Abstract
Background: The laminin-binding integrin (LBI) family are cell adhesion molecules that are essential for invasion and metastasis of human epithelial cancers and cell adhesion mediated drug resistance. We investigated whether copy number alteration (CNA) or mutations of a five-gene signature (ITGB4, ITGA3, LAMB3, PLEC, and SYNE3), representing essential genes for LBI adhesion, would correlate with patient outcomes within human epithelial-type tumor data sets currently available in an open access format. Methods: We investigated the relative alteration frequency of an LBI signature panel (integrin β4 (ITGB4), integrin α3 (ITGA3), laminin β3 chain (LAMB3), plectin (PLEC), and nesprin 3 (SYNE3)), independent of the epithelial cancer type, within publically available and published data using cBioPortal and Oncomine software. We rank ordered the results using a 20% alteration frequency cut-off and limited the analysis to studies containing at least 100 samples. Kaplan-Meier survival curves were analyzed to determine if alterations in the LBI signature correlated with patient survival. The Oncomine data mining tool was used to compare the heat map expression of the LBI signature without SYNE3 (as this was not included in the Oncomine database) to drug resistance patterns. Results: Twelve different cancer types, representing 5,647 samples, contained at least a 20% alteration frequency of the five-gene LBI signature. The frequency of alteration ranged from 38.3% to 19.8%. Within the LBI signature, PLEC was the most commonly altered followed by LAMB3, ITGB4, ITGA3, and SYNE3 across all twelve cancer types. Within cancer types, there was little overlap of the individual amplified genes from each sample, suggesting different specific amplicons may alter the LBI adhesion structures. Of the twelve cancer types, overall survival was altered by CNA presence in bladder urothelial carcinoma (p=0.0143*) and cervical squamous cell carcinoma and endocervical adenocarcinoma (p=0.0432*). Querying the in vitro drug resistance profiles with the LBI signature demonstrated a positive correlation with cells resistant to inhibitors of HDAC (Vorinostat, Panobinostat) and topoisomerase II (Irinotecan). No correlation was found with the following agents: Bleomycin, Doxorubicin, Methotrexate, Gemcitabine, Docetaxel, Bortezomib, and Shikonen. Conclusions: Our work has identified epithelial-types of human cancer that have significant CNA in our selected five-gene signature, which was based on the essential and genetically-defined functions of the protein product networks (in this case, the LBI axis). CNA of the gene signature not only predicted overall survival in bladder, cervical, and endocervical adenocarcinoma but also response to chemotherapy. This work suggests that future studies designed to optimize the gene signature are warranted. General Significance: The copy number alteration of structural components of the LBI axis in epithelial-type tumors may be promising biomarkers and rational targets for personalized therapy in preventing or arresting metastatic spread.
Keywords: Cancer, gene copy, laminin, integrin, cBioPortal, copy number alterations
Introduction
DNA copy number alterations in cancer
Human cancer is often caused by irreparable structural mutations in cells. The mutations can promote alterations in DNA copy number at very specific genomic locations [1], changing the function of the gene, and thereby producing a transformed phenotype [2]. Several developmental disorders, such as Down Syndrome, Prader Willi, and Angelman, for example, are triggered by gain or loss in a single copy of a chromosome [3]. Pollack and his team provide evidence that extensive DNA copy number alterations (CNA) can generate global deregulation in gene expression, which may be a factor in the genesis and progression of tumors [4]. Identifying and locating copy number alterations can offer an approach for linking CNA with disease phenotype and for pinpointing critical genes, all of which can be highly useful in treating tumors or in developing novel treatments.
Clinicians and researchers are now able to employ increasingly sophisticated sequencing technology (comparative genomic hybridization [CGH], single-nucleotide polymorphism [SNP] [5]) to identify copy number alterations and their relationship to tumor severity in a variety of cancers, including head neck squamous cancers [6], multiple myeloma [7], prostate cancer [8-10], primary cutaneous malignant melanomas [11], chronic lymphocytic leukemia [12], the transformation of follicular lymphoma to diffuse large cell lymphoma [13], urinary bladder cancer [14], breast cancer [15], hepatocellular carcinoma [16], and pancreatic cancer [17,18], among many others.
Access to publically available databases with user-friendly interfaces, such as the cBioPortal (http://www.cbioportal.org) [19,20], Oncomine (http://www.oncomine.com) [21,22], and STRING (http://string-db.org) [23,24], prompted the current study. Our goal was to determine if the copy number alterations of essential members of the laminin-binding integrin (LBI) axis correlated with aggressive cancer subtypes or drug-resistant phenotypes. Investigating CNA (i.e. gene amplification) as a point of regulation for the abundance of LBIs is particularly relevant since integrins are constitutively synthesized, recycled, rarely degraded, and have a biological half-life longer than the duration of a cell cycle [25,26]. CNA would be the major mechanism to increase LBI abundance on tumor cell surfaces during tumor metastasis. The essential gene products in the laminin-binding integrin axis required for tumor metastatic progression were investigated, in contrast to standard approaches investigating CNA in prostate cancer (PCa) [27].
In the study by Ross-Adams, et al. [27], candidates were selected based on transcription and gene variation data by comparing normal and cancer tissue in 259 men. Five separate patient subgroups were identified based on 100 unique genes, of which six were previously known to play a role in prostate cancer (MAP3K7, MELK, RCBTB2, ELAC2, TPD52, ZBTB4), and 94 genes were previously unlinked to PCa progression. This observation allowed the authors to reliably predict biochemical relapse. However, patients with poor prognosis (Gleason score >7) were spread across all five clusters, failing to differentiate the clinical significance of any one cluster. Furthermore, since the identified genes were specific to nucleic acid processing, transcription factor binding, and phosphorylation of proteins, the authors could only predict biochemical relapse, whereas predicting metastatic potential is much more beneficial in determining survival [28,29].
Our approach was to query the data for CNA of the LBI axis. An abundance of cell culture and experimental mouse models have investigated the role of laminin-binding integrins and their interacting proteins in cancer progression [30-33], but only sporadic reports exist with human tissue studies indicating the LBI axis as important in cancer progression [34-38]. A five-gene signature consisting of essential laminin adhesion structures known to cause human disease was created—β4 integrin (ITGB4) [39-42], α3 integrin (ITGA3) [35,43,44], laminin β3 chain (LAMB3) [45,46], plectin (PLEC) [40,47,48], and nesprin 3 (SYNE3) [49-51]. We sought to determine if CNA in this five-gene signature could be observed in human cancer by screening numerous cancer types using the open-access resources. Selection of these five-genes synthesizes many separate research strands linking two or more of these genes as critical elements in cancer progression and/or metastasis [45-56], and defines more clearly the potential role of the laminin-binding integrin axis in disease progression. Our approach may suggest alternative targeted therapies or biomarker networks based upon phenotype selection of the gene candidates, in line with emerging research efforts to subtype-classify various tumors [42,57,58].
