Abstract
Geobacillus sp. WCH70 was one of several thermophilic organisms isolated from hot composts in the Middleton, WI area. Comparison of 16 S rRNA sequences showed the strain may be a new species, and is most closely related to G. galactosidasius and G. toebii. The genome was sequenced, assembled, and annotated by the DOE Joint Genome Institute and deposited at the NCBI in December 2009 (CP001638). The genome of Geobacillus species WCH70 consists of one circular chromosome of 3,893,306 bp with an average G + C content of 43 %, and two circular plasmids of 33,899 and 10,287 bp with an average G + C content of 40 %. Among sequenced organisms, Geobacillus sp. WCH70 shares highest Average Nucleotide Identity (86 %) with G. thermoglucosidasius strains, as well as similar genome organization. Geobacillus sp. WCH70 appears to be a highly adaptable organism, with an exceptionally high 125 annotated transposons in the genome. The organism also possesses four predicted restriction-modification systems not found in other Geobacillus species.
Keywords: Geobacillus sp. WCH70, Wood compost, Thermophile, Transposons, Restriction-modification
Introduction
Originally classified as members of the genus Bacillus, Geobacillus species were reclassified into a separate genus based on properties such as 16S rRNA gene sequence analysis, lipid and fatty acid analysis, phenotypic characterization, and DNA–DNA hybridization experiments [1]. Geobacillus species have been isolated from high-temperature oilfields [2], a corroded pipeline in an extremely deep well [3], American [4, 5] African [6] and Russian [7] hot springs, marine vents [8], and the Mariana Trench [9]. In addition to these extreme environments, Geobacillus species are commonly found in composting materials [10]. Geobacillus. sp. WSUCF1 [11], G. galactosidasius [12] and G. toebii [13] were isolated from high-temperature composts. The ability of Geobacillus species to thrive in these varied and often hostile environments suggests that these species possess enzymes suitable for applications in challenging industrial environments [14, 15]. As part of a program to identify organisms, we isolated Geobacillus species from a variety of composts in Middleton, WI. We report here the isolation and genome sequence of Geobacillus sp. WCH70, isolated from high-temperature wood compost.
Organism information
Classification and features
Geobacillus sp.WCH70 is a novel thermophilic species isolated from a hot wood compost pile (~70 °C) in Middleton, WI (43.097090° latitude and -89.504730° longitude). The organism was isolated from a piece of decaying wood by enrichment and plating on YTP-2 medium (YTP-2 media contains (per liter) 2.0 g yeast extract, 2.0 g tryptone, 2.0 g sodium pyruvate, 1.0 g KCl, 2.0 g KNO3, 2.0 g Na2HPO4.7H2O, 0.1 g MgSO4, 0.03 g CaCl2, and 2.0 ml clarified tomato juice) at 70 °C. The culture is available from the Bacillus Genetic Stock Center. Cultures are routinely grown on tryptic soy broth without glucose (Difco) media and maintained on TSB agar plates. C5-6 Technologies, Lucigen, and the Joint Genome Institute have placed no restrictions on the use of the culture or sequence data. Geobacillus sp.WCH70 is a Gram-positive, rod-shaped facultative anaerobe (Table 1), with optimum growth temperature of 70 °C and maximum growth temperature of 80 °C. Geobacillus sp.WCH70 appears to grow as a mixture of single cells and large clumps in liquid culture (Fig. 1).
Table 1.
Classification and general features of Geobacillus strain WCH70 [33]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [34] | |
| Phylum Firmicutes | TAS [35] | ||
| Class Bacilli | TAS [36, 37] | ||
| Order Bacillales | TAS [38, 39] | ||
| Family Bacillaceae | TAS [39, 40] | ||
| Genus Geobacillus | TAS [1] | ||
| Species Geobacillus sp. | |||
| Strain: WCH70 | |||
| Gram stain | Positive | IDA | |
| Cell shape | Rods and chains of rods | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Subterminal spores | IDA | |
| Temperature range | 55 °C to 80 °C | IDA | |
| Optimum temperature | 70 °C | IDA | |
| pH range; Optimum | 5.8-8.0; 7.5 | IDA | |
| Carbon source | Carbohydrate or protein | IDA | |
| MIGS-6 | Habitat | Compost | IDA |
| MIGS-6.3 | Salinity | Not reported | IDA |
| MIGS-22 | Oxygen requirement | Facultative anaerobe | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Non-pathogen | IDA |
| MIGS-4 | Geographic location | Middleton, WI, USA | IDA |
| MIGS-5 | Sample collection | September 2003 | IDA |
| MIGS-4.1 | Latitude | 43.097090 | IDA |
| MIGS-4.2 | Longitude | -89.504730 | IDA |
| MIGS-4.4 | Altitude | 342 | TAS |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [41]
Fig. 1.
