Abstract
Circular RNAs (circRNAs), a class of endogenous RNAs, are characterized by covalently closed continuous loop without 5’ to 3’ polarity and polyadenylated tail. Recent studies indicated that circRNAs might play an important role in cancer. However, the function of circRNA in laryngeal squamous cell cancer tissues (LSCC) is still unknown. In this study, we investigated the expression of circRNAs in 4 paired LSCC tissues and adjacent non-tumor tissues by microarray analysis. Results showed significant upregulation (n = 302) of or downregulation (n = 396) of 698 circRNAs in LSCC tissues. We further detected hsa_circRNA_100855 as the most upregulated circRNA and hsa_circRNA_104912 as the most downregulated circRNA using qRT-PCR methods. Results showed that hsa_circRNA_100855 level was significantly higher in LSCC than in the corresponding adjacent non-neoplastic tissues. Patients with T3-4 stage, neck nodal metastasis or advanced clinical stage had higher hsa_circRNA_100855 expression. The hsa_circRNA_104912 level was significantly lower in LSCC than in corresponding adjacent non-neoplastic tissues. Patients with T3-4 stage, neck nodal metastasis, poor differentiation or advanced clinical stage had a lower hsa_circRNA_104912 expression. Overall, our data suggest that circRNAs play an important role in the tumorigenesis of LSCC and may serve as novel and stable biomarkers for the diagnosis and progress of LSCC.
Keywords: Laryngeal squamous cell cancer (LSCC), noncoding RNA, circular RNAs, progress
Introduction
Laryngeal squamous cell carcinoma (LSCC) is one of the most frequent malignant neoplasms of head and neck. Current treatments include laryngectomy, radiotherapy and chemotherapy. Despite several strategies and interventions, the 5-year overall survival rate in advanced LSCC is still unsatisfactory [1]. Therefore, better understanding of the molecular mechanisms associated with progression is critical to the development of effective therapeutic approaches against LSCC. Circular RNA (circRNA) is a novel class of endogenous noncoding RNAs (ncRNAs) that regulate the gene expression at the transcriptional or post-transcriptional level by modulating microRNAs or other molecules. CircRNAs were reported decades ago, but were mostly misconstrued as “splicing rubbish” [2]. Recent advances including high-throughput sequencing and bioinformatics, led to rediscovery of circRNAs and characterized as widespread, abundant, stable, conserved and tissue-specific molecules in mammalian cells [3-6]. Compared with linear RNAs that terminate in 5’ caps and 3’ tails, circRNAs form covalently closed loops without 5’ or 3’ polarities [7]. Therefore, circRNAs are not degraded by RNase R, which digest linear RNAs [8]. CircRNAs are important competing endogenous RNAs (ceRNAs). They function as microRNA sponges and regulate the expression of miRNA-targeted transcripts in many eukaryotes [9]. Recently, circRNAs have been reported to mediate cancer progression [10]. However, little is known about the role of circRNAs in LSCC. In the present study, we analyzed the expression of circRNAs in LSCC tissues and adjacent non-tumor tissues using microarray analysis. We found that 698 circRNAs were significantly differentially expressed. Using real-time PCR, we found that hsa_circRNA_100855 upregulation and hsa_circRNA_104912 downregulation significantly correlated with the progression of LSCC. These results provide evidence that circRNAs play an important role in LSCC and may serve as novel tumor markers for the progress of LSCC.
Material and methods
Patients and specimens
The study included patients with laryngeal cancer who underwent partial or total laryngectomy at the Department of Otorhinolaryngology of the Second Affiliated Hospital of Harbin Medical University (China) between August 2013 and December 2014. Four matched samples of LSCC tissues and corresponding adjacent non-neoplastic tissues were tested using microarray analysis. Real-time qPCR was used to test 52 matched cancerous and noncancerous tissues. The study was approved by the Ethics Committee of Harbin Medical University and informed consent was obtained. The patients had not received any cancer therapy before admission. After surgery, the matched specimens of LSCC and the corresponding adjacent non-tumor tissues obtained from patients were preserved in liquid nitrogen within 5 minutes of excision. They were transported frozen to the laboratory for storage at -80°C.
