Abstract
Chromosome 12 (Chr12) abnormalities have been described for individual patients with Philadelphia chromosome-negative myeloproliferative neoplasms (Ph-neg MPN), however the frequency, characteristics, and outcomes of such patients as a whole have not been investigated. We reviewed a database of 1787 consecutive Ph-neg MPN patients seen at our institution and determined that 2% of Ph-neg MPN patients harbored an alteration involving Chr12 by cytogenetic evaluation. Retrospective chart review revealed that patients with Chr12 abnormalities had a higher likelihood of having myelofibrosis (MF) compared to patients without a Chr12 abnormality, and were more likely to have post-polycythemia vera MF. The most common alterations in Chr12 in MF patients involved 12q13, 12q15, 12q24, and trisomy 12, and >40% of Chr12 Ph-neg MPN patients had cytogenetic evolution. Chr12 abnormalities did not significantly correlate with JAK2 status, progression to acute myeloid leukemia, or survival, however patients with 12q24 abnormalities trended towards poorer outcomes.
Keywords: Myeloproliferative neoplasms, Myelofibrosis, Polycythemia vera, Chromosome aberrations
1. Introduction
Myeloproliferative neoplasms (MPNs) result from the clonal proliferation of one or more hematopoietic cell lineages and are characterized by bone marrow fibrosis and extramedullary hematopoeisis. Philadelphia chromosome-negative (Ph-neg) MPN is a subset of MPN patients who do not have a translocation (9;22) and do not have chronic myeloid leukemia. Ph-neg MPN includes polycythemia vera (PV), essential thrombocytosis (ET), myelofibrosis (MF), mastocytosis (MST), and hypereosinophilic syndrome (HES) among others [1]. Approximately 3 per 100,000 people are diagnosed with some form of Ph-neg MPN annually [2]. Frequently Phneg MPN is a progressive illness, and some patients will transform into acute leukemia [1]. Chromosome 12 (Chr12) abnormalities have been previously reported in cases of Ph-neg MPN [3-7]. Specific structural abnormalities at 12q15 [6] and 12q24 [8] have been reported for individual patients with primary MF. Disruptions that affect the genes HMGA2, an architectural transcription factor, and SH2B3 (LNK), a multifunctional adapter protein, located at the 12q15 and 12q24 loci respectively, have been identified in a handful of cases of Ph-negative MPN [3, 4, 9, 10]. How structural abnormalities of Chr12 contribute to MPN and other cancers is not entirely understood [11-14]. The scope and effects of cytogenetic Chr12 abnormalities in Ph-neg MPN has not been fully characterized. We sought to identify such patients and test whether they had common clinical characteristics, higher rates of transformation to acute myeloid leukemia (AML), and differences in prognosis compared to other Ph-neg MPN patients without chromosome 12 abnormalities (Non-Chr12).
2. Methods
We retrospectively identified all Ph-neg MPN patients (confirmed by bone marrow histology that included karyotype) seen at MD Anderson Cancer Center between 1985 and 2012. Patients were classified and subclassified according to the World Health Organization classification of hematologic malignancies [15]. For karyotypic analysis, unstimulated marrow cells were cultured for 24 to 72 hours, followed by G-banding procedure according to standard techniques in the MD Anderson clinical cytogenetics laboratory. Read out of cytogenetic data was performed by a cytogeneticist, and where possible at least 20 methaphases were examined for each patient. Abnormal clones were defined using current and previous era International System for Human Cytogenetic Nomenclature [16]. We identified patients with aberrations involving Chr12 at the time of presentation to our institution. Abnormalities included translocation breakpoint or disruption, partial or whole deletion, or hyperploidy. We further reviewed available medical records, clinical features, and patient outcomes. When available, mutational analysis for JAK2 V617F was carried out by extracting genomic DNA from bone marrow samples, and testing according to institutional standards and as previously described [17]. For all patients in the Ph-neg MPN database, we examined type of MPN, JAK2 status, progression to AML, and overall survival (OS), which was defined as time from diagnosis to death or last follow-up. For each patient with a Chr12 abnormality, we identified demographic data, specific Chr12 abnormality, therapies, and whether they had karyotypic evolution while at our institution, defined as two successive bone marrow biopsies with differing cytogenetic abnormalities. A comparison of binomial proportions was used to detect differences in characteristics between groups. Survival estimates for Chr12 and Non-Chr12 MF patients were generated using Kaplan–Meier curves, and differences assessed using a logrank test. For all analyses, p-value < 0.05 was considered to be statistically significant.
