Abstract
Primary glioblastoma always has a fatal outcome despite maximal therapy. Identification and validation of prognostic biomarkers and novel therapeutics will be potentially powerful to transform the care of glioblastoma patients. In this study, we constructed Affymetrix gene microarrays with 14 glioma samples to screen for genes with potential prognostic value by hieratical clustering, and 83 genes including WD-repeat containing protein 1 (WDR1) were filtered out. WDR1 is a major co-factor collaborating with cofilin in actin cytoskeletal dynamics, which may play vital role in glioma proliferation and invasion. Further, The Cancer Genome Atlas (TCGA) database was utilizedto verify the expression of WDR1 and its prognostic implicationin 528 glioblastoma specimens. Survival and correlation analyses showed WDR1 expression was highly expressed and related to the prognosis of glioblastoma and the expression of signal transducer and activator of transcription 3 (STAT3), respectively (p<0.05). Finally, WDR1 expression was detected in our large cohort containing 258 glioma patients (including 100 primary glioblastomas).And univariate and multivariate analyses confirmed that high WDR1 expression was an independent prognostic factor for a shorter progression-free survival (PFS) and overall survival (OS) in primary glioblastoma patients at our center [hazard ratio (HR)=1.844, p=0.005 and HR=2.085, p=0.001, respectively]. Together, WDR1 is significantly over-expressed in primary glioblastoma. High expression of WDR1 can independently predict unfavorable clinical outcome for primary glioblastoma patients. This study identifies a novel prognostic biomarker and a potential therapeutic target for glioblastoma.
Keywords: WDR1, glioblastoma, prognosis, TCGA
Introduction
Malignant glioma is the most common and lethal primary brain tumor in adults. Glioblastoma (GBM), which accounts for approximately 60-70% of malignant glioma, has the most biologically aggressive phenotype [1]. Despite undergone aggressive surgery and adjuvant radiotherapy combined with chemotherapy, patients with GBM derive little benefit from the current standard of care. Ultimately, this disease follows a fatal course with the median overall survival (OS) of 12 to 15 months for these patients [2].
Substantial efforts have been undertaken in the identification of specific molecular markers and therapeutic targets to improve the prognosis of GBM patients. The methylation status of promoter of O-6-methylguanine-DNA methltransferase (MGMT) has been demonstrated to be correlated with response to temozolomide (TMZ) treatment, as GBM patients with methylated MGMT have a longer lifespan [3]. Recently, The Cancer Genome Atlas (TCGA) research network identified three core signaling pathways underlying GBM pathogenesis: receptor tyrosine kinase (RTK)/RAS/phosphatidylinositol 3-kinase (PI3K), p53 and retinoblastoma protein (RB) signaling pathways [4]. In addition, other canonical signaling pathways like proangiogenic pathway are important for gliomagenesis and maintainance of GBM phenotypes [5]. Several therapeutic agents against targets, such as EGFR, PDGFR and mTOR, involved in these pathways have been extensively investigated in current clinical trials for treating GBM patients [5]. Notably, bevacizumab which targetedly inhibits VEGF (a major angiogenic factor) has been granted approval by U.S. Food and Drug Administration (FDA) in the treatment of recurrent GBM [6-8]. Overall, significant progress in understanding genomic and molecular abnormalities in GBM has shifted the treatment paradigm towards use of molecularly targeted agents and opened opportunities to rationally develop more molecularly targeted therapy options and discover biomarkers for outcome prediction.
WDR1 (WD-repeat containing protein 1) also known as AIP1 (actin-interacting protein 1), is a highly conserved protein (67 kDa) containing 9 WD repeats and ubiquitously expressed in eukaryotes [9]. The protein is encoded by a gene mapping to human chromosome 4p [10]. WDR1 participates in promotion of cofilin-mediated actin filament disassembly and plays a crucial role in cytokinesis and cell migration [11]. It is indicated that WDR1 may be central for the ability of proliferation and invasion, which is needed for tumor growth [12]. Previous studies showed that WDR1 was overexpressed by several cancers such as breast cancer, thyroid neoplasia and ovarian carcinoma [13-15], and the differential expression of WDR1 in cancer tissue compared to healthy tissue suggests that WDR1 may act as a tumor-specific protein and a therapeutic target [15]. Moreover, a recent study found that signal transducer and activator of transcription 3 (STAT3) regulated WDR1 promoter activity and STAT3-induced WDR1 expression was associated with breast cancer progression [16]. It is well known that STAT3 is aberrantly activated in GBM and involved in gliomagenesis [17], which can directly binds to WDR1 promoter to regulate WDR1 transcription [16]. However, there has been no study on the expression of WDR1 in GBM tissues and its importance as a therapeutic target or prognostic predictor of GBM patients.
Searching some published microarray databases in Oncomine (www.oncomine.org), we found that several databases all showed a significantly higher expression of WDR1 in GBM tissues compared with normal brain. Thus, it is reasonable to speculate that there might be some important relationships between the expression of WDR1 and GBM.
In this study, we applied Affymetrix gene microarray to detect the expression pattern of WDR1 in glioma tissues and its relationship with clinical prognosis of glioma patients. We further analyzed WDR1’s expression combined with survival data in more than 500GBM cases of TCGA database to find WDR1’s prognostic value in GBM. At last, we used Tissue Microarray (TMA) composing a large number of glioma and normal brain tissue samples to clarify the expression pattern of WDR1 and its prognostic significance in glioma.
