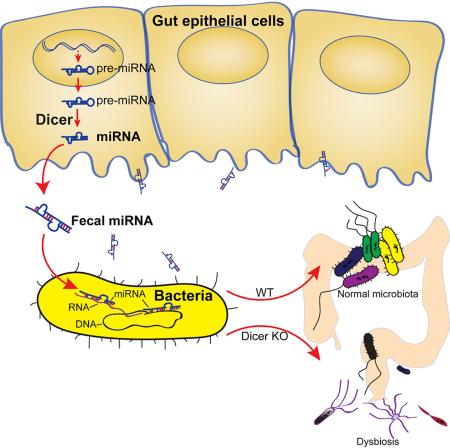
SUMMARY
The host gut microbiota varies across species and individuals but is relatively stable over time within an individual. How the host selectively shapes the microbiota is largely unclear. Here, we show that fecal microRNA (miRNA)-mediated inter-species gene regulation facilitates host control of the gut microbiota. MiRNAs are abundant in mouse and human fecal samples and present within extracellular vesicles. Cell-specific loss of the miRNA-processing enzyme, Dicer, identified intestinal epithelial cells (IEC) and Hopx-positive cells as predominant fecal miRNA sources. These miRNAs can enter bacteria, such as F. nucleatum and E. coli, specifically regulate bacterial gene transcripts and affect bacterial growth. IEC-miRNA deficient (Dicer1ΔIEC) mice exhibit uncontrolled gut microbiota and exacerbated colitis and WT fecal miRNA transplantation restores fecal microbes and ameliorates colitis. These findings identify both a physiologic role by which fecal miRNA shapes the gut microbiota and a potential strategy for manipulating the microbiome.
Graphical Abstract
INTRODUCTION
The gut hosts a complex microbiota which is initially comprised of microbes from the mother and continues to develop through feeding and other environmental contacts (Mändar and Mikelsaar, 1996). At approximately three years of age in humans, the intestinal microbiota resembles an adult-like microbiota that is relatively stable over time (Faith et al., 2013; Schloissnig et al., 2013). Many factors contribute to shaping the mammalian microbiota, including host genetics, diet and disease states (Goodrich et al., 2014; Ley et al., 2005; Turnbaugh et al., 2008). Broad trends exist within a given species but interspecies microbial composition often differs dramatically. Reciprocal transplantation of gut microbiota into germ-free zebrafish and mouse recipients revealed that the relative abundance of microbial lineages resembles the composition of the recipient host (Rawls et al., 2006), suggesting that there are selective mechanisms in the host for the maintenance of specific components of the microbiota. Since the gut microbiota play an important role in host metabolism and immunity as well as in disease (An et al., 2014; Belkaid and Naik, 2013; Hooper et al., 2012; Iida et al., 2013; Koren et al., 2011; Smith et al., 2013; Tremaroli and Bäckhed, 2012; Turnbaugh et al., 2008), it is important to understand the mechanisms by which the microbiota is regulated by the host and to identify ways in which to manipulate the microbiome (Goodrich et al., 2014).
MicroRNAs (miRNAs) are small non-coding RNAs, 18-23 nucleotides in length, synthesized in nucleus, that are processed and function in the cytoplasm. However, increasing evidence demonstrates that miRNAs exist extracellularly and circulate in body fluids (Weber et al., 2010). Isolated studies have measured RNA in human stool and identified miRNAs as potential makers of intestinal malignancy (Ahmed et al., 2009; Link et al., 2012). Whether functional fecal miRNA exists in the normal gut is unexplored. Here we identify gut miRNAs in intestinal contents and feces and demonstrate their role in modulating the gut microbiota composition.
RESULTS
Identification of miRNA in Mouse and Human Feces
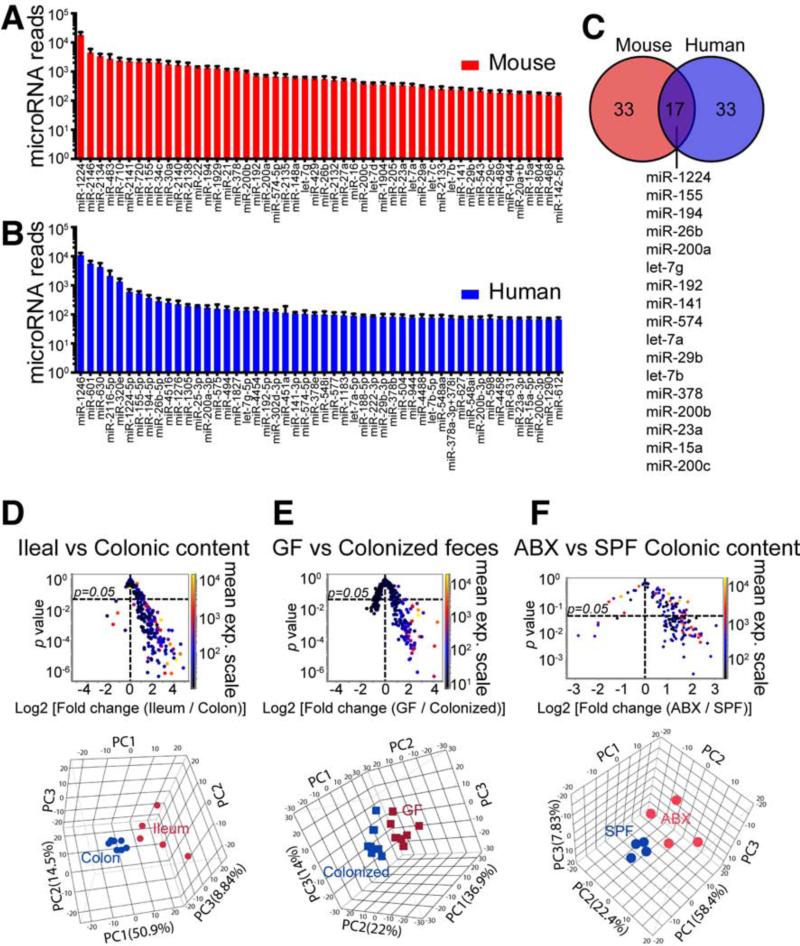
To determine whether miRNAs could be identified in the stool, we isolated RNA from both human and mouse fecal samples using the mirVana™ miRNA isolation kit and compared it to splenic RNA isolates. We found that both human and mouse fecal samples contained small RNA species in a pattern similar to extracellular exosome RNA (Figure S1A), whose size pattern lacks peaks for the 18S and 28S rRNA subunits (Valadi et al., 2007) as measured by bioanalyzer. We then performed small RNA bioanalyzer on the fecal samples (Figure S1B) and found three major RNA peaks in the small RNA region: ~20 nt, ~65 nt, and ~100 nt. The majority of these small RNAs were of miRNA size. In order to establish that specific miRNAs were present in mouse fecal samples, we carried out a pilot study and found that specific miRNAs could be identified by real time quantitative PCR (qPCR) (not shown). Based on these data, we profiled mouse fecal miRNA using the NanoString nCounter platform (Geiss et al., 2008). Of the 566 miRNAs tested in mouse feces, 283 miRNAs were detected (Table S1) with miR-1224, miR-2146, miR-2134, miR-483, miR-710, miR-2141, miR-720, miR-155 and miR-34c being the most abundant miRNAs (Figure 1A).
Figure 1. Identification of miRNA in Feces and Intestinal Luminal Contents.
(A) Mean values ± SEM for the 50 most abundant miRNAs in mouse fecal samples (n=6). See Table S1 for full list.
(B) Mean values ± SEM for the 50 most abundant miRNAs in human feces (n=10). See Table S2 for full list.