3. Results
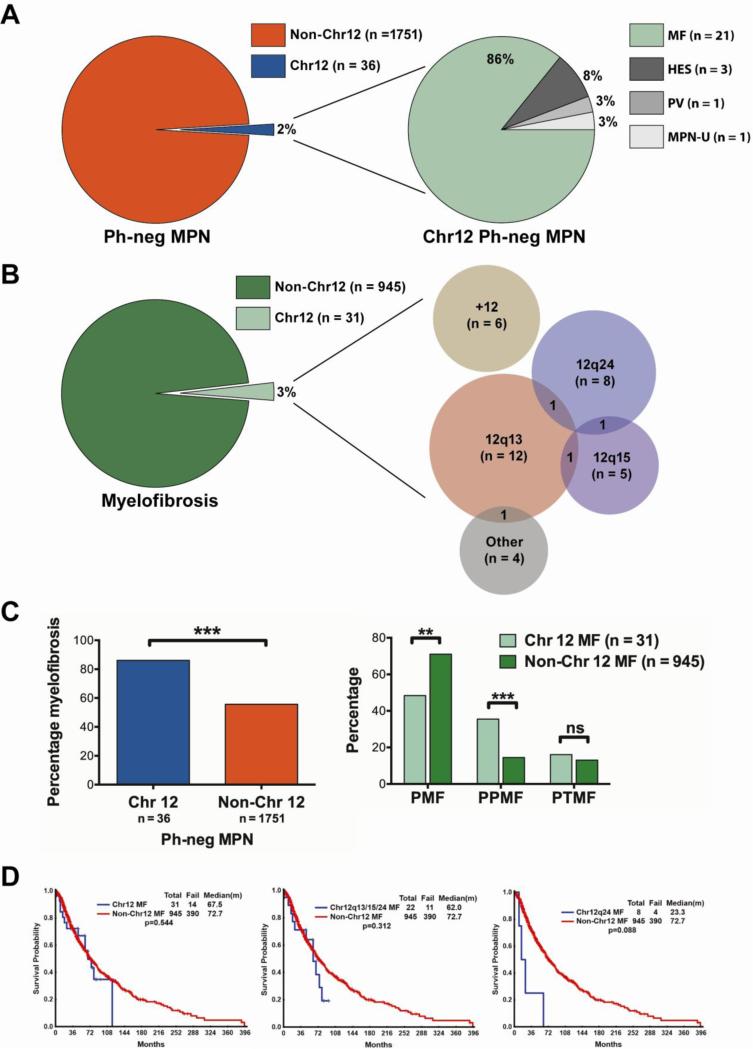
The Ph-neg MPN patient database included 1787 patients. A total of 36 patients (2%, 21 male and 15 female) had at least one cytogenetic abnormality in Chr12 (Figure 1A). Among all Non-Chr12 patients (n = 1751), 945 (54%) were diagnosed with MF, 295 (17%) with ET, 229 (13%) with PV, 126 (7%) with MST, 81 (5%) with HES, and 75 (4%) with unspecified MPN. By comparison, in the Chr12 group (n = 36), 31 (86%) were diagnosed with MF, 1 (3%) with PV, 3 (8%) with HES, and 1 (3%) with unspecified MPN (Figure 1A). Median age was 61 years (range, 36-83 years) for the Chr12 patients versus 59 years (range, 14-89 years) for the Non-Chr12 patients. A complete list of all Chr12 Ph-neg MPN patients, their clinical characteristics, and the locations of their Chr12 abnormalities is provided in Table 1.
Figure 1.