Integrins and their role in cancer
Integrins are a class of non-covalently bound, heterodimeric cell surface receptors composed of α and β subunits, and responsible for cell adhesion to the extracellular matrix (ECM), cell signaling, and cell migration [59-63]. Humans possess 18 α and 8 β integrin subunits, combining in 24 distinct heterodimers [63]. Three alpha integrins are involved in laminin-binding (α3, α6, and α7), comprising heterodimers α3β1, α6β1, α6β4, and α7β1, and representing a highly conserved class of integrins essential for normal development [62,64-66]. There is considerable evidence for the role of α6 integrin, also called CD49f, in the progression of a variety of epithelial cancers [67-70], as well as its role in the migration of normal cell processes associated with neuroblasts [71,72] and the myelination of peripheral nerve cells via Schwann cell activity [73]. In partnership with neuroligin 1 (NLGN1), a cell adhesion molecule, α6 integrin plays a crucial role in neurovascular development [74]. Compared with α6, the role of α7β1 has been so far limited to its presence in melanoma cells [75], myoblasts [76], and the skeletal neuromuscular junction [77], although there is some evidence of anti-metastatic properties for α7β1 [47,78]. Conversely, α3β1 is a basement membrane receptor that also appears to modulate adhesion, migration, and cytoskeletal organization [43,79] and, along with α6β1, is important for proper formation of the cerebral cortex [80].
The three laminin-binding alpha integrins also are implicated in the invasion steps of cancer metastasis [60,61,81-83], in part through interactions with tetraspanins [32,84,85], and the drug-resistant phenotype of metastatic disease [86-88]. Previous research from our group has detailed regulation by laminin-binding integrins and laminin ECM proteins of cell adhesion and migration (metastasis) during prostate cancer progression [61,89-91]. Das, et al. [89], and Sroka, et al. [61], have demonstrated the role of α3 and α6 in perineural invasion (PNI) as a major route for prostate cancer metastasis, while Liebig, et al. [92], offers a comprehensive overview of PNI in various cancers. Further, our group has identified a novel variant of α6β1 in prostate cancer—α6pβ1—which is unique to human cancer tissue and tumor cell lines [91,93]. The α6p variant occurs on the tumor cell surface by removal of the extracellular laminin-binding domain by the serine protease urokinase plasminogen activator (uPA) [91,94], an occurrence that may provide the most common mechanism of extracapsular spread in prostate metastasis [95].
Materials and methods
Identifying the protein components of the laminin-binding integrin (LBI) axis
The STRING analysis tool was used to determine interacting proteins using ITGB4 as the query. The β4 integrin was used because, biologically, this is the seed site for building dominant adhesion structures in normal epithelial tissues. Several known partners have been genetically verified and therefore served as the foundation for finding the other protein partners in the axis. Any proteins identified that were not specific to the LBI axis, (e.g., adapter proteins [GRB2]) were excluded from the gene signature.
Immunohistochemistry
Human cancer tissues were fixed in 10% neutral buffered formalin for 24 hours, processed, paraffin embedded, and immunohistochemistry performed using the AA6NT antibody (1:700) on a Discovery XT Automated Immunostainer (Ventana Medical Systems, Inc., Tucson, AZ). All de-identified tissues were processed through the tissue acquisition and molecular analysis support resource (TACMASR) of the UA Cancer Center.
Analysis of cBioPortal data
We utilized the ability to conduct an integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal data, an open-access resource at http://www.cbioportal.org/[96,97]. The portal reduces molecular profiling data from cancer tissues and cell lines into readily understandable genetic, epigenetic, gene expression, and proteomic events. The query interface combined with customized data storage enabled us to interactively explore genetic alterations across samples curated from national and international cancer studies and specific genes. This web-based tool was used to query five genes simultaneously: ITGB4, ITGA3, LAMB3, PLEC, and SYNE3. In the query, no cancer studies were pre-selected and approximately 91 studies were analyzed. The data type priority, selected by us, was mutation and copy number alteration (CNA). The LBI gene set was defined by the STRING analysis coupled with retaining candidate gene products genetically defined as axis partners and eliminating gene products with known interactions across multiple pathways. The resulting HUGO gene symbols were submitted together and the analysis was provided by the tool. Further data analysis was restricted by requiring at least 100 samples in the data set and at least a 20% or larger change in the CNA and mutation within the gene set.
Analysis of Oncomine data
The four LBI genes (the fifth gene in our selection, SYNE3, is not in this database) were entered into the Oncomine database (Thermo Fisher Scientific Inc., v4.5: http://www.oncomine.com) [21,22] and searched with a variety of filters until “drug sensitivity analysis” and “chemotherapy sensitivity analysis” were arrived at as most useful. All results were then filtered for top 1% of gene rank, a fold change value of 4, and a P-value of 1E-4. Each drug resistance profile was viewed through both an under- and over-expression order. Heat maps of gene expression were generated for the four LBI genes and each of the chemo drugs in the Oncomine database.
Statistical analysis
Survival curves generated by the cBioPortal were analyzed to determine whether any alterations in patient survival occurred when comparing cases that contained an alteration in the five LBI genes with those without an alteration in the five LBI genes. The results are displayed as Kaplan-Meier plots with P values from a logrank test. Similarly, with Oncomine, heat maps were generated comparing the expression of the LBI genes between drug-resistant and drug-sensitive cells. Statistical significance of the data (P-values) was provided by the program.
Results
Immunohistochemistry detection of integrin α6 in aggressive human epithelial-type tumors
Laminin-binding integrins and, in particular, the α6 integrin, have been shown to be a normal stem cell adhesion and signaling protein axis for the invasion, migration, and patterning of embryonic tissue and, in adults, regenerating tissue following injury. In human cancer, cohesive collectives of cells are found in invasive prostate cancer, cancer in circulation, and in prostate cancer within metastatic sites such as bone. In human prostate cancer tissues, α6 integrin is found typically between the tumor cells as a cohesive collection of tumor during cancer invasion and metastasis [38]. Here we surveyed by immunohistochemistry α6 protein expression in other aggressive epithelial tumors (pancreatic, breast) in bone, lymph node, and a highly infiltrative axial skeletal neoplasm with epithelial characteristics (chordoma). In these aggressive human cancer specimens, α6 integrin is predominantly expressed on the cell membrane as well as in the cytoplasm (Figure 1), suggesting active trafficking of the adhesion receptor.
Figure 1.