Micrograph of Geobacillus sp. Y412MC52 cells showing individual cells and clumps of cells. Cells were grown in TSB plus 0.4 % glucose for 18 h. at 70 °C. A 1.0 ml aliquot was removed, centrifuged, re-suspended in 0.2 ml of sterile water, and stained using a 50 μM solution of SYTO® 9 fluorescent stain in sterile water (Molecular Probes). Dark field fluorescence microscopy was performed using a Nikon Eclipse TE2000-S epifluorescence microscope at 2000× magnification using a high-pressure Hg light source and a 500 nm emission filter
A phylogenetic tree was constructed to identify the relationship of Geobacillus sp.WCH70 to other members of the Geobacillus family (Fig. 2). The phylogeny of Geobacillus sp.WCH70 was determined using one of the ten16S rRNA gene sequence (genome coordinates 10256 through 11801), as well as those of the type strains of all validly described Geobacillus spp. The 16S rRNA gene sequences were aligned using MUSCLE [16], pairwise distances were estimated using the Maximum Composite Likelihood (MCL) approach, and initial trees for heuristic search were obtained automatically by applying the Neighbour-Joining method in MEGA 5 [17]. The alignment and heuristic trees were then used to infer the phylogeny using the Maximum Likelihood method based on Tamura-Nei [18]. Comparison of 16 S rRNA sequences shows Geobacillus sp.WCH70 clades with other 42 to 45 % G + C content species including G. thermoglucosidasius, G. caldoxylolyticus,G. galactosidasius and G. toebii and is most closely related to G. galactosidasius and G. toebii. Bootstrap analysis indicates that G. galactosidasius and G. toebii are more closely related to each other than to Geobacillus sp.WCH70, suggesting Geobacillus sp.WCH70 may be a new Geobacillus sp. Essentially identical trees were obtained when the other nine Geobacillus sp.WCH70 16S rRNA gene sequences were used to generate phylogenetic trees.
Fig. 2.
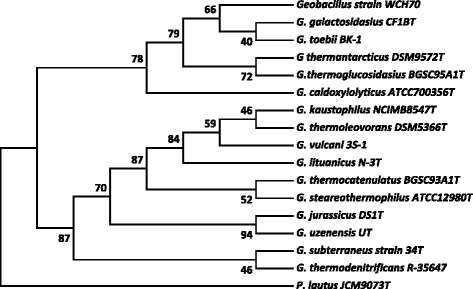
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model [18]. The bootstrap consensus tree inferred from 500 replicates [42] is taken to represent the evolutionary history of the taxa analyzed [42]. Branches corresponding to partitions reproduced in less than 50 % bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches [42]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The analysis involved 26 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1271 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [17]. The type strains of all validly described species are included (NCBI accession numbers): G. caldoxylosilyticus ATCC700356T (AF067651), G. galactosidasius CF1BT (AM408559), G. jurassicus DS1T (FN428697), G. kaustophilus NCIMB8547T (X60618), G. lituanicus N-3T (AY044055), G. stearothermophilus R-35646T (FN428694), G. subterraneus 34T (AF276306), G. thermantarcticus DSM9572T (FR749957), G. thermocatenulatus BGSC93A1T (AY608935), G. thermodenitrificans R-35647T (FN538993), G. thermoglucosidasius BGSC95A1T (FN428685), G. thermoleovorans DSM5366T (Z26923), G. toebii BK-1T (FN428690), G. uzenensis UT (AF276304) and G. vulcani 3S-1T (AJ293805). The 16S rRNA sequence of Paenibacillus lautusJCM9073T (AB073188) was used to root the tree
Genome sequencing information
Genome project history
Geobacillus sp.WCH70 was selected for sequencing on the basis of its biotechnological potential as part of the U.S. Department of Energy’s Genomic Science program (formerly Genomics:GTL). The genome sequence is deposited in the Genomes On Line Database [19, 20] (GOLD ID = Ga0028898), and in GenBank (NCBI Reference Sequence = CP001638.1). Sequencing, finishing and annotation were performed by the DOE JGI. A summary of the project information and its association with MIGS identifiers is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | 8 Kb and 40 Kb |
| MIGS 29 | Sequencing platforms | Sanger and 454 |
