Abstract
Intercalating fluorescent probes are widely used to visualize DNA in studies on DNA-protein interactions. Some require the presence of adenosine triphosphate (ATP). We have investigated the mechanical properties of DNA stained with the fluorescent intercalating dyes YOYO-1 and YOYO-3 as a function of ATP concentrations (up to 2 mM) by stretching single molecules in nanofluidic channels with a channel cross-section as small as roughly 100×100 nm2. The presence of ATP reduces the length of the DNA by up to 11 %. On the other hand, negligible effects are found if DNA is visualized with the minor groove-binding probe 4′,6-diamidino-2-phenylindole. The apparent drop in extension under nanoconfinement is attributed to an interaction of the dye and ATP, and the resulting expulsion of YOYO-1 from the double helix.
Keywords: DNA, ATP, Intercalating dye, YOYO-1, DAPI, Nanofluidics
Graphical Abstract

Nanochannel-stretched DNA (48.5 kbp) stained with YOYO-1 is sensitive to ATP concentration in buffer. Nanochannels with a cross-section of 80×80 nm2 were used to stretch DNA.
Introduction
Single-molecule fluorescence microscopy is a powerful tool to explore the statistical, mechanical, and transport properties of DNA [1–5]. It in particular enables nanofluidic methods for genome analysis that aid rapid sequence assembly and elucidate structural variations as well as haplotype [6]. Since DNA is not fluorescent, it must be stained with fluorescent dyes. One of the most commonly used dyes in such experiments is the bis-intercalator YOYO-1. YOYO-1 forms a very stable complex with double-stranded DNA (dsDNA) and undergoes about 500-fold fluorescence enhancement upon binding, thus providing high signal to noise ratio [7].
Intercalating dyes affect both the mechanical and structural properties of DNA [8–10]. Generally, YOYO-1 causes DNA elongation, and it is assumed that the contour length of dsDNA complex with YOYO-1 increases linearly with increasing staining ratio by up to 35% compared to native DNA [9, 11, 12]. This lengthening effect originates from the fact that each YOYO-1 molecule separates two pairs of neighboring DNA base pairs by ~0.4 nm [13, 14]. Currently, contradicting ideas exist about how YOYO-1 binding alters DNA rigidity, with results ranging from a decreased persistence length [9], over unchanged persistence length [12, 15, 16], to an assumed increase in persistence length [2]. The more recent studies favor a persistence length that is independent of YOYO-1 concentration.
An alternative to intercalating fluorescent dyes is DAPI (4′,6-diamidino-2-phenylindole). It attaches inside the minor groove of A-T rich DNA sequences [17]. DAPI is a useful fluorescent dye to visualize DNA molecule as it shows insignificant effect on the local structure of DNA. In particular, DAPI does not inhibit the enzymatic activities of many restriction endonucleases [18]. However, it was found that DAPI exhibits a marked effect on the higher order structure of DNA [19].
Many factors such as proteins and coenzymes can alter the stability of fluorescence dye binding and therefore exert an effect on DNA contour length. In our previous work we showed that T4 DNA ligase can change the DNA configuration in nanochannels [20]. This protein requires ATP as a cofactor. Given the near ubiquity of ATP as a coenzyme, it is useful to know how it interacts with staining agents in order to decouple that interaction from observations and extract the true phenomena of interest. Therefore, we here study the effect of ATP on YOYO-1, YOYO-3, and DAPI stained λ-DNA.
Stretching of DNA in nanochannels is applied here as the main analytical technique for following DNA configuration [21]. Such nanochannels have a cross-section on the order of ~100×100 nm2, and a length of 100s of micrometers. After DNA is brought into a nanochannel, it will assume an equilibrium configuration that is governed by the channel cross-section, the contour length of DNA, its persistence length, and its width [22]. Example data is shown in Fig. 1. The dependence on the persistence length and width make nanochannel stretching a promising probe for these parameters as a function of buffer conditions and protein binding [23]. DNA inside nanochannels fluctuates [24, 25], and can thus explore many possible configurations during a single experiment.
Figure 1.

Nanochannel-stretched DNA (48.5 kbp) stained with YOYO-1 is sensitive to ATP concentration in buffer. Nanochannels with a cross-section of 80×80 nm2 were used to stretch DNA.
