Abstract
Predicting recurrence risk and chemotherapy benefit in early-stage breast cancer is challenging. The Oncotype DX gene assay is often used. Using a database of 221 patients a simple 2-rule model was developed and validated on an independent group of 319 patients. The model categorizes patients unlikely to benefit from the test thus achieving significant avoidance of cost.
Background
Predicting recurrence risk and chemotherapy benefit in early-stage breast cancer can be challenging, and Oncotype DX (ODX) is often used to gain insight. However, it is still unclear whether ODX can benefit in all cases. To clarify ODX’s usefulness we sought to develop a model using readily available pathologic markers to help clinicians make that determination.
Patients and Methods
Clinical pathologic data from 221 hormone receptor-positive, HER2-negative invasive breast cancer patients was used to create a model. The model was then validated on a second institution’s set of 319 patients.
Results
The model has 2 simple rules: low grade and positive progesterone receptor tumors (LG+PR) are low risk, and high grade or low estrogen receptor (ER) (ER < 20%) tumors (HG/LER) are high risk. The TAILORx (Trial Assigning Individualized Options for Treatment (Rx)) trial thresholds of Recurrence Score (RS) ≤ 10, when chemotherapy is of little benefit, and RS ≥ 26 when chemotherapy might be beneficial were used to judge model performance. Impressively, the misclassifications of an HG/LER patient who has an RS ≤ 10 were 0% and 2%, and for LG+PR patients who had an RS ≥ 26 were 0% and 2.6%. In the validation set, 28% (66 of 232) of the indeterminate group (neither in the HG/LER nor the LG + PR groups) had an RS ≤ 10 or an RS ≥ 26; this group might clinically benefit from ODX.
Conclusion
A simple 2-rule model based on readily available pathologic data was developed and validated, which categorized patients into high and low risk for recurrence. Identification of patients who are unlikely to benefit from ODX testing could result in significant cost avoidance.
Keywords: Classification model, Grade, Oncotype DX, Predictive value of tests, Risk assessment, TAILORx, Tumor markers
Introduction
Adjuvant chemotherapy treatment decisions are straightforward in early-stage breast cancer patients with good prognostic findings and with poor prognostic findings. The dilemma is the patient with conflicting clinicopathologic data, for whom the benefit of giving or withholding chemotherapy is unclear. This dilemma has fueled our demand for technology that allows us to predict, with greater accuracy, tumor responsiveness, risk of recurrence, and mortality. The promise of tumor genomic signatures is to provide biological information above and beyond current clinical and pathological data and ultimately solve this problem.
The 21-gene Recurrence Score (RS) assay (Oncotype DX [ODX], Genomic Health, Redwood City, CA) is a 21-gene reverse transcriptase-polymerase chain reaction assay first introduced in 2004 to provide additional clinical information regarding the risk of recurrence of estrogen receptor (ER)-positive (ER+) breast cancers.1 Tumor expression of these genes (representing proliferation, invasion, HER2, and ER-related genes) is analyzed and calculated, using a weighted formula, into a single RS, which is classified into 1 of 3 risk categories: low, RS ≤ 17; intermediate, RS 18 to 30; or high, RS ≥31. In 2006, work was published predicting the 10-year survival benefit of chemotherapy as a function of the ODX recurrence score.2
In the series that established the RS as a valuable test, patients were included who clearly were on either end of the spectrum of favorable and unfavorable clinicopathology. It is therefore possible that the published performance of this test is driven largely by the results of patients for whom we could have easily predicted the prognosis and benefit of chemotherapy. This raises the concern that, in the very patients with conflicting clinicopathologic findings for whom additional information is most needed, the ODX test is weaker in its predictive power than implied in the published reports.