(C) Venn diagram showing 17 miRNAs from the 50 most abundant fecal miRNAs shared between human and the mouse as shown in (A) and (B).
(D-F) (Upper panels), volcano plots for miRNA level based on nanostring detection in ileal luminal contents (n=5) vs. colonic luminal contents (n=8) (D), germ-free (GF) mouse feces (n=8) vs. SPF colonized (Colonized) mouse feces (n=8) (E), and antibiotic treated (ABX) mouse feces (n=4) vs. SPF mouse feces (n=4) (F). Each dot represents one miRNA; x-Axis: fold change; y-Axis: p value comparing individual miRNAs between groups (unequal variance t test followed by Benjamini-Hochberg correction); the color of the dot indicates mean expression (exp.) level of the corresponding miRNA in both groups as shown in side color scale bar. (Down panel), PCA analyses of miRNAs based on the same nanostring data sets.
See also Figure S1.
The stability of miRNAs is robust compared to mRNA (Jung et al., 2010). Extracellular miRNAs can be released both in extracellular vesicle (EV) form (eg. microvesicle, exosome), and in an EV-free form associating with high-density lipoproteins or argonaute protein. These forms may contribute to extracellular miRNA stability (Creemers et al., 2012). It was reported that epithelial cells could release exosomes-like vesicles displaying major histocompatibility complexes (Van Niel et al., 2003). To explore whether EV exist in the feces, we examined the specimen with NanoSight and electron microscopy and observed EV in the fecal samples (Figures S1C-S1D). Furthermore, we found that the most abundant miRNAs in feces, such as miR-1224, miR-2146, miR-2134, miR-2141 and miR-34c, are abundantly present in EVs (Figure S1E).
We then performed Nanostring analyses of human fecal samples to determine which miRNAs are expressed and how they compare to the mouse. Of the 800 miRNAs tested in human feces, 181 miRNAs were detected (Table S2) with miR-1246, miR-601, miR-630, miR-2116-5p, miR-320e, miR-1224-5p, miR-155-5p and miR-194-5p being the most abundant miRNAs (Figure 1B). When we compared the 50 most abundant miRNAs in mouse and human feces, we found that 17 miRNAs were shared between these species (Figure 1C).
In order to investigate whether the miRNAs are present in different sections of the gut lumen, we collected gut luminal contents from the distal ileum and colon of C57BL/6J mice, isolated RNA and measured miRNA profiles. We observed that the miRNAs were significantly different between different regions of the intestine. More abundant miRNAs were found in the ileal lumen compared to the colon (Figure 1D). This distribution is opposite to the abundance of the gut microbes, which are abundant in the colon.
The gut microbiota shapes many aspects of gut physiology and immune system maturation (Lee and Mazmanian, 2010). To determine whether resident gut microbes affect fecal miRNA, we compared the fecal miRNA profile of germ-free (GF) mice with that of SPF colonized littermates. We found that the abundance of fecal miRNA in GF mice was higher than in SPF colonized mice and that the miRNA profiles in these two populations differed (Figure 1E). We further clarified this correlation by comparing SPF mice with antibiotic treated mice and found that removal of microbes in the gut by antibiotics resulted in significantly more luminal miRNA (Figure 1F).
Intestinal Epithelial Cells and Hopx-positive Cells are Two Main Sources of Fecal miRNA
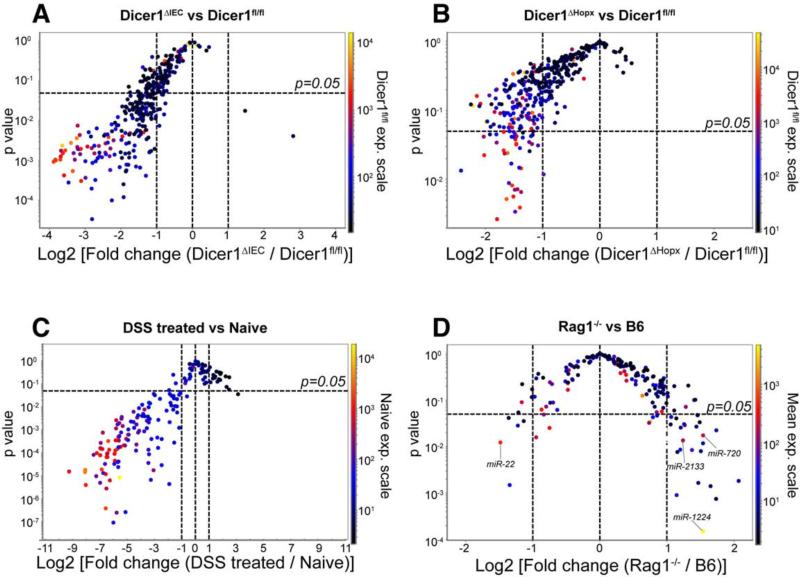
The source of fecal miRNA has not been reported. Since intestinal epithelial cells (IEC) were reported to secret exosomes (Van Niel et al., 2003) and we observed miRNA containing exosome-like EVs in the feces (Figures S1C-S1E), we investigated whether fecal miRNAs originated from IECs. Villin protein is universally expressed in IECs. Villin-cre transgenic mice (Vil-cre) express Cre recombinase under the direction of the villin 1 promoter (Madison et al., 2002). Dicer is required for the processing of miRNAs. FloxP sites on either side of the Dicer1 gene (Dicer1fl/fl) result in the Cre-expressing cell specific deletion of miRNAs (Harfe et al., 2005). We bred Vil-Cre mice with Dicer1fl/fl mice to generate mice defective in IEC-specific miRNA (Vil-CreTg/−, Dicer1fl/fl; referred to as Dicer1ΔIEC hereafter) (McKenna et al., 2010). We compared the fecal miRNA profiles of Dicer1ΔIEC mice with miRNA profiles of their wild type littermates (Vil-Cre−/−, Dicer1fl/fl; referred to as Dicer1fl/fl or WT hereafter) and found that the miRNA abundance was decreased and the profiles were altered (Figure 2A and Table S3), suggesting that intestinal epithelial cells are a major source of fecal miRNA.
Figure 2. Intestinal Epithelial Cells and Hopx-expressing Cells are Two Predominant Fecal miRNA Sources.
Volcano plot of fecal miRNA levels detected by nanostring in feces from: (A) Dicer1ΔIEC (n=6) v.s. Dicer1fl/fl (n=5) mice (See also Table S3); (B) Dicer1ΔHopx (n=4) v.s. Dicer1fl/fl (n=4) mice (See also Table S4); (C) DSS treated day 4 (n=4) v.s. naïve mice (n=4); and (D) Rag1−/− (n=5) v.s. wild type C57Bl/6J (B6) (n=5) mice. x-Axis: Log2 (fold change) of expression level between the groups as indicated; y-Axis: Benjamini-Hochberg corrected unequal variance t test p-value of the compared groups. Dotted horizontal line: p=0.05. The color of the dot indicates expression level of the corresponding miRNA in (A) and (B): Dicer1fl/fl (WT) group; (C): naïve group; (D): mean of both Rag1−/− and B6 groups.
We then asked whether Paneth cells and goblet cells also contributed to fecal miRNA. The HOP homeobox gene (Hopx) is expressed in intestinal epithelial +4 niche stem cells and in their derived cells including Paneth and goblet cells (Takeda et al., 2011). We thus bred HopxERCre mice (Takeda et al., 2011) with Dicerflox mice to generate Dicer1ΔHopx (Hopx-ERCreTg/−, Dicer1fl/fl; referred to as Dicer1ΔHopx hereafter) mice in which Hopx-expressing cells are deficient in miRNA following tamoxifen induction. We found that most of the detectable fecal miRNAs in Dicer1ΔHopx mice were decreased as compared to WT littermates and the profiles were changed (Figure 2B and Table S4), suggesting that the Hopx expressing cells are also a source of fecal miRNAs. The decreased fecal miRNAs in Dicer1ΔHopx mice were different from those affected in Dicer1ΔIEC mice (Tables S3 and S4) suggesting that epithelial cells and Hopx-expressing cells are responsible for the generation of different miRNAs in the feces.