Methods and materials
Glioma and normal brain tissue samples
The study protocol and acquisition of tissue specimens were approved by the Tissue Committee and Research Ethics Board, Second Military Medical University, Shanghai, China. Glioma tissue specimens were obtained from archived tissue samples of glioma patients who underwent surgical treatment at Changzheng Hospital, China from January, 1999 to December, 2010. Glioma was diagnosed by two experienced pathologists independently according to the 2007 WHO Classification of Tumors of the Central Nervous System. Patients were eligible for the study if they presented with a diagnosis of glioma and no history of other tumors. Additional eligibility criteria included complete demographic and clinical data, such as age, gender, clinical manifestations, tumor size, extent of resection, adjuvant therapy and data of progression and/or death, and evaluation by enhanced MRI scanning for tumor relapse or progression after surgery at least once every six months. Patients receiving chemotherapy or radiotherapy prior to surgery were excluded. Normal brain tissues were obtained from surgical resections of severe trauma patients who required a partial resection of normal brain tissue as a decompression treatment to reduce increased intracranial pressure. And all the patient’s families signed the written informed consent about human tissue acquisition and usage in this study completely compiled with the National Regulations of Clinical Samples in China.
Gene microarray
We built 14 Affymetrix microarrays (Affymetrix Human U133 plus 2.0, Affymetrix, Santa Clara, CA, USA) in 2007. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols. Briefly, 1 μg of total RNA from each sample was amplified and transcribed into fluorescent cRNA using the manufacturer’s labeling protocol. The labeled cDNAs were hybridized onto the Affmetrix U133 plus 2.0. After washing the slides, the arrays were scanned by GeneChip® Scanner 3000 (Affymetrix, Santa Clara, CA, USA). The data from the experiments was obtained by original signal value of each gene, which was used for hierarchical clustering analysis (MeV 4.9 software, Dana-Farber Cancer Institute, Boston, MA, USA). Patients with glioma were dived into two groups according to their overall survival time calculated at the end of the follow-up period. The “Good” group was defined as patients with OS more than 60 months, while the “Poor” was less than 60 months. Genes that were important for prognostic prediction of glioma were identified through Student t-test and fold change filtering.
TCGA data
We searched TCGA (www. cancaergenome.nih.gov), which provides multimodal data of more than 500GBM cases, to identify dataset suitable for the analysis. Available raw microarray gene expression data based on the Affmetrix microarrays (Human gene U133A), and clinical treatment and follow-up information were included. The expression of several genes including WDR1 was collected for each case, and WDR1 expression was classified as either High (expression value ≥7.90) or Low (expression value <7.90). OS and progression free survival (PFS) were calculated in days from the data of diagnosis to the time of death and to the time of tumor progression or recurrence, or death of the patient from GBM, respectively.
Tissue microarray and immunohistochemistry
All the paraffin-embedded tissues used for analysis were acquired from 272 gliomas and 16 trauma patients as mentioned. And the microarray was constructed as described previously in Shanghai Biochip Co., Shanghai, China [18]. Immunohistochemistry staining using rabbit polyclonal anti-WDR1 antibody (AbcamCo., Ltd, Cambridge, MA, USA) was performed according to instructions, while sections incubated with the antibody were used as negative controls. The immunohistochemical results were examined by two independent pathologists who were blinded to the demographic and clinicopathological data of all subjects. The evaluation of WDR1 expression was based on both intensity and extensiveness. The intensity of positive staining in the cytoplasm was scored on a scale of 0 to 3 (0, no immunostaining; 1, light brown color; 2, medium brown color; 3, dark brown color). The percentage of positive staining cells was scored as follows: 0, ≤10% of the entire tumor cells; 1, 11-45% of the entire tumor cells; 2, 46-80% of the entire tumor cells; 3, >80% of the entire tumor cells. Then the two scores were multiplied to arrive at the final composite score which would be classified as: strong (+++, final score ≥6), moderate (++, final score = 4~6), weak (+, final score = 1-3), and null (-, final score = 0). WDR1 expression was divided into “High” (+++) and “Low” (++ and + and -) according to the rate of stained tumor cells and cytoplasm staining intensity. Scoring discrepancies were reviewed by the two pathologists who finally arrived at a consensus.
Follow-up
All the glioma patients were followed every 6 months by telephone or outpatient, and postoperative treatment including chemotherapy or radiotherapy was suggested according to the tumor’s pathological grade, patient’s systemic condition and wishes. All the complete follow-up information was recorded. OS was calculated in months from the initial date of diagnosis to the time of death, regardless of cause. PFS was measured from the date of diagnosis to the time of tumor progression on enhanced MRI, or death of patients from glioma.
