Summary
The agr locus encodes a quorum sensing (QS) circuit required for the virulence of a spectrum of gram-positive pathogens and is, therefore, regarded as an important target for the development of chemotherapeutics. In recent years, many of the biochemical events in the Staphylococcus aureus agr circuit have been reconstituted and subject to quantitative analysis in vitro. This work, in conjunction with structural studies on several key players in the signaling circuit, has furnished mechanistic insights into the regulation and evolution of the agr quorum sensing system. Herein, we review this progress and discuss the remaining open questions in the area. We also highlight advances in the discovery of small-molecule agr modulators and how the newly available biochemical and structural information might be leveraged for the design of next generation therapeutics targeting the agr system.
Introduction
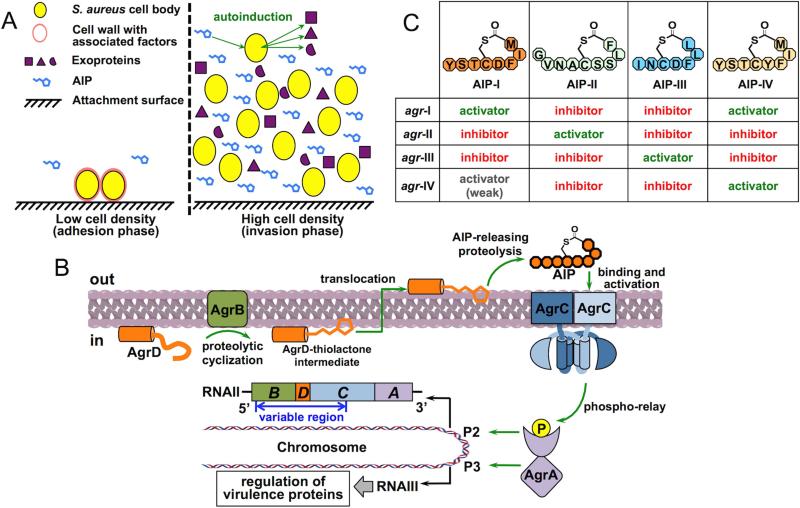
Staphylococcus aureus (S. aureus) is a commensal symbiont and an opportunistic pathogen. Once S. aureus invades host tissues, it causes both acute and chronic illnesses such as bacteremia, sepsis, endocarditis and toxic shock syndrome (Lowy, 1998). To establish and sustain its infection, this bacterium deploys a diverse arsenal of virulence factors, depending on its growth phase. During the lag and early exponential phases, S. aureus produces cell wall-associated factors that facilitate tissue attachment and evasion of the host immune system, allowing the bacteria to accumulate and form a biofilm (Kong et al., 2006) (Figure 1A, left panel). Once the bacterial population reaches the late exponential phase, it begins to secrete a spectrum of exoproteins, including proteases, hemolysins and super-antigens, and at the same time down-regulates the cell wall-associated factors, leading to dispersion of the biofilm and the spread of the infection (Dinges et al., 2000) (Figure 1A, right panel). This population density-dependent behavior essentially delineates two stages of the S. aureus life cycle, i.e., an adhesion phase and an invasion phase. The timing and expeditious transition between these two phases occurs through an intercellular communication process called quorum sensing (QS), in which the bacterium produces a diffusive molecule, termed the autoinducer (AI), as an indicator of the local population density. Detection of the AI is central to the decision making process that ultimately controls gene expression programs (Waters and Bassler, 2005).
Figure 1. Role of the agr QS circuit in virulence regulation in S. aureus.
(A) Two phases of the S. aureus life cycle featuring distinct patterns of virulence protein production. At high cell density, AIP accumulates in the extracellular environment and triggers the agr QS circuit leading to decreased production of cell wall-associated factors and a simultaneous increase in exoprotein production. (B) Schematic of the agr autoinduction circuit. (C) Structure and efficacy of AIPs from all four S. aureus subgroups.
