Abstract
In this issue of Developmental Cell, Okamoto and Nishimura (2015) identify a positive feedback loop between neuronal cells that maintains insulin signaling and growth under restricted nutritional conditions.
The early discovery that Drosophila encodes eight Drosophila insulin-like peptides (Dilps) raised many questions regarding how these ligands might exert distinct functions through a single insulin receptor (InR). Subsequent functional studies showed that the downstream insulin/insulin-like growth factor signaling pathway (IIS) is conserved in metazoans, promoting growth and metabolic homeostasis in response to nutrient availability (Kannan and Fridell, 2013; Nässel et al., 2013). Moreover, a series of papers over the past few years have revealed that individual Dilps have distinct spatial and temporal patterns of expression and can signal either locally or remotely to control growth and physiology (Kannan and Fridell, 2013; Nässel et al., 2013). Much attention has focused on Dilp2, Dilp3, and Dilp5, which are highly expressed in the neurosecretory insulin producing cells (IPCs) that act like mammalian β cells. While a number of studies have focused on the regulated secretion of Dilps from the IPCs into the circulating hemolymph, relatively little is known about the control of their transcription, and functions for Dilp3 and Dilp5 remain unclear. In this issue of Developmental Cell, Okamoto and Nishimura (2015) explore the regulation and function of Dilp5. Through a series of elegant and detailed experiments, they discover a positive feedback loop between the surface glial cells, cholinergic neurons, and IPCs in the brain that sustains dilp5 production under restricted nutritional conditions to maintain insulin signaling during larval growth.
Okamoto and Nishimura (2015) first confirmed that dilp5 expression is repressed by starvation and found that feeding animals a rich diet—or one containing primarily amino acids, but not lipids or carbohydrates—is sufficient to restore dilp5 expression. In addition, while dilp5 mutants display relatively normal larval growth on rich media, they have reduced growth when maintained under nutrient-restricted conditions. Taken together with other experiments, these results raise the intriguing model that nutrient regulation of dilp5 expression is needed to maintain larval growth under suboptimal dietary conditions. Moreover, the authors show that disruption of the TOR amino acid-sensing pathway in the IPCs has no effect on dilp5 expression, indicating that other cells must sense nutrient-derived amino acids to control dilp5 production. These observations prompted the authors to undertake a detailed study of the regulation of dilp5 and led to the discovery of a remote signaling system that maintains Dilp5 in response to dietary signals (Figure 1).
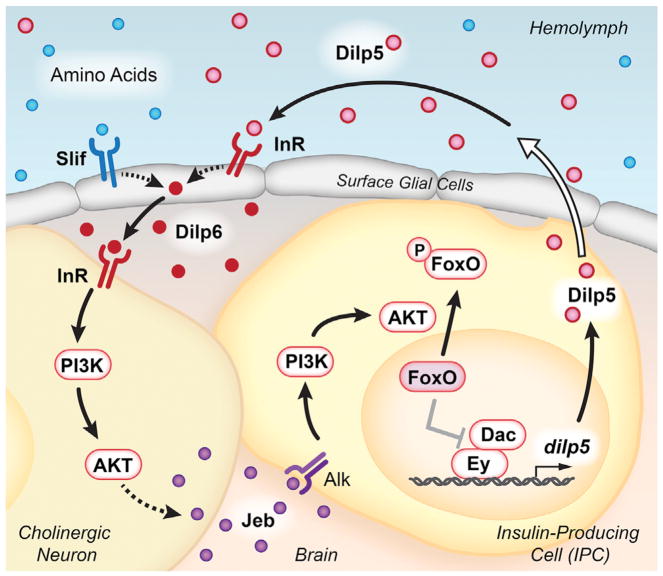
Figure 1. Intercellular Signaling Supports dilp5 Expression.
Circulating Dilps, in conjunction with dietary amino acids, promote Dilp6 production by surface glial cells. This ligand activates InR on the surface of cholinergic neurons, in close proximity to both the glia and the IPCs. IIS within cholinergic neurons leads to Jeb secretion, which activates the Alk receptor on IPCs. Downstream activation of the PI3K/AKT signaling pathway leads to phosphorylation and nuclear exclusion of Foxo. In the absence of IIS, nuclear Foxo negatively regulates the Ey and Dac transcription factors to suppress dilp5 expression. IPC-derived Dilps are secreted into the hemolymph via projections to the circulatory system (represented by the open arrow), completing the positive feedback loop.