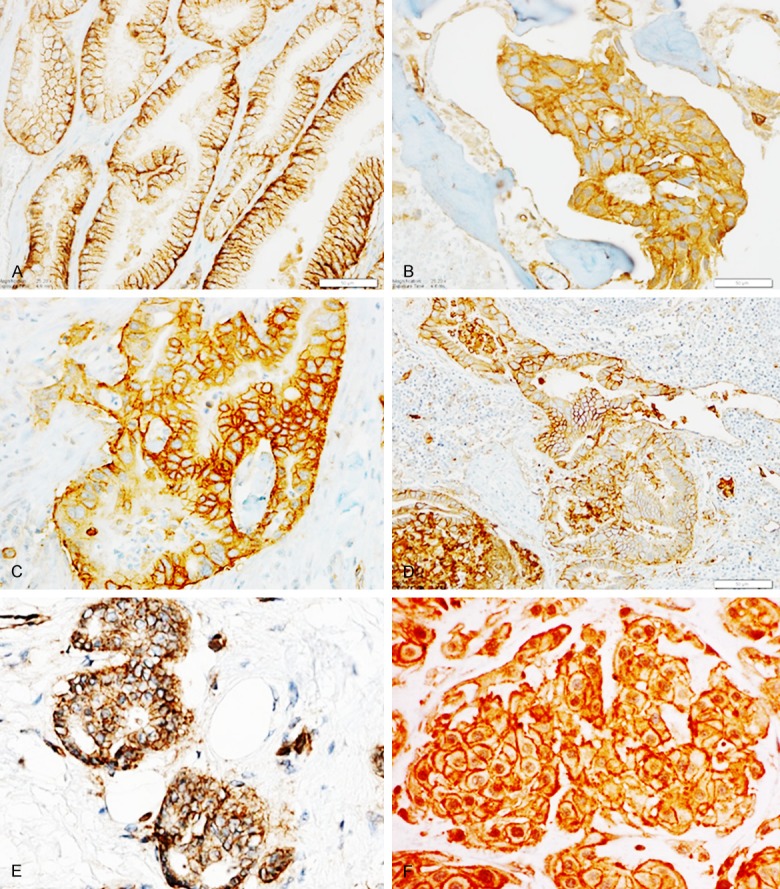
Immunohistochemistry detection of laminin-binding integrin A6 in aggressive human cancer specimens. (scale bar, as indicated): (A) prostate cancer, (B) prostate cancer bone metastasis, (C) pancreatic tumor, (D) pancreatic tumor metastatic to lymph node, (E) breast cancer, and (F) chordoma (highly infiltrative skeletal neoplasm with epithelial characteristics).
Significantly, the distribution in tumors is around the tumor cells in a pattern distinct from the polarized cell-ECM distribution that is observed in normal tissues [38]. For example, in normal prostate glands, the α6 integrin is distributed at the base of the gland, anchoring the basal cells to a basal lamina composed of laminin 332. In contrast, the tumor tissue contains the α6 integrin distributed as a cell-cell adhesion molecule, suggesting a dramatic change in function.
The α6 integrin is a laminin-binding integrin that will dominantly pair with β4 or pair with β1 when β4 is absent. Since β1 will pair with many alpha integrin subunits, the β4 subunit was used as the query to find other protein partners associated with α6β4. Our next step was to utilize a STRING program to survey potential candidates based on the eight lines of evidence used in the algorithm.
Protein components of nodes across the laminin-binding integrin axis
Using an open-access resource called STRING v10.0 (http://string-db.org), we selected the functional protein partners of integrin α6β4 using data from peer-reviewed publications and curated databases (Figure 2). The ten predicted proteins (with the corresponding gene names) include: plectin (PLEC), integrin α6 (ITGA6), collagen type XVII (COL17A1), laminin β3 (LAMB3), integrin α3 (ITGA3), laminin α3 (LAMA3), met proto-oncogene (hepatocyte growth factor receptor, MET), the adapter proteins, Src homology 2 domain, which contains (SHC1) and growth factor receptor-bound protein 2 (GRB2), and protein kinase C, alpha (PRKCA).
Figure 2.
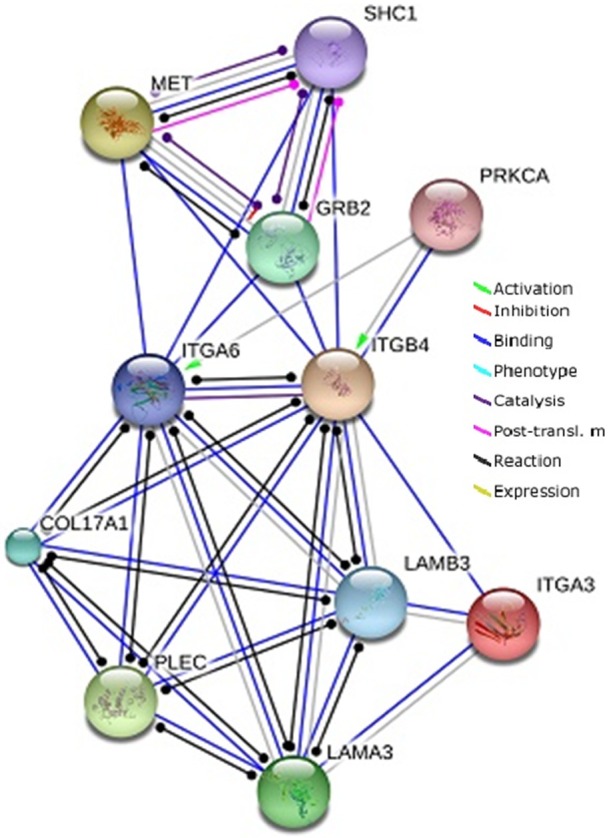
Identification of known and predicted structural proteins essential for laminin-binding integrin (ITGB4) function. Interacting nodes are displayed in colored circles using String, v10.0. Predicted functional partners of β4 integrin are shown based upon peer reviewed published data and curated database entries. [STRING v.10 (http://string-db.org)].
As Figure 2 illustrates, ITGB4 interacts with ITGA6 as expected for normal heterodimer formation and interacts with its ligands, LAMA3, LAMB3, and with PLEC, which is known to be a component of a LBI-based adhesion structure called the hemidesmosome. In considering the proteins essential for the LBI axis, proteins that were required but not specific to the LBI axis or those that were not rate limiting for its function were eliminated from further analysis. The excluded genes included GRB2, PRKCA, COL17A1, LAMA3, MET, and SHC1. Reduction from the 10 original proteins to the five used in the cBioPortal analysis (and four in the Oncomine analysis, as SYNE3 was not in their database) was based on knowing the essential genetic components for the LBI axis and the components associated with cancer invasion and metastasis.
Unbiased cross cancer subtypes correlations using cBioPortal data
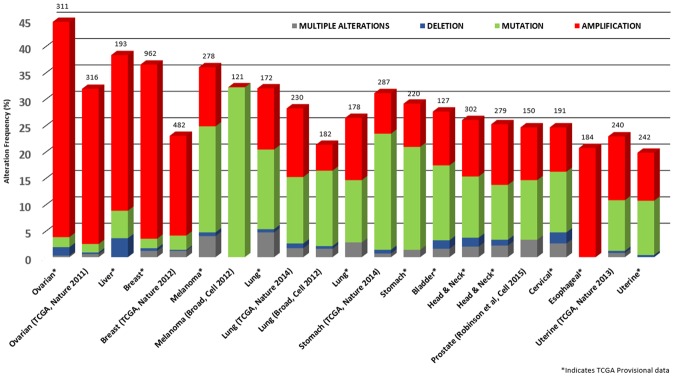
Using the five-gene query, the cBioPortal tool analyzed 91 different cancer studies for mutation or copy number alterations. The results returned 21 different cancer studies representing 5,647 samples that contained a >20% alteration frequency and at least 100 samples in the data set (Figure 3). On closer inspection, this represents approximately 12 different epithelial cancer types. Of particular interest is that the predominant pattern of amplification occurred in ovarian, liver, breast, and esophageal cancer. Evidence of mutation was most predominant in melanoma and stomach cancer. Minor changes in deletion or multiple alterations were observed in the data.