| MIGS 31.2 | Fold coverage | 13 × |
| MIGS 30 | Assemblers | Phred/Phrap/Consed |
| MIGS 32 | Gene calling method | Prodigal, GenePRIMP |
| Locus Tag | GWCH70 | |
| Genbank ID | NC_012793 | |
| GenBank Date of Release | December 1, 2009 | |
| GOLD ID | Gs0012167 | |
| BIOPROJECT | PRJNA20805 | |
| MIGS 13 | Source Material Identifier | Genome |
| Project relevance | Biotechnological |
Growth conditions and genomic DNA preparation
For preparation of genomic DNA, one liter cultures of Geobacillus sp.WCH70 were grown from a single colony in YTP-2 medium at 70 °C in flasks agitated at 200 rpm and collected by centrifugation. Culture stocks were maintained on YTP-2 agar plates grown at 70 °C. The cell concentrate was lysed using a combination of SDS and proteinase K, and genomic DNA was isolated using a phenol/chloroform extraction. The genomic DNA was precipitated, and treated with RNase to remove residual contaminating RNA. The purity and concentration of the recovered DNA was determined by gel electrophoresis in 0.7 % agarose containing ethidium bromide. Low and high molecular weight lambda DNA ladders were used as standards. The purity,and quantity of the recovered DNA was also independently confirmed by the JGI as suitable for sequencing prior to initiation of the project.
Genome sequencing and assembly
The genome of Geobacillus sp.WCH70 was sequenced at the JGI using a combination of Sanger and 454 technologies [21]. Two Sanger libraries with average insert size of 8 Kb and 40 Kb (fosmid) were generated for this genome. In addition to Sanger sequencing, 454 pyrosequencing was done to a depth of 20x coverage. Draft assemblies were based on 52,102 total reads. All three libraries provided 12.7x coverage of the genome. The Phred/Phrap/Consed software package was used for sequence assembly and quality assessment [22–24] in the following finishing process. After the shotgun stage, reads were assembled with parallel phrap (High Performance Software, LLC). Possible mis-assemblies were corrected with gapResolutioin (Cliff Han, unpublished), Dupfinisher, or sequencing cloned bridging PCR fragments with subcloning. Gaps between contigs were closed by editing in Consed, by PCR and by Bubble PCR primer walks. A total of 2,285 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The completed genome sequences of Geobacillus contains 56,142 reads, achieving an average of 13-fold sequence coverage per base with an error rate less than 1 in 100,000.
Genome annotation
Genes were identified using Prodigal [25] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [26]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. These data sources were combined to assert a product description for each predicted protein. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [26], RNAMMer [27], Rfam [28], TMHMM [29], and signalP [29].
Genome properties
The genome of Geobacillus sp.WCH70 consists of one circular chromosome (Table 3 and Fig. 3) of 3,464,618 bp and an average G + C content of 43 % and two circular plasmids of 33,899 and 10,287 bp and an average G + C content of 40 % (Table 4). There are 92 tRNA genes and 28 rRNA genes. There are 3,477 predicted protein-coding regions and 309 pseudogenes in the genome. A total of 2,373 genes (66.0 %) have been assigned a predicted function while the rest have been designated as hypothetical proteins (Table 4). The numbers of genes assigned to each COG functional category are listed in Table 5. About 39 % of the annotated genes were not assigned to a COG or have an unknown function.
Table 3.
Summary of genome: one chromosome and 2 plasmids
| Label | Size (Mb) | Topology | INSDC identifier | RefSeq ID |
|---|---|---|---|---|
| Chromosome | 3.46 | Circular | CP001638.1 | NC_012793 |
| Plasmid 1 | 0.034 | Circular | CP001639.1 | NC_012794 |
| Plasmid 2 | 0.010 | Circular | CP001640.1 | NC_012790 |
Fig. 3.
