A combination of surface plasmon resonance (SPR) spectroscopy and interaction map® (IM) analysis was used to characterize binding of the LytTR-type response regulator YpdB to promoter DNA. YpdB follows an ‘AB-BA’ mechanism involving sequential and cooperative DNA binding followed by rapid successive promoter clearance.
Keywords: interaction map® (IM) analysis, protein–DNA interaction, pyruvate sensing, nutrient scavenging, response regulator YpdB, surface plasmon resonance (SPR) spectroscopy
Abstract
Most bacterial response regulators (RRs) make contact with DNA through a recognition α-helix in their DNA-binding domains. An emerging class of RRs interacts with DNA via a relatively novel type of binding domain, called the LytTR domain, which is mainly composed of β-strands. YpdB belongs to this latter class, is part of a nutrient-sensing network in Escherichia coli and triggers expression of its only target gene, yhjX, in response to extracellular pyruvate. Expression of yhjX mainly occurs in the late exponential growth phase, and in a pulsed manner. Although the DNA-binding sites for YpdB are well defined, exactly how YpdB initiates pulsed gene expression has remained elusive. To address this question, we measured the binding kinetics of wild-type YpdB and the phosphomimetic variant YpdB-D53E to the yhjX promoter region (PyhjX) using surface plasmon resonance (SPR) spectroscopy combined with interaction map® (IM) analysis. Both YpdB and YpdB-D53E bound as monomers to the tandem-repeat sequences in the promoter, with YpdB-D53E displaying a higher maximal binding rate than YpdB. Furthermore, we identified a high-affinity (A-site) and a low-affinity binding site (B-site) within the yhjX promoter. Only YpdB-D53E utilizes an ‘AB-BA’ DNA-binding mechanism, involving sequential and cooperative promoter binding, and rapid, successive promoter clearance. We propose that response regulator phosphorylation, in combination with the cycle of cooperative DNA binding and rapid promoter clearance just described, can account for pulsed gene expression.
INTRODUCTION
Bacteria are equipped with specific signal transduction systems, which allow for optimal adaptation to changing environmental conditions. The most prominent bacterial signal transduction systems are of the histidine kinase/response regulator (HK/RR) type, also referred to as two-component systems. Upon stimulus perception, the HK autophosphorylates a highly conserved histidine, and subsequently transfers the phosphoryl group to a highly conserved aspartate in the cognate RR. The majority of RRs are DNA-binding proteins, which act as transcriptional regulators when phosphorylated [1]. The family of LytS/LytTR HK/RR systems is widespread among human and plant pathogenic bacteria, and controls production of a variety of virulence and virulence-associated factors. Among known LytS/LytTR-regulated pheno-types are toxin production in Staphylococcus aureus [2], natural competence in Streptococcus pneumoniae [3] and the biosynthesis of extracellular polysaccharides in Pseudomonas aeruginosa [4,5].
High-resolution structural analyses of LytTR-family members have revealed an uncommon DNA-binding domain. A conserved 10-stranded β-fold mediates DNA binding via three elongated β-sheets, whereas highly variable amino acids within the connecting loop-regions are responsible for binding specificity [6]. The DNA-binding sites of almost all LytTR RRs are similar in their overall structure, comprising two direct or inverse repeats (9–11 nucleotides in length) separated by 11–13 spacer nucleotides, but they do not share a common consensus binding sequence [7–9]. Moreover, the flanking DNA regions also seem to be important for binding of LytTR-type transcription factors [10–13]. The YpdA/YpdB system of Escherichia coli, a LytS/LytTR-type HK/RR system, has recently been investigated in more detail [13]. The HK YpdA responds to extracellular pyruvate and activates its cognate RR YpdB, which in turn induces yhjX expression [13]. The gene yhjX is the only direct target gene of YpdB and codes for a putative transport protein of the major facilitator superfamily. When E. coli is grown in peptone broth, pulsatile induction of yhjX is observed during the late exponential growth phase [14]. The gene product is assumed to contribute to nutrient scavenging before cells enter stationary phase [13]. The RR YpdB is composed of an N-terminal receiver domain, with a conserved aspartate at position 53, and a C-terminal LytTR effector domain with DNA-binding affinity. Replacement of aspartate 53 by glutamate (YpdB-D53E) results in constitutive yhjX expression, presumably because the variant mimics phospho-YpdB [13].
Although two studies have provided detailed insights into the structural properties of the LytTR domain of AgrA from S. aureus when bound to its cognate DNA sequence [6,15], details of the mechanism of transcriptional activation by LytTR-type transcriptional regulators are still obscure. Only Straume et al. [16] have reported cooperative DNA binding by PlnC to tandemly arranged sites in the plnA promoter. It nevertheless remains unclear how the LytTR-type transcription factor YpdB induces pulsed expression of yhjX [13]. In order to answer this question, our study used a combination of surface plasmon resonance (SPR) spectroscopy with interaction map® (IM) analyses to determine the binding kinetics of YpdB and its phosphomimetic variant YpdB-D53E to the tandem repeat sequences in the yhjX promoter.
