Forward | Reverse |
|---|---|---|
| β-actin | 5’-GCACCGTCAAGGCTGAGAAC | 5’-GGATCTCGCTCCTGGAAGATG |
| CXCL1 | 5’-AGTCATAGCCACACTCAAGAATGG | 5’-GATGCAGGATTGAGGCAAGC |
| IL-6 | 5’-TCTCCACAATTCGGTCCAGTT | 5’-CAACACCAGGAGCAGCCC |
| IL-8 | 5’-GGCAGCCTTCCTGATTTCTG | 5’-CTTGGCAAAACTGCACCTTCA |
| IL-1β | 5’-GCCTGAAGCCCTTGCTGTAGT | 5’-GCGGCATCCAGCTACGAAT |
| TNF-α | 5’-AGGGATGAGAAGTTCCCAAATG | 5’-TGTGAGGGTCTGGGCCATA |
Statistical analysis
Statistical analyses were conducted with SPSS 19.0 software (Chicago, IL, USA). Wilcoxon signed-rank test was used to compare the qualitative variables of IHC results between the CRC tissues and paired normal mucosal tissues. Pearson χ2 or correctional χ2 tests were employed to analyze the correlation between the differential expression of indicated proteins and clinicopathologic characteristics of patients. Risk factors that could affect the expression of potential protein biomarkers were analyzed by logistic regression. ROC curves were used to determine the diagnostic values of the markers.
Results
iTRAQ 2D-LC-MS/MS-based quantitative proteome analysis
Given that the 2D-LC-MS/MS method provides a great alternative to gels, a newly developed iTRAQ technique was used in this study to compare protein expressions between CRC and matched normal mucosal tissue. A total of 568 proteins were identified in the tissues. Among these proteins, 115 [45 upregulated (Table 4) and 70 downregulated (data not shown)] were identified to be differentially expressed in the tissues by the twofold cut-off value for upregulation and downregulation. Representative MS/MS and MS spectra for one peptide identified from PRDX1 are shown in Figure 1.
Table 4.
iTRAQ ratios of up-regulated proteins in CRC tissues
| No. | Name | Accession Number | Gene Name | Molecular Weight | T/Na |
|---|---|---|---|---|---|
| 1 | Vimentin | gi|340219 | VIM | 54 kDa | 2 |
| 2 | Purine-nucleoside phosphorylase | gi|157168362 | PNP | 32 kDa | 2.1 |
| 3 | Enoyl coenzyme A hydratase 1, peroxisomal | gi|16924265 | ECH1 | 36 kDa | 2.1 |
| 4 | Extracellular superoxide dismutase [Cu-Zn] precursor | gi|118582275 | SOD3 | 26 kDa | 2.1 |
| 5 | Thiosulfate sulfurtransferase | gi|17402865 | TST | 33 kDa | 2.1 |
| 6 | 6-phosphogluconolactonase | gi|6912586 | PGLS | 28 kDa | 2.1 |
| 7 | 78 kDa glucose-regulated protein precursor | gi|16507237 | HSPA5 | 72 kDa | 2.2 |
| 8 | Glutathione S-Transferase pi | gi|119595056 | GSTP1 | 19 kDa | 2.2 |
| 9 | Lamin A/C, isoform CRA_c | gi|119573383 | LMNA | 87 kDa | 2.3 |
| 10 | Prohibitin | gi|4505773 | PHB | 30 kDa | 2.3 |
| 11 | Actin-related protein 2/3 complex subunit 2 | gi|5031599 | ARPC3 | 34 kDa | 2.3 |
| 12 | Annexin A1 | gi|119582950 | ANXA3 | 40 kDa | 2.4 |
| 13 | Syndecan binding protein (syntenin), isoform CRA_a | gi|119607218 | SDCBP | 35 kDa | 2.4 |
| 14 | Plectin isoform 1e | gi|41322908 | PLEC | 514 kDa | 2.5 |
| 15 | Cathepsin B | gi|16307393 | CTSB | 38 kDa | 2.5 |
| 16 | Keratin 6B, isoform CRA_a | gi|119617032 | KRT6B | 60 kDa | 2.5 |
| 17 | Heterogeneous nuclear ribonucleoprotein A1 | gi|111306451 | HNRNPA1 | 34 kDa | 2.5 |
| 18 | GTPase activating protein 1 | gi|141797011 | IQGAP1 | 189 kDa | 2.5 |
| 19 | Glutathione peroxidase | gi|2160390 | GPX | 26 kDa | 2.5 |
| 20 | Annexin A3, isoform CRA_b | gi|119626228 | ANXA3 | 40 kDa | 2.5 |
| 21 | Fibronectin 1, isoform CRA_h | gi|119590943 | FN1 | 256 kDa | 2.6 |
| 22 | Transforming growth factor-beta-induced protein ig-h3 precursor | gi|4507467 | TGF-β IG-H3 | 75 kDa | 2.6 |
| 23 | Tropomyosin alpha-4 chain isoform 2 | gi|4507651 | TPM4 | 29 kDa | 2.6 |
| 24 | Cathepsin G preproprotein | gi|4503149 | CTSG | 29 kDa | 2.6 |
| 25 | Galectin-4 | gi|5453712 | Gal-4 | 36 kDa | 2.6 |
| 26 | Myosin heavy chain 9 | gi|12667788 | MYH9 | 227 kDa | 2.7 |
| 27 | Oncogene family, isoform CRA_c | gi|119618923 | RAB1B | 26 kDa | 2.7 |
| 28 | Dihydropyrimidinase-like 3 | gi|24659471 | DPYSL3 | 62 kDa | 2.8 |
| 29 | Rho GDP-dissociation inhibitor 1 isoform a | gi|4757768 | RhoGDIA | 23 kDa | 2.8 |
| 30 | Adaptor-related protein Complex 1, beta 1 subunit, isoform CRA_b | gi|119580203 | AP1B1 | 104 kDa | 2.8 |
| 31 | Lumican precursor | gi|4505047 | LUM | 38 kDa | 2.9 |
| 32 | Thymidine phosphorylase | gi|17390355 | TYMP | 50 kDa | 2.9 |
| 33 | Catechol-O-methyltransferase, isoform CRA_b | gi|119623416 | COMT | 34 kDa | 2.9 |
| 34 | Fibulin-1 isoform D precursor | gi|13661193 | FBLN1 | 77 kDa | 2.9 |
| 35 | Glutathione peroxidase 1 | gi|156602648 | GPX1 | 22 kDa | 2.9 |
| 36 | CAPNS1 protein | gi|15080279 | CAPNS1 | 34 kDa | 3.1 |
| 37 | Peroxiredoxin-1 | gi|4505591 | PRDX1 | 22 kDa | 3.2 |
| 38 | Thrombospondin 2 | gi|148922238 | THBS2 | 130 kDa | 3.3 |
| 39 | T200 leukocyte common antigen precursor, partial | gi|10999057 | CD45 | 128 kDa | 3.3 |
| 40 | Phosphoglycerate kinase 1 | gi|4505763 | PGK1 | 45 kDa | 3.4 |
| 41 | Peroxiredoxin-6 | gi|4758638 | PRDX6 | 25 kDa | 3.6 |
| 42 | Coagulation factor XIII A chain precursor | gi|119395709 | Factor XIII-A | 83 kDa | 4 |
| 43 | Ribosomal Protein S6 | gi|20381196 | RPS6 | 29 kDa | 4.1 |
| 44 | Chloride intracellular channel protein 1 | gi|14251209 | CLIC1 | 27 kDa | 4.1 |
| 45 | Rho Guanine Nucleotide Dissociation Inhibitor 2 | gi|56676393 | RhoGDI2 | 23 kDa | 4.2 |
T/N: The ratio of quantity analysis between CRC tissue and normal tissue.