The chromosomal locus responsible for QS in S. aureus is named agr (accessory gene regulator). The locus encodes a signaling circuit that both produces and senses the AI, a small peptide named the AIP (for autoinducing peptide), featuring a unique thiolactone linkage between the C-terminal carbonyl and the sulfur atom in a cysteine side-chain (Novick and Geisinger, 2008) (Figure 1B). Importantly, it has been known since the late 1990s that inhibition of agr-mediated signaling attenuates the spread of S. aureus infections in animal models, thus qualifying agr as a potential drug target (Mayville et al., 1999). These early discoveries have fueled interdisciplinary efforts to understand the underlying mechanisms of agr-mediated signaling. Accordingly, all of the protein components encoded by the agr locus have been extensively studied in whole cell-based systems, primarily employing mutagenesis and sequence swapping approaches. Recent years, however, have seen the emergence of in vitro reconstitution systems for studying most biochemical events in which these proteins participate. These studies have collectively addressed, or provided a promising starting point to address, many long-standing mechanistic questions regarding the regulation and evolution of the system. In this review, we integrate the results of both in-cell and in vitro studies to provide an up-to-date mechanistic description of the S. aureus QS circuit. We also discuss on-going efforts to identify agents that interfere with S. aureus QS as a potential route to treating infections and highlight opportunities in this area presented by recent biochemical breakthroughs.
Basic architecture of the agr autoinuction circuit
Analogous to other QS systems, production and sensing of the AIP in S. aureus are mutually enhancing, leading to a positive-feedback autoinduction circuit (Novick and Geisinger, 2008) (Figure 1B). In the agr locus, the P2 operon encodes a polycistronic messenger RNA (mRNA), termed RNAII, containing four open reading frames (ORFs), from which four Agr proteins involved in the autoinduction circuit are translated (Novick et al., 1995). AgrD, the precursor of the AIP, is first proteolytically processed by a membrane-bound peptidase, AgrB, to generate a thiolactone intermediate. This intermediate is then exported across the membrane and subject to a second cleavage to release the mature AIP pheromone into the extracellular space. The AIP is the activating signal of the sensing pathway, which is detected by a classic two-component signaling system (TCS) consisting of the membrane-bound receptor histidine kinase (RHK), AgrC, and the response regulator (RR), AgrA. The AIP interacts with AgrC to activate a phospho-relay cascade that leads to the phosphorylation AgrA. Upon phosphorylation, AgrA binds to the P2 promoter, up-regulates the transcription of RNAII and thereby the production of all four Agr proteins, conferring positive feedback to the AIP synthesis.
Regulation of virulence-factor production is accomplished through the AgrA-dependent P3 operon, located back-to-back with P2, that encodes RNAIII, the mRNA for δ-toxin (an exoprotein) and a pleiotropic regulatory factor (Novick et al., 1993) (Figure 1B). RNAIII primarily functions through base-pairing to the 5’-ends of virulence-factor mRNAs, suppressing the synthesis of proteins required for the adhesion phase of the life cycle, while de-repressing those involved in the invasion phase. A full discussion of the targets and mechanisms of action of RNAIII is beyond the scope of this review, and readers are referred to a recent account of this topic (Fechter et al., 2014). It is worth noting, however, that RNAIII-independent agr effectors have also been identified in recent years, with the most prominent example being phenol-soluble modulins (PSMs) that facilitate the bacterium's immune evasion (Queck et al., 2008).
Intraspecies variation of agr
One of the most intriguing features of the agr locus is its polymorphism within a single species. Within five years of the first agr locus being cloned in S. aureus, four allelic variants were reported (Jarraud et al., 2000; Ji et al., 1997). The variable region spans half the length of RNAII, covering coding region of the main body of AgrB, the entirety of AgrD and the sensor domain of AgrC (Figure 1B, marked with a blue, double-headed arrow). This setting allows each agr variant to specifically produce, and mediate autoinduction in response to, its own AIP. In S. aureus strains carrying different agr variants, the vast majority of conserved, structural genes (excluding mobile genetic elements) are predominantly identical, suggesting that the variation occurs at a sub-species level. Strains harboring each agr allele are therefore classified as a pherotype or a specificity subgroup.