MATERIALS AND METHODS
Protein purification and molecular biological techniques
For the purification of 6His-YpdB and 6His-YpdB-D53E, E. coli strain BL21(DE3) [17] was transformed with pBAD24-ypdB and pBAD24-ypdB-D53E respectively [13]. Overproduction of 6His-tagged proteins and subsequent purification by Ni-NTA-affinity chromatography was performed as described before [12], yielding approximately 95% pure protein as estimated by SDS-PAGE [18]. The oligomeric state of both proteins was analysed using size-exclusion chromatography on a calibrated Superdex 200 Increase GL HR10/300 column (GE Healthcare), equilibrated with 50 mM Tris/HCl pH 7.6, 10% (v/v) glycerol, 150 mM NaCl, 2 mM DTT.
The size of biotinylated DNA fragments used in the SPR measurements was adapted from earlier experiments initially characterizing the yhjX promoter [13] and resembles all protected nucleotides from DNaseI footprint experiments. To generate double-stranded biotinylated PyhjX, equimolar amounts of 5′-biotinylated oligonucleotides (Sigma–Aldrich) and their complementary non-biotinylated strands (flanked by four guanine-cytosine nucleotides) (Table 1) were first heated at 95°C for 5 min and subsequently annealed by slow cooling to room temperature. The additional four G-C flanking the promoter region were added to further stabilize the DNA fragment and are therefore referred to as clamp. Depending on the oligonucleotides used the motifs of interest in the biotinylated double-stranded DNAs carried purine-to-pyrimidine or pyrimidine-to-purine transversions.
Table 1. Oligonucleotides used for SPR experiments on a SA sensor chip.
Biotinylated [BTN] oligonucleotides of the native promoter as well as BTN-oligonucleotides with motif substitutions (lower case letters) were annealed, and the double stranded DNA was immobilized onto the surface of a SA sensor chip. As a control the promoter DNA fragment of PyjiY was used.
| Name | 5′ → 3′ sequence |
|---|---|
| [BTN] yhjX YpdB bs sense | [BTN]GGGGCGCGTCATTCATTCCTGAACTAAGGCATTTCATTCCGTTCTGATGGCATTTCATGCCGGGGG |
| yhjX YpdB bs antisense | CCCCCGGCATGAAATGCCATCAGAACGGAATGAAATGCCTTAGTTCAGGAATGAATGACGCGCCCC |
| [BTN] yhjX YpdB A bs sense | [BTN]GGGGCGCGTCATTCATTCCTGAACTAAttacgggacgTCCGTTCTGATGGCATTTCATGCCGGGGG |
| yhjX YpdB A bs antisense | CCCCCGGCATGAAATGCCATCAGAACGGAcgtcccgtaaTTAGTTCAGGAATGAATGACGCGCCCC |
| [BTN] yhjX YpdB B bs sense | [BTN]GGGGCGCGTCATTCATTCCTGAACTAAGGCATTTCATTCCGTTCTGATttacgggacgGCCGGGGG |
| yhjX YpdB B bs antisense | CCCCCGGCcgtcccgtaaATCAGAACGGAATGAAATGCCTTAGTTCAGGAATGAATGACGCGCCCC |
| [BTN] yjiY YehT bs sense | [BTN]GGGGCCTTTGCCGCTCAACCGCAAAACTGACCGCTTACATCCCTAAAATAACCACTCAGTTAGGGG |
| yjiY YehT bs antisense | CCCCTAACTGAGTGGTTATTTTAGGGATGTAAGCGGTCAGTTTTGCGGTTGAGCGGCAAAGGCCCC |
Surface plasmon resonance spectroscopy
SPR assays were performed in a Biacore T200 using carboxymethyl dextran sensor chips pre-coated with streptavidin (SA Sensor Chip Series S). All experiments were carried out at a constant temperature of 25°C using HBS-EP [10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) detergent P20] as running buffer. Before immobilizing the DNA fragments, the chips were equilibrated by three injections of 1 M NaCl/50 mM NaOH applied at a flow rate of 10 μl/min. Then the respective double-stranded biotinylated DNA fragment (10 nM) was injected at a flow rate of 10 μl/min for a total contact time of 420 s. The chips were then washed by injecting 1 M NaCl/50 mM NaOH/50% (v/v) propan-2-ol. Approximately 100–200 RU (response units) of the relevant DNA fragment was bound per flow cell. Analyses of the kinetics of interaction of YpdB and YpdB-D53E with the various DNA fragments were performed at a flow rate of 30 μl/min in HBS-EP buffer at 25°C. Various concentrations of the proteins (1–50 nM), dissolved in HBS-EP buffer, were passed over the flow cells for 180 s, and the complexes formed were allowed to dissociate for 300 s before the next cycle started. After each cycle, the surface was regenerated by injection of 2.5 M NaCl for 30 s, followed by 0.5% (w/v) SDS for 60 s, at a flow rate of 30 μl/min. All experiments were performed at 25°C. Sensorgrams were recorded using Biacore T200 Control Software 1.0 and analysed with Biacore T200 Evaluation Software 1.0. The surface of flow cell 1 was coated with a control DNA (PyjiY DNA, no binding) and used to obtain blank sensorgrams for subtraction of the bulk refractive index background. The referenced sensorgrams were normalized to a baseline of 0. Peaks in the sensorgrams at the beginning and the end of the injection are due to the run-time difference between the flow cells for each chip.