Figure 1.
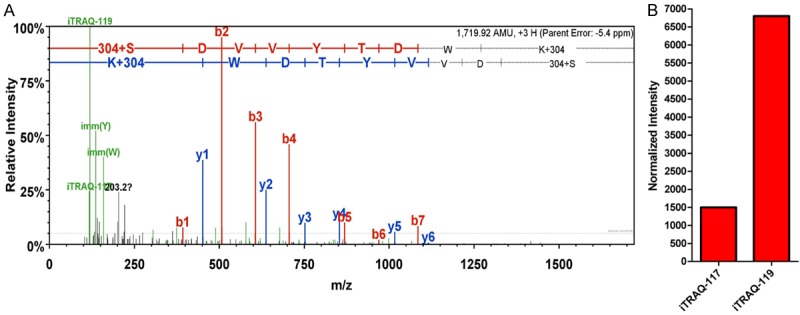
MS spectra showing PRDX1 peptides.
Bioinformatics analysis of differentially expressed proteins
According to the annotations from the UniProt Knowledgebase (Swiss-Prot/TrEMBL) and GEO database, 76.9% of the differentially expressed proteins were located in more than one cellular component. In brief, 53.8%, 53.8%, 69.2%, 69.2%, 23.1%, 38.5%, and 7.7% of the proteins were located in the cell, cell part, extracellular region, extracellular region part, macromolecular complex, organelle, and organelle part, respectively (Figure 2). Furthermore, 25.46%, 30.77%, 66.67%, and 41.3% of the proteins were located in the extracellular region, plasma membrane, cytoplasm, and nucleus of cells, respectively (Figure 2). Cytoplasmic proteins were located in the cytosol, intermediate and keratin filaments, mitochondrion, endoplasmic reticulum lumen, melanosome, and trans-Golgi network vesicles. Nucleic proteins were found in the nuclear matrix, nucleoplasm, nucleolus, nuclear speck, ribonucleoprotein complex, and nuclear inner membrane. We further analyzed the protein function using the UniProt Knowledgebase (Swiss-Prot/TrEMBL) and GEO database. The following molecular function and biological processes were observed to change more frequently: binding (84.6%), response to stimulus (84.6%), biological regulation (76.9%), cellular process (69.2%), and metabolic process (69.2%). A total of 83.6% of differentially expressed proteins were involved in more than one molecular function and biological process. A more detailed data are shown in Figure 2.
Figure 2.
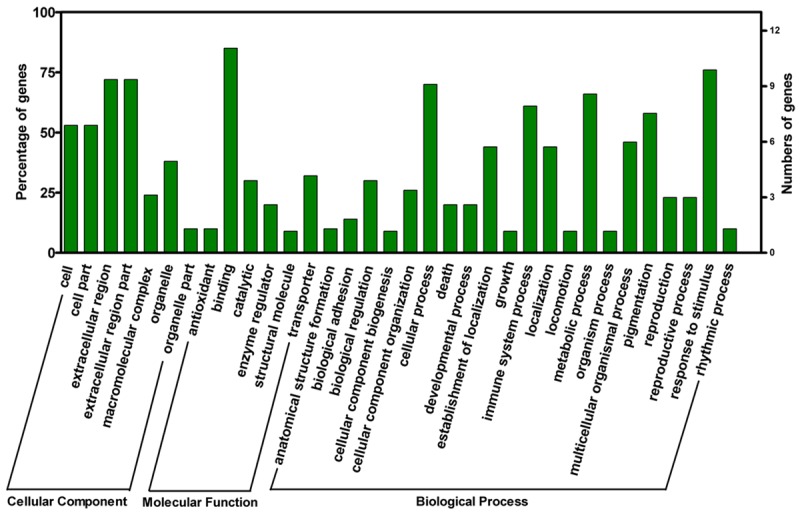
Cellular component analysis and functional distribution graph of differentially expressed proteins.
Validation of PRDX1 as a potential candidate protein marker
PRDX1 is a member of the PRDX family that is nonclassically secreted from cells and acts as a mediator of inflammation in prostate cancer [62,63]. Therefore, attention was focused on PRDX1, which showed a robust expression change in CRC tissues (3.2-fold increase, Table 4) in this study. qPCR, western blot, and IHC assays were performed to verify the iTRAQ results and assess the differential patterns of PRDX1 in CRC and paired control tissues. Figure 3A-C showed that both mRNA and protein levels of PRDX1 were increased in CRC tissues as compared with control tissues, which is consistent with the results from the iTRAQ experiments. In addition, PRDX1 expression was verified by IHC assay in 80 cases of human CRC tissues at stages I, IIa, IIb, and III and paired normal mucosa tissues (Figure 3D). The PRDX1 scores between CRC tissues and normal mucosa tissues showed statistically significant differences (P < 0.01, Figure 3E), which agrees with the data obtained by both iTRAQ and western blot analysis.
Figure 3.