Total RNA isolation and quality control (QC)
Total RNA was isolated from tumor and adjacent tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The purity and concentration of total RNA samples were quantified using the NanoDrop ND-1000 (NanoDrop Technologies/Thermo Scientific, Wilmington, DE, USA). The integrity of RNA was determined by electrophoresis using a denaturing agarose gel.
RNA labeling and hybridization
In order to enrich pure circRNAs, the total RNA in each sample was treated with RNase R (Epicentre, Inc.) to delete linear RNAs. The enriched circRNAs in each sample were amplified and transcribed into fluorescent circRNA utilizing a random priming method according to Arraystar Super RNA Labeling protocol (Arraystar, Inc). The labeled circRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled circRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. The labeled circRNAs were hybridized onto the Arraystar Human circRNA Arrays (8×15 K, Arraystar). The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven (Agilent Technologies, Santa Clara, CA, USA). After washing the slides, we scanned the arrays using the Agilent Scanner G2505C (Agilent Technologies, Santa Clara, CA, USA).
Data collection and circRNA expression
Raw data was extracted using Agilent Feature Extraction software (version 11.0.1.1). After quantile normalization performed using the R software package of the raw data, low intensity filtering was performed and the circRNAs with at least 4 out of 8 samples containing flags in Present or Marginal (“All Targets Value”) were retained for further analyses. We drew a Box Plot to quickly visualize the distribution of the intensities from all datasets after normalization. The unsupervised hierarchical clustering was performed to analyze the circRNA expression of the samples. CircRNAs expressed significantly different between the LSCC and paired non-tumor tissues were identified using fold change cut-off and Volcano Plot filtering, respectively. The Scatter-Plot was used to assess the variation in circRNA expression between the two groups of samples.
qRT-PCR
The real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using the qPCR Master Mix (Arraystar) on a ViiA 7 Real-time PCR System (Applied Biosystems) following the manufacturer’s instructions. Divergent primers of hsa_circRNA_100855 and hsa_circRNA_104912 were designed. The sequences of hsa_circRNA_100855, hsa_circRNA_104912 and GAPDH primers were as follows: 5’-GCCAAGGCTCAGCAGAAACTA-3’ (sense) and 5’-TGTTGCTCCAAGACCTTGTCC-3’ (antisense) for hsa_circRNA_100855; 5’-GAACTTCACATTCGTGCTCAC-3’ (sense) and 5’-TCCTGGTCACTGTAG TCCTCC-3’ (antisense) for hsa_circRNA_104912; and 5’-GGGAAACTGTGGCG TGAT-3’ (sense) and 5’-GAGTGGGTGTCGCTGTTGA-3’ (antisense) for GAPDH. The Ct values obtained against known concentrations of serially diluted circRNAs were used to construct a standard curve. The circRNA levels were normalized using GAPDH as the internal control. The relative expression levels of cirRNAs were calculated using T (tumor)/N (non-tumor) ratios.
Statistical analysis
All quantitative data were presented as mean ± SEM as indicated from at least three independent experiments. Significance was determined using the Student’s t test for differences of circRNAs expression. P < 0.05 was considered as statistically significant.