Statistical analysis
All the calculations were performed with the SPSS 18.0 software program (SPSS Inc, Chicago, IL, USA), and results were presented as the means ± standard deviation (SD). Independent T-test was used to compare the difference of measurement data between two groups. Wilcoxon rank-sum test was used to estimate the difference of counting data between two groups. Chi-square test was calculated the difference of rates among different groups. Linear correlation analysis was used to evaluate mathematical relationship of two variables. Variables related to cumulative survival of glioma patients were evaluated by the Kaplan-Meier method and analyzed by log-rank test. The identification of candidate prognostic factors was performed by univariate analysis, multivariate analysis and stepwise backward Cox regression model. Factors with a result of p<0.2 in the univariate analysis were able to be added into multivariate analysis. All statistical tests were two-tailed, and p<0.05 was considered statistically significant.
Results
Demographic and clinicopathological characteristics of glioma patients
We performed gene microarray of a set of glioma samples, and the clinicalpathological data of these patients are shown in Table 1. And other 258 glioma tissue samples (8 grade I, 92 grade II, 44 grade III, and 114 grade IV) and 16 normal brain tissue samples were enrolled in our study for tissue microarray, while 256 glioma and 16 normal brain tissue samples met the criteria for further evaluation (the other 2 tissue dots were lost from the TMA slide). The period of follow-up of 256 patients was from 0.3 to 119 months. And at the final follow-up, 164 patients died and the rest were still alive. The median PFS and OS were 20 months and 21.5 months, respectively. For primary GBM patients accounting for 39.1% of all TMA patients, the median PFS was 9 months and the OS rates were 47% at 1year and 18% at 2 year, respectively, with a median OS of 11 months.
Table 1.
Expression of WDR1 gene as determined by Affymetrix microarray and relevant clinical data
| Number | Patient ID | WHO Grade | WDR1 Expression | Survival | OS (months) | Group |
|---|---|---|---|---|---|---|
| 1 | 04 | 2 | 992.7 | Alive | 86 | Good |
| 2 | 12 | 2 | 834.8 | Alive | 85 | Good |
| 3 | 27 | 3 | 1738 | Dead | 5 | Poor |
| 4 | 52 | 2 | 957 | Alive | 70 | Good |
| 5 | 69 | 2 | 985.1 | Alive | 66 | Good |
| 6 | 76 | 2 | 1106.5 | Dead | 29 | Poor |
| 7 | 86 | 2 | 731.6 | Alive | 73 | Good |
| 8 | 88 | 3 | 1795.7 | Dead | 21 | Poor |
| 9 | 89 | 4 | 1319.4 | Dead | 7 | Poor |
| 10 | 90 | 4 | 1996.2 | Dead | 20 | Poor |
| 11 | 92 | 4 | 1651.4 | Dead | 11.9 | Poor |
| 12 | 94 | 3 | 716.9 | Alive | 71 | Good |
| 13 | 98 | 2 | 717.7 | Dead | 70 | Good |
| 14 | 100 | 1 | 474.6 | Alive | 70 | Good |
Gene microarray analysis reveals WDR1 has the potential of prognostic predictor in glioma
14 glioma tissue samples were cultured and the cDNA were subsequently hybridized onto the Affymetrix platform. Then, we divided these patients into two groups to study the gene expression profile relevant to prognosis. In the “Good” group, 8 patients had been lived for more than 60 months by the end of the follow-up, and the average OS was 73.9 months, whereas other 6 patients whose OS was less than 60 months were described as the “Poor”. And we calculated the fold of increase or decrease of each gene’s expression in patients with poor prognosis comparing that in patients with good prognosis, and made hierarchal clustering. Of the 54675 genes represented on the array, 83 genes were filtered out with at least a 2-fold change in expression at the p<0.001 level (Figure 1). Of these, 59 genes (WDR1 included) were upregulated, while 24 genes were downregulated.