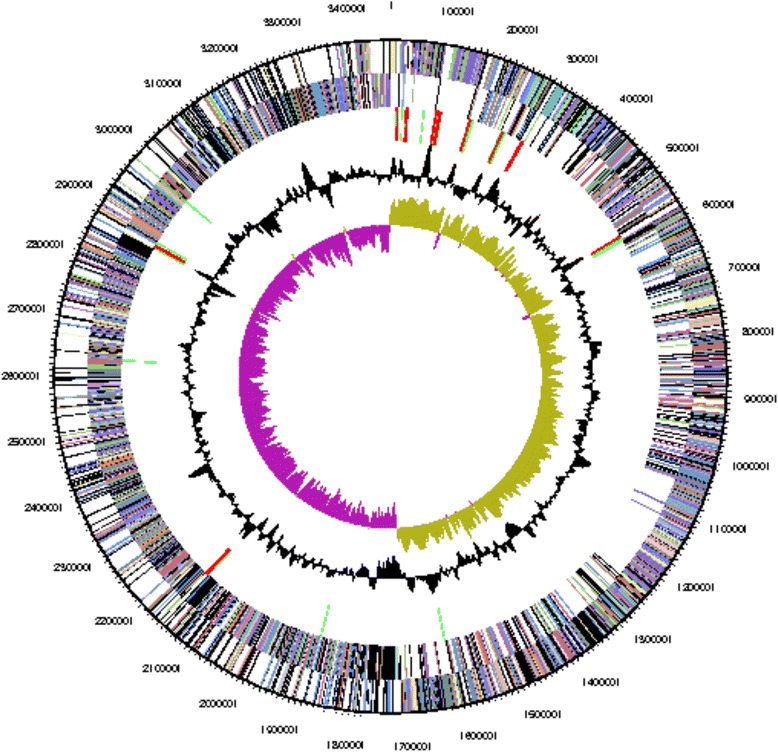
Graphical circular map of the Geobacillus sp. WCH70 chromosome. From outside to the center: Genes on forward strand (color by COG categories) Genes on reverse strand (color by COG categories) RNA genes (tRNAs green, rRNAs red, other RNAs black) GC content, GC skew
Table 4.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,508,804 | 100.0 |
| DNA coding (bp) | 3,033,424 | 86.4 |
| DNA G + C (bp) | 1,501,708 | 42.8 |
| DNA scaffolds | 3 | |
| Total genes | 3597 | 100.0 |
| Protein coding genes | 3477 | 96.7 |
| RNA genes | 120 | 3.3 |
| Pseudo genes | 309 | 8.6 |
| Genes in internal clusters | ||
| Genes with function prediction | 2373 | 66.0 |
| Genes assigned to COGs | 2201 | 61.2 |
| Genes with Pfam domains | 2946 | 81.9 |
| Genes with signal peptides | 125 | 3.5 |
| Genes with transmembrane helices | 805 | 22.4 |
| CRISPR repeats | 6 |
Table 5.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 195 | 8.0 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 143 | 5.8 | Transcription |
| L | 94 | 3.8 | Replication, recombination and repair |
| B | 1 | 0.1 | Chromatin structure and dynamics |
| D | 102 | 4.2 | Cell cycle control, Cell division, chromosome partitioning |
| V | 65 | 2.6 | Defense mechanisms |
| T | 104 | 4.2 | Signal transduction mechanisms |
| M | 102 | 4.2 | Cell wall/membrane biogenesis |
| N | 62 | 2.5 | Cell motility |
| U | 33 | 1.4 | Intracellular trafficking and secretion |
| O | 97 | 4.0 | Posttranslational modification, protein turnover, chaperones |
| C | 140 | 5.7 | Energy production and conversion |
| G | 128 | 5.2 | Carbohydrate transport and metabolism |
| E | 222 | 9.1 | Amino acid transport and metabolism |
| F | 71 | 2.9 | Nucleotide transport and metabolism |
| H | 158 | 6.5 | Coenzyme transport and metabolism |
| I | 99 | 4.0 | Lipid transport and metabolism |
| P | 131 | 5.3 | Inorganic ion transport and metabolism |
| Q | 45 | 1.8 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 194 | 7.9 | General function prediction only |
| S | 157 | 6.4 | Function unknown |
| - | 1396 | 38.8 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
The genome sequence of Geobacillus sp.WCH70 was compared to Geobacillus species with sequenced genomes. The lack of genome sequence information for G. galactosidasius and G. toebii prevents direct comparisons with these two organisms that are most closely related to Geobacillus sp.WCH70 based on rRNA gene sequences. Geobacillus sp.WCH70 Average Nucleotide Identity values [30] were 86.5 to 86.7 % to five G. thermoglucosidasius strains, 85.2 % to G. stearothermophilus NUB3621, and 84.9 and 85.0 % for two G. caldoxylosilyticus strains. ANI values ranged from 75.3 to 76.3 % for 20 other Geobacillus strains including G. stearothermophilusATCC 7953, G. thermodenitrificansDSM 465, G. subterraneus PSS2, and G. kaustophilus HTA426. These values mirror the relationships of Geobacillus sp.WCH70 to other species seen in the phylogenetic tree based on rRNA. In addition to being closely related to G. thermoglucosidasius strains based on these two criteria, synteny plots reveal highly similar genome organizations in Geobacillus sp.WCH70 and G. thermoglucosidasius C56-YS93 (Fig. 4).