While S. aureus strains from all four subgroups are capable of qualitatively similar autoinduction when cultured alone, the effect of AIPs on the induction of a heterologous agr system is, in most cases, strongly inhibitory (Ji et al., 1997; Lyon et al., 2002; Mayville et al., 1999) (Figure 1C). The only exception lies between the two most closely related groups, I and IV – AIPs from these groups have 7 identical residues out of 8 positions (Figure 1C). Clinical isolates of S. aureus from one infection site rarely exhibit variegation in the agr locus, primarily because an agr-heterologous cell population cannot achieve cooperative autoinduction to support the fitness of all participant subgroups (Traber et al., 2008).
A long-standing puzzle of agr polymorphism concerns the evolutionary advantage offered by individual agr alleles. A correlation has been observed between agr variants and infection types (Traber et al., 2008). For instance, group-III strains are overrepresented in menstrual toxic shock syndromes, while the exfoliatin-producing strains causing scalded skin syndrome are predominantly group-IV. In an insightful study performed by Geisinger et al., all four agr alleles were introduced, one at a time, into an agr-null background strain through chromosomal insertion at an identical site (Geisinger et al., 2012). Comparison of these alleles on an isogenic background revealed major differences in the temporal control of autoinduction: induction was achieved earliest with group-I and group-IV and latest with group-III alleles. This observation argues for a model in which each agr variant has a different schedule for autoinduction. Conceivably, such differences in induction timing might underlie, at least in part, correlations between subgroup type and the site of infection. This idea merits further investigation as does the detailed mechanism underlying the differential autoinduction timing and dynamics mediated by the different agr groups.
Biochemistry of agr autoinduction
Several steps in the agr autoinduction circuit have been investigated using well-defined reconstitution systems. These studies have provided a number of mechanistic insights, but have also pointed to the involvement of other, as yet uncharacterized, molecular players in the autoinduction circuit. In this section, we focus primarily on the biochemical mechanisms that this circuit harnesses to achieve timing, specific and restricted autoinduction.
Translation of Agr proteins
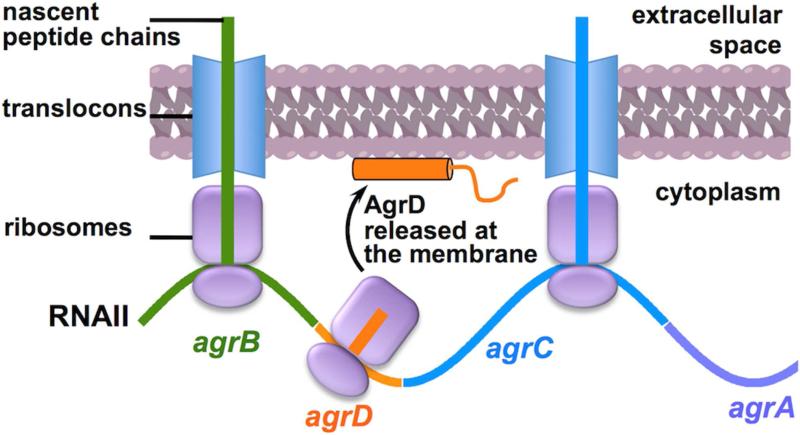
Although the coding sequence of all four Agr proteins are co-transcribed in the polycistronic RNAII, this transcript is exquisitely designed to finely coordinate their translation. For instance, the ribosomal binding sequences (RBSs) and initiating codons governing the translation of AgrD and AgrA are more favorable for translation efficiency than their counterparts in the AgrB and AgrC ORFs (Novick et al., 1995). This arrangement likely accounts for the desired stoichiometry between the AgrB-AgrD and AgrC-AgrA enzyme-substrate pairs. In addition, since the synthesis of AgrB and AgrC is coupled to co-translational insertion into the cell membrane, the intervening AgrD ORF presumably localizes in close proximity of the bilayer (Libby et al., 2012) (Figure 2). Because the 46-aa AgrD is highly hydrophobic and likely unstructured unless associated with the lipid bilayer (vide infra), it might be an important protective measure to synthesize AgrD close to the cell membrane to prevent its aggregation and degradation.
Figure 2. Membrane localization of the RNAII polysome.
Schematic shows the site of AgrD synthesis relative to the cell membrane as a consequence of the co-translational insertion of the flanking RNAII-encoded membrane proteins (AgrB and AgrC).