Results
CircRNA expression profiles in LSCC and normal tissues
Using the circRNA microarray, we assessed the circRNA profiles in four paired LSCC and normal tissues. We drew a Box plot to quickly visualize the distribution of the intensities from all the datasets after normalization and found that the distribution of log2 ratios was similar in all the tested samples (Figure 1). To hypothesize the relationships between the samples, we used unsupervised hierarchical clustering for analysis of circRNA expression. Cluster analysis segregates samples into groups based on differences in their expression levels, and hypothetic relationships among the samples. Results of hierarchical clustering show distinct circRNA expression profiling among the samples (Figure 2). The Scatter plot may be used for visual assessment of variation in circRNA expression between LSCC and non-tumor tissues. The values spotted in the X and Y axes represent the average of normalized signals of the two groups of samples (log2 scaled). The middle green line represents a fold change of 1, indicating no difference between the two groups of samples. The circRNAs above the top green line and below the bottom green line indicate more than 2.0 fold change in circRNAs between the LSCC and non-tumor tissue samples. The scatter plot was used to derive the general data involving 2.0 fold change in samples (Figure 3). The Volcano plot was performed to visualize the differential expression between LSCC and non-tumor tissues. The significant differences (fold change > 2.0, P value < 0.05) between the two different conditions are illustrated in the Volcano plot (Figure 4). The microarray data showed the significant differential expression of the 698 circRNAs (fold change > 2.0, P value < 0.05). They include 302 upregulated and 396 downregulated circRNAs in tumor tissues, respectively. Forty-two circRNAs exhibited greater than 5-fold increase, 29 greater than 5-fold decrease, 20 greater than 10-fold increase, and 2 more than 10-fold decrease. Here, we first detected the expression of hsa_circRNA_100855 (fold change 110.07 up; P < 0.001) and hsa_circRNA_104912 (fold change 21.97 down; P < 0.001), which show the highest or the lowest expression in LSCC compared with the adjacent non-tumor samples.
Figure 1.
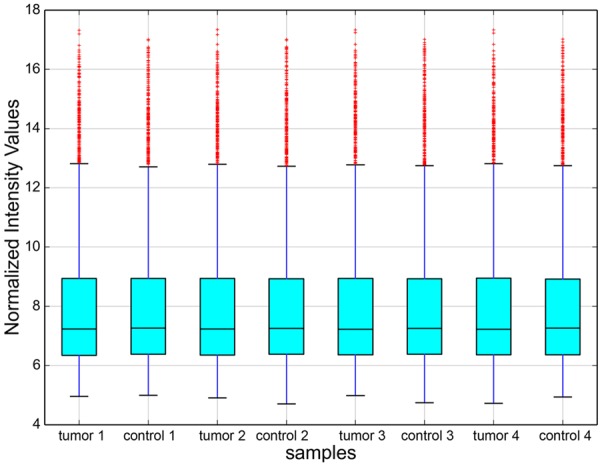
Box plot. X-axis: 8 tissue samples; Y-axis: normalized intensity values. All the 8 samples in the databases were normalized. The distribution of circRNAs was almost similar in all samples.
Figure 2.
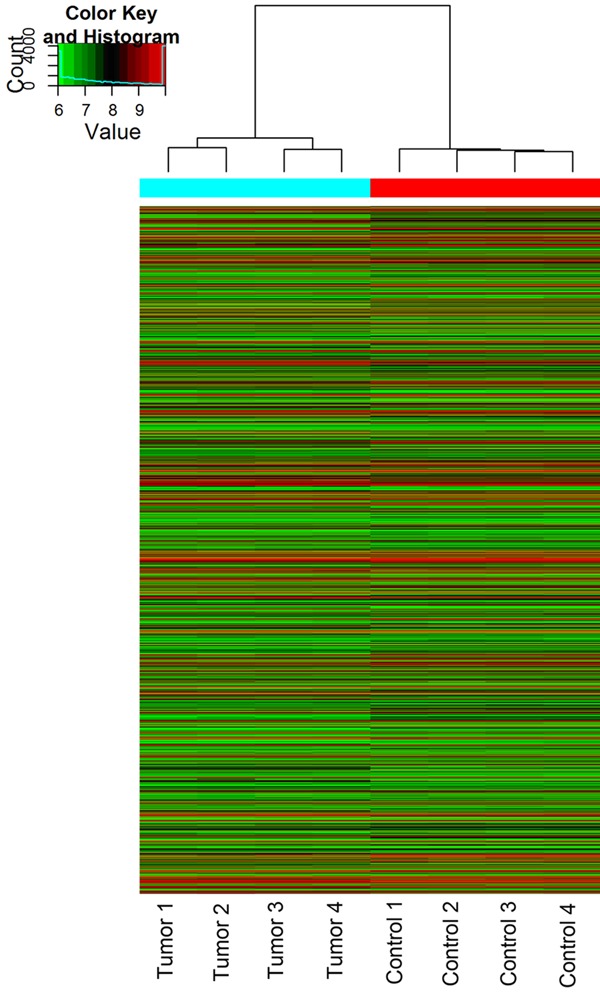
Unsupervised hierarchical cluster: unique circRNA expression profile of the 8 samples in red (low relative expression) or green (high relative expression) scale.