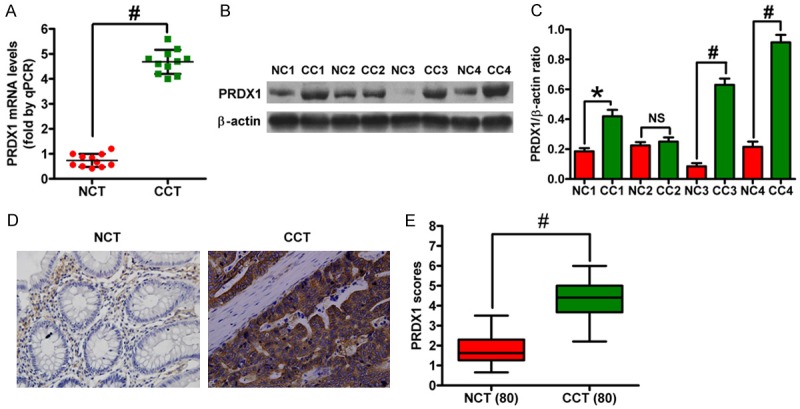
mRNA and protein levels of PRDX1 in CRC and paired control tissues were assessed by qPCR, western blot, and IHC. A. Expression of PRDX1 mRNA in the four pairs of CRC and control tissue samples by qPCR. B. Expression of PRDX1 protein in the four pairs of CRC and control tissue samples by western blot analysis. β-actin was used as a loading control. C. Relative density of PRDX1 normalized to β-actin. D. Representative images showing the expression of PRDX1 in tissues by IHC. E. Scores of PRDX1-positive staining. All figures are representative of three independent experiments performed in triplicate. Data are expressed as means ± SD. Statistical significance: *P < 0.05 vs NCT groups; #P < 0.01 vs NCT groups. (NCT: non-cancerous tissue; CCT: colon cancer tissue).
Diagnostic value of PRDX1
Cut-off values for PRDX1 were determined as scores 2 and 3 to distinguish CRC from normal mucosa tissues by comparing the sums of sensitivity and specificity of various diagnostic criteria. These criteria were set as I, II, III, IV, V, VI, and VII (scores ≥ 0, 1, 2, 3, 4, 5 and 6, respectively). The area under the ROC curves of PRDX1 was 0.847, which showed relatively high diagnostic accuracies (Figure 4).
Figure 4.
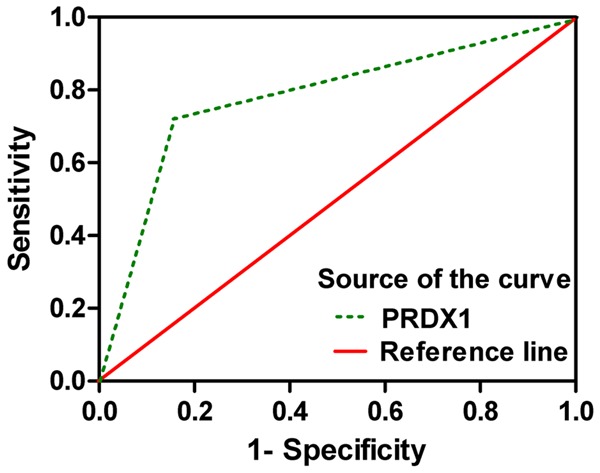
Area under the ROC curve for PRDX1 detection in CRC.
Oxidative stress causes upregulation of PRDX1 in NCM460 cells
PRDX1 expression could be elevated under oxidative stress in A549 cells [64]. PRDX1 has been reported to be secreted from tumor cells [65]. Therefore, we examined whether PRDX1 concentration is upregulated in the culture medium in NCM460 cells under H2O2. We exposed NCM460 cells to a dose range of H2O2 (0 µM-300 µM) for 0, 12, or 24 h and found that both mRNA and protein levels of PRDX1 were increased in a dose-dependent manner (Figure 5A-C). Immunocyto-chemical staining was consistent with the prior dose-dependent effect on the PRDX1 expression (Figure 5D). PRDX1 concentration in the media by ELISA increased in a dose-dependent manner after H2O2 treatment as compared with the control (Figure 5E). These results indicate that exposure of normal colon cells to H2O2 enhanced the expression level of PRDX1.
Figure 5.
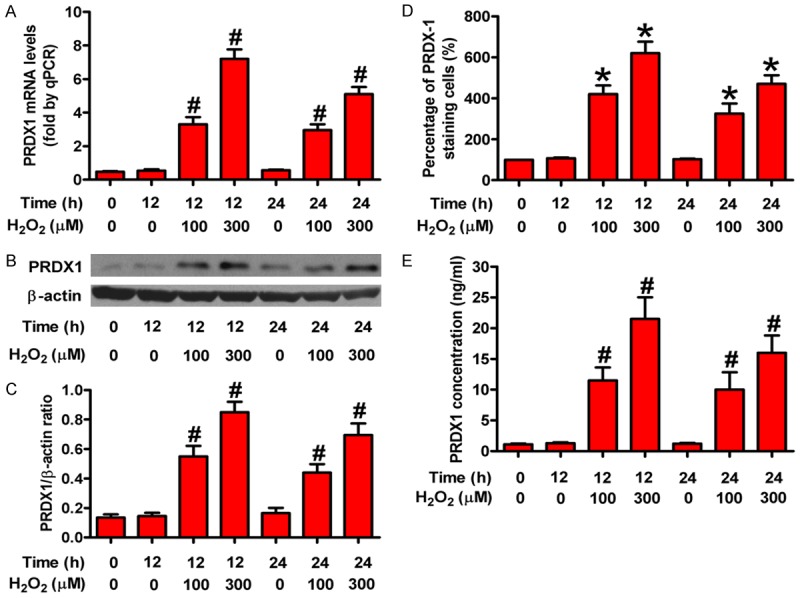
Oxidative stress caused PRDX1 upregulation in vitro. NCM460 cells were incubated with different doses of H2O2 (0 μM, 100 μM, and 300 μM) for 0 h, 12 h, and 24 h, respectively. Expression changes of PRDX1 were determined by qPCR, western blot, immunocytochemical staining, and ELISA. (A) qPCR analysis of PRDX1 mRNA expression in NCM460 cells treated with various doses of H2O2 at indicated time points. (B) Representative results of PRDX1 expression in each group analyzed by western blot. (C) Relative quantity of PRDX1 normalized to β-actin. (D and E) PRDX1 expression was examined in NCM460 cells treated with H2O2 compared with mock-treated NCM460 by immunocytochemical staining (D) and ELISA (E). All figures are representative of three independent experiments performed in triplicate. Data are expressed as means ± SD. Statistical significance: *P < 0.05 vs H2O2-free groups; #P < 0.01 vs H2O2-free groups.