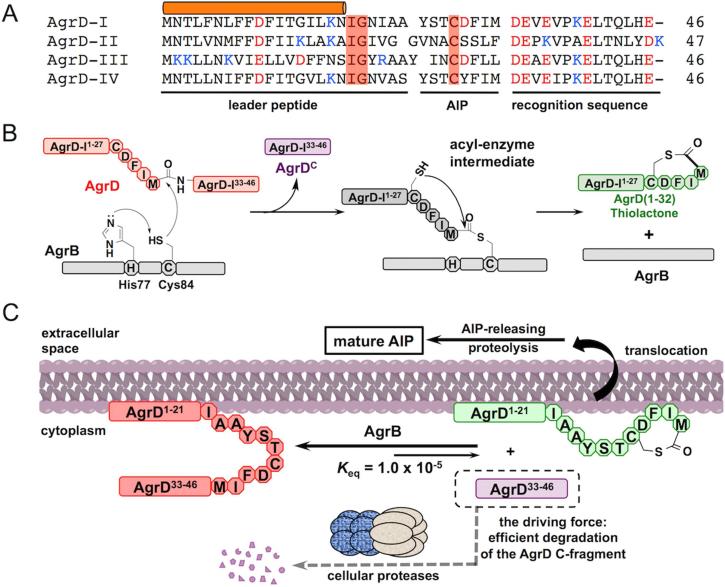
Formation of the AIP thiolactone
One of the most fascinating features of the agr system is the thiolactone structure within the AIP, closing the 16-atom macrocycle. The thiolactone is generated by a single proteolysis reaction involving AgrB and AgrD (Qiu et al., 2005; Thoendel and Horswill, 2009). The substrate, AgrD, contains the mature AIP sequence sandwiched between an N-terminal leader peptide and a C-terminal recognition sequence (Figures 1B and 3A). AgrB-catalyzed proteolysis clips off the recognition sequence as a linear peptide and concomitantly installs the thiolactone within the remaining N-fragment, herein referred to as the thiolactone intermediate. The recognition sequence is enriched in acidic residues and highly conserved in staphylococcal AgrDs. The significance of the sequence conservation is not yet fully understood (Thoendel and Horswill, 2009). The N-terminal 18 residues of the leader form an amphipathic helix that anchors AgrD to the cell membrane, putatively by lateral association (Zhang et al., 2004) (Figures 1B and 3A). This form of localization is required for AgrD processing, as the substitution of its N-terminal region with an artificial amphipathic helical sequence, but not a hydrophobic trans-membrane domain, is tolerated (Zhang et al., 2004). In all staphylococcal AgrDs, the amphipathic sequence is followed by an “IG” motif that putatively act as a helicity breaker to facilitate the proteolytic release of the mature AIP from the thiolactone intermediate (Kavanaugh et al., 2007).
Figure 3. Formation of the AIP thiolactone.
(A) Sequence alignment of the four AgrD variants from S. aureus: acidic and basic residues are highlighted in red and blue, respectively. Residues playing explicit roles in AgrD processing are highlighted with red boxes. The cylinder indicates the amphipathic α-helical sequence. (B and C) Schematics showing (B) the catalytic mechanism and (C) the thermodynamic driving force of the AgrB-mediated proteolytic cyclization of AgrD, exemplified by the formation of the group-I AgrD thiolactone intermediate.