The type of patients for whom ODX is ordered is evolving. Experience is allowing oncologists to identify patients for whom it is unlikely to provide additional information to aid in treatment decisions. This experience can be captured and enhanced in a model. We sought to develop such a model that can be used to guide clinicians as to when not to order the test (at significant savings of $4400), because the chances that the RS would contribute more information about the patient’s prognosis and chemotherapy responsiveness would be minimal.
Patients and Methods
All ER+, lymph node-negative breast cancer patients with complete clinicopathologic data who underwent ODX testing at Anne Arundel Medical Center (AAMC) between 2006 and July 2013 were included in our study. A developmental set of 221 patients (evaluated using the same staining and computer-aided slide-reading methodology, with proven high interpathologist reliability) was used to develop the current model. It was then tested on a “superset” which included an additional 108 patients with nonuniform tissue processing. Multiple tumor variables were retrospectively analyzed including ODX RS, tumor type, Nottingham score and grade, ER and progesterone receptor (PR) levels, HER2, and Ki-67.
Multiple statistical models were initially investigated using multiple variables including age, Ki-67, and others. The more complicated models added little to the accuracy and the final model required only 3 variables: tumor grade, ER percentage, and PR percentage. The following rules were established for the model: rule 1, high grade or low ER (defined as ER < 20%) tumors are likely to be high-risk and herein called the HG/LER group; and rule 2, low grade and positive PR patients are likely to be low-risk and called the LG+PR group (Figure 1).
Figure 1.

Rules for Classifying Early Estrogen Receptor (ER)-Positive Breast Cancer Patients Into Groups Likely to Either Benefit or Not Benefit From Adjuvant Chemotherapy
For our RS thresholds of low- and high-risk, we used those defined by the TAILORx (Trial Assigning Individualized Options for Treatment (Rx)) trial (RS < 11 and RS ≥26)3 as an example of the consensus standard of care for either withholding or offering chemotherapy.
Last, we validated our model with an independent data set from a second institution (Johns Hopkins Hospital [JHH]). This independent data set of patients was divided into 2 groups, similar to the AAMC developmental set and superset. The JHH MAIN set included only specimens evaluated by JHH campus pathologists, who review most of the breast pathology, and the JHH superset included all specimens, including cases that originally were evaluated by satellite center pathologists or non-Hopkins pathologists and then forwarded to JHH campus pathologists for review. Institutional review board approval was obtained from both institutions.
Results
Anne Arundel Medical Center Data Set Results
In the developmental set (n =221), 17% (n =37) were high-grade or low ER (the HG/LER group) and 33% (n = 73) were low-grade and positive PR (the LG+PR group). The remaining 50% (n = 111), had grade, ER, and PR results that rendered them indeterminate according to our model’s rules.
All 37 HG/LER patients had an RS > 11, with most (31 of 37; 84%) with an RS ≥ 26 (Table 1). Similarly, all LG+PR patients had RS < 26, with most (60 of 73; 82%) with RS < 18. The indeterminate patients (those neither in the HG/LER group nor the LG+PR group) were distributed across the spectrum, with most (70%; 76 of 111) resulting in an intermediate RS between 11 and 25, 17% (19 of 111) resulting in an RS < 11, and 14% (16 of 111) resulting in an RS > 25.
Table 1.