Effect of PRDX1 depletion on ROS, NO, and ONOO- production in colon cancer cells
A PRDX1 sequence-specific siRNA mimics was transfected into colon cancer SW480 cells to explore the effect of PRDX1 on CRC development. Figure 6A showed that the PRDX1 mRNA level in siPRDX1 group was significantly decreased compared with the mock and siScramble groups. The results from western blot, immunocytochemical staining, and ELISA assays also showed that siPRDX1 efficiently inhibited PRDX1 expression (Figure 6B-E). PRDX1 belongs to a family of PRDX with six isoforms that protect cells against oxidative stress [62,63]. High levels of ROS are produced to activate redox-sensitive transcription factors and downstream genes regulated in colon cancer cells [66]. In this study, SW480 cells highly expressed ROS and PRDX1 knockdown significantly increased the production of ROS, NO, and ONOO- in SW480 cells (Figure 6F-H). These data suggest that PRDX1 functioned as a negative regulator of oxidative stress in colon cancer cells.
Figure 6.
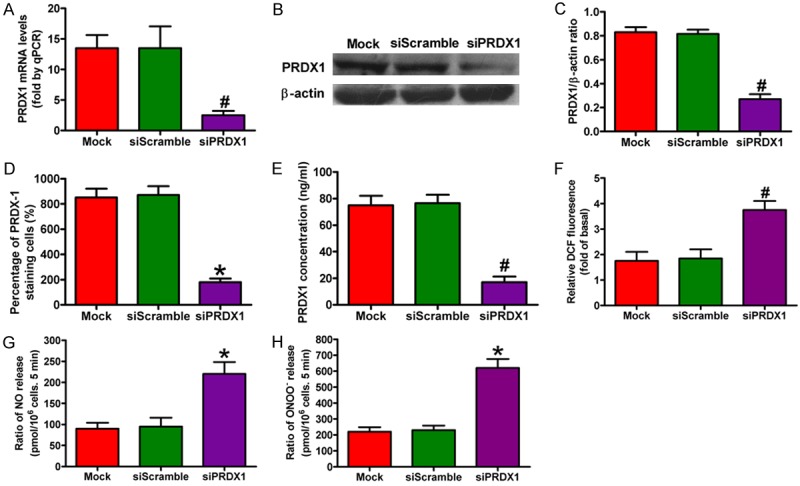
PRDX1 silencing inhibited ROS, NO, and ONOO- production in colon cancer cells. SW480 cells were transfected with PRDX1 or scramble siRNA for 24 h. (A) Expression of PRDX1 mRNA in siPRDX1-treated SW480 cells was examined by qPCR. β-actin was used as the internal control. (B) PRDX1 protein levels in siPRDX1-treated SW480 cells were analyzed by western blot. (C) Relative quantity of PRDX1 normalized to β-actin. (D) Immunocytochemical staining showing PRDX1 expression levels in all groups. (E) Secreted PRDX1 levels in media from different groups as determined by ELISA. The production of ROS (F), NO (G), and ONOO- (H) in various groups was calculated. All figures are representative of three independent experiments performed in triplicate. Data are expressed as means ± SD. Statistical significance: *P < 0.05 vs Mock and siScramble groups; #P < 0.01 vs Mock and siScramble groups.
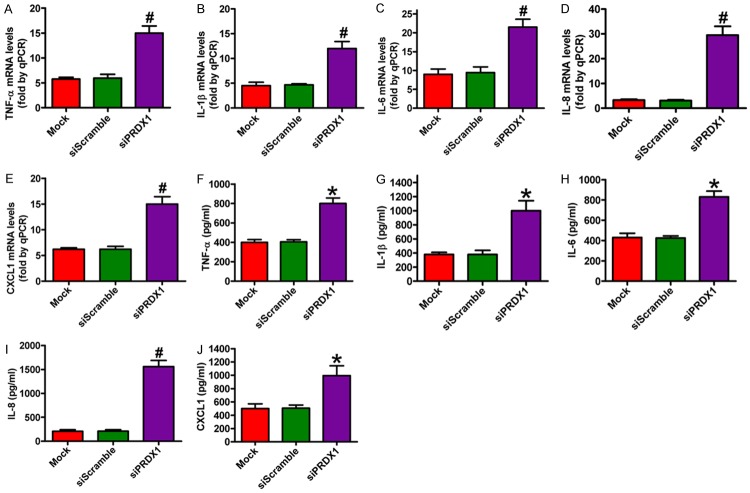
PRDX1 silencing promoted inflammation in colon cancer cells
The NF-κB signaling pathway plays a critical role in regulating proinflammatory cytokine and chemokine production [67]. Nucleic and cytosolic fractions of the cell lysates were extracted and the expression of NF-κB p65 protein was analyzed by western blot analysis to investigate the effect of PRDX1 on the nuclear translocation of NF-κB p65. As shown in Figure 7A (lower panel) and 7C, the depletion of PRDX1 caused considerable increase of nuclear NF-κB p65 levels and simultaneous significant decrease of cytosolic NF-κB p65 levels [Figure 7A (upper panel), 7B] in SW480 cells compared with the siScramble- and mock-treated groups. The effects of siPRDX1 on mRNA and protein expressions of proinflammatory cytokines and chemokines (TNF-α, IL-1β, IL-6, IL-8, and CXCL1) in SW480 cells were also investigated by qPCR and ELISA. As shown in Figure 8, PRDX1 knockdown in SW480 cells caused a significant increase in TNF-α, IL-1β, IL-6, IL-8, and CXCL1 expressions compared with the siScramble- and mock-treated group. These findings suggest that PRDX1 expression modulated the expression of proinflammatory cytokines and chemokines as well as regulated NF-κB p65 activation in colon cancer cells.
Figure 7.
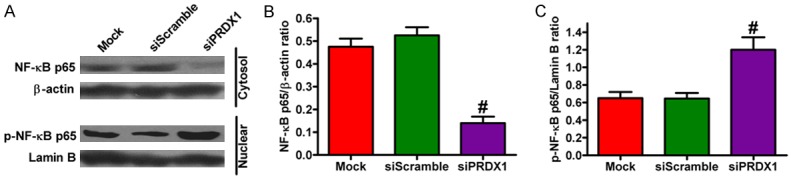
PRDX1 depletion promoted NF-κB p65 activation in SW480 cells. After 24 h of siPRDX1 or siScramble transfection, the protein expressions of NF-κB p65 in cytosol and in nuclear fragments of SW480 cells were analyzed by western blot. A. Protein levels of NF-κB p65 in cytosol extracts and p-NF-κB p65 in nuclear extracts. β-actin and lamin B were used as loading controls. B. Relative density of NF-κB p65 normalized to β-actin. C. Relative quantity of p-NF-κB p65 normalized to lamin B. All figures are representative of three independent experiments performed in triplicate. Data are expressed as means ± SD. Statistical significance: *P < 0.05 vs Mock and siScramble groups; #P < 0.01 vs Mock and siScramble groups.