To further investigate the gut epithelium as a source of fecal miRNAs, we studied dextran sulfate sodium (DSS)-induced colitis in which DSS treatment results in epithelial cell destruction and mild goblet cell loss (Solomon et al., 2010). Destruction of intestinal epithelial cells secondary to DSS treatment caused a marked decrease in fecal miRNAs (Figure 2C) supporting a key role for gut epithelial cells in the generation of fecal miRNAs. In order to distinguish whether the identified miRNAs are secreted from epithelial cells or are derived from sloughed epithelial cells, we compared the miRNA profile of epithelial cells and luminal content. Although, as expected, many miRNAs were found in both luminal content and epithelial cells, we also found many miRNAs such as miR-1224, miR-155, miR-710, miR-2138, at a much higher concentration in the gut luminal contents compared to epithelial cells (Figure S1F), suggesting that these miRNAs are specifically secreted into the gut lumen.
To investigate the degree to which fecal miRNAs are derived from lymphocytes that may be present in the gut, we analyzed fecal miRNA in Rag1−/− mice which have no mature B cells or T cells (Mombaerts et al., 1992). We found no major differences in the abundance of fecal miRNAs as compared to WT mice (Figure 2D), suggesting that these cells are not a major source of fecal miRNA.
Increased Gut Microbiota Dissimilarity in IEC miRNA-deficient Mice
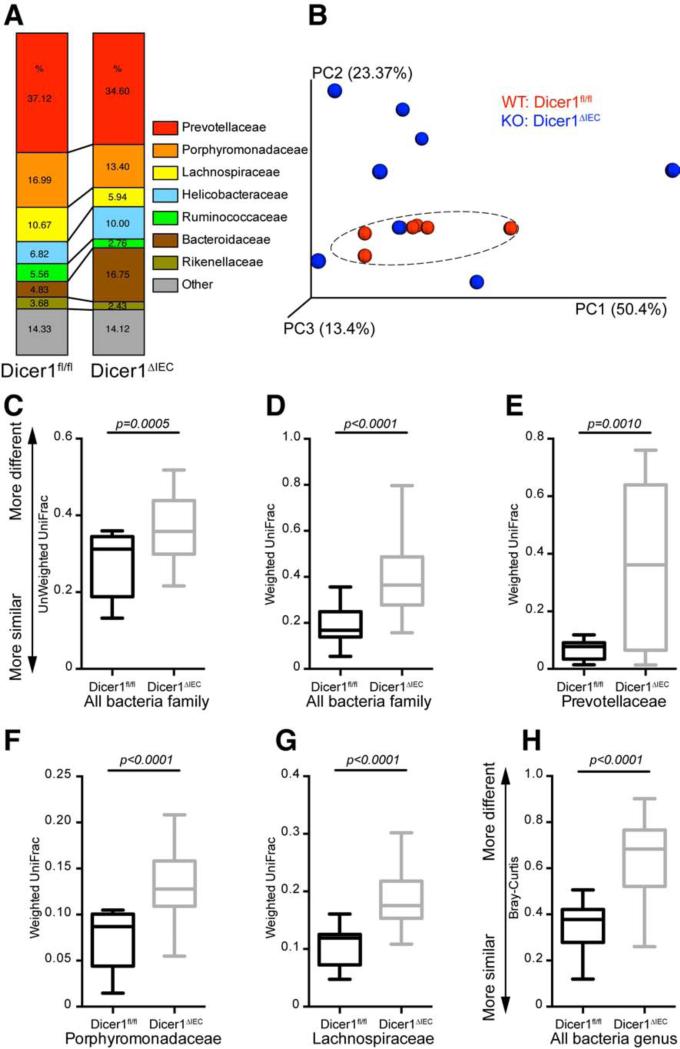
To evaluate whether fecal miRNA affects the gut microbiota, we surveyed the fecal bacterial composition of Dicer1ΔIEC and Dicer1fl/fl littermate mice by sequencing the V4 region of 16S rRNA gene using an Illumina MiSeq platform and analyzing with the QIIME software following an established protocol (Caporaso et al., 2010). We generated over 250,000 sequences for each sample, unique sequences were classified and grouped into 1,895 operational taxonomic units based on 97% nucleotide sequence identity (97%ID OTUs). Taxonomic classification revealed a shift of the dominant bacterial phyla Firmicutes and Proteobacteria in Dicer1ΔIEC mice compare to Dicer1fl/fl mice (Figure S2A). At the family level, we found an increase of Bacteroidaceae and Helicobacteraceae, and decrease of Prevotellaceae, Porphyromonadaceae, Lachnospiraceae and Ruminococcaceae in Dicer1ΔIEC mice (Figure 3A). Furthermore, UniFrac metric β-diversity-based principal coordinate analysis (PCoA) showed a phylogenetic architecture that was more dissimilar within Dicer1ΔIEC mice compared to WT littermates (Figure 3B), as indicated both by unweighted UniFrac analyses (Figure 3C), which measures the phylogenetic similarity between microbial communities, and by weighted UniFrac analyses (Figure 3D), which is an abundance-based metric (Goodrich et al., 2014).
Figure 3. Deficiency of Intestinal Epithelial Cell miRNA Increases the Dissimilarity of the Gut Microbiota.
Bacterial 16S rDNA sequence-based surveys were performed on the feces of 16 mice (n=7 Dicer1fl/fl, 9 Dicer1ΔIEC mice). (A) Relative abundance of bacteria was classified at a family-level taxonomy. (B) Principal coordinates analysis (PCoA) based on weighted UniFrac metrics. Dashed circle indicates the clustering of Dicer1fl/fl samples. (C-H) Box and whiskers plots of β-diversity distances between microbial communities comparing individuals within Dicer1fl/fl mice and between Dicer1ΔIEC individual mice. (C-G) β-diversity at family level: (C and D) of the whole microbiota; (E) the bacterial family Prevotellaceae; (F) the bacterial family Porphyromonadaceae; (G) the bacterial family Lachnospiraceae. (H) β-diversity at the genus level. (C-H) the specific distance metric used in each analysis is indicated on the axes. Values are: box: median, whiskers: min to max, p-value: non-parametric t-test).
We then constrained the distance metric analyses to the three most dominant bacteria families detected in WT mice: Prevotellaceae, Porphyromonadaceae and Lachnospiraceae (Figure 3A). We found marked dissimilarity between Dicer1ΔIEC individuals compared to WT individuals using the weighted UniFrac metric within the Prevotellaceae family, Porphyromonadaceae, and Lachnospiraceae family (Figures 3E-3G). Within the Porphyromonadaceae and Lachnospiraceae family, we observed greater dissimilarity between Dicer1ΔIEC individuals compared to WT individuals using unweighted UniFrac. However, no significant β-diversity differences were observed within Prevotellaceae families using unweighted UniFrac (Figures S2B-S2D). This indicates that the increased β-diversity within the Dicer1ΔIEC mice was either from changed bacterial species or the changed abundance of particular bacteria. Furthermore, we confirmed the marked dissimilarity within Dicer1ΔIEC individuals at the genus level by analyzing the overall OTU counts using Bray-Curtis β-diversity analyses (Figure 3H). Due to high β-diversity in the Dicer1ΔIEC mice, we were not able to identify major individual OTU differences between Dicer1ΔIEC mice and WT littermates. However, we did find 10 OTUs which had lower abundance and 2 OTUs with higher abundance in Dicer1ΔIEC mice compared to WT mice (Figure S2E and Table S5).