Chr12 abnormalities in Philadelphia-negative myeloproliferative neoplasms and myelofibrosis. (A) Frequency of Chr12 abnormality in Ph-neg MPN and (expanded) type of MPN among Chr12 patients. (B) Frequency of Chr12 abnormality in MF and (expanded) Venn diagram of specific type of Chr12 abnormality in MF. Area of circle represents the relative frequency of abnormality. (C) Comparison of proportions of Chr12 and Non-Chr12 Ph-neg MPN patients who had MF, and MF patients who had PMF, PPMF, and PTMF. **p-value < 0.01; ***p-value < 0.001. (D) Comparison between survival of patients with and without Chr12 abnormalities, between combined patients with 12q13, 12q15 and 12q24 abnormalities and Non-Chr12 patients, and between 12q24 patients only versus Non-Chr12 patients. Ph-neg MPN, Philadelphia-chromosome negative myeloproliferative neoplasm; MF, myelofibrosis; HES, hypereosinophilic syndrome; PV, polycythemia vera; MPN-U, unspecified MPN; PMF, primary myelofibrosis; PPMF, post-polycythemia vera myelofibrosis; PTMF, post-thrombocytosis myelofibrosis.
Table 1.
Ph-neg MPN patients with Chr12 abnormalities.
| Patient N | Age | Sex | Diagnosis | PV/ET | Chr12 loci involved | JAK2 | No. prior txs | CG Evolution |
|---|---|---|---|---|---|---|---|---|
| 1 | 58 | M | MF | none | 12q24, 12q15 | Unk | 5 | Y |
| 2 | 83 | M | MF | PV | 12q24 | + | 1 | N |
| 3 | 66 | M | MPN-U | none | 12q24 | + | 4 | N |
| 4 | 49 | F | MF | none | 12p13, 12q10 | Unk | 2 | N |
| 5 | 75 | M | MF | none | 12q13 | Unk | 3 | Y |
| 6 | 52 | F | MF | none | 12q24 | Unk | 3 | N |
| 7 | 70 | F | MF | none | 12q24 | Unk | 2 | Y |
| 8 | 58 | M | PV | PV | 12q21 | + | 3 | N |
| 9 | 62 | M | HES | none | 12q21 | Unk | 2 | Y |
| 10 | 54 | F | MF | none | 12q15 | Neg | 4 | Y |
| 11 | 66 | M | HES | none | 12q10 | Unk | 5 | N |
| 12 | 53 | F | MF | none | 12q13 | Unk | 0 | N |
| 13 | 71 | M | MF | PV | 12q13, 12q15 | Unk | 4 | N |
| 14 | 57 | F | MF | ET | 12q13, −12, 12p12, 12q21, 12q23 | + | 6 | Y |
| 15 | 59 | M | MF | none | 12q13 | + | 1 | Y |
| 16 | 76 | M | MF | PV | −12 | + | 1 | N |
| 17 | 61 | F | MF | PV | 12q15 | + | 4 | Y |
| 18 | 21 | M | MF | PV | 12q13 | + | 2 | Y |
| 19 | 57 | M | MF | PV | 12q24 | + | 1 | N |
| 20 | 40 | M | HES/CEL | none | 12q24 | Neg | 3 | N |
| 21 | 50 | F | MF | none | +12 | + | 1 | Y |
| 22 | 61 | F | MF | PV | 12q13 | + | 3 | N |
| 23 | 61 | M | MF | ET | 12q15 | + | 2 | N |
| 24 | 35 | F | MF | PV | +12 | + | 1 | N |
| 25 | 58 | F | MF | ET | +12 | + | 1 | N |
| 26 | 56 | F | MF | none | +12 | Neg | 1 | Y |
| 27 | 52 | M | MF | PV | +12 | + | 4 | Y |
| 28 | 32 | M | MF | none | +12 | + | 2 | Y |
| 29 | 24 | M | MF | ET | 12q12 | Neg | 2 | Y |
| 30 | 60 | F | MF | none | 12q13 | + | 1 | N |
| 31 | 46 | M | MF | PV | 12q13, 12q24 | + | 0 | N |
| 32 | 36 | F | MF | none | 12q13 | Neg | 4 | N |
| 33 | 47 | F | MF | PV | 12q13 | + | 2 | N |
| 34 | 63 | M | MF | none | 12q24 | Neg | 0 | N |
| 35 | 51 | M | MF | PV | 12q24 | + | 0 | N |
| 36 | 63 | M | MF | ET | 12q13 | Neg | 3 | N |
Patient N, Patient number; PV, Polycythemia vera; ET, Essential thrombocytosis; No. prior txs, Number of prior thearpies; CG, Cytogenetic; MPN-U, Unspecified myeloproliferative neoplasm; HES, Hypereosinophilic syndrome; CEL, Chronic eosinophilic leukemia.