Figure 8.
PRDX1 knockdown increased proinflammatory cytokine and chemokine production in SW480 cells. After 24 h of siPRDX1 or siScramble transfection, the mRNA expression of TNF-α (A), IL-1β (B), IL-6 (C), IL-8 (D), and CXCL1 (E) were assessed by qPCR assay in SW480 cells subjected to different treatments. And the production of TNF-α (F), IL-1β (G), IL-6 (H), IL-8 (I), and CXCL1 (J) were detected by ELISA assay. All figures are representative of three independent experiments performed in triplicate. Data are expressed as means ± SD. Statistical significance: *P < 0.05 vs Mock and siScramble groups; #P < 0.01 vs Mock and siScramble groups.
Discussion
Proteomics provides unique tools for the high-throughput screening of biomarkers and therapeutic targets that have potential for preventing and curing cancer. Therefore, proteomics can possibly help translate basic science discoveries into the clinical practice of personalized medicine [68]. The high-throughput proteomic technique iTRAQ can be used to label four or eight samples simultaneously for cancer biomarker identification; thus, iTRAQ emerges as a great methodology for discovering disease biomarkers [69,70]. In this study, we compared protein expression levels in CRC tissues and matched normal mucosa samples by using the iTRAQ-based proteomic method. Proteomic analysis revealed 115 differentially expressed proteins, of which 45 were upregulated (Gal-4, PGK1, ribosomal protein S4, ribosomal protein S5, annexin A5, molecular chaperone GRP78, PRDX1, PRDX6, etc.) and 70 were downregulated (FABP1, apolipoprotein A-IV precursor, cytochrome b-c1 complex subunit 2, MHC class I antigen, HSP90, cytokeratin 9, etc.) in CRC tissues as compared to the normal control. Functional and positional analyses showed that the aforementioned differentially expressed proteins were mainly associated with biological processes of binding (84.6%), response to stimulus (84.6%), biological regulation (76.9%), cellular process (69.2%), and metabolic process (69.2%) through the Swiss-Prot and GEO databases. Notably, overexpression of the inflammation-related molecule PRDX1 was verified by western blot, qPCR, and IHC analysis. An in vitro study using the NCM460 cell model showed that PRDX1 expression was upregulated under H2O2 exposure. Moreover, PRDX1 depletion enhanced ROS, NO, and ONOO- production in SW480 cells. PRDX1 knockdown also increased NF-κB p65 activation and the production of proinflammatory cytokines and chemokines in SW480 cells. Findings from the current study provide a novel insight into the mechanisms underlying inflammation-associated CRC.
Chronic inflammation has been reported to increase the risk for developing more severe CRC, indicating the significance of inflammation in CRC [71]. In this study, changes in 45 proteins that were upregulated involved in cellular metabolism and the pathogenesis of CRC (Table 4). For example, Gal-4 is proved to be an important molecule in elucidating the biology of CRC; it has a tumor suppressive effect in CRC cells in vitro through its inhibitory ability of Wnt signaling pathway [72]. Glutathione peroxidases (GPXs) are selenium-dependent antioxidant enzymes, which can detoxify hydrogen peroxide and many lipid hydroperoxides [73]. Decreased activity of these GPXs may increase oxidative stress and damage several biomolecules, which may promote neoplastic transformation in affected tissues [74]. Excess ROS or active nitrogen species (RNS) may also trigger and accelerate chronic inflammation, which is considered a risk factor for CRC [75]. Whether the other proteins that showed differential expression play roles in CRC pathogenesis remains unknown and needs further investigation.
PRDX1 was the significantly overexpressed protein in CRC tissues compared with the normal control as examined by iTRAQ. Given this finding, we explored the specific role of PRDX1 in CRC development. We further validated the overexpression of PRDX1 by qPCR, western blot, and IHC in CRC tissues. PRDX1 belongs to the PRDX family that reduces peroxides, such as H2O2 [76]. Interestingly, PRDX1-/- mice develop several cancers at increased frequency [77]. PRDX1 is not a traditional tumor inhibitor because re-expression of PRDX1 in cancer cells fails to induce cell death [78]. PRDX may act as a messenger of oxidative damage in normal tissues that are exposed to increased metabolic oxidative stress [79,80]. Various oxidative stress stimuli, including H2O2 and sulfhydryl reactive agents, can upregulate the PRDX1 expression in mouse peritoneal macrophages [81,82]. Two oxidative stress-sensitive transcription factors, namely, NF-E2 related factor (Nrf) 1 and Nrf2, interact with MafG to upregulate the transcription of many antioxidant genes, including PRDX1 when activated by oxidative stress [83-85]. In the present study, we investigated whether the expression of PRDX1 could be increased in NCM460 under H2O2 exposure. Our results verified that PRDX1 expression upregulated in a dose-dependent manner at 12 or 24 h. Notably, the level of PRDX1 in conditioned media was higher after 12 h H2O2 treatment than that after 24 h H2O2 treatment. Thus, we speculate that the quantity of PRDX1 secreted from the cells may be time dependent, but this speculation needs further investigation. Collectively, upregulation of PRDXI expression in NCM460 cells by H2O2 treatment was considered a cellular response to oxidative stress.
PRDX1 plays a rheostat role in regulating gene transcription by controlling cellular ROS in maintaining homeostasis. ROS also participated in various cellular functions, including cell proliferation, differentiation, and apoptosis [86]. ROS accumulation promotes nuclear translocation of NF-κB p65 and then activates the NF-κB p65 signaling pathway, resulting in a series of pro-inflammatory gene transcription [87]. We depleted PRDX1 in SW480 cells to investigate the effect of PRDX1 on inflammatory processes in CRC progression. Some results showed that PRDX1 knockdown increased the production of ROS, NO, and ONOO-. We also observed that PRDX1 depletion in SW480 cells promoted NF-κB p65 nuclear translocation and increased the production of pro-inflammatory cytokines and chemokines (TNF-α, IL-1β, IL-6, IL-8, and CXCL1). These results indicate that PRDX1 inhibited inflammatory processes possibly via the NF-κB p65 signaling pathway. This study provided novel insights into the anti-inflammatory mechanism of PRXD1 in CRC and revealed that PRDX1 may be a biomarker for detection of cancer progression.