Concentration-free calibration analysis (CFCA) was performed using a standard 5 μM solution of purified YpdB-D53E (calculated from absorbance-based determination of protein concentration using a molar absorption coefficient for YpdB of 3.24×105 M−1·cm−1), which was diluted stepwise (1:2, 1:5, 1:10 and 1:20). Each protein dilution was injected twice, first at 5 μl/min, then at 100 μl/min. PyhjX DNA was used for YpdB-D53E binding in the active flow cell, and PyjiY DNA (control DNA) in the reference cell. CFCA basically relies on mass transport, which is a diffusion phenomenon that describes the movement of molecules between the solution and the surface. The CFCA therefore relies on the measurement of the observed binding rate during sample injection under partially or completely mass transport-limited conditions. In general, the initial binding rate (dR/dt), which is dependent on the diffusion constant of the protein, is measured at two different flow rates. The diffusion coefficient of YpdB-D53E was calculated by the Biacore diffusion constant calculator and converter webtool (https://www.biacore.com/lifesciences/Application_Support/online_support/Diffusion_Coefficient_Calculator/index.html), based on the assumption that the protein is globular in shape. In this way, the diffusion coefficient of YpdB-D53E was determined to be D=9.94×10−11 m2·s−1. The initial rates for those dilutions that differed by a factor of at least 1.5 were utilized to calculate the ‘active’ concentration, which was determined as 5×10−8 M (1% of the absorbance-based measured protein concentration) for YpdB-D53E. The active protein concentration was then used for calculation of the kinetic binding constants.
Interaction map® analysis
IM calculations were performed on the Ridgeview Diagnostic Server (Ridgeview Diagnostics, Uppsala, Sweden). For this purpose, the SPR sensorgrams were exported from the Biacore T200 Evaluation Software 1.0 as *.txt files and imported into TraceDrawer Software 1.5 (Ridgeview Instruments). IM files were created using the IM tool within the software, generating files that were sent via e-mail to the server (im@ridgeviewdiagnostics.com), where the IM calculations were performed [19]. The resulting files were then evaluated for spots in the TraceDrawer 1.5 Software, and the IM spots were quantified.
RESULTS AND DISCUSSION
Purified 6His-YpdB and 6His-YpdB-D53E are monomeric in solution
YpdB-mediated expression of yhjX in E. coli cells grown in LB medium follows upon the accumulation of extracellular pyruvate and results in a pulse-shaped burst of transcription upon entry into stationary phase. Replacement of the highly conserved aspartate residue at position 53 within the receiver domain of the RR by glutamate (YpdB-D53E) causes yhjX expression to become constitutive and independent of growth phase [13]. Aspartate-to-glutamate substitutions have been shown to simulate phosphorylation in several RR classes [20], and this property, which often increase their DNA-binding affinities [7–9], has allowed the characterization of the active states of RRs [21].
In the present study, we have compared the properties of the constitutively active YpdB-D53E with those of the inactive wild-type protein to gain insights into the mechanism of promoter activation by this LytTR-type transcriptional activator.
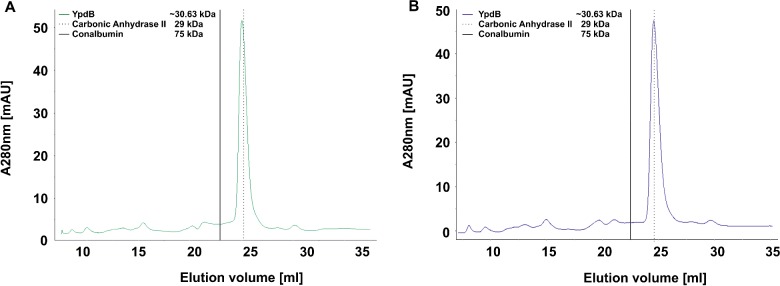
For in vitro studies, 6His-YpdB and 6His-YpdB-D53E were overproduced and purified by Ni-NTA-affinity chromatography as described before. Subsequent size-exclusion chromatography showed that both species (with an estimated molecular mass of 30.6 kDa) are exclusively monomeric in solution (Figures 1A and 1B).
Figure 1. Absorbance spectra obtained after size exclusion chromatography of purified 6His-YpdB and 6His-YpdB-D53E.