Classification of Patients Using the Model’s Rules and ODX/TAILORx Risk Groups
| Data Set | ODX Low Risk | ODX Intermediate | ODX High Risk | Total | ||
|---|---|---|---|---|---|---|
| TX NC | TAILORx Intermediate | TX Chemo | ||||
| RS <11 | 11–17 | 18–25 | 26–30 | RS >30 | ||
| AAMC Dev (Development) | ||||||
| HG/LER | 0 | 2 (5%) | 4 (11%) | 6 (16%) | 25 (68%) | 37 |
| Indeterminate | 19 (17%) | 41 (37%) | 35 (32%) | 8 (7%) | 8 (7%) | 111 |
| LG+PR | 27 (37%) | 33 (45%) | 13 (18%) | 0 | 0 | 73 |
| Total | 46 (21%) | 76 (34%) | 52 (24%) | 14 (6%) | 33 (15%) | 221 |
| AAMC Superset (Consults Included) | ||||||
| HG/LER | 1 (2%) | 5 (10%) | 7 (15%) | 8 (17%) | 27 (56%) | 48 |
| Indeterminate | 37 (19%) | 72 (38%) | 54 (28%) | 14 (7%) | 13 (7%) | 190 |
| LG+PR | 31 (34%) | 42 (46%) | 18 (20%) | 0 | 0 | 91 |
| Total | 69 (21%) | 119 (36%) | 79 (24%) | 22 (7%) | 40 (12%) | 329 |
| JHH MAIN (Main Hospital Only) | ||||||
| HG/LER | 1 (2.6%) | 8 (21%) | 11 (29%) | 6 (16%) | 12 (32%) | 38 |
| Indeterminate | 33 (19%) | 63 (36%) | 59 (34%) | 12 (7%) | 8 (5%) | 175 |
| LG+PR | 7 (28%) | 12 (48%) | 6 (24%) | 0 | 0 | 25 |
| Total | 41 (17%) | 83 (35%) | 76 (32%) | 18 (8%) | 20 (8%) | 238 |
| JHH Superset (Consults Included) | ||||||
| HG/LER | 1 (2%) | 10 (20%) | 13 (27%) | 7 (14%) | 18 (37%) | 49 |
| Indeterminate | 39 (17%) | 88 (38%) | 78 (34%) | 16 (7%) | 11 (5%) | 232 |
| LG+PR | 11 (29%) | 17 (45%) | 9 (24%) | 0 | 1 (2.6%) | 38 |
| Total | 51 (16%) | 115 (36%) | 100 (31%) | 23 (7.2%) | 30 (9%) | 319 |
Abbreviations: AAMC = Anne Arundel Medical Center; Chemo = chemotherapy; HG/LER = high grade or low estrogen receptor; JHH = Johns Hopkins Hospital; LG + PR = low grade and progesterone receptor-positive; NC = no chemotherapy; ODX = Oncotype DX (Genomic Health, Redwood City, CA); TAILORx = Trial Assigning Individualized Options for Treatment (Rx); TX = treatment.
When we applied our model to the AAMC superset of 329 patients, the breakdown was similar with 48 of 329 (15%) stratified as HG/LER, 190 of 329 (58%) as indeterminate, and 91 of 329 (28%) as LG+PR. The ability of the model to predict the RS group was equally accurate, showing all but 1 (47 of 48; 98%) of the HG/LER patients with an RS ≥ 11. Similarly, all LG+PR patients (91 of 91) had an RS < 26. Most indeterminate patients had an intermediate or low RS (126 of 190 [66%] with an RS of 11–25; 37 of 190 [19%] with an RS < 11); and 27 of 190 (14%) had an RS > 25.
Johns Hopkins Hospital Model Validation Results
The JHH MAIN set included 238 patients. Of these, 38 of 238 (16%) were stratified as HG/LER patients, 175 of 238 (74%) as indeterminate, and 25 of 238 (11%) as LG+PR. Of those stratified as HG/LER, 37 of 38 (97%) had an RS ≥ 11. Of those stratified as LG+PR, all tumors (25 of 25) resulted in an RS < 26. Most indeterminate tumors resulted in an intermediate RS (122/175, 70%), 33 of 175 (19%) were RS < 11, and 20 of 175 (12%) were RS ≥ 26.
The JHH superset included 319 patients. Of these, 49 of 319 (15%) were stratified as HG/LER patients, 232 of 319 (73%) indeterminate, and 38 of 319 (12%) LG+PR. Of those stratified as HG/LER, 48 of 49 (98%) had an RS ≥ 11. Of those stratified as LG+PR, 37 of 38 (97%) had an RS < 26. Again, most indeterminate tumors resulted in an intermediate RS between 11 and 25 (166 of 232; 72%), 39 of 232 (17%) had an RS < 11, and 27 of 232 (12%) had an RS > 25.