Figure 3.
Copy number alteration of laminin-binding integrin genes and cancer subtypes. The alteration frequency of a five-gene signature (ITGA3, ITGB4, LAMB3, PLEC, and SYNE3) was determined using the cBioPortal (http://www.cbioportal.org). Only cancer types containing >100 samples and an alteration frequency of >20% are shown. The alteration frequency included deletions (blue), amplification (red), multiple alterations (grey) or mutation (green). The total number of samples for each cancer type are indicated by the numbers at the top of each column.
The frequency of alteration ranged from 38.3% to 19.8% with the rank order (highest to lowest) as ovarian serous cystadenocarcinoma, liver hepatocellular carcinoma, breast invasive carcinoma, skin cutaneous melanoma, lung adenocarcinoma, lung squamous cell carcinoma, stomach adenocarcinoma, bladder urothelial carcinoma, head and neck squamous cell carcinoma, metastatic prostate cancer, cervical squamous cell carcinoma, endocervical carcinoma, and uterine corpus endometrial carcinoma.
Oncoprints of three epithelial cancer subtypes
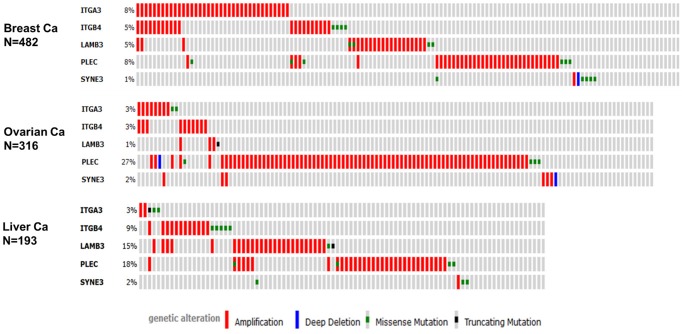
Within the LBI gene query and across all twelve cancer types, PLEC was the most commonly altered followed by LAMB3, ITGB4, ITGA3, and SYNE3. Although specific mutations occurred in PLEC, amplification was the most common feature. Since the major changes for copy number alteration were found in ovarian, breast, and liver cancer, we used the Oncoprint feature of the tool to determine the specific alterations in each gene of the signature in the data set for each cancer type.
In the analysis depicted in Figure 4 each row represents a gene and each column represents a tumor sample. The PLEC gene was amplified predominantly in all three cancer types. Inspecting the data across the grey vertical bars, which represent unique samples interrogated for the gene alteration, shows that individual samples in the majority of cases are not altered in every gene of the signature. Within cancer types, there was little overlap of the individual amplified genes within a specific case, suggesting different specific amplicons may alter the LBI adhesion structures. Stated another way, the Oncoprint shows different mechanisms of altering the LBI axis across a set of cancer samples based on a query of the five genes. Further analysis of the specific mutations listed did not reveal any “hot spots” of mutation within any of the genes in the set (data not shown). Further analysis also revealed that amplification of the genes was correlated with increased levels of mRNA in the samples (data not shown).
Figure 4.
Epithelial cancer types frequently amplify PLEC. We used the Oncoprint feature of the cBioPortal (http://www.cbioportal.org) to determine the copy number alteration frequency of each individual gene in the LBI signature within selected cancer subtypes. Grey bars along a vertical line represent the same sample interrogated for amplification (red), deep deletion (blue), missense mutation (green), truncating mutation (black) or in-frame mutation (brown).
cBioPortal LBI and CNA survival curves: two cancer types
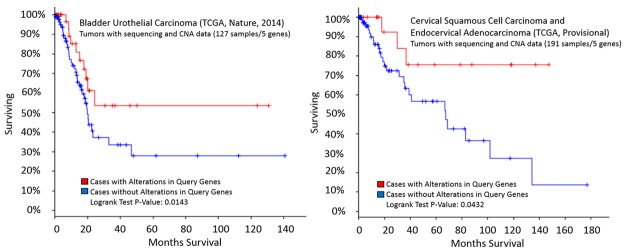
Of the twelve cancer types, a significant alteration in overall survival (Figure 5) was indicated in bladder urothelial carcinoma (p=0.0143*), as well as cervical squamous cell carcinoma and endocervical adenocarcinoma (p=0.0432*).
Figure 5.
Kaplan-Meier survival curves generated by the cBioPortal (http://www.cbioportal.org). The overall fraction of subjects surviving over time following treatment was measured among cases with and without alterations in the five-gene signature (ITGA3, ITGB4, LAMB3, PLEC, and SYNE3). A significant difference in survival between groups was observed among cases involving bladder urothelial carcinoma (p=0.0143*), cervical squamous cell carcinoma, and endocervical adenocarcinoma (p=0.0432*).
Oncomine LBI expression signature and drug resistance
The drug resistance profile from 28 chemotherapeutic agents were independent of the LBI CNA signature, including Bleomycin, Bosutinib, Carboplatin, Cisplatin, Docetaxel, Doxorubicin, Fluorouracil, Gemcitabine, Methotrexate, Mitomycin, Paclitaxel, Shikonin, and Vinblastine, among others (data not shown). Querying the drug resistance profiles with the LBI axis signature unexpectedly resulted in a positive correlation with cells resistant to two inhibitors of histone deacetylase (HDAC) (Vorinostat, Panobinostat) and topoisomerase II (Irinotecan) (Figure 6).
Figure 6.
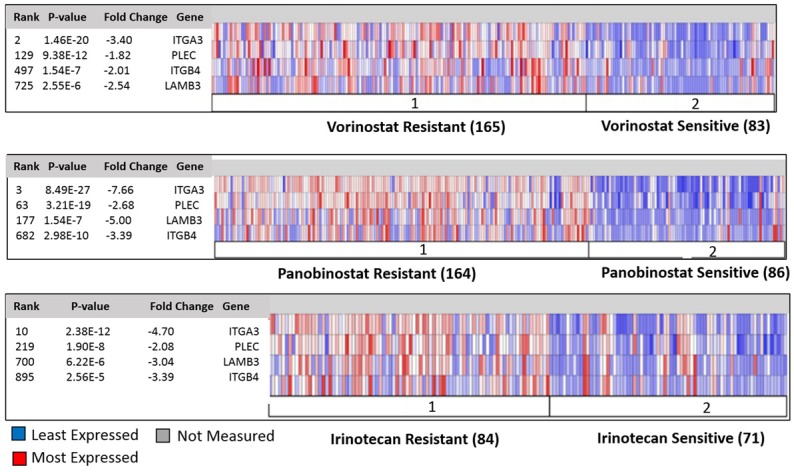
LBI expression signature and drug resistance. The Oncomine data mining tool (v4.5: http://www.oncomine.com) was used to compare the heat map expression pattern of the four gene signature (ITGA3, PLEC, ITGB4, LAMB3) in the Garnett cell lines to inhibitors of HDAC (Vorinostat, Panobinostat) and Topoisomerase II (Irinotecan).