Fig. 4.
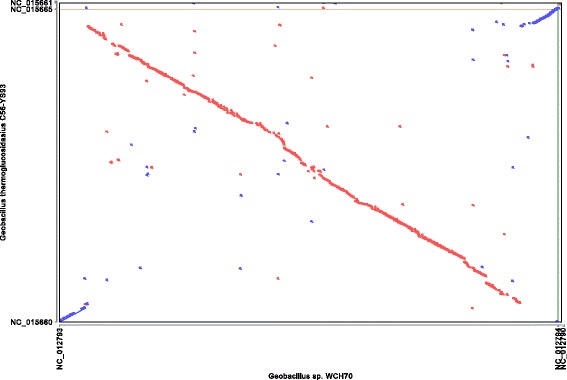
Synteny plot of Geobacillus sp. WCH70 versus G. thermoglucosidasius C56-YS93
Geobacillus sp.WCH70 possesses a number of unusual features when compared to other Geobacillus species. A major feature of Geobacillus sp.WCH70 is the presence of 125 insertion-sequence (IS) elements predicted to code for transposons, significantly more elements than are found in sequenced strains of either G. thermoglucosidasius or G. caldoxylosilyticus (Table 6). In addition to these IS elements, Geobacillus sp.WCH70 possesses four predicted restriction-modification gene clusters not found in other Geobacillus species. Genes GWCH70_1298 through GWCH70_1302 code for a predicted Type I restriction system most closely related to a system in B. cereus VD021, while GWCH70_2032 through GWCH70_2034 and GWCH70_3440 through GWCH70_3444 code for predicted Type I restriction systems most closely related to systems in B. coagulans XZL4. Genes GWCH70_2067 through GWCH70_2069 code for a predicted Type III restriction system most closely related to a system in Thermincola ferriaceticaDSM 14005™. Genes GWCH70_1385 and GWCH70_1386 code for restriction system proteins most closely related to proteins in Streptosporangium roseumDSM 43021™. These restriction systems may facilitate transfer of DNA to and from other organisms in the compost microbiome.
Table 6.