Because a large number of Chr12 patients had MF, we focused on the MF group in greater detail. Of all MF patients in the Ph-neg MPN database (n = 976), 31 (3%) had an abnormality of Chr12 (Figure 1B). Cytogenetic Chr12 abnormalities occurred at 12q13 (most frequently), 12q15, and 12q24 among others. Six patients had trisomy 12, and 3 patients had a structural abnormality at 12q13 in addition to another Chr12 abnormality (Figure 1B). Other Chr12 aberrations (some occurring in the same patient) included monosomy 12 (n = 2), and structural breakpoints at 12p13 (n = 1), 12q10 (n = 2), 12q12 (n = 1), 12q21 (n = 1), and 12q23 (n = 1).
We investigated whether the existence of a Chr12 abnormality correlated with a diagnosis of MF, and found that a significantly higher proportion of Chr12 Ph-neg MPN patients were diagnosed with MF compared to Non-Chr12 patients (Figure 1C, Table 2, p-value < 0.001). In addition, a significantly higher proportion of patients with Chr12 abnormality also had post-PV MF (PPMF, Figure 1C, Table 2, p-value < 0.001). Among Chr12 MF patients, 11/31 (35%) had PPMF, whereas among Non-Chr12 MF patients 137/945 (14%) had PPMF. There was no significant difference in the proportion of patients who had post-ET MF (PTMF), although a higher fraction (16% versus 13%) had PTMF in the Chr12 group (p-value = 0.11, Figure 1C, Table 2). In contrast, a significantly higher fraction of patients in the Non-Chr12 group had primary MF (PMF, p-value < 0.01, Figure 1C, Table 2). Interestingly, 5/12 (42%) patients who had an abnormality at 12q13 had PPMF, which was a significantly higher fraction than in the Non-Chr12 group (p-value = 0.01).
Table 2.
Incidence of primary myelofibrosis, post-PV MF, post-ET MF, JAK2 status, and transformation to leukemia for Chr12 and Non-Chr12 patients.
| Characteristic | Chr. 12 (n=31) | Non-Chr. 12 (n=945) | p-value |
|---|---|---|---|
| PMF, N (%) | 15 (48) | 685 (72) | <0.01 |
| PPMF, N (%) | 11 (35) | 137 (14) | <0.001 |
| PTMF, N (%) | 5 (16) | 123 (13) | 0.11 |
| JAK2-positive, N (%) [median allele burden % |range|] | 18 (58) [69.7 (10.5-95.9)] | 471 (50) [57.3 (1-99.1)] | 0.55 |
| JAK2-negative, N (%) | 7 (23) | 239 (25) | |
| JAK2-unknown, N (%) | 6 (19) | 235 (25) | |
| Transformation to leukemia, N (%) | 1 (3) | 76 (8) | 0.660 |
| Months to transformation to leukemia (range) | 70.2 | 33.6 (1.1-315) |
PMF, Primary myelofibrosis; PPMF, Post-polycythemia vera myelofibrosis; PTMF, Post-thrombocytosis myelofibrosis
Age, JAK2 status and incidence of leukemic transformation from MF were not significantly different between Chr12 and Non-Chr12 groups (Table 2). The two groups had 80% and 75% of patients tested for JAK2 mutation, respectively; among patients who were tested, 72% of Chr12 MF patients had the V617F mutation and 66% of Non-Chr12 patients had the mutation (difference p-value = 0.55, Table 2). We additionally found that Chr12 status did not correlate with progression from MF to AML, and only 1 of the 31 Chr12 MF patients transformed into AML (Table 2). Further, we looked at how frequently Chr12 patients had clonal evolution. Fourteen out of 36 (39%) Chr12 MPN patients, and 13 of 31 (42%) Chr12 MF patients had cytogenetic evolution. There was no correlation between clonal evolution and disease, loci, JAK2 status, or number of prior therapies in Chr12 patients.