After affinity chromatography via an N-terminal 6His-tag, the oligomeric state of (A) YpdB (green) and (B) YpdB-D53E (blue) was analysed by size-exclusion chromatography using a calibrated Superdex 200 Increase GL HR10/300 column. The solid line represents the retention volume of 75 kDa protein conalbumin whereas the dotted line refers to 29 kDa protein carbonic anhydrase II, both applied in the same buffer used for 6His-YpdB/6His-YpdB-D53E purification.
YpdB-D53E binds to the PyhjX promoter with high affinity
In the next step we determined the kinetics of binding of YpdB and YpdB-D53E to the native promoter (PyhjX) using SPR spectroscopy.
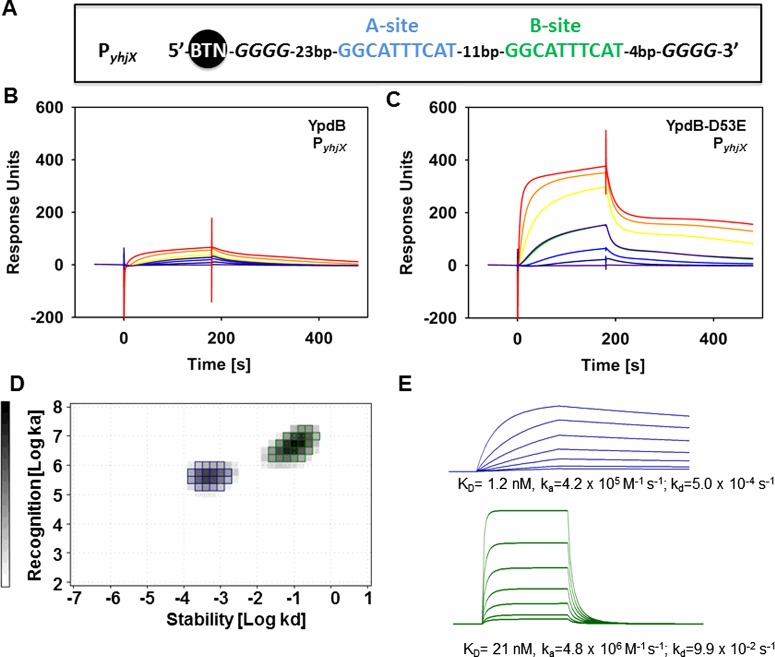
The corresponding 5′-biotinylated double-stranded DNA (Figure 2A) was immobilized onto an SA Sensor Chip, and increasing concentrations (1–50 nM) of YpdB or YpdB-D53E were passed over the chip surface for 180 s (association), followed by a 300 s dissociation phase. We found that the maximal binding capacity (Rmax RU) of YpdB-D53E to PyhjX (Figure 2C) was approxi-mately 8-fold higher than that of wild-type YpdB (Figure 2B). In addition, the sensorgrams differ in their overall association rates, which are significantly higher for YpdB-D53E than for YpdB to PyhjX.
Figure 2. Binding of YpdB and YpdB-D53E to the promoter region of yhjX (PyhjX).
(A) Schematic presentation of the binding sites A (blue) and B (green) within the biotinylated yhjX promoter fragment used in the consecutive SPR analyses. (B and C) The biotin-labelled DNA fragment comprising PyhjX was captured onto a SA sensor chip, and solutions containing purified YpdB at concentrations of 1 nM (violet line), 2.5 nM (blue line), 5 nM [dark blue and green line (internal reference)], 10 nM, 20 nM (orange line) and 50 nM (red line) (B) or YpdB-D53E (C) were passed over the chip. (D) IM analyses of the YpdB-D53E-PyhjX interaction. The green and blue spots both represent interactions of YpdB-D53E with the PyhjX DNA. (E) Sensorgrams were inferred for each specific KD value calculated from the IM analyses. The blue sensorgram corresponds to the blue spot and the green sensorgram to the green spot from ON/OFF rate map. The calculated affinities, as well as the ON/OFF rates, are indicated below the sensorgrams.
Furthermore, simple affinity calculations based on maximal binding rates (excluding ON and OFF rates) would result in an overall affinity of YpdB-D53E for PyhjX of 10 nM. This value is approximately 10 times higher than the KD value previously determined from gel-retardation experiments [13]. This discrepancy can be explained by the fact that the new calculation is based on the concentration of ‘active’ YpdB-D53E (as determined by CFCA) (for details please see Materials and Methods) and not on the total protein concentration.