Host miRNA Affects the Growth of Gut Bacteria
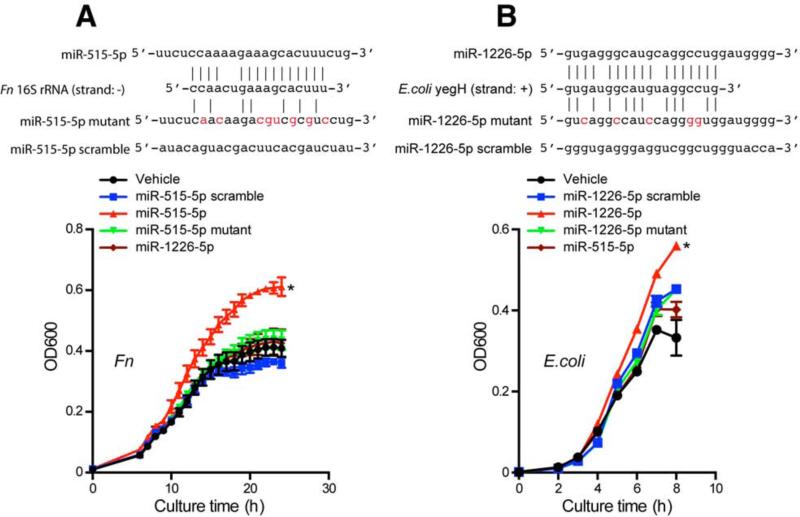
We next asked whether host miRNA affects individual microbial species in the gut. To determine whether specific miRNAs can affect bacteria directly in vitro, we examined two gut bacteria: the anaerobic species Fusobacterium nucleatum (Fn) and the facultative anaerobic species Escherichia coli (E. coli). Prior to in vitro culturing, we asked whether bacteria had any nucleic acid sites that could be targeted by miRNAs based on sequence similarity. We input seven nucleic acid sequences of Fn, E. coli and segmental filamentous bacteria (SFB), another bacteria species that is important for gut immune development (Lee and Mazmanian, 2014), to miRbase (Griffiths-Jones et al., 2008) to search for potential targets of miRNAs. We found that each bacterial nucleic acid sequence was predicted to be targeted by many miRNAs (Figure S3A and Table S6). Of note, these miRNAs come from both lower species such as worm (Caenorhabditis elegans) and fly (Drosophila melanogaster), and higher species including mouse and humans (Figure S3A and Table S6). The miRNA could align either to the plus or minus strand, and thus potentially act at the DNA level to affect gene expression or directly on RNA. Among these miRNAs, we found that miR-101, hsa-miR-515-5p, miR-876-5p, hsa-miR-325 and hsa-miR-1253 could potentially target Fn nucleic acid sequences; hsamiR-4747-3p, hsa-miR-1224-5p, hsa-miR-1226-5p and hsa-miR-623 could potentially target E. coli nucleic acid sequences (Figures S3B-S3G). The abundance of these miRNAs in human feces was determined by qPCR and is shown in Figure S4A. Accordingly, we cultured Fn and E. coli with synthesized miRNA mimics of these miRNAs in vitro and found that hsa-miR-515-5p promoted the growth of Fn (Figure 4A and Figure S3C) whereas hsa-miR-1226-5p promoted the growth of E. coli (Figure 4B and Figure S3F). These results demonstrate that miRNAs directly affect bacterial growth. As a control we used miRNAs that were synthesized by changing the predicted miRNA-target pairing sites of the miRNA. These mutated miRNAs did not confer the growth promoting effect on target bacteria (Figures 4A-4B, Figures S3D and S3G), suggesting the effect is sequence specific.
Figure 4. Host miRNA Directly Affects the Growth of Gut Bacteria.
Based on pilot culture experiments (see Figure S3C, Figure S3F), (A) Fn was grown in media with 1.25 μM miRNA mimics hsa-miR-515-5p, mutated hsa-miR-515-5p, scrambled control and hsa-miR-1226-5p. Growth was monitored as absorbance at 600 nm (OD600) once per hour for 24 hours. Representative growth curves of 5 independent experiments with triplicates are presented. See Figure S3D for additional growth curves. (B) E. coli was grown in media with 2 μM miRNA mimics hsa-miR-1226-5p, mutated hsa-miR-1226-5p, scrambled control and hsa-miR-515-5p. Growth was monitored as absorbance at 600 nm (OD600) once per hour for 8 hours. Representative growth curves of 5 independent experiments with duplicates are presented. See Figure S3G for additional growth curves. Upper panels show target site sequence alignment of hsa-miR-515-5p and mutant (mutant site highlighted) vs Fn 16S rRNA (A) and hsa-miR-1226-5p and mutant vs E. coli yegH sequence (B). *: Growth differs from other groups in 5 consecutive experiments. Related to Figure S3 and Table S6.
Host miRNA Enters Bacteria and Regulates Bacterial Gene Transcripts
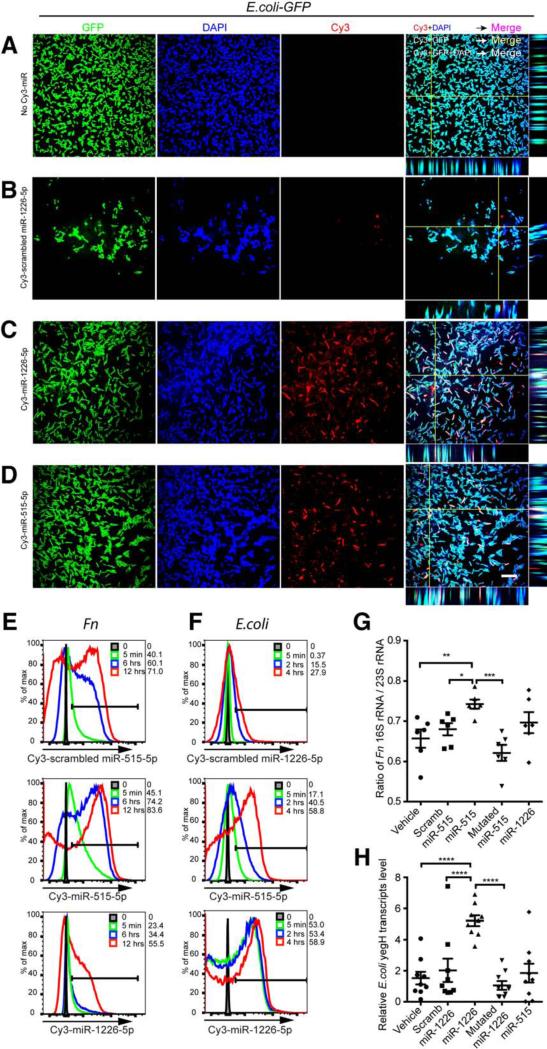
It has recently been reported that cellular miRNA can enter mitochondria and regulate mitochondrial gene expression (Zhang et al., 2014). It has also been well accepted that mitochondria originated evolutionarily from bacteria (Thrash et al., 2011) providing a theoretical basis for regulation of bacteria by miRNA. Thus, to elucidate how host miRNA affects bacteria growth, we first asked whether host miRNA was able to enter bacteria. We cultured a GFP-expressing E. coli strain (E. coli-GFP) with different synthesized fluorescence (Cy3)-conjugated miRNAs and examined the E. coli-GFP by confocal microscopy. We found that miRNAs entered the bacteria and co-localized with bacterial nucleic acids (Figures 5A-5D, Movie S1). Additionally, Cy3-conjugated miRNAs were also observed to co-localize with nucleic acids in Fn (Figures S4B-S4E, Movie S2). Furthermore, we observed the dynamic accumulation of miRNA in bacteria by flow cytometry during culture (Figures 5E-5F), providing a temporal and spatial basis for miRNA-bacterial nucleic acid interaction. Of note, the different capability of different miRNAs to enter bacteria, may in part explain different miRNA effects on bacteria gene transcripts and growth.