We present an investigation of the influence of ATP on the configuration of long DNA strands under nanoconfinement. We introduced λ-DNA molecules into nanochannels and observed extended DNA configurations fluctuating around the equilibrium point. A contraction of bis-intercalator stained DNA in the presence of ATP is found. In contrast, DAPI-stained DNA is not affected by ATP.
Experimental
λ-DNA (Roche Diagnostics GmbH, 1 μg·mL−1, www.roche.com) was stained with YOYO-1 or YOYO-3 (Life Technologies, www.lifetechnologies.com) at a ratio of 1 dye per 10 base pairs. We also investigated DNA stained with DAPI (10 μM). The contour length of unstained λ-DNA (48.5 kbp) is approximately 16 μm. After staining, that length can increase by about 35% [11], where a linear interpolation would predict about 14% extension for our staining. DNA was suspended in ½ x TBE buffer (pH 8) at room temperature. BSA (0.1 mg·mL−1, New England Biolabs, www.neb.com) was added to prevent sticking of DNA to the nanochannels. Adenosine triphosphate (ATP) at concentrations of 0.5 mM, 1 mM, and 2 mM were investigated. For preparing “quick-stained” DNA, DNA and YOYO-1 were incubated for 30 minutes at room temperature. For preparation of an “equilibrated” stained DNA, solutions were incubated for 48 hours at 4°C.
All experiments used mixed micro- and nanofluidic devices made from fused silica, which were prepared by methods discussed elsewhere [26]. Nanofluidic channels with 80×80 nm2, 100×100 nm2, 130×130 nm2, and 150×150 nm2 cross-sections and a length of 200 μm were placed between microchannels [27]. Each DNA molecule is driven from microchannel to nanochannel by a hydrostatic pressure gradient. After the DNA molecule has entered the nanochannel, the pressure gradient was removed and the dynamics of molecules were observed. Molecules were observed using a fluorescence microscope with a 1.3-N.A. oil-immersion microscope objective under illumination from a metal halide lamp, and images were recorded by an emCCD camera (Andor, www.andor.com).
For calculating the end-to-end extended length of DNA molecules, we fitted the brightness along the molecule backbone to
Here I(x) is the intensity along nanochannel, σ is the slope of the edge, I0, xc, l, and Ib are the brightness, center position, length, and background signal, respectively. Only hairpin-free molecules were considered for analysis.
Results and discussion
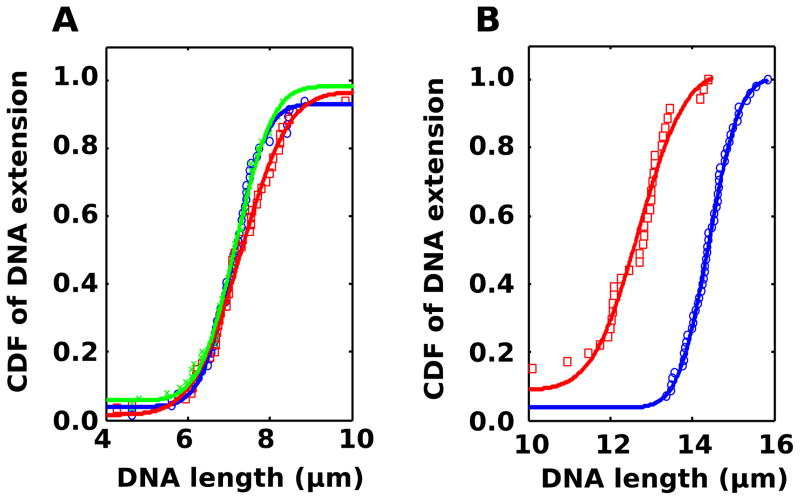
In Fig. 2a we show the extension of YOYO-1 “quick-stained” DNA inside nanochannels with and without added ATP, at the same conditions as in our previous publication [20]. We observed a significant shortening of DNA upon addition of 1 mM ATP (Tab. 1). This shortening could be an effect peculiar to quick stained DNA, because quick staining produces inhomogeneous fluorescence along the length of the DNA. In comparison, longer equilibration times promotes uniform stating, which indicates that dye-interaction is a dynamic process [28].