Figure 3.
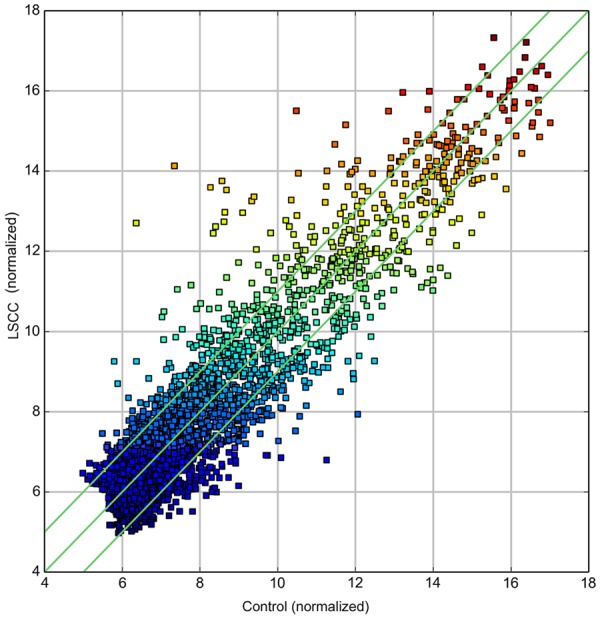
Scatter plot. X-axis: adjacent non-tumor tissues (normalized), Y-axis: LSCC tissues (normalized). The green lines represent fold change. The circRNAs above the top green line and below the bottom green line indicate more than 2.0 fold change in circRNAs between the two groups of samples.
Figure 4.
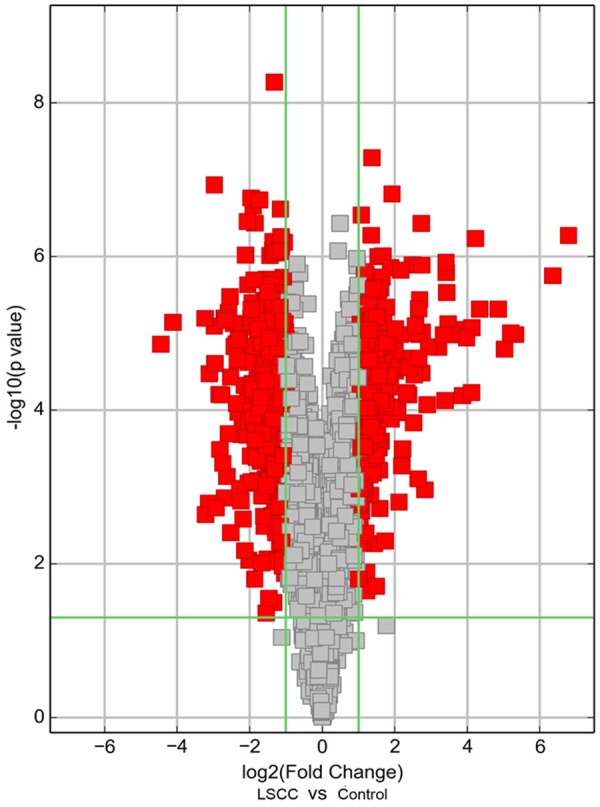
Volcano plot. X-axis: log2 (fold change); Y-axis: -log10 (P-value). The vertical green lines correspond to 2.0-fold up and down, and the horizontal green line represents a p-value of 0.05. The red points in the plot represent circRNAs expressed differentially with statistical significance.