AgrB, the peptidase responsible for proteolysis of AgrD, is a multi-pass membrane protein. Two residues invariable in all known AgrB homologs, one cysteine and the other histidine, have been identified as the catalytic diad (Qiu et al., 2005) (Figure 3B). An elegant peptidyl-transfer mechanism has been proposed for this reaction (Thoendel and Horswill, 2009), in which AgrB first attacks the scissile bond using its active-site cysteine to form an acyl-enzyme thioester intermediate with the concomitant release of the linear AgrD C-fragment (Figure 3B). The intermediate is then resolved by transferring the peptidyl group of the AgrD N-fragment to the side-chain thiol group of its internal cysteine residue (C28), which leads to the formation of the thiolactone macrocycle. This ‘proteolytic cyclization’ process is unusual from a thermodynamic perspective in that it results in the net conversion of a stable peptide bond into a high-energy thioester bond. Reconstitution of this reaction, employing highly purified AgrD peptides and liposome-incorporated AgrB, has confirmed that the thiolactone N-fragment is indeed a kinetically favorable product (Wang et al., 2015). Intriguingly, the proteolytic cyclization exhibited a dynamic equilibrium behavior in the in vitro system. The equilibrium constant determined therein revealed that, in order to maintain a sufficient intracellular pool of the thiolactone intermediate to support the rapid production of AIP, S. aureus cells have to efficiently degrade the C-terminal cleavage fragment of AgrD, limiting its half-life to the order of 10 seconds (Wang et al., 2015) (Figure 3C). In other words, the bacterium follows Le Châtelier's principle, harnessing the favorable free energy from the hydrolytic degradation of one cleavage fragment to power the installation of a high-energy thiolactone motif in the other. In line with this notion, over-production of the AgrD C-fragment in S. aureus causes a decrease of AIP production. Because of its thermodynamic contribution, degradation of the AgrD C-fragment should be added as an essential step to the AIP production pathway.
Translocation and maturation of the AIP
After being processed by AgrB, the thiolactone intermediate undergoes a second proteolysis event to release the freely diffusive AIP pheromone from the membrane-anchoring N-terminal leader peptide. An active or facilitated translocation event is required for the successful secretion of the AIP due to its presumed lack of membrane permeability. Intriguingly, the agr locus does not encode designated proteins to account for these steps. Consequently, AgrB had been surmised to also export and/or cleave the thiolactone intermediate (Zhang et al., 2002). Biochemical studies have since ruled out a role for AgrB in the second proteolysis step (Qiu et al., 2005; Wang et al., 2015) and, while an involvement in AgrD translocation has not been formally ruled out, the absence of an ATP-binding cassette within the enzyme would make such a translocation activity quite extraordinary.
In principle, peptide translocation could occur either before or after the second proteolysis event, although a few lines of evidence favor the former scenario. In 2007, Kavanaugh and coworkers showed that the general signal peptidase, SpsB, cleaves a heptapeptide mimicking the leader peptide-AIP junction of AgrD-I at the expected site (Kavanaugh et al., 2007). Because the catalytic domain of SpsB localizes in the extracellular space, the authors argued in favor of the translocation-first model. It should be noted that only indirect evidence is provided in this study for the ability of SpsB to cleave the native thiolactone intermediate. More recently, large quantities of the AgrD leader peptide have been detected in the S. aureus extracellular matrix (Gonzalez et al., 2014; Schwartz et al., 2014). This result also supports the translocation-first model, as the alternative, proteolysis-first model would entail separate translocation of both leader and AIP fragments, which would be substantially less economical. The transporter responsible for AIP production, regardless of the substrate being exported, is yet to be identified.
The second proteolysis of AgrD sets the length of the exocyclic tail region within the AIP. Intriguingly, depending upon the S. aureus agr group, the AIP tail can vary between 2-4 amino acids in length (see Figure 1.4), with even larger variation found in other gram-positive species (Olson et al., 2014; Sturme et al., 2005). Nonetheless, this proteolysis event is remarkably specific within a given agr group – the growth media of over a dozen AIP-producing gram-positive strains have been subjected to mass-spectrometric analysis and in no case has a single agr locus been found to produce chemically heterogeneous AIPs (Gray et al., 2013). The sequence contexts of the scissile bonds are so diverged among their AgrD precursors that it would be nothing short of shocking if the orthologs of a single protease family carried out the AIP-releasing function in all species.