HDAC inhibitors are a group of compounds that disrupt the function of histone deacetylase [73,74], and topoisomerase II (Top2) inhibitors [75,76] are enzymes that regulate DNA structural changes during the cell cycle. Recent work has suggested a link between drug resistance from gemcitabine and increased sensitivity to HDAC inhibitors [98].
Discussion
The cBioPortal analysis program identified 12 epithelial types of human cancer that have significant CNA in the chosen five-gene signature (ITGA3, ITGB4, LAMB3, PLEC, and SYNE3). The LBI signature was created representing the essential and genetically verified functional components of laminin-binding integrins and their adhesion structures.
Although CNA increased significantly in specific cancer subtypes, there was not uniform increase in all genes of the signature. The increase in the LBI using several different genes within the cluster, independently, suggests that the phenotype is selected and warrants further study to identify the essential elements of the LBI axis. It is important to note that the LBI structural proteins were good candidates for genes altered in copy number since they are essential “housekeeping genes” found in normal tissue, continually expressed in cancer (Figure 1) and used in cancer for metastasis. Human essential genes, similar to those in the LBI axis, are retained as duplicates to serve as “backed up copies” and normally are under stringent dosage regulation [99]. Currently, mass spectrometry approaches are identifying new targets in integrin structural and signaling complexes [100,101] that could be genetically tested for function in the LBI axis.
While some significant alterations in overall survival were indicated when considering the LBI signature using the current data (Figure 5), future work will be to monitor the trend with additional data as it becomes publically available. Another limitation of the work is that RNA transcription signatures were not sufficiently available to determine if the CNA across all the genes in the data sets correlated with increased transcription.
We note with interest that of the many drug resistance profiles in Oncomine, only the HDAC and topoisomerase inhibitor resistance correlated with the increase in laminin-binding integrin copy number expression. While potentially interesting for understanding regulation of the LBI axis, it is noted that the preferred drug-based management of epithelial tumors, such as prostate [102], are Cabazitaxel [103,104], Docetaxel and/or Mitoxantrone [105,106], and Cyclophosphamide [107,108], many of which were not in the database. Neither were the anti-androgen receptor (AR) signaling inhibitors and antagonists, Enzalutamide (2nd gen), Flutamide, Bicalutamide, Nitulamide (1st Gen), and Galterone (3rd Gen). These drugs are not considered chemotherapeutic agents for killing cancer cells but, rather, are anti-growth agents via binding to AR and displacing androgen, or down-regulating expression of the androgen-dependent genes, such as PSA and TMPRSS2. Future open-access data detailing sensitivity and response of currently tested agents with copy number analysis and mutation data will likely be useful for additional analysis.
Conclusions
The open access databases cBioPortal and Oncomine both contain user-friendly interfaces to query data across genes and cancer types from many clinical studies that are independently curated. The programs identified other epithelial types that likely will have detectable immunohistochemistry signatures of the LBI and will prompt new studies in other epithelial cancer types. The copy number alterations of specific structural components of the LBI axis in epithelial tumors may be promising targets to prevent or manage metastatic spread.
Acknowledgements
The use of the Tissue Acquisition and Molecular Analysis Support Service (TACMASR) of the UA Cancer Center was essential for this work. Of particular note was the expert technical assistance of Ed Abril. The work was supported in part by CA P30 23074, CA164484 and CA159406.
Disclosure of conflict of interest
None.
References
- 1.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 2.Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, Chin K, Devries S, Feiler H, Gray JW, Waldman F, Pinkel D, Albertson DG. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 4.Pollack JR, Sørlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Børresen-Dale A-L, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. PNAS. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BS, Barretina J, Socci ND, DeCarolis P, Ladanyi M, Meyerson M, Singer S, Sander C. Functional Copy-Number Alterations in Cancer. PLoS One. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, Lang W, Issa JP, Lee JJ, Mao L. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J. Clin. Oncol. 2007;25:2164–2170. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 7.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, Attal M, Marit G, Mathiot C, Facon T, Moreau P, Anderson KC, Campion L, Munshi NC, Minvielle S. Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. 2009;27:4585–4590. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, Xu J. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, Turner AR, Kim JW, Meyers DA, Zheng SL, Isaacs WB, Xu J. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosom Cancer. 2006;45:1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 10.Schlomm T, Kirstein P, Iwers L, Daniel B, Steuber T, Walz J, Chun FH, Haese A, Kollermann J, Graefen M, Huland H, Sauter G, Simon R, Erbersdobler A. Clinical Significance of Epidermal Growth Factor Receptor Protein Overexpression and Gene Copy Number Gains in Prostate Cancer. Clin Cancer Res. 2007;13:6579–6584. doi: 10.1158/1078-0432.CCR-07-1257. [DOI] [PubMed] [Google Scholar]
- 11.Rákosy Z, Vízkeleti L, Ecsedi S, Vokó Z, Bégány Á, Barok M, Krekk Z, Gallai M, Szentirmay Z, Ádány R, Balázs M. EGFR gene copy number alterations in primary cutaneous malignant melanomas are associated with poor prognosis. Int J Cancer. 2007;121:1729–1737. doi: 10.1002/ijc.22928. [DOI] [PubMed] [Google Scholar]
- 12.Gunnarsson R, Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N, Rasmussen M, Staaf J, Lundin J, Norin S, Buhl AM, Smedby KE, Hjalgrim H, Karlsson K, Jurlander J, Juliusson G, Rosenquist R. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2009;24:211–215. doi: 10.1038/leu.2009.187. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Climent JA, Alizadeh AA, Segraves R, Blesa D, Rubio-Moscardo F, Albertson DG, Garcia-Conde J, Dyer MJ, Levy R, Pinkel D, Lossos IS. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101:3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- 14.Simon R, Richter J, Wagner U, Fijan A, Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knönagel H, Rist M, Wilber K, Anabitarte M, Hering F, Hardmeier T, Schönenberger A, Flury R, Jäger P, Fehr JL, Schraml P, Moch H, Mihatsch MJ, Gasser T, Sauter G. High-Throughput Tissue Microarray Analysis of 3p25 (RAF1) and 8p12 (FGFR1) Copy Number Alterations in Urinary Bladder Cancer. Cancer Res. 2001;61:4514–4519. [PubMed] [Google Scholar]