Figure 5. Host miRNA Enters Bacteria and Specifically Regulates Bacterial Gene Transcripts.
(A-D) E. coli GFP (Green) was cultured in the presence of 2 μM Cy3-labeled (red) hsamiR-1226-5p, scrambled hsa-miR-1226-5p control or hsa-miR-515-5p for 4 hours and washed with PBS, and fixed in 2% PFA, followed by nucleic acid staining with DAPI (Blue). Images were acquired by confocal microscopy with a 100x objective. Merged channel and orthogonal view were processed with Fiji/ImageJ. Scale bars, 10 μm. Representative of 2 experiments (See also Movie S1).
(E) Fn was cultured in the presence of 1.25 μM Cy3-labeled (red) hsa-miR-515-5p, scrambled hsa-miR-515-5p control or hsa-miR-1226-5p for 0, 5 min, 6 hrs and 12 hrs and terminated on ice, washed once with cold PBS and fixed with 2% PFA; followed by flow cytometry detection of Cy3 in the bacteria. The percentage of Cy3-miR positive Fn is shown. Representative of 2 experiments.
(F) E. coli GFP was cultured in the presence of 2 μM Cy3-labeled (red) hsa-miR-1226-5p, scrambled hsa-miR-1226-5p control or hsa-miR-515-5p for 0, 5 min, 2 hrs and 4 hrs and terminated on ice, washed once with cold PBS and fixed with 2% PFA; followed by flow cytometry detection of Cy3 in the GFP+ E. coli. The percentage of Cy3-miR positive E. coli is shown. Representative of 2 experiments.
(G) Fn was cultured in the presence of vehicle, 1.25 μM scrambled hsa-miR-515-5p control, hsa-miR-515-5p, mutated hsa-miR-515-5p, or hsa-miR-1226-5p for 16 hours. RNA was isolated and the ratio of Fn 16S rRNA/ 23S rRNA transcript level of was quantified by qPCR.
(H) E. coli was cultured in the presence of vehicle, 1.25 μM scrambled hsa-miR-1226-5p control, hsa-miR-1226-5p, mutated hsa-miR-1226-5p, or hsa-miR-515-5p for 4 hours. RNA was isolated and transcript levels of E. coli yegH were quantified by qPCR.
(G-H) Values are mean ± SEM, One-way ANOVA followed by Dunnett's multiple comparison tests. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Data summarize 3 independent experiments.
See also Figure S4.
To further examine the effect of the interaction of host miRNA with bacteria, we asked whether the bacterial gene expression could be specifically affected by the introduced miRNA. We cultured Fn with human miR-515-5p and quantified the 16S rRNA transcripts by qPCR. As we predicted (Figure 4A), the ratio of Fn 16S rRNA / 23S rRNA transcripts was increased (Figure 5G). Similarly, E. coli yegH mRNA was increased by miR-1226-5p (Figure 5H), RNaseP was increased by miR-4747-3p (Figure S4F), rutA mRNA was decreased by miR-1224-5p (Figure S4G) and fucO decreased by miR-623 (Figure S4H). In order to investigate whether the regulation is sequence-specific, we disrupted the predicted base pairing by using mutated miRNAs and determining gene regulation and growth effects. Because the most profound bacterial growth interference effect was miR-515-5p on Fn (Figure S3C) and miR-1226-5p on E. coli (Figure S3F), we mutated the predicted pairing sites of miR-515-5p and miR-1226-5p (Figure 4). These alterations not only impaired the gene regulation of the miRNAs (Figures 5G-5H) but also impaired their growth enhancing effects (Figure 4, Figures S3D and S3G).
WT Fecal miRNA Transplantation Restores the Fecal Microbes in IEC miRNA-deficient Mice
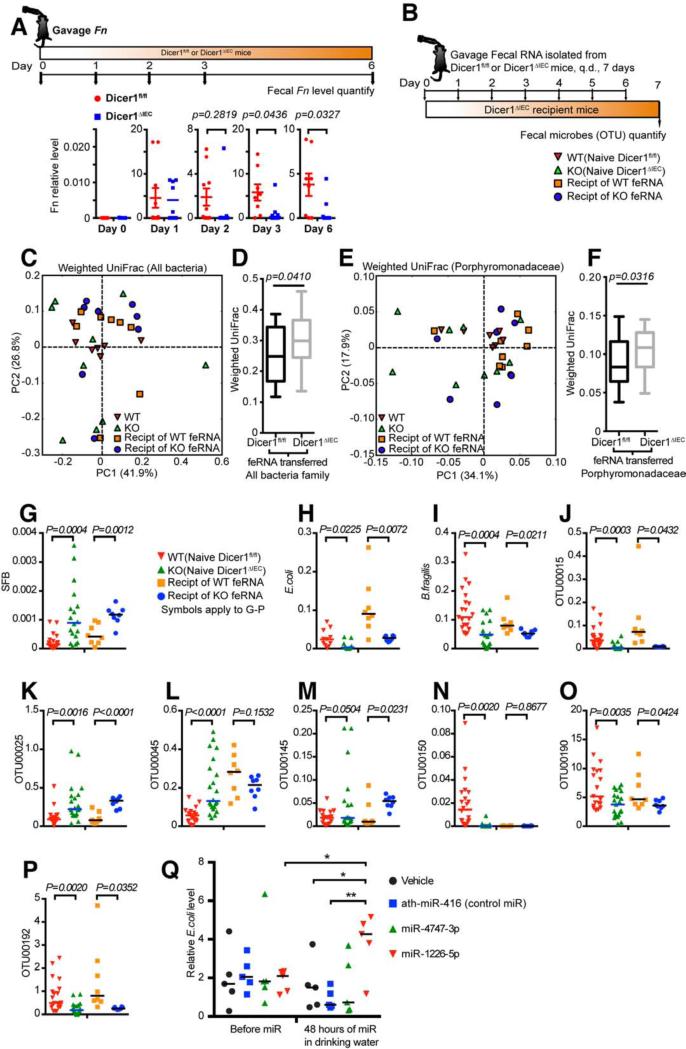
Our in vitro bacteria culture data suggest that miRNA regulate bacterial genes and a bacterial gene could be targeted by different miRNAs. We thus hypothesized that a set of host fecal miRNAs could contribute to shaping the composition of the gut microbiota. To investigate whether miRNAs in the gut affect the growth of exogenously introduced bacteria, we gavaged Dicer1ΔIEC mice and their WT littermates with Fn, which is not normally present in mouse feces. We found a significantly higher level of Fn in the feces of WT mice compared to feces from Dicer1ΔIEC mice (Figure 6A).
Figure 6. WT Fecal RNA Transplantation Restores the Fecal Microbes in IEC miRNA-deficient Mice.