Finally, we performed a survival analysis of MF patients with versus without Chr12 abnormalities (Figure 1D). There was no statistical difference in the survival probability between the two groups (p-value = 0.54). If Chr12 patients with an abnormality at 12q13, 12q15, or 12q24 were grouped, the difference in survival was also not significant (p-value = 0.31). We investigated each Chr12 abnormality individually, however there were no significant differences between the groups identified and Non-Chr12 patients. The 8 patients with an abnormality at 12q24 trended towards worse prognosis (p-value = 0.09).
4. Discussion
In total, approximately 2% of patients with Ph-neg MPN, and 3% with MF harbor a Chr12 abnormality. Our results suggest that Ph-neg MPN patients with Chr12 abnormalities are more likely to have MF than Non-Chr12 patients. Additionally, Chr12 patients have an increased likelihood of MF that has progressed from PV. We did not detect a higher proportion of patients with PV who had Chr12 abnormality, however many patients at M. D. Anderson Cancer Center are evaluated as tertiary referrals. Taken together, these results suggest that a patient with PV who has a Chr12 abnormality is more likely to progress to MF. An alternative explanation is that patients with PPMF have a propensity to develop a chromosomal 12 cytogenetic abnormality, as many patients had cytogenetics only after development of PPMF. This data also suggests that Chr12 MF patients are not more likely to progress to AML than those without an abnormality of Chr12. Over 40% of Chr12 MF patients have cytogenetic evolution, demonstrating that some of these patients had chromosomal structural instability. Interestingly, clonal and mutational complexity has been reported previously in a patient with LNK mutations at Chr12q24 [18]. Instability of chromosomal structure is a known factor in myeloid leukemogenesis [19]. Whether Chr12 patients in particular have a predilection for chromosomal instability cannot be determined by the current study. Previous reports have indicated that patients who possess LNK mutations have a loss of negative feedback on JAK-STAT signaling that can result in MPN, JAK2-negative erythrocytosis, and post-PMF AML [10, 18, 20, 21].
There were no statistically significant differences in survival between Chr12 MF patients (as a whole, and based on site/type of abnormality) and Non-Chr12 MF patients, however a Chr12q24 aberration was associated with a non-significant trend towards a worse prognosis. Limitations of the current study include the small number of patients with Chr12 abnormality and lack of complete mutational genotyping. This analysis does demonstrate that cytogenetic information and specific chromosomal abnormalities continue to offer important information about an individual patient's Ph-neg MPN.
Acknowledgements
We are grateful to University of Texas M. D. Anderson Cancer Center for support, technical assistance, and data analysis. This research is supported in part by the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship
C.B. designed research, performed research, analyzed data, wrote the manuscript. M.T. performed research, analyzed data. C.W. analyzed data, assisted with the manuscript. S.P and L.Z. contributed essential tools, performed research, analyzed data. J.C. and H.K. contributed essential tools, analyzed data. S.V. contributed essential tools, designed research, analyzed data.
Conflict-of-interest
The authors declare no competing financial interests.