In summary, our findings provide data on global alteration in the proteome of CRC tissues and confirm that PRDX1 is a potential biomarker of inflammation-associated CRC. PRDX1 expression was significantly upregulated in NCM460 cells exposed by H2O2 in a dose-dependent fashion. PRDX1 depletion in SW480 cells enhanced ROS, NO, and ONOO- production, and increased the mRNA and protein levels of pro-inflammatory factors (TNF-α, IL-1β, IL-6, IL-8, and CXCL1), and partly activated NF-κB p65 in CRC cells. This discovery has an important implication in elucidating the molecular mechanisms in inflammation-associated CRC and suggests that PRDX1 is a potential therapeutic target against CRC.
Acknowledgements
We thank Yang Li and Xu Zhan (Beijing Protein institute, China) for their expertise in iTRAQ and 2D-LC-MS/MS. We thank Yangkun Wang and Nianlong Meng (Department of Pathology, The 150th Central Hospital of PLA) for their help in clinical tissue collections.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Oliphant R, Nicholson GA, Horgan PG, Molloy RG, McMillan DC, Morrison DS. West of Scotland Colorectal Cancer Managed Clinical Network. Deprivation and colorectal cancer surgery: longer-term survival inequalities are due to differential postoperative mortality between socioeconomic groups. Ann Surg Oncol. 2013;20:2132–2139. doi: 10.1245/s10434-013-2959-9. [DOI] [PubMed] [Google Scholar]
- 3.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 4.Ohshima H, Tazawa H, Sylla BS, Sawa T. Prevention of human cancer by modulation of chronic inflammatory processes. Mutat Res. 2005;591:110–122. doi: 10.1016/j.mrfmmm.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 5.Rhodes JM, Campbell BJ. Inflammation and colorectal cancer: IBD-associated and sporadic cancer compared. Trends Mol Med. 2002;8:10–16. doi: 10.1016/s1471-4914(01)02194-3. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese S, Mantovani A. Inflammatory bowel disease and intestinal cancer: a paradigm of the Yin-Yang interplay between inflammation and cancer. Oncogene. 2010;29:3313–3323. doi: 10.1038/onc.2010.109. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Itzkowitz SH. Cancer in inflammatory bowel disease. World J Gastroenterol. 2008;14:378–389. doi: 10.3748/wjg.14.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potack J, Itzkowitz SH. Colorectal cancer and dysplasia in inflammatory bowel disease. Gut Liver. 2008;2:61–73. doi: 10.5009/gnl.2008.2.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 11.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 12.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 13.Song M, Wu K, Ogino S, Fuchs CS, Giovannucci EL, Chan AT. A prospective study of plasma inflammatory markers and risk of colorectal cancer in men. Br J Cancer. 2013;108:1891–1898. doi: 10.1038/bjc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe M, Takemasa I, Kaneko N, Yokoyama Y, Matsuo E, Iwasa S, Mori M, Matsuura N, Monden M, Nishimura O. Clinical significance of circulating galectins as colorectal cancer markers. Oncol Rep. 2011;25:1217–1226. doi: 10.3892/or.2011.1198. [DOI] [PubMed] [Google Scholar]
- 16.Naguib A, Cooke JC, Happerfield L, Kerr L, Gay LJ, Luben RN, Ball RY, Mitrou PN, McTaggart A, Arends MJ. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC Cancer. 2011;11:123. doi: 10.1186/1471-2407-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XH, Zheng HC, Takahashi H, Masuda S, Yang XH, Takano Y. PTEN expression and mutation in colorectal carcinomas. Oncol Rep. 2009;22:757–764. doi: 10.3892/or_00000497. [DOI] [PubMed] [Google Scholar]
- 18.Luo Y, Wang L, Wang J. Developing proteomics-based biomarkers for colorectal neoplasms for clinical practice: opportunities and challenges. Proteomics Clin Appl. 2013;7:30–41. doi: 10.1002/prca.201200071. [DOI] [PubMed] [Google Scholar]
- 19.González-González M, Garcia JG, Montero JA, Fernandez LM, Bengoechea O, Muñez OB, Orfao A, Sayagues JM, Fuentes M. Genomics and proteomics approaches for biomarker discovery in sporadic colorectal cancer with metastasis. Cancer Genomics Proteomics. 2013;10:19–25. [PubMed] [Google Scholar]
- 20.Yan S, Zhou C, Lou X, Xiao Z, Zhu H, Wang Q, Wang Y, Lu N, He S, Zhan Q, Liu S, Xu N. PTTG overexpression promotes lymph node metastasis in human esophageal squamous cell carcinoma. Cancer Res. 2009;69:3283–3290. doi: 10.1158/0008-5472.CAN-08-0367. [DOI] [PubMed] [Google Scholar]
- 21.Gronemeyer T, Wiese S, Ofman R, Bunse C, Pawlas M, Hayen H, Eisenacher M, Stephan C, Meyer HE, Waterham HR, Erdmann R, Wanders RJ, Warscheid B. The proteome of human liver peroxisomes: identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS One. 2013;8:e57395. doi: 10.1371/journal.pone.0057395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan NJ, Gao CF, Wang CS, Zhao G, Lv JJ, Wang XL, Chu GH, Yin J, Li DH, Chen X, Yuan XT, Meng NL. Identification of the up-regulation of TP-alpha, collagen alpha-1(VI) chain, and S100A9 in esophageal squamous cell carcinoma by a proteomic method. J Proteomics. 2012;75:3977–3986. doi: 10.1016/j.jprot.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 24.Jin GZ, Li Y, Cong WM, Yu H, Dong H, Shu H, Liu XH, Yan GQ, Zhang L, Zhang Y, Kang XN, Guo K, Wang ZD, Yang PY, Liu YK. iTRAQ-2DLC-ESIMS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res. 2011;10:3418–3428. doi: 10.1021/pr200482t. [DOI] [PubMed] [Google Scholar]
- 25.Hou Q, Tan HT, Lim KH, Lim TK, Khoo A, Tan IB, Yeoh KG, Chung MC. Identification and functional validation of caldesmon as a potential gastric cancer metastasis-associated protein. J Proteome Res. 2013;12:980–990. doi: 10.1021/pr3010259. [DOI] [PubMed] [Google Scholar]
- 26.Yang LS, Xu XE, Liu XP, Jin H, Chen ZQ, Liu XH, Wang Y, Huang FP, Shi Q. iTRAQ-based quantitative proteomic analysis for identification of oligodendroglioma biomarkers related with loss of heterozygosity on chromosomal arm 1p. J Proteomics. 2012;77:480–491. doi: 10.1016/j.jprot.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 27.Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci U S A. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weitzman SA, Gordon LI. Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood. 1990;76:655–663. [PubMed] [Google Scholar]