(A) WT (Dicer1fl/fl) or Dicer1ΔIEC Mice were gavaged Fn. The relative abundance of Fn in the mouse feces was monitored by qPCR at gavage (day 0), day 1, day 2, day 3 and day 6-post gavage. Data are mean ± SEM, t-test, n≥8 mice and 6 littermate pairs per group, summary of two independent experiments. (B) Schematic diagram of fecal RNA transplantation that applies to (C-P): Donor fecal RNA (feRNA) was isolated from Dicer1fl/fl or Dicer1ΔIEC mice and was transferred by gavage once daily (q.d.) for 7 days to Dicer1ΔIEC recipient mice (Recipt), one donor to one recipient. (C-F) Bacterial 16S rDNA sequence-based UniFrac similarity matrix of the 18 recipient mice (n=8 Dicer1fl/fl fecal RNA recipients, 8 Dicer1ΔIEC fecal RNA recipients) was performed on the feces and compared with naïve mice as presented in Figure 3 of the Dicer1fl/fl and Dicer1ΔIEC mice. (C and E) Principal coordinates analysis (PCoA) of weighted UniFrac values of 4 groups. Each point represents one mouse sample and each sample is colored according to gene background or treatment. (C) Weighted UniFrac PCoA clustering results for all bacteria. (E) Weighted UniFrac PCoA clustering results for the family of Porphyromonadaceae. (D and F) Box and whiskers plots of β-diversity unifrac values between individuals within Dicer1fl/fl fecal RNA recipients and between Dicer1ΔIEC fecal RNA recipients of all bacteria (D) and the bacterial family Porphyromonadaceae (F). (Values are: box: median, whiskers: min to max, p-value: non-parametric t-test).
(G-P) Symbiotic bacteria SFB (G), E. coli (H), B. fragilis (I) loads in the naïve Dicer1fl/fl and Dicer1ΔIEC mouse feces, as well as in the feces of Dicer1ΔIEC mice received fecal RNA, were determined and the loads of the bacterial OTU00015 (J), OTU00025 (K), OTU00045 (L), OTU00145 (M), OTU00150 (N), OTU00190 (O) and OTU00192 (P) as tagged in Figure S2E were determined by qPCR (naïve group, n≥11, fecal RNA transplanted group, n=8). (Values are mean ± SEM, t-test).
(Q) C57BL/6J mice were administrated 200 nM indicated synthesized miRNA mimics in drinking water for 48 hours and the relative abundance of E. coli in the feces was determined by qPCR (n=5, scatter dot plot with line at median, One-way ANOVA followed by Dunnett's multiple comparison tests. *p<0.05, **p<0.01).
We then asked whether the endogenous microbes in the gut would also be directly shaped by fecal miRNAs. We performed a compensation assay. In this assay, RNA was isolated from WT or Dicer1ΔIEC mouse feces and then administrated to Dicer1ΔIEC recipient mice by gavage, one donor to one recipient (Figure 6B). Seven days after fecal miRNA transplantation, the microbiota profile of the recipient mice was examined by 16S rDNA sequencing. We found that transfer of WT fecal RNA, as compared to transfer of Dicer1ΔIEC fecal RNA, more profoundly reduced the gut microbiota β-diversity in KO recipients to be more similar to that of WT as evaluated by weighted UniFrac analyses of the whole microbiota (Figures 6C-6D) and of the family Porphyromonadaceae (Figures 6E-6F). The restoration effect was confirmed by a qPCR platform detecting the level of three species known to influence host immune cell function (Fritz et al., 2012; Vaishnava et al., 2011) and metabolism (Chen et al., 2014) including SFB, E. coli and B. fragilis, as well as seven highly detected OTUs. We found that SFB and OTU00025 were more abundant in naïve Dicer1ΔIEC mice, whereas WT fecal RNA transfer reduced their abundance (Figures 6G and 6K). E. coli, B. fragilis, OTU00015, OTU00190 and OTU00192 were increased in abundance in Dicer1ΔIEC mice gavaged with WT fecal RNA transfer. Three of the 10 detected species/OTUs were not restored by WT fecal RNA transfer (Figures 6H-6P).
To exclude the possibility that the changed microbiota in Dicer1ΔIEC mice was due to direct effect of antimicrobial components, i.e. IgA and RegIII-γ (Hooper et al., 2012) in the host, we measured free IgA concentration in the feces and found no difference between WT and Dicer1ΔIEC mice (Figure S5A). RegIII-γ expression in the small intestine was also not changed (Figure S5B), though it was increased in the colon (Figure S5C). As SFB colonize the small intestine of mice, these data collectively suggests that these host components do not contribute to compositional changes in fecal RNA transferred Dicer1ΔIEC mice. We also measured inflammatory cytokines and receptors in the colonic tissue of both naïve mice and fecal RNA transferred recipients. We found a decrease of Il-15 and Tnfsf13 and increase of Tnf in naïve Dicer1ΔIEC mice (Figure S5D), but found no difference between the WT and Dicer1ΔIEC fecal RNA transferred recipients (Figure S5E), suggesting that the microbiota restoration effects were not mediated by antimicrobial or inflammatory components.
In order to further test whether specific miRNAs could change specific bacteria in vivo, we synthesized miR-4747-3p and miR-1226-5p that enhanced E. coli in vitro (Figure 4B) and placed them in the drinking water of WT mice for 48 hours. We found that supplying miR-1226-5p increased the E. coli abundance in the feces (Figure 6Q).
WT Fecal miRNA Transplantation Rescued DSS Colitis in Dicer1ΔIEC Mice
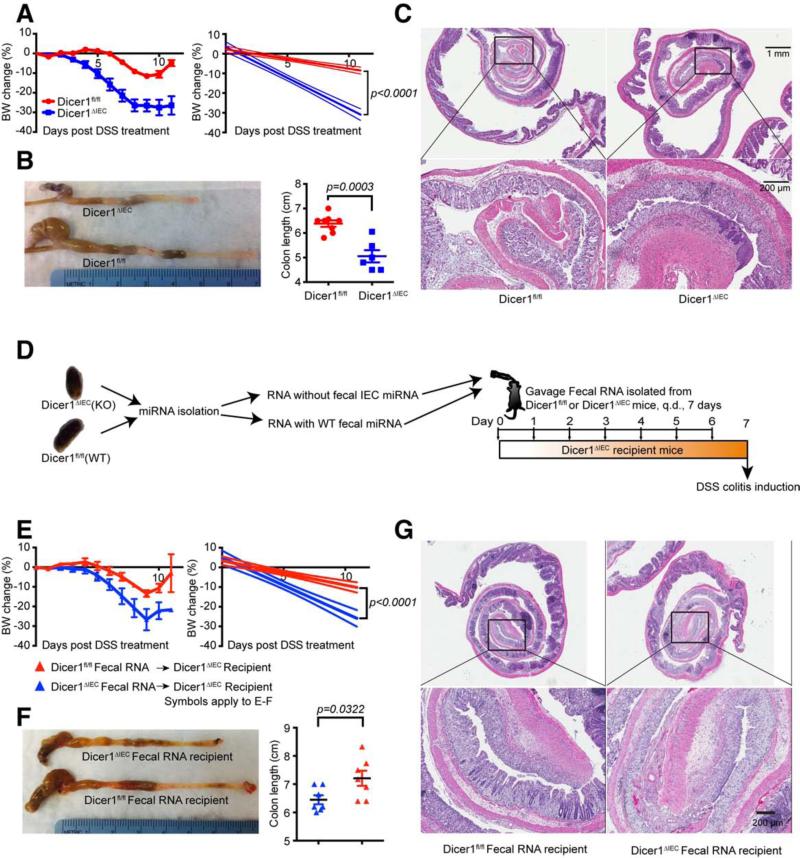
Given our finding that the composition of the microbiota is affected by fecal miRNA, we asked if there was a physiologic consequence in Dicer1ΔIEC mice. We found that the expression of MHCII in intestinal lymphoid tissue inducer cells (LTi) (Longman et al., 2014) was reduced in Dicer1ΔIEC mice (Figure S6A) and this was associated with decreased expression of lt-β, ifn-γ and tgf-β (Figure S6B) in the ileum, decreased expression of ifn-γ and increased expression of il-17 in the colon (Figure S6C). Resistin-like molecules are critical for the maintenance of colonic barrier integrity and their expression is regulated by symbiotic bacteria (Banerjee and Lazar, 2001; Hogan et al., 2006). We found that resistin-like molecules Relm-α and Relm-β were decreased in the Dicer1ΔIEC mice (Figures S6D-S6E). We further determined by qPCR the expression of epithelial tight junction molecules (Zo-1, occludin-1, claudin-1, claudin-2, and claudin-5) that were demonstrated to be regulated by symbiotic bacteria (Braniste et al., 2014). We found a significant reduction of ZO-1, claudin-1 and occludin-1 in the ileum (Figure S6F), and reduction of all of the tight junction protein transcripts except for claudin-2 in the colon (Figure S6G). These changes in the Dicer1ΔIEC mice increased susceptibility to colitis. Therefore, we induced colitis in mice using DSS. We found that Dicer1ΔIEC mice, as compared to WT mice, exhibited greater body weight loss (Figure 7A) and shortening of the colon (Figure 7B), as well as higher cellular infiltration in the colon and extensive loss of colonic tissue integrity (Figure 7C). We next determined whether the pathology was caused by the loss of secreted fecal miRNAs or by intrinsic cellular malfunction due to deficiency of epithelial miRNA. We administered fecal RNA from WT or Dicer1ΔIEC mice to Dicer1ΔIEC recipient mice by gavage for seven days prior to DSS treatment (Figure 7D). Transfer of WT fecal RNA to Dicer1ΔIEC mice resulted in less body weight loss (Figure 7E), longer colon length (Figure 7F) and less severe colonic damage (Figure 7G). These data suggest that intestinal luminal miRNAs are important in protecting the integrity of the intestinal epithelial barrier.