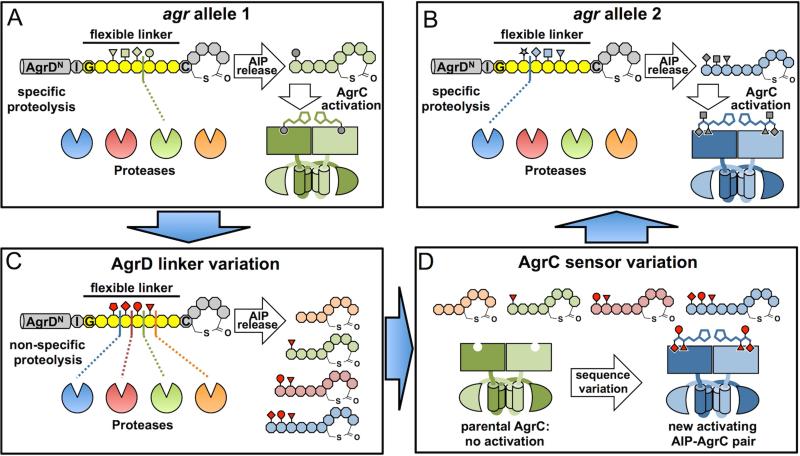
How might such variation within AIP length have arisen? Here we present a model that reconciles the absence of a designated AIP-releasing protease to both the homogeneity of the cleavage site within each native AIP, as well as the tail-length variation among AIPs from closely-related the agr loci. The AgrD-thiolactone intermediate consists of two structurally rigid and putatively protease-resistant elements, i.e., an amphipathic α-helix and a thiolactone macrocycle, flanking an 8-aa linker starting from the conserved, helicity-breaking glycine and ending at the ring-forming cysteine (Figures 4A and 4B). Because all known AIP-releasing sites are located within this linker, we postulate that this region exhibits considerable conformational flexibility so that each peptide bond is potentially cleavable if exposed to a protease that recognizes the relevant sequence context. As a consequence, the cleavage site is dictated by a competition among all proteases with access to this linker (Figures 4A and 4B). In wild-type agr systems, the exquisite specificity of the AIP-releasing proteolysis would come from the selective pressure imposed by the need for highly efficient conversion of AgrD to the native AIP pheromone. The protease of choice thereby overwhelms all other competitor proteases in terms of both efficiency and specificity (Figures 4A and 4B). Conceivably, however, sequence changes occurring within or adjacent to the AgrD linker region through random mutagenesis or DNA recombination may loosen the proteolysis specificity (Figure 4C). The mutant strain may thus produce a series of AIPs with variable tail lengths (Figure 4C), which significantly increases the likelihood for a new, activating AIP-AgrC pair to emerge through a second mutational event affecting the sensor domain sequence of AgrC (Figure 4D). Importantly, because autoinduction is by nature a collective behavior of bacterial populations, selection pressure against the AgrD mutant would be low if it stays within its parental population (Schuster et al., 2013). This may allow persistence of the mutant and also facilitate the co-evolution between AgrD and AgrC. Once bacteria containing this nascent AIP-AgrC pair are isolated from a parental population, they would be again selected for AIP-production efficiency, leading to the co-evolution between the new AgrD-protease pair and hence, the re-establishment of AIP-releasing specificity (from Figure 4D to 4B). We imagine that this model should be experimentally testable through a combination of AgrD mutagenesis and peptidomics. Ultimately, identification of the AIP-releasing proteases in all S. aureus subgroups will provide a key starting point to investigate the evolutionary trajectory of these agr variants.
Figure 4. A Hypothetical trajectory for hyper-variation of agr alleles.
(A and B) Specific AIP-releasing proteolysis and AIP-AgrC recognition in two hypothetical, wild-type agr variants from the same bacterial species. Amino-acid residues in the AgrD thiolactone intermediate are shown in octagons, with the proteolysis-susceptible linker highlighted in yellow. AIP, AgrC, the AIP-releasing protease and side-chains of four linker residues in the thiolactone intermediate are colored for group specificity (green in panel A, blue in panel B). (C) Loosened AIP-releasing specificity after sequence variation occurs within the AgrD linker region depicted in (A). Side-chains of four linker residues subject to changes are highlighted in red. (D) Further sequence variation in the AgrC sensor domain gives rise to a new, activating AgrC-AIP pair (blue). This nascent, functional agr allele would then be selected for releasing specificity of the new AIP to become the wild-type allele depicted in (B).