Two-step discordances were calculated for each model rule and each data set (Table 2). The definition of a 2-step discordance is the number of cases that were not correctly predicted by 2 steps. For example, the 2-step discordance of those predicted to be HG/LER would count only those patients that were in the low-risk (RS < 11) group but not the 1-step (RS 11–25) group. For each model rule and data set, the 2-step discordance was < 3%. For the model’s first rule of HG/LER, the 2-step discordances for the AAMC developmental set, AAMC superset, JHH MAIN, and JHH superset were 0%, 2.1%, 2.6%, and 2.0%, respectively. For the model’s second rule of LG+PR, the 2-step discordances were 0%, 0%, 0%, and 2.6%, respectively.
Table 2.
The Model’s 2-Step Discordances for the Different Patient Populatons
| Two-Step Discordance | Rule 1: HG/LER Group | Rule 2: LG+PR Group |
|---|---|---|
| AAMC Development Set | 0% | 0% |
| AAMC Superset Set | 2.1% | 0% |
| JHH MAIN Set | 2.6% | 0% |
| JHH Superset Set | 2.0% | 2.6% |
Abbreviations: AAMC = Anne Arundel Medical Center; HG/LER = high grade or low estrogen receptor; JHH = Johns Hopkins Hospital; LG+PR = low grade and progesterone receptor-positive.
Discussion
The model presented herein reliably classified many patients into a group for which the ODX test would provide little actionable information. ODX testing of HG/LER tumors consistently resulted in an RS that has a proven benefit of adding chemotherapy (RS ≥ 26) or a value at which chemotherapy benefit is indeterminate (RS 11–25). The same finding can be found in LG+PR tumors; in this population, tumors consistently resulted in an RS that has no proven benefit of adding chemotherapy (RS < 11) or in the intermediate range (RS 11–25). For high and low grade tumors, an intermediate RS score is unlikely to provide additional clinical information regarding the potential benefit of adjuvant chemotherapy because grade itself is prognostic, independent of RS, demonstrated by equal hazard ratios of RS and grade for distant recurrences in the 2004 multivariate Cox proportional-hazards model described by Paik et al.1 An interesting and perhaps underappreciated finding of Paik et al is that when the variables considered are: age at time of surgery, tumor grade, HER2 status, and ER-receptor protein level in a proportional-hazard model only tumor grade 3 versus grades 1 and 2, and RS were significant as predictors of distant recurrence (see Table 2 in Paik et al, page 2822).1 Both of these significant variables had similar P values (P < .001), and the hazard ratio of grade 3 versus grades 1 and 2 was 5.14 compared with a hazard ratio of 2.81 relative to an increment of 50 units in RS.
Results of other studies in which the usefulness of ODX testing over routine histopathologic analysis was investigated4–15 are consistent with our findings. The pioneering report by pathologists Dabbs, Bhargava, and colleagues12 at the Magee-Women’s Hospital in Pittsburgh showed the power of a linear regression model to make predictions of ODX’s RS using nuclear grade, mitotic count, ER and PR, and HER2 status. Their pilot study was based on a sample size of 42 patients. In 2013, they expanded this presenting work with 3 linear regression equations8 derived using samples from approximately 250 patients. The most appealing of the “Magee equations” uses Nottingham score, tumor size, ER and PR, and HER2 status and is available on the Web to compute the estimated RS (“Magee equation 2”16). The estimated RS determined using any of the “Magee equations” can be used to rule out patients for whom the ODX test will provide little actionable information because it is unlikely to provide information to change the recommendation for chemotherapy. The 2-step discordance in using any of the “Magee equations” to classify a patient’s risk group compared with the ODX test classification is approximately the same as the rule in this report. A possible drawback of the “Magee equations” is their use of “H-scores” to quantify the ER and PR immunohistochemical (IHC) test results, (not the more commonly used percentage of stained cells of an IHC test), however, at least 1 study has shown that replacing the “H-score” with the percentage of stained cells multiplied by intensity does not significantly compromise the equations’ performance.17 Others have developed linear regression models to predict ODX’s RS using other methods for scoring the ER and PR test or additional IHC markers. Geradts et al use the Allred scoring system in their model.13 Tang et al presented a linear regression equation using the IHC markers cytokeratin (CK) 5/6 and epidermal growth factor receptor (EGFR) in addition to the commonly used IHC markers.14 There is no significant difference in performance among these regression models in their ability to classify a patient into 1 of the ODX risk groups. The advantage of ours is its simplicity.