Figure 1.
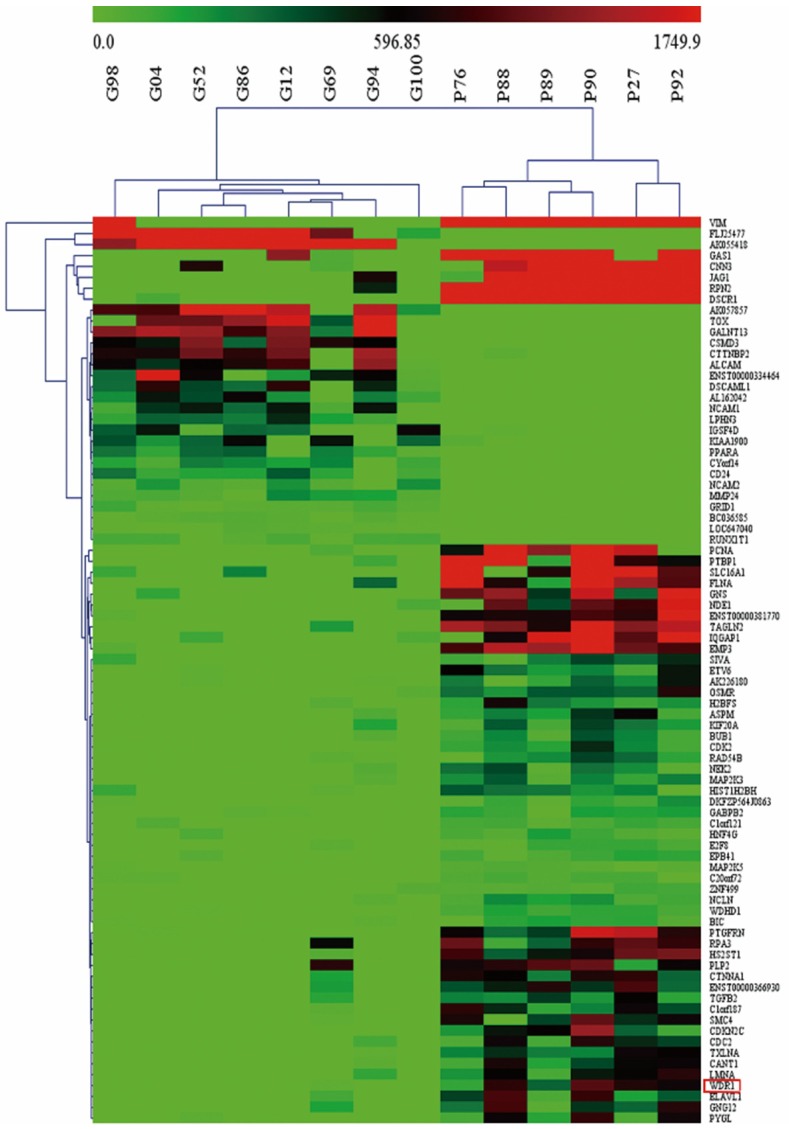
WDR1 is included in a gene set differentiating 14 gliomas with different prognoses by hierarchal cluster analysis. The cutoffs of fold change ≥2 and t-test p<0.001 based on expression profile data by Affymetrix microarrays were used to screen differently expressed genes between “Good” and “Poor” groups. 83 genes were filtered out and associated with the prognosis of glioma patient. As shown in the heat map, each column represents a tumor sample and each row is for a single gene. And color bar indicates gene with expression intensity, high level of mRNA is shown in red and low level is shown in green. WDR1 is highlighted by a red box, which is upregulated in the “Poor” group comparing to the “Good”.
TCGA dataset analysis shows WDR1 is highly expressed and acts as a risk factor for GBM survival
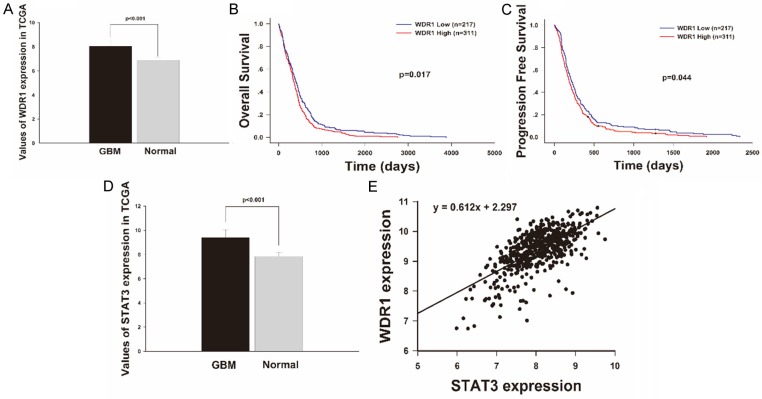
By searching the TCGA dataset, we have collected more than 500GBM cases with clinical follow-up information and got 528 GBM cases and 10 normal brains with level 3 gene expression data based on the Affymetrix microarrays (Human Gene U133A). We then used Wilcoxon rank-sum test to study the differentially expressed WDR1 and calculated the fold change of WDR1 expression in GBM patients versus normal brain cases. Figure 2A shows WDR1 is significantly highly expressed in GBM (p<0.001).
Figure 2.
WDR1 expression levels are elevated in GBMs of TCGA database and associated with patients’ survival and progression and related to the expression of STAT3. A. WDR1 mRNA expression level were detected in 528 GBM samples and 10 normal brain tissues obtained by TCGA. The values represent average mRNA levels of GBMs (8.043±0.618) and normal brains (6.876±0.230), and their difference was statistically significant (p<0.001). B, C. Kaplan-Meier survival analysis in 528 GBMs stratified by WDR1 expression showed that high level of WDR1 expression predicted shorter OS (p=0.017) and PFS (p=0.044). D. STAT3 mRNA expression level was higher in GBM samples than in normal brain tissues obtained by TCGA (9.3890±0.662 and 7.856±0.324, respectively; p<0.001). E. Both levels of STAT3 and WDR1 expression in 528 GBMs were extracted from the TCGA database and analyzed by liner correlation, the result showed a moderate positive correlation between expressions of the two genes (p<0.001, r=0.612).
By matching the clinical information data and gene expression profile data of 528 GBM patients, we did Kaplan-Meier analysis (log-rank test) to estimate the relationship between WDR1 expression and patient survival. Results were displayed in Figure 2B and 2C, WDR1 expression was significantly related to the OS and PFS of GBM patients (p=0.017 and p=0.044, respectively). Median OS and PFS in the WDR1 Low expression group were 216±17.903 days and 394±31.678 days, respectively. As for WDR1 High expression group, the median OS was 345±21.052 days and the median PFS was 186±16.120 days. So the WDR1 expression High group has an obviously shorter OS and poor prognosis than the Low group.