Figure 7. IEC miRNA-deficiency is Associated with Exacerbated DSS Colitis and is Rescued Following WT Fecal RNA Transplantation.
(A to C) 6-wk-old gender-matched Dicer1fl/fl and Dicer1ΔIEC littermates were treated with 3% DSS in drinking water for 7 days. (A) Percentile change of body weight (BW). Linear regression curves of the BW change are shown at right panel (Values are mean ± SEM, p< 0.0001 between groups); (B) Colonic length (Values are mean ± SEM, p= 0.0003 between groups, t-test); (C) Histologic analysis (H&E) at day 9 post DSS administration (n = 8, represents two independent experiments). (D-G), (D) Schematic diagram of fecal RNA transfer and colitis induction: Donor fecal RNA was isolated from Dicer1fl/fl (n=8) or Dicer1ΔIEC (n=8) mice feces and was administrated by gavage to Dicer1ΔIEC recipient mice, once daily (q.d.) for 7 days. Colitis was then induced by applying 3% DSS in drinking water for another 7 days. In the recipients, (E) Body weight change (Values are mean ± SEM, p< 0.0001 between groups); (F) Colonic length (Values are mean ± SEM, p= 0.0322 between groups, t-test); and (G) histologic analysis (H&E) at day 9 post DSS administration were analyzed.
Related to Figure S6.
DISCUSSION
The gut harbors approximately 10-100 trillion microorganisms which includes 100-200 different bacterial species and approximately 2 to 4 million genes (Faith et al., 2013). How the microbes are selected and whether the host specifically regulates microbial gene expression is not clear. Here we identified fecal miRNAs and found that they directly regulate specific bacterial gene expression and affect gut microbial growth.
Fecal miRNAs have not been characterized in normal human and animal feces. We found that miRNAs are a normal component in feces both in mice and humans and identified gut epithelial cells and +4 niche derived Hopx-expressing cells as two main sources of the fecal miRNAs. We found that fecal miRNAs are present in extracellular vesicles. However, since miRNAs are stable compared to other RNAs (Jung et al., 2010), whether fecal miRNAs could exist in EV-free forms such as associating with high-density lipoproteins or argonaute protein (Creemers et al., 2012), or in a completely free form needs further investigation.
Using miRbase (Kozomara and Griffiths-Jones, 2014), we identified that fecal miRNAs could base pair with specific bacterial genes (Table S6). By using E. coli and Fn, a species that has been reported to promote colorectal cancer (Rubinstein et al., 2013), as models, we observed that miRNAs were able to enter bacteria and co-localize with bacterial nucleic acids. This provides a temporal and spatial basis for miRNA-bacteria gene interaction. We observed that different miRNAs had different capacities to enter bacteria. This may in partial explain their different regulatory effects. However, the mechanisms controlling the entry of miRNAs into bacteria, as well as the mechanisms by which miRNAs are processed after they enter bacteria requires future investigation. By using different miRNAs and mutants, we showed that specific bacterial gene transcripts were regulated by specific miRNAs in culture. Since the miRNA could align either to the plus or minus target strand and thus may act at the DNA level to affect gene expression or directly on RNA. The detailed mechanism by which this occurs requires further study. The bacterial regulation we describe is different from traditional miRNA regulation in eukaryotic cell posttranscriptional repression, which includes cleaving of mRNA, destabilization of mRNA and reducing the translation efficiency (Bartel, 2009; Fabian et al., 2010). In our case, the host miRNA regulation of bacterial targets extended to rRNA (16S rRNA) and ribozyme (RNaseP), and the effect included not only a decrease of but also enhancement of the transcripts. However, we only observed miR-515-5p and miR-1226-5p promoted the growth of Fn and E. coli, respectively. We did not observe a suppressive effect on growth. How miRNA regulation of gene expression affects bacteria growth may rely on the function of the gene targeted by the miRNA.
The DSS-induced colitis model is independent of T and B lymphocytes (Dieleman et al., 1994), but dependent on symbiotic microbiota. Conventional microbiota provide a protective role in this model (Kitajima et al., 2001). By microbial 16S rRNA gene sequencing, we observed that a deficiency of epithelial originated miRNAs resulted in a more diverse gut microbiota and changed the intestinal barrier integrity. To investigate the consequence of a changed microbiota secondary to a gut miRNA-deficiency, we compared DSS-induced colitis in Dicer1ΔIEC versus WT mice and found marked exacerbation of colitis in the Dicer1ΔIEC mice. Importantly, we further found that fecal miRNA transplantation could help to restore the gut microbiota, which may have therapeutic applications (Goodrich et al., 2014).
In conclusion, we identified fecal miRNA as a normal component of the gut lumen. We identified host intestinal epithelial cells and +4 niche derived cells as two main sources of fecal miRNA. We further demonstrated that fecal miRNA specifically targets bacterial genes and thus regulates the gut microbiota. Given the importance of gut microbiota in host physiology and pathology (Honda and Littman, 2012), our findings reveal a host defense mechanism and highlight miRNA as a strategy for specific manipulation of microbiome for the health of the host.
EXPERIMENTAL PROCEDURES
(For detailed procedure see Extended Experimental Procedures)
Animals, Tissue and Feces Sampling
Animals were handled according to protocols approved by the Harvard Medical Area (HMA) Standing Committee on Animals. Mouse strains B6.Cg-Tg(Vil-cre)997Gum/J (Van Niel et al., 2003) and Hopxtm2.1(cre/ERT2)Joe/J (Takeda et al., 2011) were cross-bred with strain B6.Cg-Dicer1tm1Bdh/J (Harfe et al., 2005). Germ-free conventionalization was performed by oral gavage SPF mouse caecal material. Fecal specimens and intestinal luminal contents were collected, snap frozen, and stored at −80 °C for microbiota and miRNA analysis.
Antibiotic Treatment
Mice were given a mixture of antibiotics (ampicillin 1 mg/ml, vancomycin 500 mg/ml, neomycin 1 mg/ml, metronidazole 1 mg/ml, and streptomycin 1 mg/ml (Sigma-Aldrich)) in drinking water for 1 week according to established protocols (Benjamin et al., 2013).