Figure 2.
A) Cumulative distribution function (CDF) of extensions of “quick-stained”, YOYO-1 bearing DNA without ATP (blue ○), and with 1 mM (red □). Measurements were performed in channels with 100×100 nm2 cross-section and 60 molecules for each condition. B) CDF of DNA with equilibrated YOYO-1 stain at 0 mM ATP (blue ○), 0.5 mM ATP (red □), 1 mM ATP (green ×), 2 mM ATP (cyan ◇). There are over 100 molecules for each condition, and the channel cross-section is 130×130 nm2.
Table 1.
Numerical results of fitting cumulative distribution functions in all figures to a single error function model.
| Fig. # | Dye | [ATP] (mM) | Channel cross-section (nm2) | l (μm)* |
|---|---|---|---|---|
| 2a | YOYO-1 | 0.0 | 100×100 | 11.28 ±0.06 |
| 1.0 | 10.27 ±0.05 | |||
|
| ||||
| 2b | YOYO-1 | 0.0 | 130×130 | 7.46 ±0.03 |
| 0.5 | 7.37 ±0.05 | |||
| 1.0 | 7.17 ±0.05 | |||
| 2.0 | 6.71 ±0.03 | |||
|
| ||||
| 3 | YOYO-1 | 0.0 | 100×100 | 10.84 ±0.05 |
| 2.0 (ATP) | 9.67 ±0.03 | |||
| 2.0 (AMP) | 9.42 ±0.04 | |||
| 2.0 (GTP) | 9.24 ±0.05 | |||
|
| ||||
| 4a | DAPI | 0.0 | 150×150 | 7.14 ±0.08 |
| 1.0 | 7.31±0.12 | |||
| 2.0 | 7.08 ±0.1 | |||
|
| ||||
| 5b | YOYO-3 | 0.0 | 80×80 | 14.38 ±0.08 |
| 2.0 | 12.75 ±0.09 | |||
l is the extension defined in equation (1).
To investigate whether the shortening effect in Fig. 2a is specific to quick-stained DNA or whether it is a general property, we exposed DNA with an equilibrated YOYO-1 stain to a range of ATP concentrations (Fig. 2b). We observed that concentrations as low as 0.5 mM alter the DNA length (Tab. 1). Furthermore, as we increased the concentration, the apparent shortening became more dramatic, i.e. the strength of the ATP effect depends on its concentration. Thus it appears that ATP alters both quick and equilibrated stained DNA, but that its effect on quick-stained DNA is stronger. This would suggest that ATP’s effect is stronger when the dye has not had time to fully thermalize and that it may be linked to the some redistribution of dye along the DNA. The extension observed in Fig. 2b was slightly smaller than that in Fig. 2a because wider nanochannels (130 × 130 nm2) were used.
ATP’s molecular structure consists of a phosphate chain, a sugar, and a nucleoside base. To explore which part of ATP is interacting with DNA or YOYO-1, we repeated the same experiment with GTP (Guanosine triphosphate) and AMP (adenosine monophosphate). GTP has the same triphosphate chain as ATP but a different nucleoside, while AMP has the same nucleoside as ATP but only one phosphate group (Fig. 3). It is apparent that all of those nucleosides alter DNA length (Tab. 1). In particular the effects of GTP and ATP are quite similar.
Figure 3.
Cumulative distribution function of extensions of DNA without nucleotide triphosphate (blue ○), 2 mM ATP (red □), 2 mM AMP (green ×), 2m M GTP (cyan ◇). 60 molecules each condition. Channel cross-section was 80×80 nm2.
In contrast, the effect of AMP is less pronounced, pointing to a dependence on the size of the phosphate chain. We would naively anticipate an ionic strength of ~15 mM in the absence of ATP, and of ~25 mM in the presence of 2 mM ATP, which according to Reisner et al. led to a reduction of up to 8% in extension along the channel axis [23]. Given the complex behaviour of ATP and AMP, the prediction of the buffer strength effect is fraught with considerable uncertainty, but it is safe to assume that AMP should carry at most half the contribution of ATP. However, the differences between the lengths are far less than half the amplitude of the peak shift, and so it does not appear that the ionic strength effect makes the leading contribution.