STAT3 expression is relevant to the expression of WDR1 in GBM in TCGA
In order to study the interaction of STAT3 and WDR1 in GBM, we searched the two molecules’ expression in TCGA dataset. All 528 cases experimented on the Affymetrix microarrays (Human Gene U133A) were available for further analysis. Wilcoxon rank-sum test also showed that STAT3 was significantly highly expressed in GBM comparing to normal brain tissues (p<0.001) (Figure 2D). Based on linear correlation analysis, we found WDR1 expression was linear associated with the expression of STAT3 (p<0.001) (Figure 2E). And the linear coefficient r was 0.612, which revealed a moderate correlation pattern.
Large sample TMA proves WDR1 is overexpressed in primary GBM
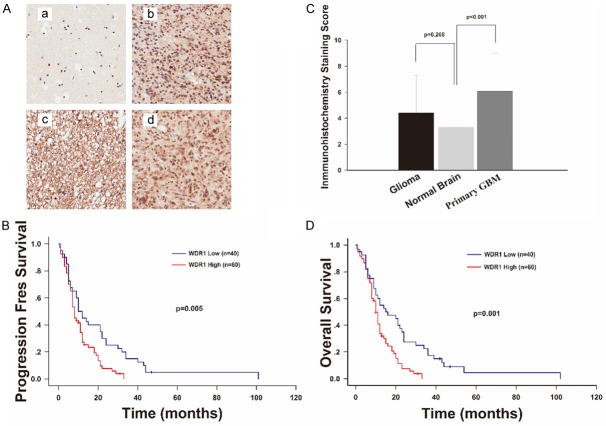
We further explored the WDR1 expression in a large sample TMA of glioma to confirm the results in gene microarray and TCGA database study. A total of 256 glioma specimens and 16 normal brain tissues were performed immunohistochemical staining in the TMA. We found WDR1 almost expressed in the cytoplasm of glioma cells (Figure 3A). The scores of WDR1 expression in normal brain and glioma were 3.313±2.182 and 3.302±2.902, respectively. As shown, WDR1 expression was not obviously different between these two groups (p=0.208). Notably, the expression of WDR1 in primary GBM (n=100, 6.070±2.934) was significantly higher than normal brain (p<0.001) (Figure 3C).
Figure 3.
WDR1 is upregulated in primary GBMs based on the TMA data and associated with patients’ survival and progression. (A) Representative staining images in normal brain and primary GBMs were shown in (a-d), respectively. WDR1 protein almost expressed in the cytoplasm as indicated. The intensity of positive staining in the cytoplasm in primary GBM, as well as the percentage of positive staining cells, was much higher than that in normal brain. (C) Immunohistochemical staining scores of WDR1 in 256 gliomas and 100 primary GBMs WDR1 were compared with the score in 16 normal brain tissues, respectively. Score of WDR1 expression in primary GBM (6.070±2.934) was significantly higher than that in normal brain (3.313±2.182) (p<0.001), while score of WDR1 in whole glioma group (3.302±2.902) was not (p=0.208). (B and D) Kaplan-Meier survival analysis in 100 primary GBMs stratified by WDR1 expression showed that high level of WDR1 expression was associated with shorter OS (p=0.001) and PFS (p=0.005).
Correlation of WDR1 expression with other clinical features of primary GBM patients in TMA
The clinicopathological characteristics of 100 primary GBM patients with WDR1 expression are shown in Table 2. We then tested the correlation of WDR1 expression and other clinical features and found that high WDR1 correlated with unclear border of tumor on MRI (p=0.033), but did not correlate with the rest characteristics in Table 2. It is known that blurred boundary is an important marker of invasive capability of malignancy [12], which indicated that higher WDR1 expression might be related to poorer prognosis of GBM.
Table 2.
Demographic and clinicopathological characteristics of the patients with primary GBM
| Characteristics | Number | WDR1 Expression | |
|---|---|---|---|
|
| |||
| Low | High | ||
| Age(years) | |||
| <55 | 53 (53%) | 21 | 32 |
| ≥55 | 47 (47%) | 19 | 28 |
| Gender | |||
| Male | 67 (67%) | 26 | 41 |
| Female | 33 (33%) | 14 | 19 |
| Seizure | |||
| Yes | 11 (11%) | 3 | 8 |
| No | 89 (89%) | 37 | 52 |
| IICP | |||
| Yes | 44 (44%) | 14 | 30 |
| No | 56 (56%) | 26 | 30 |
| Cysticdegeneration | |||
| Yes | 26 (26%) | 10 | 16 |
| No | 74 (74%) | 30 | 44 |
| Necrosis on MRI | |||
| Yes | 16 (16%) | 6 | 10 |
| No | 84 (84%) | 34 | 50 |
| Border on MRI | |||
| Clear | 18 (18%) | 3 | 15 |
| Unclear | 82 (82%) | 37 | 45 |
| MTD(cm) | |||
| <4 | 45 (45%) | 17 | 28 |
| ≥4 | 55 (55%) | 23 | 32 |
| Resection degree | |||
| Total | 77 (77%) | 31 | 46 |
| Subtotal | 20 (20%) | 8 | 12 |
| Partial | 3 (3%) | 1 | 2 |
| Chemotherapy | |||
| Yes | 72 (72%) | 27 | 45 |
| No | 28 (28%) | 13 | 15 |
| Radiotherapy | |||
| Yes | 70 (71.6%) | 30 | 40 |
| No | 30 (28.4%) | 10 | 20 |
| Survival status | |||
| Live | 6 (6%) | 2 | 4 |
| Dead | 94 (94%) | 38 | 56 |
| Recurrence | |||
| No | 5 (5%) | 1 | 4 |
| Yes | 95 (95%) | 39 | 56 |
Abbreviations: IICP, increased intracranial pressure; MTD, mean tumor diameter.