Binding of YpdB-D53E to the DNA is complex
A more precise evaluation of the YpdB-D53E binding curves revealed that none of the sensorgrams for the phosphomimetic RR followed a hyperbolic curve, which is expected for 1:1 binding events. This result indicates that the interaction of YpdB-D53E with its binding sites in PyhjX does not reflect equal binding of the RR to each single site, but supports the possibility of more complex binding interactions [16]. To calculate reliable binding constants and kinetic parameters, a computational approach was employed to evaluate the sensorgrams. In this approach, the experimental SPR curves are considered as the sum of individual binding curves, each representing a monovalent interaction [22]. Consequently, we generated a so-called interaction map® from the YpdB-D53E sensorgrams in order to determine and quantify the individual binding events represented by the SPR curves. In brief, the algorithm behind IM splits an experimental SPR data set into se-veral theoretically 1:1 binding curves, which need to be summed up to match the initially observed experimental data. By plotting the unique combination of the association rate ka and dissociation rate kd within a 2D distribution (surface plot with the function of ka and kd), it is possible to display heterogeneous binding data as weight peaks, in which each peak corresponds to one component that contributes to the cumulative binding curve [19]. Fused peaks within this IM plot represent interactions with similar ON and OFF rates and can be taken into account with a peak weight proportion of more than 10%, whereas peaks below 10% most likely represent bulk effects. Based on the SPR sensorgrams for YpdB-D53E, IM analyses identified two clearly separated peaks (Figure 2D). The first peak (blue), with a peak weight of 33.5% (Figure 2D), displays an interaction with an average ON rate of 4.2×105 M−1 · s−1 and an OFF rate of 5.0×10−4 s−1. Referred to a monovalent binding curve (Figure 2E, upper panel) it results in an overall affinity of 1.2 nM. The second peak (green), with a peak weight of 51.6% (Figure 2D), is characterized by a 10-fold higher ON rate, whereas the OFF rate for this interaction was determined to be 9.9×10−2 s−1, which is even 50-fold higher compared with the interaction derived from the first peak (blue). The monovalent binding curve of this interaction (green) results in a KD of 21 nM (Figure 2E, lower panel).
In summary, the two YpdB-binding sites in PyhjX displayed the following features: One is characterized by a high affinity and has slow ON and slow OFF rates, whereas the second has a 10-fold lower affinity, with fast ON and fast OFF rates.
Binding of YpdB-D53E to site B requires prior occupation of site A
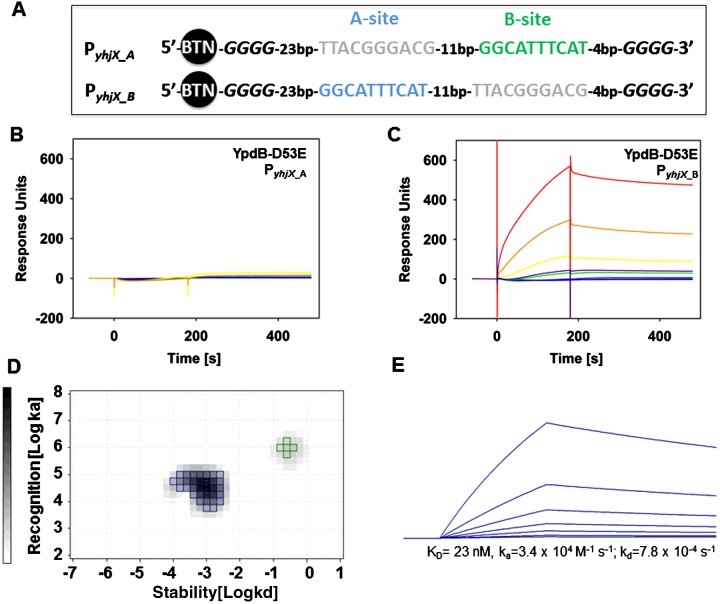
In order to probe these two interaction modes further, we examined the two YpdB-binding sites in more detail. YpdB binds to a well characterized direct repeat of the nucleotide motif GGCATTTCAT in PyhjX [13], hereinafter referred to as sites A and B (Figure 2A). We therefore generated two biotinylated DNA fragments in which either the A- (PyhjX_A) or the B-site (PyhjX_B) was inactivated by introducing transversion mutations (purines to pyrimidines and vice versa) (Figure 3A).
Figure 3. Binding of YpdB-D53E to the PyhjX_A and PyhjX_B.
(A) Schematic presentation of the two biotinylated yhjX promoter fragments carrying substitutions within binding site A or binding site B, respectively. (B, and C), SPR analyses. Biotin-labelled DNA fragments of the yhjX promoter region carry substitutions that inactivate site A (PyhjX_A) (B) or site B (PyhjX_B) (C), respectively. These fragments were captured onto SA sensor chips, and solutions of purified YpdB-D53E at concentrations of 1 nM (violet line), 2.5 nM (blue line), 5 nM [dark blue and green line (internal reference)], 10 nM (yellow line), 20 nM (orange line) and 50 nM (red line) were passed over the chip. (D) IM analyses of the YpdB-D53E-PyhjX_B interaction. The blue spot represents the YpdB-D53E interaction with the PyhjX_B. The green spot with peak weight of <5% does not correspond to a binding event (see text). (E) Calculated sensorgram for the interaction of YpdB-D53E with PyhjX_B. The calculated affinity, as well as the ON and OFF rates, are indicated below the sensorgram.