One of the possibilities for the apparent shortening could be an interaction between ATP and YOYO-1 in solution leading to less free dye available for staining of DNA. Such an interaction could be driven by the high positive and negative charges, respectively. The drop in the staining ratio would result in shorter molecules.
To further support our hypothesis of ATP-YOYO-1 interaction instead of an ATP-DNA interaction, we repeated the measurement on DNA stained with DAPI, a minor groove binder (Fig. 4a). It is apparent that the DAPI- stained DNA is not affected by ATP to the same extent that YOYO-1 stained DNA is (Tab. 1). This strongly suggests that the hypothesis of ionic strength-driven contraction is not correct.
Figure 4.
A) Cumulative distribution function of extension of DNA stained with DAPI without ATP (blue ○), 1mM ATP (red □), and with 2 mM ATP (green ×). Measured in channels with 150×150 nm2 cross-section and 50 molecules for each condition. B) CDF of DNA with equilibrated YOYO-3 stain at 0 mM ATP (blue ○), and with 2 mM ATP (red □). 55 molecules each condition, and channel cross-section of 80×80 nm2.
In order to confirm that ATP does affect bis-intercalated dyes other than YOYO-1, we repeated the same experiment with YOYO-3, which is also a bis-intercalating dye. We observed an effect similar to what we observed on YOYO-1 stained DNA Fig. 4b, (Tab. 1).
Conclusions
We have investigated the physical behavior of nanoconfined, bis-intercalator-stained DNA in the presence of ATP. In particular, the extensions of DNA molecules reduced when more than 1 mM ATP was added to YOYO-1 and YOYO-3 stained DNA. By comparing ATP, AMP, and GTP, we concluded that that the action of ATP is through an action of the nucleoside on the dye that is intercalated into the DNA. On the other hand DAPI- stained DNA is not affected by ATP to the same extent as YOYO-1 stained DNA. We believe that this result is important because it could lead to a considerable artifact when studying the interaction of DNA and ATP-dependent proteins on YOYO-1 stained DNA.
Acknowledgments
We acknowledge support from the National Institutes of Health (R01GM107559), and thank Keith Weninger for fruitful discussions.
References
- 1.Wirtz D. Direct measurement of the transport properties of a single DNA molecule. Phys Rev Lett. 1995;75:2436–2439. doi: 10.1103/PhysRevLett.75.2436. [DOI] [PubMed] [Google Scholar]
- 2.Bakajin O, Duke TAJ, Chou C, et al. Electrohydrodynamic Stretching of DNA in Confined Environments. Phys Rev Lett. 1998;80:2737–2740. doi: 10.1103/PhysRevLett.80.2737. [DOI] [Google Scholar]
- 3.Perkins TT. Single Polymer Dynamics in an Elongational Flow. Science (80- ) 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 4.Maier B, Rädler J. Conformation and Self-Diffusion of Single DNA Molecules Confined to Two Dimensions. Phys Rev Lett. 1999;82:1911–1914. doi: 10.1103/PhysRevLett.82.1911. [DOI] [Google Scholar]
- 5.Turner S, Cabodi M, Craighead H. Confinement-Induced Entropic Recoil of Single DNA Molecules in a Nanofluidic Structure. Phys Rev Lett. 2002;88:128103. doi: 10.1103/PhysRevLett.88.128103. [DOI] [PubMed] [Google Scholar]
- 6.Lam ET, Hastie A, Lin C, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glazer A, Rye H. Stable dye-DNA intercalation complexes as reagents for high-sensitivity fluorescence detection. Nature. 1992;359:859– 861. doi: 10.1038/359859a0. [DOI] [PubMed] [Google Scholar]