Expression of hsa_circRNA_100855 and hsa_circRNA_104912 in LSCC tissues
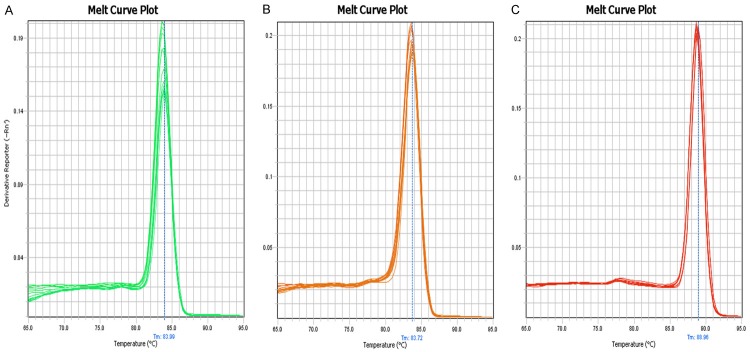
A melting curve analysis showed that the qPCR amplified product yielded a single peak, characteristic of hsa_circRNA_100855 and hsa_circRNA_104912 (Figure 5). We found that hsa_circRNA_100855 levels in LSCC tissues were significantly higher (10.486-fold) than in the corresponding nontumor tissues. Hsa_circRNA_100855 expression was related to T grade, neck nodal metastasis, primary location and clinical stage of LSCC (Table 1; Figure 6). Tumors with grade T3 to T4, lymph node metastasis, supraglottic location or advanced clinical stages expressed higher levels of hsa_circRNA_100855. Hsa_circRNA_104912 level in LSCC tissues was significantly lower (0.211-fold) than in the corresponding non-tumor tissues. Hsa_circRNA_104912 expression was statistically related to T grade, differentiation, neck nodal metastasis, and clinical stage of LSCC (Table 2; Figure 7). Tumors with grade T3 to T4, lymph node metastasis, poor differentiation or advanced clinical stages expressed lower levels of hsa_circRNA_104912.
Figure 5.
Melting curve. The qPCR-amplified product yielded a single peak, indicating the specific amplifications. A: Hsa_circRNA_100855; B: hsa_circRNA_104912; C: GAPDH.
Table 1.
Relationship between hsa_circRNA_100855 expression and clinicopathological features of LSCC
| Characteristics (n) | hsa_circRNA_100855 level | P |
|---|---|---|
| Gender | 0.113 | |
| Male (37) | 10.095±3.385 | |
| Female (15) | 11.451±3.632 | |
| Age | 0.121 | |
| ≥ 58 (26) | 9.918±3.287 | |
| < 58 (26) | 11.054±3.632 | |
| T classification | 0.006 | |
| T1-2 (32) | 9.468±2.906 | |
| T3-4 (20) | 12.114±3.761 | |
| Differentiation | 0.487 | |
| G1 (35) | 10.467±2.045 | |
| G2 (17) | 10.495±4.021 | |
| Lymph node metastasis | 0.003 | |
| Negative (34) | 9.403±2.622 | |
| Positive (18) | 12.531±4.019 | |
| Primary location | 0.007 | |
| Supraglottis (23) | 11.846±3.811 | |
| Glottis (29) | 9.407±2.810 | |
| Clinical stage | 0.001 | |
| I-II (31) | 9.230±2.614 | |
| III-IV (21) | 12.341±3.810 |
Figure 6.