- 15.Zhang Y, Martens JW, Yu JX, Jiang J, Sieuwerts AM, Smid M, Klijn JG, Wang Y, Foekens JA. Copy Number Alterations that Predict Metastatic Capability of Human Breast Cancer. Cancer Res. 2009;69:3795–3801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 16.Kim TM, Yim SH, Shin SH, Xu HD, Jung YC, Park CK, Choi JY, Park WS, Kwon MS, Fiegler H, Carter NP, Rhyu MG, Chung YJ. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008;123:2808–2815. doi: 10.1002/ijc.23901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidenblad M, Lindgren D, Veltman JA, Jonson T, Mahlamaki EH, Gorunova L, van Kessel AG, Schoenmakers EF, Hoglund M. Microarray analyses reveal strong influence of DNA copy number alterations on the transcriptional patterns in pancreatic cancer: implications for the interpretation of genomic amplifications. Oncogene. 2005;24:1794–1801. doi: 10.1038/sj.onc.1208383. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Auletta T, Dovirak O, Hutter C, Kuntz K, El-ftesi S, Kendall J, Han H, Von Hoff DD, Ashfaq R, Maitra A, Iacobuzio-Donahue CA, Hruban RH, Lucito R. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J. Integrin traffic - the update. J Cell Sci. 2015;128:839–852. doi: 10.1242/jcs.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witkowski CM, Bowden GT, Nagle RB, Cress AE. Altered surface expression and increased turnover of the alpha6beta4 integrin in an undifferentiated carcinoma. Carcinogenesis. 2000;21:325–330. doi: 10.1093/carcin/21.2.325. [DOI] [PubMed] [Google Scholar]
- 27.Ross-Adams H, Lamb AD, Dunning MJ, Halim S, Lindberg J, Massie CM, Egevad LA, Russell R, Ramos-Montoya A, Vowler SL, Sharma NL, Kay J, Whitaker H, Clark J, Hurst R, Gnanapragasam VJ, Shah NC, Warren AY, Cooper CS, Lynch AG, Stark R, Mills IG, Grönberg H, Neal DE CamCaP Study Group. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine. 2015;2:1133–44. doi: 10.1016/j.ebiom.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman RE. Clinical Features of Metastatic Bone Disease and Risk of Skeletal Morbidity. Clin Cancer Res. 2006;12:6243s–6249s. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 29.Coleman RE. Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat Rev. 2001;27:165–176. doi: 10.1053/ctrv.2000.0210. [DOI] [PubMed] [Google Scholar]
- 30.Sadej R, Grudowska A, Turczyk L, Kordek R, Romanska HM. CD151 in cancer progression and metastasis: a complex scenario. Lab Invest. 2014;94:41–51. doi: 10.1038/labinvest.2013.136. [DOI] [PubMed] [Google Scholar]
- 31.van der Linde MR, Crijns HJ, de Koning J, Hoogkamp-Korstanje JA, de Graaf JJ, Piers DA, van der Galien A, Lie KI. Range of atrioventricular conduction disturbances in Lyme borreliosis: a report of four cases and review of other published reports. Br Heart J. 1990;63:162–168. doi: 10.1136/hrt.63.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 33.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teller IC, Beaulieu JF. Interactions between laminin and epithelial cells in intestinal health and disease. Expert Rev Mol Med. 2001;3:1–18. doi: 10.1017/S1462399401003623. [DOI] [PubMed] [Google Scholar]
- 35.Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. α3β1 integrin–CD151, a component of the cadherin–catenin complex, regulates PTPμ expression and cell–cell adhesion. J cell biol. 2003;163:1351–1362. doi: 10.1083/jcb.200306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshioka T, Otero J, Chen Y, Kim YM, Koutcher JA, Satagopan J, Reuter V, Carver B, de Stanchina E, Enomoto K, Greenberg NM, Scardino PT, Scher HI, Sawyers CL, Giancotti FG. β4 Integrin signaling induces expansion of prostate tumor progenitors. J Clin Invest. 2013;123:682–699. doi: 10.1172/JCI60720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmelz M, Cress AE, Scott KM, Burger F, Cui H, Sallam K, McDaniel KM, Dalkin BL, Nagle RB. Different phenotypes in human prostate cancer: alpha6 or alpha3 integrin in cellextracellular adhesion sites. Neoplasia. 2002;4:243–254. doi: 10.1038/sj.neo.7900223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagle RB, Cress AE. Metastasis Update: Human Prostate Carcinoma Invasion via Tubulogenesis. Prostate Cancer. 2011;2011:249290. doi: 10.1155/2011/249290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nisticò P, Di Modugno F, Spada S, Bissell MJ. β1 and β4 integrins: from breast development to clinical practice. Breast Cancer Res. 2014;16:459. doi: 10.1186/s13058-014-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nievers MG, Kuikman I, Geerts D, Leigh IM, Sonnenberg A. Formation of hemidesmosomelike structures in the absence of ligand binding by the (alpha) 6 (beta) 4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the (beta) 4 integrin subunit. J cell sci. 2000;113:963–973. doi: 10.1242/jcs.113.6.963. [DOI] [PubMed] [Google Scholar]
- 41.Cress A, Rabinovitz I, Zhu W, Nagle R. The α6β1 and α6β4 integrins in human prostate cancer progression. Cancer and Metast Rev. 1995;14:219–228. doi: 10.1007/BF00690293. [DOI] [PubMed] [Google Scholar]
- 42.Lu S, Simin K, Khan A, Mercurio AM. Analysis of integrin beta4 expression in human breast cancer: association with basal-like tumors and prognostic significance. Clin Cancer Res. 2008;14:1050–1058. doi: 10.1158/1078-0432.CCR-07-4116. [DOI] [PubMed] [Google Scholar]
- 43.DiPersio CM, Hodivala-Dilke KM, Jaenisch R, Kreidberg JA, Hynes RO. α3β1 integrin is required for normal development of the epidermal basement membrane. J Cell Biol. 1997;137:729–742. doi: 10.1083/jcb.137.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bijnsdorp IV, Geldof AA, Lavaei M, Piersma SR, van Moorselaar RJ, Jimenez CR. Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuruta D, Kobayashi H, Imanishi H, Sugawara K, Ishii M, Jones JC. Laminin-332-Integrin Interaction: A Target For Cancer Therapy? J Med Chem. 2008;15:1968–1975. doi: 10.2174/092986708785132834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon O, Park J, Kim M, Kim J, Lee H, Kim H, Noh S, Song K, Yoo H, Paik S. Aberrant upregulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem Biophys Res Commun. 2011;406:539–545. doi: 10.1016/j.bbrc.2011.02.082. [DOI] [PubMed] [Google Scholar]