Comparison of predicted transposons
| Function Name | COG id | WCH70 | CIC9a | NBRCb | YUc | YS93d | GT20e | M10EXGf |
|---|---|---|---|---|---|---|---|---|
| Transposase, IS605 family | COG0675 | 62 | 3 | 2 | 0 | 1 | 0 | 0 |
| REP element-mobilizing transposase RayT | COG1943 | 8 | 0 | 1 | 0 | 0 | 0 | 0 |
| Transposase | COG3316 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Transposase, mutator type | COG3328 | 15 | 0 | 1 | 4 | 7 | 4 | 3 |
| Transposase, IS66 family | COG3436 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
| Transposase, IS204 family | COG3464 | 9 | 0 | 0 | 0 | 0 | 0 | 1 |
| Transposase, IS116 family | COG3547 | 11 | 0 | 1 | 0 | 5 | 0 | 1 |
| Transposase | COG5421 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| Transposase | Not in WCH70 | 0 | 4 | 10 | 13 | 13 | 13 | 19 |
| Total | 125 | 7 | 15 | 17 | 26 | 17 | 24 |
a Geobacillus caldoxylosilyticus CIC9, b Geobacillus caldoxylosilyticus NBRC 107762, c Geobacillus thermoglucosidans YU, d Geobacillus thermoglucosidasius C56-YS93, e Geobacillus thermoglucosidasius GT20, f Geobacillus thermoglucosidasius M10EXG, Geobacillus thermoglucosidasius NBRC 107763
Surprisingly, the genome of Geobacillus sp.WCH70 is lacking many of the predicted polysaccharide degradation clusters seen in other Geobacillus species [5], including the metabolic cluster for degrading hemicellulose [31]. The organism may utilize starch and other alpha-glucans based on the presence of a eleven-gene cluster GWCH70_0695 through GWCH70_0704 that is predicted to code for two, three-gene ABC carbohydrate transport systems, three alpha-amylase catalytic regions, an alpha-glucosidase, and a LacI family transcriptional regulator.
Conclusions
Geobacillus sp.WCH70 is a thermophilic gram-positive, spore-forming organism isolated from hot wood compost in the Middleton, WI area. Comparison of 16 S rRNA sequences showed the strain may be a new species, and is most closely related to G. galactosidasius and G. toebii. The genome of Geobacillus has an average G + C content of 43 %, similar to that reported for G. toebii (43.9 %) [13]. G. galactosidasius is reported to possess a 53.5 % average G + C content [12] significantly higher than the value for Geobacillus sp.WCH70. Six G. thermoglucosidasius strains have 43.8 to 44.0 % average G + C content based on genomic sequence [32], similar to the value obtained for Geobacillus sp.WCH70. These G + C content values are lower than the 53 to 54 % obtained using chemical analyses [1, 12]. Genomic sequencing of G. galactosidasius and G. toebii is necessary to clarify the relationships among Geobacillus sp.WCH70, G. galactosidasius and G. toebii, and G. thermoglucosidasius.
The presence of 125 insertion-sequence (IS) elements predicted to code for transposons along with multiple restriction-modification systems suggests Geobacillus sp.WCH70 possesses a highly mutable chromosome, able to add or delete non-essential genes and gene clusters depending on the environmental conditions. Genomic sequencing of other Geobacillus species may help clarify if this mutability is a common element in other organisms in composts, or unique to Geobacillus sp.WCH70.
Acknowledgements
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494). Sequencing work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396.
Abbreviations
- IMG
integrate microbial genomes database
- JGI
joint genome institute
Footnotes
Competing interests
Great Lakes Bioenergy Research Center, C5•6 Technologies, and Lucigen Corporation provided support in the form of salaries for authors PB, & DM, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
Authors’ contributions
PJB isolated and characterized the organism and wrote the manuscript, DAM managed the DNA preparation and submission to JGI for sequencing, and MLL did the genome annotation and document editing. All authors read and approved the final manuscript.
References
- 1.Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, et al. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int J Syst Evol Microbiol. 2001;51(Pt 2):433–46. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 2.Kuisiene N, Raugalas J, Chitavichius D. Geobacillus lituanicus sp. nov. Int J Syst Evol Microbiol. 2004;54(Pt 6):1991–5. doi: 10.1099/ijs.0.02976-0. [DOI] [PubMed] [Google Scholar]
- 3.Popova NA, Nikolaev Iu A, Turova TP, Lysenko AM, Osipov GA, Verkhovtseva NV, et al. Geobacillus uralicus, a new species of thermophilic bacteria. Mikrobiologiia. 2002;71(3):391–8. [PubMed] [Google Scholar]
- 4.Brumm P, Land M, Hauser LJ, Jeffries C, Chang YJ, Mead D. Complete genome sequence of Geobacillus strain Y4.1MC1, a novel CO-utilizing Geobacillus thermoglucosidasius strain isolated from Bath Hot Spring in Yellowstone National Park. Bioenerg Res. 2015;8(3):1039–1045.