References
- 1.Campbell PJ, Green AR. The myeloproliferative disorders. New England Journal of Medicine. 2006;355:2452–66. doi: 10.1056/NEJMra063728. [DOI] [PubMed] [Google Scholar]
- 2.Johansson P, Kutti J, Andréasson B, Safai-Kutti S, Vilén L, Wedel H, et al. Trends in the incidence of chronic Philadelphia chromosome negative (Ph-) myeloproliferative disorders in the city of Göteborg, Sweden, during 1983-99. Journal of internal medicine. 2004;256:161–5. doi: 10.1111/j.1365-2796.2004.01357.x. [DOI] [PubMed] [Google Scholar]
- 3.Etienne A, Carbuccia N, Adélaïde J, Bekhouche I, Rémy V, Sohn C, et al. Rearrangements involving 12q in myeloproliferative disorders: possible role of HMGA2 and SOCS2 genes. Cancer genetics and cytogenetics. 2007;176:80–8. doi: 10.1016/j.cancergencyto.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Aliano S, Cirmena G, Garuti A, Fugazza G, Bruzzone R, Rocco I, et al. HMGA2 overexpression in polycythemia vera with t(12;21)(q14;q22). Cancer genetics and cytogenetics. 2007;177:115–9. doi: 10.1016/j.cancergencyto.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Storlazzi CT, Albano F, Locunsolo C, Lonoce A, Funes S, Guastadisegni MC, et al. t(3;12)(q26;q14) in polycythemia vera is associated with upregulation of the HMGA2 gene. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006;20:2190–2. doi: 10.1038/sj.leu.2404418. [DOI] [PubMed] [Google Scholar]
- 6.Andrieux J, Demory J-L, Morel P, Plantier I, Dupriez B, Caulier MT, et al. Frequency of structural abnormalities of the long arm of chromosome 12 in myelofibrosis with myeloid metaplasia. Cancer genetics and cytogenetics. 2002;137:68–71. doi: 10.1016/s0165-4608(02)00554-x. [DOI] [PubMed] [Google Scholar]
- 7.Tefferi A, Mesa RA, Schroeder G, Hanson CA, Li CY, Dewald GW. Cytogenetic findings and their clinical relevance in myelofibrosis with myeloid metaplasia. British journal of haematology. 2001;113:763–71. doi: 10.1046/j.1365-2141.2001.02796.x. [DOI] [PubMed] [Google Scholar]
- 8.Ohyashiki K, Tauchi T, Kuroda M, Kodama A, Ohyashiki JH. Recurrent chromosomal aberration at 12q15 in chronic idiopathic myelofibrosis with or without JAK2(V617F) mutation. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007;21:1578–80. doi: 10.1038/sj.leu.2404700. [DOI] [PubMed] [Google Scholar]
- 9.Martin SE, Sausen M, Joseph A, Kingham BF, Martin ES. Identification of a HMGA2-EFCAB6 gene rearrangement following next-generation sequencing in a patient with a t(12;22)(q14.3;q13.2) and JAK2V617F-positive myeloproliferative neoplasm. Cancer genetics. 2012;205:295–303. doi: 10.1016/j.cancergen.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–92. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nature Reviews Cancer. 2007 doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 12.Devallière J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochemical pharmacology. 2011;82:1391–402. doi: 10.1016/j.bcp.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda K, Mason PJ, Bessler M. 3'UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood. 2011;117:5860–9. doi: 10.1182/blood-2011-02-334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bersenev A, Wu C, Balcerek J, Jing J, Kundu M, Blobel GA, et al. Lnk constrains myeloproliferative diseases in mice. Journal of Clinical Investigation. 2010;120:2058–69. doi: 10.1172/JCI42032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. IARC Press; Lyon, France: 2008. p. 439. [Google Scholar]
- 16.Shaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013) Karger; Basel: 2013. [Google Scholar]
- 17.McClure R, Mai M, Lasho T. Validation of two clinically useful assays for evaluation of JAK2 V617F mutation in chronic myeloproliferative disorders. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006;20:168–71. doi: 10.1038/sj.leu.2404007. [DOI] [PubMed] [Google Scholar]
- 18.Lasho TL, Tefferi A, Finke C, Pardanani A. Clonal hierarchy and allelic mutation segregation in a myelofibrosis patient with two distinct LNK mutations. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011;25:1056–8. doi: 10.1038/leu.2011.45. [DOI] [PubMed] [Google Scholar]
- 19.Vajen B, Thomay K, Schlegelberger B. Induction of Chromosomal Instability via Telomere Dysfunction and Epigenetic Alterations in Myeloid Neoplasia. Cancers. 2013;5:857–74. doi: 10.3390/cancers5030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardanani A, Lasho T, Finke C, Oh ST, Gotlib J, Tefferi A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:1713–8. doi: 10.1038/leu.2010.163. [DOI] [PubMed] [Google Scholar]
- 21.Oh ST. When the Brakes are Lost: LNK Dysfunction in Mice, Men, and Myeloproliferative Neoplasms. Therapeutic advances in hematology. 2011;2:11–9. doi: 10.1177/2040620710393391. [DOI] [PMC free article] [PubMed] [Google Scholar]