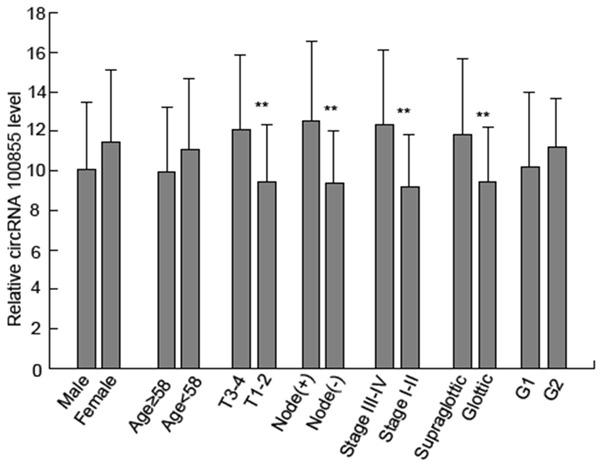
Expression of hsa_circRNA_100855 in LSCC tissues. T3-4 grade tumors, with supraglottic location, lymph node metastasis or advanced clinical stage expressed higher levels of hsa_circRNA_100855. *P < 0.05; **P < 0.01.
Table 2.
Relationship between hsa_circRNA_104912 expression and clinicopathologic features of LSCC
| Characteristics (n) | hsa_circRNA_104912 level | P |
|---|---|---|
| Gender | 0.280 | |
| Male (37) | 0.216±0.108 | |
| Female (15) | 0.198±0.080 | |
| Age | 0.160 | |
| ≥58 (26) | 0.197±0.097 | |
| <58 (26) | 0.226±0.113 | |
| T classification | 0.010 | |
| T1-2 (32) | 0.237±0.108 | |
| T3-4 (20) | 0.170±0.088 | |
| Differentiation | 0.039 | |
| G1 (35) | 0.227±0.114 | |
| G2 (17) | 0.178±0.076 | |
| Lymph node metastasis | 0.020 | |
| Negative (34) | 0.236±0.109 | |
| Positive (18) | 0.174±0.093 | |
| Primary location | 0.109 | |
| Supraglottis (23) | 0.191±0.089 | |
| Glottis (29) | 0.227±0.115 | |
| Clinical stage | 0.008 | |
| I-II (31) | 0.239±0.109 | |
| III-IV (21) | 0.171±0.086 |
Figure 7.
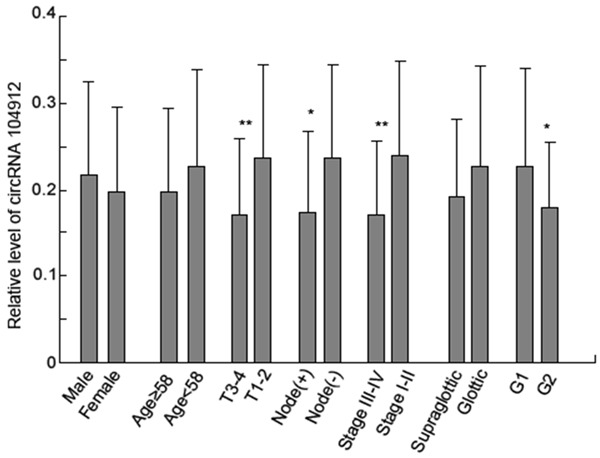
Expression of hsa_circRNA_104912 in LSCC tissues. Advanced tumors with poor differentiation, T3-4 grade or lymph node metastasis expressed lower levels of hsa_circRNA_104912. *P < 0.05; **P < 0.01.
Discussion
Evidence confirms that the mammalian genome encodes thousands of ncRNAs that are pervasively transcribed. Although ncRNAs have frequently been disregarded as transcriptional ‘noise’, substantial evidence suggests a functional role. The most studied ncRNAs to date are miRNAs and long noncoding RNAs, which have been implicated in a variety of biological processes. Dysregulated miRNAs and lncRNAs represent potential biomarkers and therapeutic targets for cancer [11,12]. Our previous studies also demonstrated that miRNAs and lncRNAs play an important role in the carcinogenesis of LSCC [13,14].