- 47.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. doi: 10.1017/S1462399409001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McInroy L, Määttä A. Plectin regulates invasiveness of SW480 colon carcinoma cells and is targeted to podosome-like adhesions in an isoform-specific manner. Exp Cell Res. 2011;317:2468–2478. doi: 10.1016/j.yexcr.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Morgan JT, Pfeiffer ER, Thirkill TL, Kumar P, Peng G, Fridolfsson HN, Douglas GC, Starr DA, Barakat AI. Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Mol Biol Cell. 2011;22:4324–4334. doi: 10.1091/mbc.E11-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilhelmsen K, Litjens SHM, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 52.Koster J, Geerts D, Favre B, Borradori L, Sonnenberg A. Analysis of the interactions between BP180, BP230, plectin and the integrin α6β4 important for hemidesmosome assembly. J Cell Sci. 2003;116:387–399. doi: 10.1242/jcs.00241. [DOI] [PubMed] [Google Scholar]
- 53.Yamakawa N, Kaneda K, Saito Y, Ichihara E, Morishita K. The Increased Expression of Integrin α6 (ITGA6) Enhances Drug Resistance in EVI1(high) Leukemia. PLoS One. 2012;7:e30706. doi: 10.1371/journal.pone.0030706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng Q, Lin HC, He XH, Li JJ, Yao M. Integrative analyses identify osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for lung cancer. PLoS One. 2013;8:e55714. doi: 10.1371/journal.pone.0055714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Tzu J, Marinkovich MP. Bridging structure with function: structural, regulatory, and developmental role of laminins. Int J Biochem Cell Biol. 2008;40:199–214. doi: 10.1016/j.biocel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagata M, Noman AA, Suzuki K, Kurita H, Ohnishi M, Ohyama T, Kitamura N, Kobayashi T, Uematsu K, Takahashi K, Kodama N, Kawase T, Hoshina H, Ikeda N, Shingaki S, Takagi R. ITGA3 and ITGB4 expression biomarkers estimate the risks of locoregional and hematogenous dissemination of oral squamous cell carcinoma. BMC Cancer. 2013;13:410–410. doi: 10.1186/1471-2407-13-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penney KL, Sinnott JA, Tyekucheva S, Gerke T, Shui IM, Kraft P, Sesso HD, Freedman ML, Loda M, Mucci LA, Stampfer MJ. Association of prostate cancer risk variants with gene expression in normal and tumor tissue. Cancer Epidemiol Biomarkers Prev. 2015;24:255–260. doi: 10.1158/1055-9965.EPI-14-0694-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choudhury Y, Wei X, Chu YH, Ng LG, Tan HS, Koh V, Thike AA, Poon E, Ng QS, Toh CK, Kanesvaran R, Tan PH, Tan MH. A Multigene Assay Identifying Distinct Prognostic Subtypes of Clear Cell Renal Cell Carcinoma with Differential Response to Tyrosine Kinase Inhibition. Eur Urol. 2015;67:17–20. doi: 10.1016/j.eururo.2014.06.041. [DOI] [PubMed] [Google Scholar]
- 59.Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 61.Sroka IC, Anderson TA, McDaniel KM, Nagle RB, Gretzer MB, Cress AE. The laminin binding integrin alpha6beta1 in prostate cancer perineural invasion. J Cell Physiol. 2010;224:283–288. doi: 10.1002/jcp.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishiuchi R, Takagi J, Hayashi M, Ido H, Yagi Y, Sanzen N, Tsuji T, Yamada M, Sekiguchi K. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin–integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 2006;25:189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Takagi J. Structural basis for ligand recognition by integrins. Curr Opin Cell Biol. 2007;19:557–564. doi: 10.1016/j.ceb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 65.Lowell CA, Mayadas TN. Overview: studying integrins in vivo. Methods Mol Biol. 2012;757:369–397. doi: 10.1007/978-1-61779-166-6_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes AL. Evolution of the integrin alpha and beta protein families. J Mol Evol. 2001;52:63–72. doi: 10.1007/s002390010134. [DOI] [PubMed] [Google Scholar]
- 67.Landowski TH, Gard J, Pond E, Pond GD, Nagle RB, Geffre CP, Cress AE. Targeting Integrin alpha6 Stimulates Curative-Type Bone Metastasis Lesions in a Xenograft Model. Mol Cancer Ther. 2014;13:1558–1566. doi: 10.1158/1535-7163.MCT-13-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demetriou MC, Kwei KA, Powell MB, Nagle RB, Bowden GT, Cress AE. Integrin A6 Cleavage in Mouse Skin Tumors. Open Cancer J. 2008;2:1–4. doi: 10.2174/1874079000802010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kacsinta AD, Rubenstein CS, Sroka IC, Pawar S, Gard JM, Nagle RB, Cress AE. Intracellular modifiers of integrin alpha 6p production in aggressive prostate and breast cancer cell lines. Biochem Biophys Res Commun. 2014;454:335–40. doi: 10.1016/j.bbrc.2014.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, Rich JN. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley JG, Hagg T. α6β1 integrin directs migration of neuronal precursors in adult mouse forebrain. Exp Neurol. 2003;183:273–285. doi: 10.1016/s0014-4886(03)00209-7. [DOI] [PubMed] [Google Scholar]
- 72.Flanagan LA, Rebaza LM, Derzic S, Schwartz PH, Monuki ES. Regulation of Human Neural Precursor Cells by Laminin and Integrins. J Neurosci Res. 2006;83:845–856. doi: 10.1002/jnr.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKee KK, Yang DH, Patel R, Chen ZL, Strickland S, Takagi J, Sekiguchi K, Yurchenco PD. Schwann cell myelination requires integration of laminin activities. J Cell Sci. 2012;125:4609–4619. doi: 10.1242/jcs.107995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samarelli AV, Riccitelli E, Bizzozero L, Silveira TN, Seano G, Pergolizzi M, Vitagliano G, Cascone I, Carpentier G, Bottos A, Primo L, Bussolino F, Arese M. Neuroligin 1 induces blood vessel maturation by cooperating with the alpha6 integrin. J Biol Chem. 2014;289:19466–19476. doi: 10.1074/jbc.M113.530972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kramer RH, Vu MP, Cheng YF, Ramos DM, Timpl R, Waleh N. Laminin-binding integrin alpha 7 beta 1: functional characterization and expression in normal and malignant melanocytes. Cell Regul. 1991;2:805–817. doi: 10.1091/mbc.2.10.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von der Mark H, Dürr J, Sonnenberg A, von der Mark K, Deutzmann R, Goodman S. Skeletal myoblasts utilize a novel beta 1-series integrin and not alpha 6 beta 1 for binding to the E8 and T8 fragments of laminin. J Biol Chem. 1991;266:23593–23601. [PubMed] [Google Scholar]
- 77.Martin PT, Kaufman SJ, Kramer RH, Sanes JR. Synaptic Integrins in Developing, Adult, and Mutant Muscle: Selective Association of α1, α7A, and α7B Integrins with the Neuromuscular Junction. Devel Biol. 1996;174:125–139. doi: 10.1006/dbio.1996.0057. [DOI] [PubMed] [Google Scholar]