The authors inactivate TOR signaling in a range of tissues and show that this pathway is required in surface glial cells, but not in the fat body, intestine, or neurons, to maintain dilp5 expression in the IPCs. The surface glia, which are exposed to the hemolymph and in close contact with the larval IPCs, are known to regulate neuroblast proliferation through Dilp6 signaling in response to circulating nutrients (Chell and Brand, 2010; Sousa-Nunes et al., 2011). Intriguingly, specific loss of dilp6 within the surface glia leads to reduced dilp5 expression in IPCs, whereas dilp6 overexpression in glia is sufficient to induce dilp5 even under fasting conditions, indicating that Dilp6 provides a critical link between nutritional signals and dilp5 expression. The authors also show that both TOR and insulin signaling are required in surface glia for dilp6 expression, suggesting that these cells sense both circulating Dilps and nutrients in the hemolymph. Consistent with this, ectopic expression of dilp5 in the fat body is sufficient to restore dilp6 expression in the brains of dilp2,3,5 triple mutants. These results suggest that dietary amino acids in conjunction with circulating Dilps induce dilp6 in the surface glia, which in turn remotely induces dilp5 expression in IPCs. The resulting signal amplification and positive feedback loop provides a focal point for this study, defining Dilp5 and Dilp6 as central factors in maintaining growth upon dietary restriction (Figure 1).
Unexpectedly, although the PI3K/AKT pathway is required in the IPCs for dilp5 expression, the InR is not. By screening the known Drosophila receptor tyrosine kinases using RNA interference, Okamoto and Nishimura (2015) identified anaplastic lymphoma kinase (Alk) as the receptor in this pathway. In addition, the Alk ligand, Jellybelly (Jeb), is both necessary and sufficient in cholinergic neurons for dilp5 expression in IPCs. These neurons underlie the surface glia and surround the IPCs, providing direct cellular contacts that can facilitate signaling. Importantly, overexpression of Jeb in cholinergic neurons lacking InR is sufficient to induce dilp5 expression, placing Dilp6 activation of IIS in cholinergic neurons upstream from Alk activation and dilp5 transcription in IPCs (Figure 1). The authors point out that the dependence on Alk signaling in IPCs maintains sensitivity to a range of nutritional levels. This is because Alk levels are unaffected by nutritional status, while InR is negatively regulated by nutrition and IIS. Thus, the employment of Jeb-Alk signaling by IPCs allows it to maintain a positive feedback loop between secreted Dilps and dilp5 expression, independent of nutritional state.
The authors complete the loop by conducting a series of detailed studies of dilp5 transcriptional regulation, building off their earlier work, showing that the Ey and Dac transcription factors directly promote dilp5 expression in IPCs (Okamoto et al., 2012). Nuclear Foxo can directly interact with Ey, disrupting the Ey-Dac protein complex and thereby down-regulating dilp5 expression under starvation conditions. Thus, the maintenance of cytoplasmic Foxo by Jeb-Alk signaling can sustain dilp5 expression under limited nutritional conditions.
This study by Okamoto and Nishimura (2015) provides a model to explain how larvae maintain their growth under restricted nutritional conditions (Figure 1). As the authors point out, dilp2 expression remains relatively constant under changing nutritional conditions, while Dilp2 peptide is rapidly secreted by IPCs in response to nutritional signals, providing a quick response mechanism on top of the sustained levels of dilp5 expression. Future experiments that examine Dilp5 secretion by IPCs can address the degree to which this might contribute to its functions. In addition, further work is needed to better define Jeb expression and secretion by cholinergic neurons (Okamoto and Nishimura, 2015). Finally, the authors point out that a receptor other than InR has been proposed to explain the ability of β cells to sustain IIS under changing nutritional conditions, and the sequence similarity between mammalian Alk and insulin/IGF-I receptors suggests that Alk may fulfill this role (Rhodes et al., 2013). In addition, cholinergic neurons are known to regulate β cells, suggesting that the functional interactions discovered by the authors are conserved through evolution and may provide new directions for understanding β cell physiology (Gilon and Henquin, 2001).
References
- Chell JM, Brand AH. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilon P, Henquin JC. Endocr Rev. 2001;22:565–604. doi: 10.1210/edrv.22.5.0440. [DOI] [PubMed] [Google Scholar]
- Kannan K, Fridell YW. Front Physiol. 2013;4:288. doi: 10.3389/fphys.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Kubrak OI, Liu Y, Luo J, Lushchak OV. Front Physiol. 2013;4:252. doi: 10.3389/fphys.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Nishimura T. Dev Cell. 2015;35:295–310. doi: 10.1016/j.devcel.2015.10.003. this issue. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Nishimori Y, Nishimura T. Proc Natl Acad Sci USA. 2012;109:2406–2411. doi: 10.1073/pnas.1116050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes CJ, White MF, Leahy JL, Kahn SE. Diabetes. 2013;62:2157–2163. doi: 10.2337/db13-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R, Yee LL, Gould AP. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]