- 29.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 30.Cerutti PA. Oxy-radicals and cancer. Lancet. 1994;344:862–863. doi: 10.1016/s0140-6736(94)92832-0. [DOI] [PubMed] [Google Scholar]
- 31.Chae HZ, Kim IH, Kim K, Rhee SG. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268:16815–16821. [PubMed] [Google Scholar]
- 32.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiolspecific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chae HZ, Uhm TB, Rhee SG. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc Natl Acad Sci U S A. 1994;91:7022–7026. doi: 10.1073/pnas.91.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poynton RA, Hampton MB. Peroxiredoxins as biomarkers of oxidative stress. Biochim Biophys Acta. 2014;1840:906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Karihtala P, Mäntyniemi A, Kang SW, Kinnula VL, Soini Y. Peroxiredoxins in breast carcinoma. Clin Cancer Res. 2003;9:3418–3424. [PubMed] [Google Scholar]
- 36.Zhang B, Wang Y, Su Y. Peroxiredoxins, a novel target in cancer radiotherapy. Cancer Lett. 2009;286:154–160. doi: 10.1016/j.canlet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 37.Cha MK, Suh KH, Kim IH. Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J Exp Clin Cancer Res. 2009;28:93. doi: 10.1186/1756-9966-28-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke-Gaffney A, Callister ME, Nakamura H. Thioredoxin: friend or foe in human disease? Trends Pharmacol Sci. 2005;26:398–404. doi: 10.1016/j.tips.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Chen MF, Lee KD, Yeh CH, Chen WC, Huang WS, Chin CC, Lin PY, Wang JY. Role of peroxiredoxin I in rectal cancer and related to p53 status. Int J Radiat Oncol Biol Phys. 2010;78:868–878. doi: 10.1016/j.ijrobp.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 40.LES Netto, Chae HZ, Kang SW, Rhee SG, Stadtman ER. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties: TSA possesses thiol peroxidase activity. J Biol Chem. 1996;271:15315–21. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- 41.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 42.Ishii T, Yamada M, Sato H, Matsue M, Taketani S, Nakayama K, Sugita Y, Bannai S. Cloning and characterization of a 23-kDa stress-induced mouse peritoneal macrophage protein. J Biol Chem. 1993;268:18633–18636. [PubMed] [Google Scholar]
- 43.Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 2001;21:2085–2090. [PubMed] [Google Scholar]
- 44.Kinnula VL, Lehtonen S, Sormunen R, Kaarteenaho-Wiik R, Kang SW, Rhee SG, Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 45.Chang JW, Jeon HB, Lee JH, Yoo JS, Chun JS, Kim JH, Yoo YJ. Augmented expression of peroxiredoxin I in lung cancer. Biochem Biophys Res Commun. 2001;289:507–512. doi: 10.1006/bbrc.2001.5989. [DOI] [PubMed] [Google Scholar]
- 46.Verdoucq L, Vignols F, Jacquot JP, Chartier Y, Meyer Y. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- 47.Wen ST, Van Etten RA. The PAG gene product, a stressinduced protein with antioxidant properties, is an Abl SH3-binding protein and a physiological inhibitor of c-Abl tyrosine kinase activity. Genes Dev. 1997;11:2456–2467. doi: 10.1101/gad.11.19.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mu ZM, Yin XY, Prochownik EV. Pag, a putative tumor suppressor, interacts with the Myc box II domain of c-Myc and selectively alters its biological function and target gene expression. J Biol Chem. 2002;277:43175–43184. doi: 10.1074/jbc.M206066200. [DOI] [PubMed] [Google Scholar]
- 49.Sobin LH, Fleming ID. TNM classification of malignant tumors, fifth edition (1997). Union internationale contre le cancer and the american joint committee on cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 50.Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preiser CM, Fléjou JF, Geboes K, Hattori T, Hirota T, Itabashi M, Iwafuchi M, Iwashita A, Kim YI, Kirchner T, Klimpfinger M, Koike M, Lauwers GY, Lewin KJ, Oberhuber G, Offner F, Price AB, Rubio CA, Shimizu M, Shimoda T, Sipponen P, Solcia E, Stolte M, Watanabe H, Yamabe H. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin J, Park J, Kim K, Kang Y, Park SG, Kim JH, Park KS, Jun H, Kim Y. Detection of differential proteomes of human beta-cells during islet-like differentiation using iTRAQ labeling. J Proteome Res. 2009;8:1393–1403. doi: 10.1021/pr800765t. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Singleton PA, Rowshan A, Gucek M, Cole RN, Graham DR, Van Eyk JE, Garcia JG. Quantitative proteomics analysis of human endothelial cell membrane rafts: evidence of MARCKS and MRP regulation in the sphingosine 1-phosphate-induced barrier enhancement. Mol Cell Proteomics. 2007;6:689–696. doi: 10.1074/mcp.M600398-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Mol Cell Proteomics. 2008;7:1434–1451. doi: 10.1074/mcp.M700478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Jia X, Feng Y, Peng X, Zhang Z, Zhou W, Zhang Z, Ma F, Liu X, Zheng Y, Yang P, Yuan Z. Plasma membrane proteome analysis of the early effect of alcohol on liver: implications for alcoholic liver disease. Acta Biochim Biophys Sin. 2011;43:19–29. doi: 10.1093/abbs/gmq108. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, Li YP, Chen JH, Yuan SF, Wang L, Zhang JL, Yao Q, Li NL, Bian JF, Fan J, Yi J, Ling R. Prognostic significance of USP22 as an oncogene in papillary thyroid carcinoma. Tumour Biol. 2013;34:1635–1639. doi: 10.1007/s13277-013-0696-0. [DOI] [PubMed] [Google Scholar]
- 57.Deliconstantinos G, Villiotou V, Fassitsas C. Ultraviolet-irradiated human endothelial cells elaborate nitric oxide that may evoke vasodilatory response. J Cardiovasc Pharmacol. 1992;20:63–65. doi: 10.1097/00005344-199204002-00019. [DOI] [PubMed] [Google Scholar]
- 58.Deliconstantinos G, Villiotou V, Stavrides JC. Scavenging effects of hemoglobin and related heme containing compounds on nitric oxide, reactive oxidants and carcinogenic volatile nitrosocompounds of cigarette smoke. A new method for protection against the dangerous cigarette constituents. Anticancer Res. 1994;14:2717–2726. [PubMed] [Google Scholar]