- 78.Ren B, Yan PY, Tseng GC, Wu C, Chen K, Rao UN, Nelson J, Michalopoulos GK, Luo JH. Analysis of integrin α7 mutations in prostate cancer, liver cancer, glioblastoma multiforme, and leiomyosarcoma. J Nat Cancer Inst. 2007;99:868–880. doi: 10.1093/jnci/djk199. [DOI] [PubMed] [Google Scholar]
- 79.Kreidberg JA. Functions of α3β1 integrin. Curr Opin Cell Biol. 2000;12:548–553. doi: 10.1016/s0955-0674(00)00130-7. [DOI] [PubMed] [Google Scholar]
- 80.Danen EH, Sonnenberg A. Integrins in regulation of tissue development and function. J Pathol. 2003;200:471–480. doi: 10.1002/path.1416. [DOI] [PubMed] [Google Scholar]
- 81.Ziober BL, Lin CS, Kramer RH. Lamininbinding integrins in tumor progression and metastasis. Semin Cancer Biology. 1996;7:119–128. doi: 10.1006/scbi.1996.0017. [DOI] [PubMed] [Google Scholar]
- 82.Vogelmann R, Kreuser ED, Adler G, Lutz MP. Integrin α6β1 role in metastatic behavior of human pancreatic carcinoma cells. Int J Cancer. 1999;80:791–795. doi: 10.1002/(sici)1097-0215(19990301)80:5<791::aid-ijc25>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 83.Lv G, Lv T, Qiao S, Li W, Gao W, Zhao X, Wang J. RNA interference targeting human integrin alpha6 suppresses the metastasis potential of hepatocellular carcinoma cells. Eur J Med Res. 2013;18:52. doi: 10.1186/2047-783X-18-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 85.Sterk LM, Geuijen CA, van den Berg JG, Claessen N, Weening JJ, Sonnenberg A. Association of the tetraspanin CD151 with the lamininbinding integrinsα 3β1, α6β1, α6β4 and α7β1 in cells in culture and in vivo. J Cell Sci. 2002;115:1161–1173. doi: 10.1242/jcs.115.6.1161. [DOI] [PubMed] [Google Scholar]
- 86.Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood. 1999;93:1658–1667. [PMC free article] [PubMed] [Google Scholar]
- 87.Bauman PA, Dalton WS, Anderson JM, Cress AE. Expression of cytokeratin confers multiple drug resistance. Proc Natl Acad Sci U S A. 1994;91:5311–5314. doi: 10.1073/pnas.91.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 89.Das L, Anderson TA, Gard J, Sroka IC, Strautman SR, Nagle RB, Morrissey C, Knudsen BS, Cress AE. Loss of A3 Integrin Expression Promotes Increased A6 Integrin Internalization to Rab4 Vesicles and Increased Migration of Human Prostate Cancer Cells. J Biol Chem (In press) [Google Scholar]
- 90.King TE, Pawar SC, Majuta L, Sroka IC, Wynn D, Demetriou MC, Nagle RB, Porreca F, Cress AE. The role of alpha 6 integrin in prostate cancer migration and bone pain in a novel xenograft model. PLoS One. 2008;3:e3535. doi: 10.1371/journal.pone.0003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Demetriou MC, Pennington ME, Nagle RB, Cress AE. Extracellular alpha 6 integrin cleavage by urokinase-type plasminogen activator in human prostate cancer. Exp Cell Res. 2004;294:550–558. doi: 10.1016/j.yexcr.2003.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer. Cancer. 2009;115:3379–3391. doi: 10.1002/cncr.24396. [DOI] [PubMed] [Google Scholar]
- 93.Davis TL, Rabinovitz I, Futscher BW, Schnölzer M, Burger F, Liu Y, Kulesz-Martin M, Cress AE. Identification of a novel structural variant of the α6 integrin. J Biol Chem. 2001;276:26099–26106. doi: 10.1074/jbc.M102811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Demetriou MC, Cress AE. Integrin clipping: A novel adhesion switch? J Cell Biochem. 2004;91:26–35. doi: 10.1002/jcb.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ayala GE, Dai H, Ittmann M, Li R, Powell M, Frolov A, Wheeler TM, Thompson TC, Rowley D. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 2004;64:6082–6090. doi: 10.1158/0008-5472.CAN-04-0838. [DOI] [PubMed] [Google Scholar]
- 96.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Samulitis BK, Pond KW, Pond E, Cress AE, Patel H, Wisner L, Patel C, Dorr RT, Landowski TH. Gemcitabine resistant pancreatic cancer cell lines acquire an invasive phenotype with collateral hypersensitivity to histone deacetylase inhibitors. Cancer Biol Ther. 2015;16:43–51. doi: 10.4161/15384047.2014.986967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Acharya D, Mukherjee D, Podder S, Ghosh TC. Investigating different duplication pattern of essential genes in mouse and human. PLoS One. 2015;10:e0120784. doi: 10.1371/journal.pone.0120784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ajeian JN, Horton ER, Astudillo P, Byron A, Askari JA, Millon-Fremillon A, Knight D, Kimber SJ, Humphries MJ, Humphries JD. Proteomic analysis of integrin-associated complexes from mesenchymal stem cells. Proteomics Clin Appl. 2016;10:51–7. doi: 10.1002/prca.201500033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones MC, Humphries JD, Byron A, Millon-Fremillon A, Robertson J, Paul NR, Ng DH, Askari JA, Humphries MJ. Isolation of integrin-based adhesion complexes. Curr Protoc Cell Biol. 2015;66:9.8.1–9.8.15. doi: 10.1002/0471143030.cb0908s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh P, Algotar AM, Bracamonte ER. Prostatic Small Cell Carcinoma: Diagnosis and Management. J Cancer Ther. 2013;4:804–810. [Google Scholar]
- 103.Paller CJ, Antonarakis ES. Cabazitaxel: a novel second-line treatment for metastatic castration-resistant prostate cancer. Drug Des Devel Ther. 2011;5:117–124. doi: 10.2147/DDDT.S13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dritselis A, Galsky MD, Kirkpatrick P, Oh WK. Cabazitaxel. Nat Rev Drug Discov. 2010;9:677–678. doi: 10.1038/nrd3254. [DOI] [PubMed] [Google Scholar]
- 105.Michels J, Montemurro T, Murray N, Kollmannsberger C, Nguyen Chi K. Firstand second-line chemotherapy with docetaxel or mitoxantrone in patients with hormone-refractory prostate cancer. Cancer. 2006;106:1041–1046. doi: 10.1002/cncr.21695. [DOI] [PubMed] [Google Scholar]
- 106.Berry DL, Moinpour CM, Jiang CS, Ankerst DP, Petrylak DP, Vinson LV, Lara PN, Jones S, Taplin ME, Burch PA, Hussain MH, Crawford ED. Quality of life and pain in advanced stage prostate cancer: results of a Southwest Oncology Group randomized trial comparing docetaxel and estramustine to mitoxantrone and prednisone. J. Clin. Oncol. 2006;24:2828–2835. doi: 10.1200/JCO.2005.04.8207. [DOI] [PubMed] [Google Scholar]
- 107.Nelius T, Rinard K, Filleur S. Oral/metronomic cyclophosphamide-based chemotherapy as option for patients with castration-refractory prostate cancer-Review of the literature. Cancer Treat Rev. 2011;37:444–455. doi: 10.1016/j.ctrv.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 108.Nicolini A, Mancini PA, Ferrari P, Anselmi L, Tartarelli G, Bonazzi V, Carpi A, Giardino R. Oral low-dose cyclophosphamide in metastatic hormone refractory prostate cancer (MHRPC) Biomed Pharmacother. 2004;58:447–450. doi: 10.1016/j.biopha.2004.08.006. [DOI] [PubMed] [Google Scholar]