Then DNA fragments corresponding to PyhjX_A and PyhjX_B, respectively, were captured onto an SA sensor chip, and SPR experiments using different concentrations of YpdB-D53E were performed as described above. Unexpectedly, we found that bin-ding of YpdB-D53E was completely abolished when site A was inactivated (Figure 3B). In contrast, YpdB-D53E was able to bind normally to PyhjX_B, in which binding site B is mutated (Figure 3C). Moreover, in contrast with the binding of YpdB-D53E to the intact PyhjX, the sensorgrams for PyhjX_B recorded a single interaction event (Figure 3C). Calculation of the corresponding IM confirmed this observation (blue peak–weight of 68.2%) (Figure 3D). The additional small peak depicted in green with a peak weight of only 3.5% did not reflect a defined binding event, and was assumed to correspond to bulk effects. The ON (3.4×104 M−1·s−1) and OFF (7.8×10−4 s−1) rates correspond to an affinity constant of 23 nM (Figure 3E). Compared with PyhjX the ON rate for YpdB-D53E binding to PyhjX_B was 10 times lower, whereas the OFF rate remained unaffected. Based on this observation the binding kinetics from the calculated binding curves of YpdB-D53E binding to PyhjX (Figure 2E) is in very good agreement with the monovalent interaction described by the blue peak, which is reflected in the upper panel of Figure 2E. Furthermore, we observed that the calculated peak in the IM peak (Figure 3D) was less sharply defined (see Figure 2D). This points to a broader range of variation for ka and kd values, which suggests that the YpdB-D53E-PyhjX_B interaction is less stable or might be affected by additional unspecific complex formation in the absence of the second binding site. Another explanation might be a disarrangement of the RR molecules that is otherwise prevented by the presence of an intact B-site. We therefore hypothesize that efficient binding of a second YpdB-D53E molecule to site B enhances protein–DNA interaction at site A by an increased overall stability.
The ‘AB-BA’ binding model
In general, activation and subsequent dimerization of RR molecules is the predominant mode of action utilized for bacterial signal transduction by two-component systems [23]. Perception of the relevant signal induces the phosphorylation of a highly conserved aspartate in the receiver domain of the RR, which induces conformational changes and in turn results in activation of the protein. The relationship between activation and dimerization has been described for several RRs, such as VraR from S. aureus [24] and PhoB from E. coli [25]. Furthermore, structural studies of isolated receiver-domain variants of the LytTR-type RR ComE from S. pneumoniae have demonstrated that activation is tightly linked to the monomer-to-dimer transition [26].
We found that both 6His-YpdB and its constitutively active variant 6His-YpdB-D53E are monomeric in solution. The activation of YpdB, which is mimicked by the aspartate-to-glutamate substitution at position 53, enhances the protein's affinity for the PyhjX promoter region and the interaction follows bipartite binding kinetics. This suggests that phosphorylation of YpdB is not only important for initial DNA binding but also facilitates gene expression by enabling dimerization of two activated RR molecules on the DNA surface.
Importantly, YpdB-D53E was completely unable to bind to PyhjX when site A was inactivated, whereas the protein could still bind to PyhjX when site B was mutated. These results suggest a two-step binding process, in which a molecule of YpdB-D53E must first bind to site A and, in so doing, stimulates binding of a second molecule to site B (Figure 4).
Figure 4. Schematic depiction of the ‘AB-BA’ mechanism of binding of activated YpdB (YpdB-D53E) to the yhjX promoter.