- 8.Smith SB, Finzi L, Bustamante C. Direct Mechanical Measurements of the Elasticity of Single DNA-Molecules by Using Magnetic Beads. Science (80- ) 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 9.Sischka A, Toensing K, Eckel R, et al. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys J. 2005;88:404–11. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murade CU, Subramaniam V, Otto C, Bennink ML. Interaction of oxazole yellow dyes with DNA studied with hybrid optical tweezers and fluorescence microscopy. Biophys J. 2009;97:835–43. doi: 10.1016/j.bpj.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuter M, Dryden DTF. The kinetics of YOYO-1 intercalation into single molecules of double-stranded DNA. Biochem Biophys Res Commun. 2010;403:225–9. doi: 10.1016/j.bbrc.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Doyle P, Ladoux B, Viovy J-L. Dynamics of a Tethered Polymer in Shear Flow. Phys Rev Lett. 2000;84:4769–4772. doi: 10.1103/PhysRevLett.84.4769. [DOI] [PubMed] [Google Scholar]
- 13.Spielmann HP, Wemmer DE, Jacobsen JP. Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy. Biochemistry. 1995;34:8542–53. doi: 10.1021/bi00027a004. [DOI] [PubMed] [Google Scholar]
- 14.Johansen F, Jacobsen JP. 1H NMR studies of the bis-intercalation of a homodimeric oxazole yellow dye in DNA oligonucleotides. J Biomol Struct Dyn. 1998;16:205–22. doi: 10.1080/07391102.1998.10508240. [DOI] [PubMed] [Google Scholar]
- 15.Kundukad B, Yan J, Doyle PS. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter. 2014;10:9721–9728. doi: 10.1039/c4sm02025a. [DOI] [PubMed] [Google Scholar]
- 16.Günther K, Mertig M, Seidel R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010;38:6526–6532. doi: 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapuscinski J. DAPI: a DNA-specific fluorescent probe. Biotech Histochem. 1995;70:220–33. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- 18.Meng X, Cai W, Schwartz DC. Inhibition of restriction endonuclease activity by DNA binding fluorochromes. J Biomol Struct Dyn. 1996;13:945–51. doi: 10.1080/07391102.1996.10508909. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzawa Y, Yoshikawa K. Change of the Higher Order Structure in a Giant DNA Induced by 4′, 6-Diamidino-2-phenylindole as a Minor Groove Binder and Ethidium Bromide as an Intercalator. Nucleosides and Nucleotides. 1994;13:1415–1423. doi: 10.1080/15257779408012161. [DOI] [Google Scholar]
- 20.Roushan M, Kaur P, Karpusenko A, et al. Probing transient protein-mediated DNA linkages using nanoconfinement. Biomicrofluidics. 2014;8:034113. doi: 10.1063/1.4882775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegenfeldt JO, Prinz C, Cao H, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc Natl Acad Sci U S A. 2004;101:10979–83. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tree DR, Wang Y, Dorfman KD. Extension of DNA in a Nanochannel as a Rod-to-Coil Transition. Phys Rev Lett. 2013;110:208103. doi: 10.1103/PhysRevLett.110.208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisner W, Beech J, Larsen N, et al. Nanoconfinement-Enhanced Conformational Response of Single DNA Molecules to Changes in Ionic Environment. Phys Rev Lett. 2007;99:058302. doi: 10.1103/PhysRevLett.99.058302. [DOI] [PubMed] [Google Scholar]
- 24.Reisner W, Morton KJ, Riehn R, et al. Statics and dynamics of single DNA molecules confined in nanochannels. Phys Rev Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 25.Karpusenko A, Carpenter JH, Zhou C, et al. Fluctuation modes of nanoconfined DNA. J Appl Phys. 2012;111:24701–247018. doi: 10.1063/1.3675207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riehn R, Reisner W, Tegenfeldt JO, et al. Nanochannels for Genomic DNA Analysis: The Long and Short of It. In: Liu RH, Lee AP, editors. Integr Biochips DNA Anal. 1. Landes Bioscience; Austin, Texas: 2007. pp. 151–186. [Google Scholar]
- 27.Riehn R, Lu MC, Wang YM, et al. Restriction mapping in nanofluidic devices. Proc Natl Acad Sci U S A. 2005;102:10012–10016. doi: 10.1073/pnas.0503809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlsson C, Jonsson M, Akerman B. Double bands in DNA gel electrophoresis caused by bis-intercalating dyes. Nucleic Acids Res. 1995;23:2413–20. doi: 10.1093/nar/23.13.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]