Activation of the AgrCA TCS
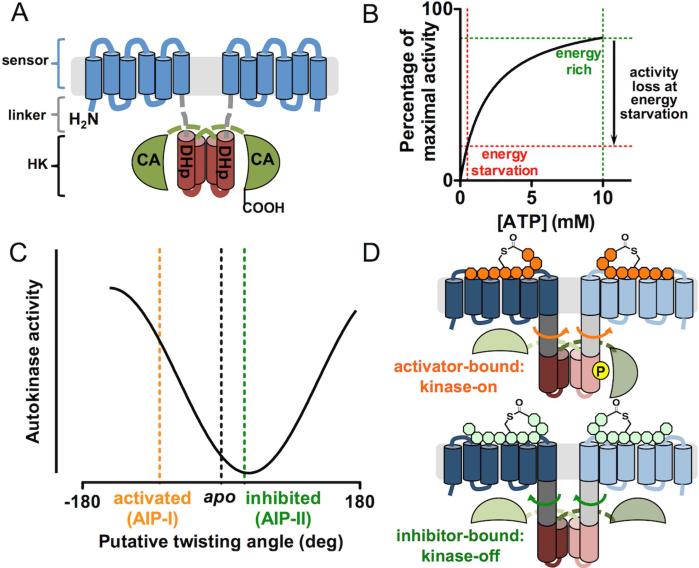
By sequence homology, AgrC and AgrA form a TCS. AgrC adopts the modular architecture commonly seen for RHKs, consisting of an N-terminal, membrane-integrated sensor module that detects AIP and a C-terminal histidine kinase (HK) module that carries out enzymatic functions (Figure 5A) (Lina et al., 1998). These two modules are connected via a short, α-helical interdomain linker (Wang et al., 2014). The HK domain contains two subdomains (Gao and Stock, 2009). Proximal to the sensor is the dimerization and histidine phosphorylation (DHp) subdomain, which folds into an α-helical hairpin and dimerizes through the formation of a four-helix bundle. Consequently, AgrC, as do most RHKs known to date, forms an obligate dimer. The distal, C-terminal subdomain is the so called catalytic and ATP-binding (CA) subdomain. In an AgrC homodimer, the CA subdomain of one subunit binds to the ATP and catalyzes the in trans phosphorylation at a histidine residue on the DHp subdomain of the opposite subunit (Cisar et al., 2009) (Figure 5A). Upon phosphorylation, AgrC transfers the phosphoryl group to AgrA to turn on its activity as a transcription factor.
Figure 5. Activation of the AgrC-AgrA TCS.
(A) Domain architecture of AgrC-I. The protein is shown as a homodimer with the sensor, DHp and CA modules colored in blue, brown and green, respectively. The interdomain linker is depicted as gray dashed lines. (B) A Michaelis-Menten plot of AgrC based on a Km value of 2 mM (Wang et al., 2014). The loss of kinase activity at energy starvation ([ATP] = 0.5 mM) relative to energy-rich conditions ([ATP] = 10 mM) is highlighted. Cellular ATP levels are inferred from measurements performed in E. coli (Tran and Unden, 1998). (C) The gradual responsiveness of the autokinase activity of the AgrC HK domain to rotational movements imposed at the interdomain linker pair. Conformational inputs in full-length AgrC-I under native ligand states are marked with dashed lines. (D) Schematic showing the opposite direction of linker rotation triggered upon the binding of an activator (top) or an inhibitor (bottom) AIP.
Although conforming to the fundamental roles of RHKs, AgrC also possesses some distinct sequence features and is classified as a member of the “HPK10” competence kinase subfamily of RHKs (Grebe and Stock, 1999). A recent survey of the protein database expanded this subfamily to more than 300 non-redundant sequences that are exclusively from low-GC gram-positive bacteria (Firmicutes) (Wang et al., 2014). In all HPK10 sequences, an Asn residue substitutes for the conserved “G1-box” Asp, which normally hydrogen-bonds to the N-6 amino group on the adenine base of the nucleotide. A recent crystal structure of the AgrC CA domain indicates that this Asn residue indeed takes the place of the canonical Asp (Srivastava et al., 2014). Therefore, the substitution is likely responsible for the exceptionally weak affinity between full-length AgrC and ATP: the Km is about 2 mM (Wang et al., 2014) (Figure 5B). This property renders the kinase activity of AgrC, and perhaps all HPK10-subfamily members, strongly dependent upon the cellular ATP level, which reflects the energy condition of the bacterium (Figure 5B). In particular, when energy starvation drives down the cellular ATP level, the AgrC kinase activity will be diminished even in the presence of AIP activators. This mechanism may account, in part, for the down-regulation of agr autoinduction in Staphylococcus and competence induction in Streptococcus (both mediated by HPK10 subfamily members) in the stationary growth phase (Claverys et al., 2006; Wright et al., 2005).