Recently, another type of ncRNA, circRNA received extensive attention of researchers with the development of transcriptome sequencing technologies. CircRNAs were observed in RNA viruses more than thirty years ago and were predominantly seen in several human gene isoforms in 2012 [2,15]. CircRNAs are abundant, stable, evolutionarily conserved and cell-type specific. Some circRNAs have miRNA response elements (MREs) and exhibit important miRNA activities, increasing the complexity of RNA regulatory networks and playing a role in gene expression [16]. These circRNAs are also called miRNA sponges [9]. CiRS-7 (also termed CDR1as), act as specific miR-7 inhibitors, and harbor more than 70 conventional miR-7 binding sites for suppression of miR-7 activities [17]. As miR-7 directly downregulates the expression of major oncogenes, ciRS-7 displays superior anti-cancer abilities and miR-7/ciRS-7 axis probably plays a critical role in cancer processes [10]. Another circRNA, cir-ITCH also functions as miRNA (miR-7, miR-17, and miR-214) sponge and plays an inhibitory role in esophageal squamous cell carcinoma [18]. Further, a few circRNAs may represent a new type of tumor biomarkers. Hsa_circ_002059 expression is lower in gastric cancer tissues and relates to distal metastasis, TNM stage, gender and age [19]. Hsa_circ_0001649 was downregulated and correlated with tumor emboli in hepatocellular carcinoma [20]. In this study, we performed microarray sequencing of four pairs of primary LSCC tissues and adjacent normal tissues. We found for the first time that there were 698 differentially expressed circRNAs in LSCC tissues. Among them, 302 were significantly upregulated and 396 were significantly downregulated. Consistent with microarray results, our qRT-PCR revealed that hsa_circRNA_100855 and hsa_circRNA_104912 levels were dysregulated in LSCC. Hsa_circRNA_100855 was upregulated in LSCC specimens compared with the control tissues. In particular, patients with grade T3 to T4, lymph node metastasis, supraglottic location or advanced clinical stages expressed higher levels of hsa_circRNA_100855. Conversely, hsa_circRNA_104912 was downregulated in LSCC and patients with grade T3 to T4, lymph node metastasis, poor differentiation or advanced clinical stages expressed lower levels of hsa_circRNA_104912. These data suggest that circRNAs may play a role in carcinogenesis and progress of LSCC. To further understand the biological role of circRNAs in LSCC, both in vitro and in vivo assays should be performed in the future after successfully engineering circRNAs.
In conclusion, we found that circRNAs are dysregulated in LSCC. Hsa_circRNA_100855 and hsa_circRNA_104912 may serve as potential biomarkers and therapeutic targets for laryngeal cancer. CircRNAs provide new insights into disease mechanisms. Further study to unravel the functional mechanisms of circRNAs in LSCC and other cancers is essential.
Acknowledgements
This study was supported by grants from the National science Foundation of china (No. 81272965, 81241085, 81402257) and Foundation of Health Department of Heilongjiang Province (No. 2013115).
Disclosure of conflict of interest
None.
References
- 1.Rudolph E, Dyckhoff G, Becher H, Dietz A, Ramroth H. Effects of tumour stage, comorbidity and therapy on survival of laryngeal cancer patients: a systematic review and a meta-analysis. Eur Arch Otorhinolaryngol. 2011;268:165–179. doi: 10.1007/s00405-010-1395-8. [DOI] [PubMed] [Google Scholar]
- 2.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 3.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 4.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 5.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 6.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase Rdigested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA Processing and Human Cancer. J Clin Med. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva A, Bullock M, Calin G. The Clinical Relevance of Long Non-Coding RNAs in Cancer. Cancers (Basel) 2015;7:2169–2182. doi: 10.3390/cancers7040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren J, Zhu D, Liu M, Sun Y, Tian L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of Laryngeal squamous cell carcinoma. Eur J Cancer. 2010;46:3409–3416. doi: 10.1016/j.ejca.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 15.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 18.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/betacatenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–9. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]