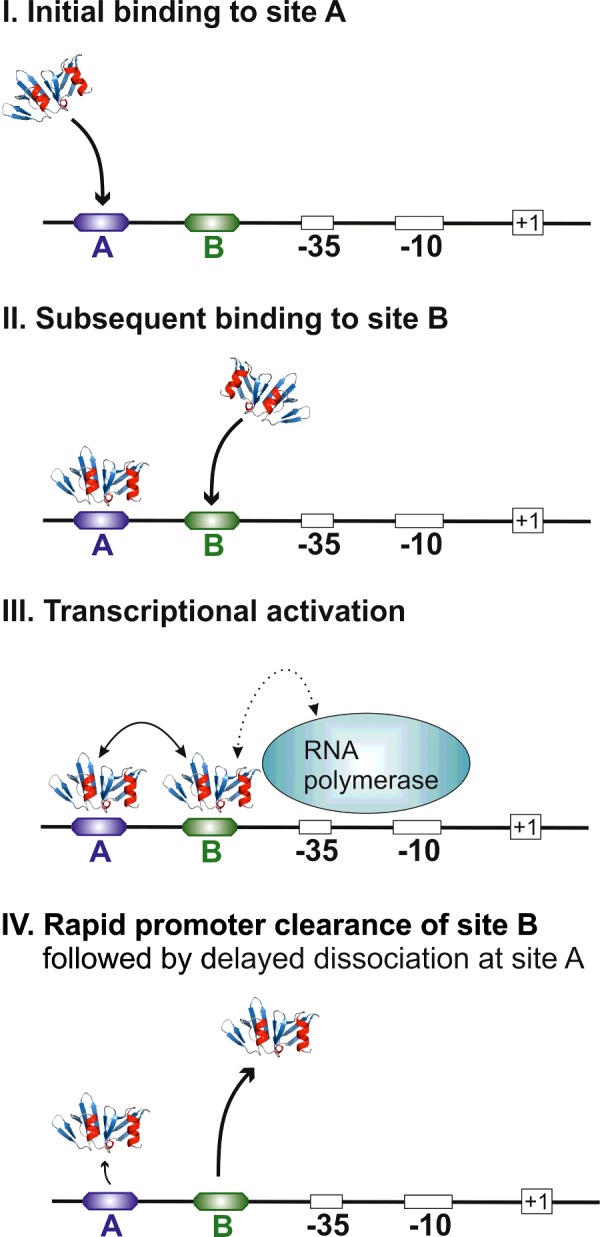
The RR is represented by the structural model of its DNA-binding domain derived by homology with known crystal structures. Initial binding of activated YpdB to yhjX promoter DNA occurs at site A. Subsequently, a second activated YpdB molecule occupies site B, and enables RNA polymerase recruitment and thus yhjX transcription. Occupation of site B is only transient, and initiates rapid and sequential promoter clearance.
Furthermore, the decrease observed in the ON rate for binding of YpdB-D53E at the A-site when the B-site is inactivated indicates that two bound YpdB-D53E molecules stabilize each other. Thus, this two-step activation might serve to trigger conformational changes in the DNA structure. Structural studies of the LytTR-binding domain of AgrA from S. aureus have revealed strong bending of the DNA upon binding of two RR molecules [6]. Interestingly, DNA analyses based on the oligonucleotide sequence used for SPR experiments (Table 1) predicted an average curvature for the A-site of between 4 and 6°, whereas the B-site curvature ranged from 8 and 11° [27]. These values already point to clear structural differences between the two sequence motifs, obviously due to flanking nucleotides, which have been shown to influence expression in vivo as well [13]. Furthermore, binding of the second RR molecule could be stabilized by intermolecular protein interactions, as has been shown for the RR KdpE [28]. Besides DNA bending, binding of the second RR to the B-site will also provide the required protein interface for recruitment of RNA polymerase. After the induction of yhjX expression, promoter clearance is initiated by rapid dissociation of the (second) RR from the B-site. This should effectively prevent (excessive) gene expression and might serve, together with a slower dissociation of the first RR from the A-site, as a tight control mechanism to generate a pulsed or switch-like expression pattern [29].
In light of the sequential nature of the binding and clearance of activated YpdB to and from the A- and B-sites within PyhjX, we refer to this mode of operation as the ‘AB-BA’ mechanism (Figure 4).
YpdB belongs to the LytTR family of RRs, and the promoter motifs of their respective target genes are similar to those of the yhjX promoter, in that they all contain A- and B-sites of comparable lengths, which are separated by a similar distance. One can therefore postulate that ‘AB-BA’ might well be a general mechanism used by LytTR-type RRs to induce pulsed expression of genes that are particularly important for tightly regulated processes, such as toxin production or pathogenicity.
Acknowledgments
SPR experiments were performed in the Bioanalytics Core Facility at the LMU Biocenter. We are grateful to Dr Karl Andersson (Ridgeview Instruments, Uppsala, Sweden) and Dr Anja Drescher (GE Healthcare, Munich, Germany) for support and helpful discussions.
Abbreviations
- BTN
biotin
- CFCA
concentration-free calibration analysis
- HK/RR
histidine kinase/response regulator
- IM
interaction map®
- ka
association rate constant
- kd
dissociation rate constant
- KD
equilibrium dissociation constant (steady-state affinity)
- PyhjX
promoter of the gene yhjX
- RU
response units
- SPR
surface plasmon resonance
AUTHOR CONTRIBUTION
Stefan Behr, Ralf Heermann and Kirsten Jung designed the concept of the study. Stefan Behr performed the protein affinity purification and size exclusion chromatography. Ralf Heermann designed and performed the surface plasmon resonance spectroscopy measurements, and analysed the resultant data [concentration-free calibration analysis (CFCA), interaction map® technology (IM)]. Stefan Behr, Ralf Heermann and Kirsten Jung wrote the manuscript.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft [grant number Exc114/2, DFGJU270/12-2 (to K.J.)].