Others have developed decision tree models similar in structure to our model as shown in Figure 2. Using a random Forest approach, Ingoldsby et al described a model to classify breast cancers into 1 of the 3 ODX risk groups with the biomarker for Survivin being a key variable.7 Auerbach et al presented a decision model with a rule that patients who are PR-negative and have a mitotic count > 1 should be in the group that might benefit from an ODX test.11 Using a classification tree analysis on a test set of 104 cases and a validation set of 69 cases, Allison et al reached a conclusion (page 413)5 similar to ours in that “a subset of cancers with a high likelihood of having a low RS (0–18) was identified with the following characteristics: grade 1, strong PR expression (Allred ≥ 5) and (Ki-67 ≤ 10%). No cases with these characteristics had a high RS (≥ 31) and 73% had a low RS. Cancers highly likely to have a high RS were grade 3, with low to absent PR expression (Allred < 5) and Ki-67 > 10%. Eighty percent of cases with these characteristics had a high RS and no cases had a low RS.”
Figure 2.
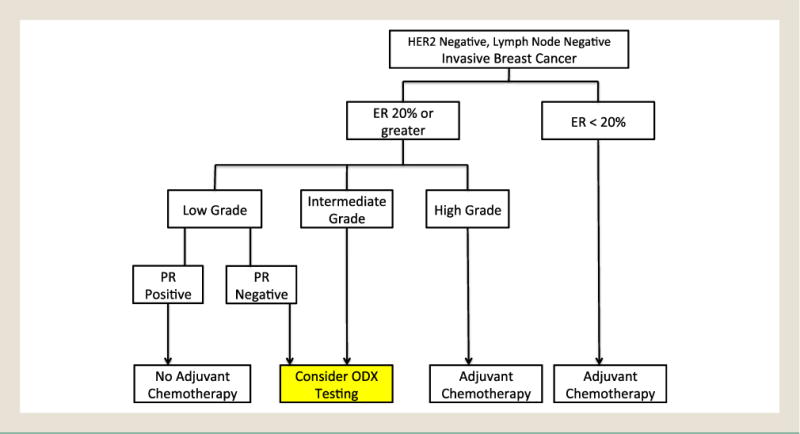
The Model’s Guidelines for Determining When Oncotype DX (ODX) Testing Might Provide Useful Information in Making the Decision for Adjuvant Chemotherapy
Abbreviations: ER = Estrogen Receptor; PR = Progesterone Receptor.