- 5.Brumm P, De Maayer P, Cowan DA, MEAD DA. Genomic analysis of six new Geobacillus strains reveals highly conserved carbohydrate degradation architectures and strategies. Front Microbiol. 2015;6:430. doi: 10.3389/fmicb.2015.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawumba JF, Theron J, Brozel VS. Thermophilic protease-producing Geobacillus from Buranga hot springs in Western Uganda. Curr Microbiol. 2002;45(2):144–50. doi: 10.1007/s00284-001-0116-3. [DOI] [PubMed] [Google Scholar]
- 7.Nazina TN, Lebedeva EV, Poltaraus AB, Tourova TP, Grigoryan AA, Sokolova D, et al. Geobacillus gargensis sp. nov., a novel thermophile from a hot spring, and the reclassification of Bacillus vulcani as Geobacillus vulcani comb. nov. Int J Syst Evol Microbiol. 2004;54(Pt 6):2019–24. doi: 10.1099/ijs.0.02932-0. [DOI] [PubMed] [Google Scholar]
- 8.Maugeri TL, Gugliandolo C, Caccamo D, Stackebrandt E. Three novel halotolerant and thermophilic Geobacillus strains from shallow marine vents. Syst Appl Microbiol. 2002;25(3):450–5. doi: 10.1078/0723-2020-00119. [DOI] [PubMed] [Google Scholar]
- 9.Takami H, Nishi S, Lu J, Shimamura S, Takaki Y. Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles. 2004;8(5):351–6. doi: 10.1007/s00792-004-0394-3. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Li L, Huang R, Sun Y, Mei X, Shen B, et al. Variations of culturable thermophilic microbe numbers and bacterial communities during the thermophilic phase of composting. World J Microbiol Biotechnol. 2014;30(6):1737–46. doi: 10.1007/s11274-013-1593-9. [DOI] [PubMed] [Google Scholar]
- 11.Bhalla A, Kainth AS, Sani RK. Draft Genome sequence of lignocellulose-degrading thermophilic bacterium Geobacillus sp. Strain WSUCF1. Genome Announc. 2013;1(4):e00595-13. [DOI] [PMC free article] [PubMed]
- 12.Poli A, Laezza G, Gul-Guven R, Orlando P, Nicolaus B. Geobacillus galactosidasius sp. nov., a new thermophilic galactosidase-producing bacterium isolated from compost. Syst Appl Microbiol. 2011;34(6):419–23. doi: 10.1016/j.syapm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Sung MH, Kim H, Bae JW, Rhee SK, Jeon CO, Kim K, et al. Geobacillus toebii sp. nov., a novel thermophilic bacterium isolated from hay compost. Int J Syst Evol Microbiol. 2002;52(Pt 6):2251–5. doi: 10.1099/00207713-52-6-2251. [DOI] [PubMed] [Google Scholar]
- 14.de Miguel BT, Barros-Velazquez J, Villa TG. Industrial applications of hyperthermophilic enzymes: a review. Protein Pept Lett. 2006;13(7):645–51. doi: 10.2174/092986606777790548. [DOI] [PubMed] [Google Scholar]
- 15.Bergquist PL, Morgan HW, Saul D. Selected enzymes from extreme thermophiles with applications in biotechnology. Curr Biotechnol. 2014;3:45–59. doi: 10.2174/2211550102999131230150918. [DOI] [Google Scholar]
- 16.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 19.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC. The Genomes On Line Database (GOLD) v. 2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34(Database issue):D332–4. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38(Database issue):D346–54. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–94. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 23.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 24.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 25.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31(1):439–41. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 30.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–51. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 31.De Maayer P, Brumm PJ, Mead DA, Cowan DA. Comparative analysis of the Geobacillus hemicellulose utilization locus reveals a highly variable target for improved hemicellulolysis. BMC Genomics. 2014;15(1):836. doi: 10.1186/1471-2164-15-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42(Database issue):D560–7. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol. 1978;28:1–6. doi: 10.1099/00207713-28-1-1. [DOI] [Google Scholar]
- 36.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. Bergey’s Manual of Systematic Bacteriology. 2009;3:19–20. [Google Scholar]
- 37.Editor L. List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol. 2010;60:469–72. doi: 10.1099/ijs.0.022855-0. [DOI] [PubMed] [Google Scholar]
- 38.Prevot AR. Dictionnaire des Bactéries Pathogènes. 1953. pp. 1–692. [Google Scholar]
- 39.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 40.Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik. 1895;27:1–163. [Google Scholar]
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;10:512–26. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]