References
- 1.Heermann R., Jung K. Stimulus perception and signaling in histidine kinases. In: Krämer R., Jung K., editors. Bacterial Signaling. Weinheim: Wiley-VCH; 2010. pp. 135–161. [Google Scholar]
- 2.Bronner S., Monteil H., Prévost G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 3.de Saizieu A., Gardes C., Flint N., Wagner C., Kamber M., Mitchell T.J., Keck W., Amrein K.E., Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 2000;182:4696–4703. doi: 10.1128/JB.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lizewski S.E., Schurr J.R., Jackson D.W., Frisk A., Carterson A.J., Schurr M.J. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 2004;186:5672–5684. doi: 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morici L.A., Carterson A.J., Wagner V.E., Frisk A., Schurr J.R., zu Bentrup K.H., Hassett D.J., Iglewski B.H., Sauer K., Schurr M.J. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 2007;189:7752–7764. doi: 10.1128/JB.01797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidote D.J., Barbieri C.M., Wu T., Stock A.M. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung J.K., Rood J.I. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 2000;182:57. doi: 10.1128/JB.182.1.57-66.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep D.B., Myhre R., Johnsborg O., Aakra A., Nes I.F. Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol. Microbiol. 2003;47:483–494. doi: 10.1046/j.1365-2958.2003.03310.x. [DOI] [PubMed] [Google Scholar]
- 9.Koenig R.L., Ray J.L., Maleki S.J., Smeltzer M.S., Hurlburt B.K. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 2004;186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Papa M.F., Perego M. Enterococcus faecalis virulence regulator FsrA binding to target promoters. J. Bacteriol. 2011;193:1527. doi: 10.1128/JB.01522-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risøen P., Johnsborg O., Diep D., Hamoen L., Venema G., Nes I. Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Genet. Genomics. 2001;265:198–206. doi: 10.1007/s004380000397. [DOI] [PubMed] [Google Scholar]
- 12.Kraxenberger T., Fried L., Behr S., Jung K. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J. Bacteriol. 2012;194:4272–4284. doi: 10.1128/JB.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fried L., Behr S., Jung K. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J. Bacteriol. 2013;195:807–815. doi: 10.1128/JB.02051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behr S., Fried L., Jung K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J. Bacteriol. 2014;196:2023–2029. doi: 10.1128/JB.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicod S.S., Weinzierl R.O., Burchell L., Escalera-Maurer A., James E.H., Wigneshweraraj S. Systematic mutational analysis of the LytTR DNA binding domain of Staphylococcus aureus virulence gene transcription factor AgrA. Nucleic Acids Res. 2014;42:12523–12536. doi: 10.1093/nar/gku1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straume D., Johansen R.F., Bjoras M., Nes I.F., Diep D.B. DNA binding kinetics of two response regulators, PlnC and PlnD, from the bacteriocin regulon of Lactobacillus plantarum C11. BMC Biochem. 2009;10:17. doi: 10.1186/1471-2091-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Altschuh D., Björkelund H., Strandgård J., Choulier L., Malmqvist M., Andersson K. Deciphering complex protein interaction kinetics using interaction map. Biochem. Biophys. Res. Commun. 2012;428:74–79. doi: 10.1016/j.bbrc.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Schaller G.E., Shiu S.-H., Armitage J.P. Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 2011;21:R320–R330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 21.Siam R., Marczynski G.T. Glutamate at the phosphorylation site of response regulator CtrA provides essential activities without increasing DNA binding. Nucleic Acids Res. 2003;31:1775–1779. doi: 10.1093/nar/gkg271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barta P., Björkelund H., Andersson K. Circumventing the requirement of binding saturation for receptor quantification using interaction kinetic extrapolation. Nucl. Med. Commun. 2011;32:863–867. doi: 10.1097/MNM.0b013e3283483e1c. [DOI] [PubMed] [Google Scholar]
- 23.Jung K., Fried L., Behr S., Heermann R. Histidine kinases and response regulators in networks. Curr. Opin. Microbiol. 2012;15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Leonard P.G., Golemi-Kotra D., Stock A.M. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8525–8530. doi: 10.1073/pnas.1302819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creager-Allen R.L., Silversmith R.E., Bourret R.B. A link between dimerization and autophosphorylation of the response regulator PhoB. J. Biol. Chem. 2013;288:21755–21769. doi: 10.1074/jbc.M113.471763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudes M., Sanchez D., Graille M., van Tilbeurgh H., Durand D., Quevillon-Cheruel S. Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae. Nucleic Acids Res. 2014;42:5302–5313. doi: 10.1093/nar/gku110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlahoviček K., Kajan L., Pongor S. DNA analysis servers: plot. it, bend. it, model. it and IS. Nucleic Acids Res. 2003;31:3686–3687. doi: 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan A., Kumar S., Evrard A.N., Paul L.N., Yernool D.A. An asymmetric heterodomain interface stabilizes a response regulator–DNA complex. Nat. Commun. 2014;5:3282. doi: 10.1038/ncomms4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisel N., Gerland U. Physical limits on cooperative protein–DNA binding and the kinetics of combinatorial transcription regulation. Biophys. J. 2011;101:1569–1579. doi: 10.1016/j.bpj.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]