High expression of WDR1 correlates with poorer survival and is an independent prognostic factor of primary GBM patients
Results from our gene microarray and TCGA dataset analysis consistently showed that WDR1 expression in groups of glioma patients with poor prognosis were significantly higher than that in groups of glioma patients with favorable prognosis. To confirm the value of WDR1 in the prognostic prediction of primary GBM patients, we made univariate survival analysis that was stratified by WDR1 expression, as well as some other clinical factors (including gender, age, seizure, intracranial hypertension, tumor size, boundary, cystic change and necrosis, extension of resection, postsurgical radiotherapy or chemotherapy), with Kaplan-Meier estimates in TMA glioma patients. Age, WDR1 expression (HR=1.844, p=0.005) (Figure 3B) and radiotherapy were found to be risk factors for PFS of primary GBM patients. And age, WDR1 expression (HR=2.085, p=0.001) (Figure 3D), radiotherapy and resection degree were significant risk factors for OS of primary GBM patients (Table 3). Furthermore, Cox proportional hazards regression model was used to analyze variables filtered from the result of Kaplan-Meier estimates (p<0.2). It was found that high WDR1 expression was an independent prognostic factor for PFS in primary GBM patients (median PFS: 8 vs. 10 months, HR=1.912, p=0.005), while post-surgical radiotherapy and age less than 55y were protective factors. As for OS of primary GBM patients, the multivariable survival analysis indicated that high level of WDR1 expression was also a strong risk factor (median OS: 10 vs. 15 months, HR=2.147, p=0.001), as well as age over 55 y, and post-surgical radiotherapy acted as a protective factor (Table 4).
Table 3.
Univariate analysis of factors associated with survival and progression of primary GBM patients
| Variable | Primary GBM | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| OS | PFS | |||||
|
|
|
|||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (≥55 vs. <55 y) | 1.773 | 1.169-2.690 | 0.005 | 1.691 | 1.120-2.555 | 0.009 |
| WDR1 (High vs. Low) | 2.085 | 1.314-3.308 | 0.001 | 1.844 | 1.173-2.897 | 0.005 |
| Chemotherapy (Yes vs. No) | 0.681 | 0.433-1.069 | 0.083 | 0.779 | 0.497-1.221 | 0.256 |
| IICP (Yes vs. no) | 0.855 | 0.567-1.289 | 0.439 | 0.849 | 0.564-1.278 | 0.414 |
| MTD (≥4 vs. <4 cm) | 1.142 | 0.754-1.731 | 0.517 | 1.401 | 0.928-2.116 | 0.094 |
| Cystic Degeneration (Yes vs. No) | 0.636 | 0.394-1.027 | 0.054 | 0.676 | 0.423-1.078 | 0.085 |
| Necrosis (Yes vs. No) | 1.137 | 0.651-1.988 | 0.641 | 1.323 | 0.770-2.276 | 0.292 |
| Radiotherapy (Yes vs. No) | 0.605 | 0.388-0.943 | 0.020 | 0.637 | 0.410-0.991 | 0.036 |
| Resection Degree | 0.034 | 0.094 | ||||
| Total vs. Partial | 0.223 | 0.068-0.725 | 0.013 | 0.291 | 0.090-0.939 | 0.039 |
| Subtotal vs. Partial | 0.196 | 0.056-0.678 | 0.010 | 0.256 | 0.075-0.878 | 0.030 |
| Seizure (Yes vs. No) | 1.334 | 0.689-1.582 | 0.376 | 1.391 | 0.718-2.694 | 0.308 |
| Gender (Female vs. Male) | 0.789 | 0.508-1.224 | 0.273 | 0.728 | 0.471-1.127 | 0.137 |
| Border on MRI (Clear vs. Unclear) | 1.049 | 0.611-1.801 | 0.859 | 1.042 | 0.607-1.788 | 0.877 |
Abbreviations: IICP, increased intracranial pressure; MTD, mean tumor diameter; HR, Hazard ratio; OS, overall survival; PFS, progression-free survival.
Table 4.
Multivariate analysis of factors associated with survival and progression of primary GBM patients
| Survival* | Median Survival (months, 95% CI) | HR | 95% CI | p | |
|---|---|---|---|---|---|
| OS | |||||
| Age (≥55 vs. <55 y) | 10 (8.351-11.649) | 15 (8.284-21.716) | 1.782 | 1.175-2.702 | 0.007 |
| WDR1 (High vs. Low) | 10 (8.545-11.455) | 15 (6.325-27.396) | 2.147 | 1.349-3.419 | 0.001 |
| Radiotherapy (Yes vs. No) | 12 (8.637-15.363) | 9 (6.858-11.142) | 0.616 | 0.395-0.960 | 0.032 |
| PFS | |||||
| Age (≥55 vs. <55 y) | 7 (5.084-8.916) | 12 (9.754-14.246) | 1.844 | 1.212-2.806 | 0.004 |
| WDR1 (High vs. Low) | 8 (6.741-9.259) | 10 (6.901-13.099) | 1.912 | 1.211-3.019 | 0.005 |
| Radiotherapy (Yes vs. No) | 10 (7.142-12.858) | 8 (5.322-10.678) | 0.528 | 0.327-0.852 | 0.009 |
Abbreviations: HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Variables were adopted for their prognostic significance by univariateanalysis (p≤0.2).