Our analysis and those of others show the importance of IHC results of the PR in evaluating breast cancer prognosis and using PR percentage information in aiding the chemotherapy decision. Prat et al proposed that the IHC-based definition of a luminal A breast cancer be one that is hormone-positive/HER2-negative with Ki-67 < 14% and PR > 20%.18
Conclusion
The potential effect of the implementation of our model would be a significant savings in the cost of breast cancer care. Since its introduction in 2004 to December of 2007, ODX has been ordered on > 175,000 patients by > 7500 physicians19 at the cost of approximately $4000 per test20 Ordering ODX as recommended by Genomic Health (“newly diagnosed patients with node-negative or node-positive, ER-positive, HER2-negative invasive breast cancer”19) and/or using the National Comprehensive Cancer Network’s guidelines (primary tumors characterized as 0.6–1.0 cm with unfavorable features or > 1 cm, and node-negative, hormone receptor-positive, and HER2-negative21) might lead to significant overordering. To investigate this further, we investigated the AAMC Breast Center’s population of invasive breast cancer patients who were evaluated by our pathologists between 2011 and 2013. We found that more than half (55%) of the patients who met the criteria for ODX testing, which we defined as ER-positive, lymph node-negative, and HER2-negative, were identified using our model as being in 1 of the groups for which the ODX test is likely to provide no additional actionable information. Use of our model would have potentially led to a $222,000 cost avoidance in ODX testing per 100 invasive breast cancer patients. The fact that only 42% (139 out of 329) of the patients in the AAMC cohort presented in this study, and 27% (87 out of 319) of the patients in the JHH cohort were classified as either HG/LER or LG+PR, is evidence that many physicians who order the ODX test are influenced by considerations similar to those that gave rise to our model.
Based on the potential effect of this model, other institutions are encouraged to evaluate their own ODX patient data, including ER, PR, tumor grade, and subsequent RS outcome. Physicians who fear the subjective variability in evaluation of grade might find that a cost-effective solution is repeat testing by a central laboratory of grade and IHC markers of the tumor specimen before ordering an ODX test.
Although there has been acceptance of these newer genomic analyses and an assumption that they must be superior to standard pathology, there have not been any published long-term, prospective studies regarding their superiority over routine histopathologic testing. Until further data are available, clinicians should choose wisely in ordering these tests, and consider use of the model described herein to select out patients for whom the ODX test is unlikely to provide additional information.
Clinical Practice Points.
The 21-gene RS assay provided by the ODX test has been shown to be of prognostic significance and predictive for the benefit of chemotherapy in patients with ER+ early breast cancer. Use of the ODX test has been included in guidelines of major medical societies.
The model presented herein reliably identified several groups of patients for whom the expensive ODX test will provide little actionable information; these are patients with high grade or low ER tumors, and patients with low grade and positive PR tumors (Figure 2).
We estimate that these new guidelines will lead to > $200,000 cost avoidance in ODX testing per 100 invasive early breast cancer patients until the information provided by the TAILORx trial becomes available.
Acknowledgments
The authors thank Dr Carol Tweed and Dr Thomas Sanders for their reviews of this report. We also appreciate the AAMC Oncology and Hematology Group for their aid identifying patients who received the ODX test. Additionally we thank an anonymous reviewer for bringing to our attention the work on the importance of the IHC results for PR in characterizing a breast cancer patient’s prognosis.
Footnotes
Disclosure
The authors have stated that they have no conflicts of interest.