- 59.Deliconstantinos G, Villiotou V, Stavrides JC. Modulation of particulate nitric oxide synthase activity and peroxynitrite synthesis in cholesterol enriched endothelial cell membranes. Biochem Pharmacol. 1995;49:1589–1600. doi: 10.1016/0006-2952(95)00094-g. [DOI] [PubMed] [Google Scholar]
- 60.Ye R, Kong X, Yang Q, Zhang Y, Han J, Zhao G. Ginsenoside Rd attenuates redox imbalance and improves stroke outcome after focal cerebral ischemia in aged mice. Neuropharmacology. 2011;61:815–824. doi: 10.1016/j.neuropharm.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Riddell JR, Bshara W, Moser MT, Spernyak JA, Foster BA, Gollnick SO. Peroxiredoxin 1 controls prostate cancer growth through tolllike receptor 4-dependent regulation of tumor vasculature. Cancer Res. 2011;71:1637–1646. doi: 10.1158/0008-5472.CAN-10-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang JW, Lee SH, Lu Y, Yoo YJ. Transforming growth factor-β1 induces the non-classical secretion of peroxiredoxin-I in A549 cells. Biochem Biophys Res Commun. 2006;345:118–123. doi: 10.1016/j.bbrc.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 64.Du Y, Zhang H, Zhang X, Lu J, Holmgren A. Thioredoxin 1 is inactivated due to oxidation induced by peroxiredoxin under oxidative stress and reactivated by the glutaredoxin system. J Biol Chem. 2013;288:32241–32247. doi: 10.1074/jbc.M113.495150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riddell JR, Wang XY, Minderman H, Gollnick SO. Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J Immunol. 2010;184:1022–1030. doi: 10.4049/jimmunol.0901945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palozza P, Serini S, Torsello A, Di Nicuolo F, Piccioni E, Ubaldi V, Pioli C, Wolf FI, Calviello G. Beta-carotene regulates NF-kappaB DNA-binding activity by a redox mechanism in human leukemia and colon adenocarcinoma cells. J Nutr. 2003;133:381–388. doi: 10.1093/jn/133.2.381. [DOI] [PubMed] [Google Scholar]
- 67.Biswas SK, Lewis CE. NF-κB as a central regulator of macrophage function in tumors. J Leukoc Biol. 2010;88:877–884. doi: 10.1189/jlb.0310153. [DOI] [PubMed] [Google Scholar]
- 68.Koomen JM, Haura EB, Bepler G, Sutphen R, Remily-Wood ER, Benson K, Hussein M, Hazlehurst LA, Yeatman TJ, Hildreth LT, Sellers TA, Jacobsen PB, Fenstermacher DA, Dalton WS. Proteomic contributions to personalized cancer care. Mol Cell Proteomics. 2008;7:1780–1794. doi: 10.1074/mcp.R800002-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9:133–148. doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su YJ, Xu F, Yu JP, Yue DS, Ren XB, Wang CL. Up-regulation of the expression of S100A8 and S100A9 in lung adenocarcinoma and its correlation with inflammation and other clinical features. Chin Med J. 2010;123:2215–2220. [PubMed] [Google Scholar]
- 71.Song M, Wu K, Ogino S, Fuchs CS, Giovannucci EL, Chan AT. A prospective study of plasma inflammatory markers and risk of colorectal cancer in men. Br J Cancer. 2013;108:1891–1898. doi: 10.1038/bjc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satelli A, Rao PS, Thirumala S, Rao US. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int J Cancer. 2011;129:799–809. doi: 10.1002/ijc.25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toppo S, Flohé L, Ursini F, Vanin S, Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 74.Brigelius-Flohé R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim Biophys Acta. 2009;1790:1555–1568. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 75.Moore MM, Chua W, Charles KA, Clarke SJ. Inflammation and cancer: causes and consequences. Clin Pharmacol Ther. 2010;87:504–508. doi: 10.1038/clpt.2009.254. [DOI] [PubMed] [Google Scholar]
- 76.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 77.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 78.Neumann CA, Fang Q. Are peroxiredoxins tumor suppressors? Curr Opin Pharmacol. 2007;7:375–380. doi: 10.1016/j.coph.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–7142. doi: 10.1158/0008-5472.CAN-05-4446. [DOI] [PubMed] [Google Scholar]
- 80.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 81.Chen MF, Keng PC, Shau H, Wu CT, Hu YC, Liao SK, Chen WC. Inhibition of lung tumor growth and augmentation of radiosensitivity by decreasing peroxiredoxin I expression. Int J Radiat Oncol Biol Phys. 2006;64:581–591. doi: 10.1016/j.ijrobp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Riddell JR, Maier P, Sass SN, Moser MT, Foster BA, Gollnick SO. Peroxiredoxin 1 stimulates endothelial cell expression of VEGF via TLR4 dependent activation of HIF-1α. PLoS One. 2012;7:e50394. doi: 10.1371/journal.pone.0050394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SU, Park YH, Min JS, Sun HN, Han YH, Hua JM, Lee TH, Lee SR, Chang KT, Kang SW, Kim JM, Yu DY, Lee SH, Lee DS. Peroxiredoxin I is a ROS/p38 MAPK-dependent inducible antioxidant that regulates NF-κB-mediated iNOS induction and microglial activation. J Neuroimmunol. 2013;259:26–36. doi: 10.1016/j.jneuroim.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 84.Kang SW, Chae HZ, Seo MS, Kim K, Baines IC, Rhee SG. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generatedin response to growth factors and tumor necrosis factor-α. J Biol Chem. 1998;273:6297–6302. doi: 10.1074/jbc.273.11.6297. [DOI] [PubMed] [Google Scholar]
- 85.Ishii T, Warabi E, Yanagawa T. Novel roles of peroxiredoxins in inflammation, cancer and innate immunity. J Clin Biochem Nutr. 2012;50:91–105. doi: 10.3164/jcbn.11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giles GI. The redox regulation of thiol dependent signaling pathways in cancer. Curr Pharm Des. 2006;12:4427–4443. doi: 10.2174/138161206779010549. [DOI] [PubMed] [Google Scholar]
- 87.Chen AC, Arany PR, Huang YY, Tomkinson EM, Sharma SK, Kharkwal GB, Saleem T, Mooney D, Yull FE, Blackwell TS, Hamblin MR. Lowlevel laser therapy activates NF-κB via generation of reactive oxygen species in mouse embryonic fibroblasts. PLoS One. 2011;6:e22453. doi: 10.1371/journal.pone.0022453. [DOI] [PMC free article] [PubMed] [Google Scholar]