Discussion
In the present study, we utilized 14 tumor samples from different grade glioma patients with clinical data to build Affymetrix microarrays, analyzed the dataset using hierarchical clustering based on post-surgery prognosis, and found that WDR1 included in a set of genes could distinguish among different prognoses of glioma patients. Then an approach was set up using extensive bioinformatics data mining process, in which public expression profile and clinical data of more than 500GBM patients were enrolled. And analysis of TCGA showed WDR1 was highly expressed in GBM samples and correlated with patients’ survival. The number of this dataset is higher than in any previous study thereby providing a robust evidence for our investigation. Finally, we examined the expression of WDR1 by immunohistochemistry using tissue microarray and found significantly higher WDR1 expression in primary GBM than in normal brain. And strong correlation between high WDR1expression and poor PFS and OS of 100 primary GBM patients was validated in the clinical cohort of our institute. Here, our study firstly provides the evidence that WDR1 expression is dysregulated in primary GBM and correlates with survival of primary GBM patients.
It is known that GBM as the highest malignant type of glioma is characterized by considerable cellularity, mitotic activity and high invasiveness [19]. Discovering the intricacies of genetic and molecular alterations involved in these processes may help us in developing a better understanding of GBM. For cancer invasion, malignant cells including GBM can utilize their migratory ability to invade adjacent tissue aggressively. And the formation of membrane protrusions is widely considered to be the initial step of migration when cells response to migratory and chemotactic stimuli [20]. Further research indicates that the driving force for membrane protrusion is localized polymerization of submembrane actin filaments [21]. Cofilin has been studied extensively that it is an essential regulator of actin dynamics at the plasma membrane during cell migration and invasion through its ability to sever actin filaments. And the suppression of cofilin expression with siRNA could reduce the invasion of carcinoma cells, while the over expression of cofilin protein could increase the velocity of cell migration in human GBM cells [22,23]. Further, WDR1 functioning as an activator of cofilin-mediated actin depolymerization has been demonstrated. All this indicates that WDR1 and cofilin work together on the increase of actin monomer pool to allow continues and robust actin polymerization to support diverse cellular processes including cell migration and invasion [24]. Coincidently, our study has shown that WDR1 was related to GBM unclear border, and a recent research enhanced the validation that upregulation of WDR1 in the interface zone of breast cancer and its relevance to cancer invasion was revealed by proteomic analysis [25].During mitosis, cells exhibit a drastic morphologic change, from essentially flat to round [26]. Mitotic cell rounding is thought to facilitate organization within the mitotic cell and be required for the geometric requirements of division [26]. In cancers, mitosis is also frequently observed and mitotic cells frequently appear round and swollen, which helps cancer cells adapt to challenges during division, including aneuploid chromosomes, multipolar spindles, and environments of high mechanical stress [27]. Thus, factors enhancing mitotic rounding are suggested to be highly favored by cancer cells. As demonstrated by a previous study, WDR1 did support mitotic cell rounding through promoting the actin cytoskleton dynamics, which ultimately regulated cancer cell proliferation [28]. It is suggested that WDR1 is likely to be involved in cancer cell growth signaling, and targeting this protein appears to be a possible therapeutic strategy.
In addition, STAT3, a latent transcriptional factor, was found to regulate WDR1 expression in breast cancer, and significantly positive correlation existed between expressions of the two genes in GBM samples of the TCGA database, which might provide molecular insight into this potential signaling pathway in GBM. Currently, researches have shown that STAT3 overexpression and hyperactivation is a feature of GBMand activated STAT3 is dimerized and accumulated in the nucleus, where it drives gene transcription by binding to the DNA promoter region [29,30]. Furthermore, there is evidence to suggest that STAT3 takes part in the regulation of cell movement mainly by cytoskeleton reorganization [31]. Intriguingly, as mentioned above, WDR1 is essential for actin cytoskeleton dynamics and possibly regulates cancer cell invasion. Thus, it is reasonable to speculate that STAT3 may act as an up steam regulator of WDR1 and control WDR1 transcription in GBM cell to affect tumor invasion and proliferation. Further investigations regarding this molecular mechanism governing GBM are warranted in this field.
In conclusion, we demonstrate that high WDR1 expression is associated with shorter OS and PFS after surgery and WDR1 can work as an independent risk factor to help define the subset of primary GBM patients. And understanding the molecular pathways involved by WDR1 will facilitate the development of designing novel targeted therapies for GBM patients.
Acknowledgements
This work was supported by the National Natural Science Foundation (81272781 and 81572501), Program for Academic Leaders in Health Sciences of Shanghai (XBR2011030), and “Pu Jiang Talent” Project of Shanghai (PJ[2014]0002617).
Disclosure of conflict of interest
None.