References
- 1.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 2.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 3.Sparano JA. TAILORx: Trial Assigning Individualized Options for Treatment (Rx) Clin Breast Cancer. 2006;7:347–50. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- 4.Milburn M, Rosman M, Mylander C, Tafra L. Is oncotype DX recurrence score (RS) of prognostic value once HER2-positive and low-ER expression patients are removed? Breast J. 2013;19:357–64. doi: 10.1111/tbj.12126. [DOI] [PubMed] [Google Scholar]
- 5.Allison KH, Kandalaft PL, Sitlani CM, Dintzis SM, Gown AM. Routine pathologic parameters can predict Oncotype DX recurrence scores in subsets of ER positive patients: who does not always need testing? Breast Cancer Res Treat. 2012;131:413–24. doi: 10.1007/s10549-011-1416-3. [DOI] [PubMed] [Google Scholar]
- 6.Mattes MD, Mann JM, Ashamalla H, Tejwani A. Routine histopathologic characteristics can predict oncotype DX(TM) recurrence score in subsets of breast cancer patients. Cancer Invest. 2013;31:604–6. doi: 10.3109/07357907.2013.849725. [DOI] [PubMed] [Google Scholar]
- 7.Ingoldsby H, Webber M, Wall D, Scarrott C, Newell J, Callagy G. Prediction of Oncotype DX and TAILORx risk categories using histopathological and immunohistochemical markers by classification and regression tree (CART) analysis. Breast. 2013;22:879–86. doi: 10.1016/j.breast.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Klein ME, Dabbs DJ, Shuai Y, et al. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol. 2013;26:658–64. doi: 10.1038/modpathol.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang G, Cuzick J, Costantino JP, et al. Risk of recurrence and chemotherapy benefit for patients with node-negative, estrogen receptor-positive breast cancer: recurrence score alone and integrated with pathologic and clinical factors. J Clin Oncol. 2011;29:4365–72. doi: 10.1200/JCO.2011.35.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamal AH, Loprinzi CL, Reynolds C, et al. Breast medical oncologists’ use of standard prognostic factors to predict a 21-gene recurrence score. Oncologist. 2011;16:1359–66. doi: 10.1634/theoncologist.2011-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auerbach J, Kim M, Fineberg S. Can features evaluated in the routine pathologic assessment of lymph node-negative estrogen receptor-positive stage I or II invasive breast cancer be used to predict the Oncotype DX recurrence score? Arch Pathol Lab Med. 2010;134:1697–701. doi: 10.5858/2009-0439-OAR.1. [DOI] [PubMed] [Google Scholar]
- 12.Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R. Histopathologic variables predict Oncotype DX recurrence score. Mod Pathol. 2008;21:1255–61. doi: 10.1038/modpathol.2008.54. [DOI] [PubMed] [Google Scholar]
- 13.Geradts J, Bean SM, Bentley RC, Barry WT. The oncotype DX recurrence score is correlated with a composite index including routinely reported pathobiologic features. Cancer Invest. 2010;28:969–77. doi: 10.3109/07357907.2010.512600. [DOI] [PubMed] [Google Scholar]
- 14.Tang P, Wang J, Hicks DG, et al. A lower Allred score for progesterone receptor is strongly associated with a higher recurrence score of 21-gene assay in breast cancer. Cancer Invest. 2010;28:978–82. doi: 10.3109/07357907.2010.496754. [DOI] [PubMed] [Google Scholar]
- 15.Rosman M, Mylander WC, Tafra L. What is the value of the 21 gene recurrence score in HER2-negative patients? J Clin Oncol. 2010;28:e647. doi: 10.1200/JCO.2010.31.2280. –author reply e648. [DOI] [PubMed] [Google Scholar]
- 16.University of Pittsburgh. Department of Pathology. Estimating Oncotype DX recurrence score. Available at: http://path.upmc.edu/onlineTools/MageeEquations.html Accessed March 26, 2015.
- 17.Turner BM, Skinner K, Huston K, et al. Validation of modified Magee equations for predicting the oncotype DS recurrence score: a cost-effective alternative for estimating the risk of distant recurrence in receptor-positive/node-negative breast cancer patients. University of Rochester Medical Center; Presented as a poster (P2-11-12) at the San Antonio Breast Cancer Symposium; San Antonio, Texas. December 10–14, 2013. [Google Scholar]
- 18.Prat A, Chon UM, Martin M, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2012;31:203–9. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genomic Health Oncotype DX Web Site. Available at: http://breast-cancer.oncotypedx.com/en-US/Professional-Invasive/Resources/FAQs.aspx Accessed May 29, 2015.
- 20.The current list price of the Oncotype DX invasive breast cancer test is $417. Available at: http://breast-cancer.oncotypedx.com/en-US/Managed-Care/Health-Economics/Financial-Impact.aspx Accessed January 6, 2015.
- 21.NCCN Clinical Practice Guidelines in Oncology (version 1.2014) 2012:1–180. Available at: http://www.nccn.org Accessed April 4, 2014.