Human Fecal Samples
Human fecal specimens were collected from 10 healthy subjects according to a protocol approved by the Institutional Review Board at Brigham and Women's Hospital.
Fecal RNA Isolation
Total RNA (including miRNAs) was extracted from stool specimens using mirVana™ miRNA isolation kit (Ambion®) following the manufacturer's procedure for total RNA isolation with modifications and with additional purification with Amicon® Ultra-0.5 Centrifugal Filter Devices-3k (Millipore).
Quantitative NanoString nCounter Fecal miRNA Analysis
nCounter® mouse miRNA Assay Kit and nCounter® human miRNA Assay Kit (NanoString Technologies) were used to detect miRNA in fecal RNA isolates following the manufacturer's protocol. Assay and spike-in controls were used for normalization based on identical amount of input RNA.
Fecal Microbes Quantification by qPCR
DNA was extracted from fecal pellets using a QIAamp Fast DNA Stool Mini Kit (Qiagen). qPCR analysis was conducted using TaqMan® Universal PCR Master Mix (Applied Biosystems).
Fecal Microbiota Diversity and Phylogenetic Analyses
16S rRNA gene V4 region amplicons from fecal DNA isolates as described were sequenced on the Illumina MiSeq 2 × 250 bp platform. Quality filtering and analysis were performed using the QIIME software pipeline following established protocols (Caporaso et al., 2010).
MiRNA Target Prediction
Nucleic acid sequences of particular bacteria were predicted for miRs targets by alignment using online miRBase tool (http://mirbase.org) (Griffiths-Jones et al., 2008).
In vitro Bacteria Growth Measurements
Bacteria strains Fusobacterium nucleatum (ATCC® 10953™) and E. coli (ATCC® 47016™) were cultured with Mission® miRNA mimics (Sigma-Aldrich) in culture medium and monitored as absorbance at 600 nm (OD600).
Detection of miRNA Entering Bacteria by Confocal and Flow Cytometry
Fn and E. coli GFP (ATCC 25922GFP) were cultured in the presence of synthesized Cy3-labeled hsa-miR-515-5p, hsa-miR-1226-5p or Cy3-labeled scrambled miRNA control (GE dharmacon) and observed with the flow cytometry and confocal microscope.
Fecal RNA Transplantation
Fecal RNA isolates from Dicer1fl/fl or Dicer1ΔIEC mice were administrated by gavage to Dicer1ΔIEC recipient mice at the dosage of 22.5 μg/day for 7 consecutive days. To investigate the effect of specific miRNAs on specific bacteria, 200 nM synthesized miRNA mimics were placed in the drinking water of WT mice for 48 hours.
Bacterial Gene Transcript Quantification by qPCR
Bacterial RNA from Fn and E. coli was extracted using TRIzol® Max™Bacterial RNA isolation Kit (Ambion). cDNA was made and qPCR was performed using Taqman probes. Outer membrane protein gene gyrA and 23S rRNA was used to normalize the transcript level of E. coli and Fn respectively.
DSS Colitis
Gender matched 6-week old Vil-cre+/− -Dicer1fl/fl and Vil-cre−/−-Dicer1fl/fl littermates were treated with 3% Dextran sodium sulfate (DSS, MW 36,000-50,000, MP Biomedicals) in the drinking water ad libitum for 7 days.
Statistical Analysis
Unless otherwise indicated, statistical significance was determined by a two-tailed Student's t-test with appropriate multiple comparison correction. p-value of <0.05 was regarded as significant. Except for specified in the context, results are expressed as mean ± SEM.
Supplementary Material
ACKNOWLEDGMENTS
We thank Oleg Butovsky and Duane Wesemann for providing GF and SPF colonized mice feces, technical assistance from Biopolymers Facility at Harvard Medical School for Bioanalyzer, Partners HealthCare Translational Genomics Core for sequencing, Center for Computational Cancer Biology (Dana-Farber Cancer Institute) for sequencing data analyses, DF/HCC research pathology core for histopathology, EM Facility for electron microscopy, Deneen E. Kozoriz for sorting and Francisco J. Quintana and Vijay K. Kuchroo for helpful discussions and reading the manuscript. The research was supported by NIH grant R01 AI43458 to H.L.W. and Susan Furbacher Conroy Fellowship to S.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.L. and H.L.W. conceived the concept and designed the study. S.L. generated transgenic mice and carried out all experiments. A.P.C. and R.M.R. were involved in cell isolation and flow cytometry. Z.W. performed extracellular vesicle isolation and nanoSight. L.B. performed GF mice colonization. L.E.C. was involved in anaerobic bacteria culture. R.G. provided human stool samples. S.L. and R.C. analyzed data. S.L. wrote the manuscript. H.L.W. supervised the study and edited the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, six tables and two movies.
REFERECES
- Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–295. [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a Symbiotic Microbe Regulate Homeostasis of Host Intestinal Natural Killer T Cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee RR, Lazar MA. Dimerization of resistin and resistin-like molecules is determined by a single cysteine. J. Biol. Chem. 2001;276:25970–25973. doi: 10.1074/jbc.M103109200. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin JL, Sumpter R, Levine B, Hooper LV. Intestinal Epithelial Autophagy Is Essential for Host Defense against Invasive Bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, Korecka A, Bakocevic N, Guan NL, Kundu P, et al. The gut microbiota influences blood-brain barrier permeability in mice. Science Translational Medicine. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, Morris LC, Matafonova E, Stien X, Kang L, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. The Journal of Clinical Investigation. 2014;124:3391–3406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- Dieleman LA, Ridwan BU, Tennyson GS, Beagley KW, Bucy RP, Elson CO. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. 1994;107:1643–1652. doi: 10.1016/0016-5085(94)90803-6. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439–1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, Rubino S, Robertson SJ, Larijani M, Gosselin J, Ivanov II, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan SP, Seidu L, Blanchard C, Groschwitz K, Mishra A, Karow ML, Ahrens R, Artis D, Murphy AJ, Valenzuela DM, et al. Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J Allergy Clin Immunol. 2006;118:257–268. doi: 10.1016/j.jaci.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annual Review of Immunology. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (New York, NY) 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (New York, NY) 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences 108 Suppl. 2011;1:4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. Has the Microbiota Played a Critical Role in the Evolution of the Adaptive Immune System? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. Microbial Learning Lessons: SFB Educate the Immune System. Immunity. 2014;40:457–459. doi: 10.1016/j.immuni.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A, Becker V, Goel A, Wex T, Malfertheiner P. Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS ONE. 2012;7:e42933. doi: 10.1371/journal.pone.0042933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Mändar R, Mikelsaar M. Transmission of mother's microflora to the newborn at birth. Biol. Neonate. 1996;69:30–35. doi: 10.1159/000244275. [DOI] [PubMed] [Google Scholar]
- McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–64. 1664, e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Rawls JFJ, Mahowald MAM, Ley RER, Gordon JIJ. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127:11–11. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E- cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (New York, NY) 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon L, Mansor S, Mallon P, Donnelly E, Hoper M, Loughrey M, Kirk S, Gardiner K. The dextran sulphate sodium (DSS) model of colitis: an overview. Comp Clin Pathol. 2010;19:235–239. [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappé MS, Giovannoni SJ. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:13. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2008;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The Antibacterial Lectin RegIII Promotes the Spatial Segregation of Microbiota and Host in the Intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Van Niel G, Mallegol J, Bevilacqua C, Candalh C, Brugière S, Tomaskovic-Crook E, Heath JK, Cerf-Bensussan N, Heyman M. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52:1690–1697. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al. MicroRNA Directly Enhances Mitochondrial Translation during Muscle Differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.