References
- 1.Patrick Y, Wen SK. Malignant Gliomas in Adults. N Engl J Med. 2008:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Xu T, Jiang Y, Xu H, Yan Y, Fu D, Chen J. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia. 2015;17:239–255. doi: 10.1016/j.neo.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen MH, Shen YL, Keegan P, Pazdur R. FDA drug approval summary: bevacizumab (Avastin) as treatment of recurrent glioblastoma multiforme. Oncologist. 2009;14:1131–1138. doi: 10.1634/theoncologist.2009-0121. [DOI] [PubMed] [Google Scholar]
- 7.Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J. Clin. Oncol. 2009;27:740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, Yung WK, Paleologos N, Nicholas MK, Jensen R, Vredenburgh J, Huang J, Zheng M, Cloughesy T. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J. Clin. Oncol. 2009;27:4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 9.Chu D, Pan H, Wan P, Wu J, Luo J, Zhu H, Chen J. AIP1 acts with cofilin to control actin dynamics during epithelial morphogenesis. Development. 2012;139:3561–3571. doi: 10.1242/dev.079491. [DOI] [PubMed] [Google Scholar]
- 10.Adler HJ, Winnicki RS, Gong TW, Lomax MI. A gene upregulated in the acoustically damaged chick basilar papilla encodes a novel WD40 repeat protein. Genomics. 1999;56:59–69. doi: 10.1006/geno.1998.5672. [DOI] [PubMed] [Google Scholar]
- 11.Kato A, Kurita S, Hayashi A, Kaji N, Ohashi K, Mizuno K. Critical roles of actin-interacting protein 1 in cytokinesis and chemotactic migration of mammalian cells. Biochem J. 2008;414:261–270. doi: 10.1042/BJ20071655. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Kim DH, Bae J, Lee JW, Kim SY, Kim YH, Bae JY, Yi JK, Yu MH, Noh DY, Lee C. Proteomic analysis of breast cancer tissue reveals upregulation of actin-remodeling proteins and its relevance to cancer invasiveness. Proteomics Clin Appl. 2009;3:30–40. doi: 10.1002/prca.200800167. [DOI] [PubMed] [Google Scholar]
- 14.Izawa S, Okamura T, Matsuzawa K, Ohkura T, Ohkura H, Ishiguro K, Noh JY, Kamijo K, Yoshida A, Shigemasa C, Kato M, Yamamoto K, Taniguchi S. Autoantibody against WD repeat domain 1 is a novel serological biomarker for screening of thyroid neoplasia. Clinical Endocrinology. 2013;79:35–42. doi: 10.1111/cen.12121. [DOI] [PubMed] [Google Scholar]
- 15.Haslene-Hox H, Oveland E, Woie K, Salvesen HB, Wiig H, Tenstad O. Increased WD-repeat containing protein 1 in interstitial fluid from ovarian carcinomas shown by comparative proteomic analysis of malignant and healthy gynecological tissue. Biochim Biophys Acta. 2013;1834:2347–2359. doi: 10.1016/j.bbapap.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Cho N. 262: STAT3-induced WDR1 expression is associated with breast cancer cell migration. European Journal of Cancer. 2014:S61. [Google Scholar]
- 17.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 19.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 20.Bailly M, Condeelis J. Cell motility: insights from the backstage. Nature Cell Biology. 2002;4:E292–E294. doi: 10.1038/ncb1202-e292. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil Cytoskeleton. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- 24.Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang S, Kim MJ, An H, Kim BG, Choi YP, Kang KS, Gao MQ, Park H, Na HJ, Kim HK, Yun HR, Kim DS, Cho NH. Proteomic Molecular Portrait of Interface Zone in Breast Cancer. J Proteome Res. 2010;9:5638–5645. doi: 10.1021/pr1004532. [DOI] [PubMed] [Google Scholar]
- 26.Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2011;469:226–230. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- 27.Cadart C, Zlotek-Zlotkiewicz E, Le Berre M, Piel M, Matthews HK. Exploring the Function of Cell Shape and Size during Mitosis. Developmental Cell. 2014;29:159–169. doi: 10.1016/j.devcel.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Fujibuchi T, Abe Y, Takeuchi T, Imai Y, Kamei Y, Murase R, Ueda N, Shigemoto K, Yamamoto H, Kito K. AIP1/WDR1 supports mitotic cell rounding. Biochem Biophys Res Commun. 2005;327:268–275. doi: 10.1016/j.bbrc.2004.11.156. [DOI] [PubMed] [Google Scholar]
- 29.Kang SH, Yu MO, Park KJ, Chi SG, Park DH, Chung YG. Activated STAT3 Regulates Hypoxia-Induced Angiogenesis and Cell Migration in Human Glioblastoma. Neurosurgery. 2010;67:1386–1395. doi: 10.1227/NEU.0b013e3181f1c0cd. [DOI] [PubMed] [Google Scholar]
- 30.Gray GK, McFarland BC, Nozell SE, Benveniste EN. NF-kappa B and STAT3 in glioblastoma: therapeutic targets coming of age. Expert Rev Neurother. 2014;14:1293–1306. doi: 10.1586/14737175.2014.964211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siveen KS, Sikka S, Surana R, Dai XY, Zhang JW, Kumar AP, Tan BKH, Sethi G, Bishayee A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845:136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]