Until recently, all mechanistic studies on the AgrCA TCS employed cell-based assays, which, while informative, precluded detailed biophysical and biochemical characterizations (Novick and Geisinger, 2008; Thoendel et al., 2011). The successful reconstitution of the AgrCA TCS thus stands as an exciting breakthrough in this field (Wang et al., 2014). Key to this achievement was the use nanoscale lipid bilayer discs, or nanodiscs (Ritchie et al., 2009), to afford an active preparation of full-length AgrC. This new system has already led to some remarkable insights. For instance, maximal activation of AgrC-I only occurs when the receptor is embedded in a highly anionic lipid bilayer that approximates the native lipid composition of the S. aureus cell membrane. Within this membrane environment, AIP-I non-cooperatively binds to the receptor in a 2:2 stoichiometry, while the inhibitor, AIP-II, competes for both sites where AIP-I binds. Dissociation constants (KD) of both ligands were found to be in the mid-nanomolar range. Interestingly, experiments employing this reconstitution system ruled out a previous model in which the AIP binding results in acylation of AgrC via opening the chemically labile thiolactone (Mayville et al., 1999). AgrC-I possesses a baseline level of autokinase activity, which is strongly activated when bound to the agonist, AIP-I. Interestingly, engagement with the AIP-II inhibitor leads to reduction in baseline AgrC activity, indicative of inverse agonism, while the AIP-III inhibitor has no effect on basal AgrC activity, consistent with neutral antagonism (Geisinger et al., 2009). Kinetic studies on the reconstituted AgrCA TCS indicate that autophosphorylation, rather than phosphoryl transfer, is the rate-determining step. Unlike most RHKs that have been characterized in vitro, AgrC-I lacks phosphatase activity on AgrA in all ligand states tested. Perhaps not coincidently, AgrA features one of the fastest rates for spontaneous chemical dephosphorylation (t1/2 = 3.9 min at 37 °C) among transcription-factor RRs (Thomas et al., 2008). Thus, a “kinase-off, phosphatase-on” ligand state of AgrC may be dispensable for the rapid inactivation of AgrA, potentially allowing shutdown of the agr signaling in response to, for instance, energy starvation even in the presence of activator AIPs.
Perhaps the most intriguing finding from the reconstituted agr TCS is the signaling plasticity of AgrC-I (Wang et al., 2014): the RHK exhibited four distinct levels of autokinase activity when bound to AIP-I, -II, -III and a non-native partial activator, the truncated AIP-I (Lyon et al., 2002). This plasticity contradicts the generally accepted two-state model of RHK autokinase activation (Wang et al., 2013). Employing a chimeric-protein strategy in which the entire sensor domain was replaced by a stable coiled-coil motif, rotational perturbations were systematically introduced to the α-helical interdomain linkers preceding the AgrC-I HK domain. Autokinase analysis of these chimera proteins revealed that the kinase activity of the AgrC-I HK domain changes gradually with the magnitude of twisting movement applied to the linkers (Figure 5C). This result is in stark contrast with a previous report arguing for a model in which HK domains are inactive unless subjected to highly specific conformational inputs (Moglich et al., 2009). We note that HK domains exhibiting gradual input-response properties, when recombined with non-cognate sensor domains during the course of evolution, should enjoy a better chance of generating a signaling-competent new RHK (Capra and Laub, 2012). Furthermore, full-length AgrC-I appears to harness the signaling plasticity of its HK domain: cysteine-specific crosslinking data is consistent with a model in which AIP-I or AIP-II binding rotates the interdomain linker in different directions and thus confers activation or inhibition to the kinase activity (Figures 5C and 5D). These findings provide the first view on molecular motions triggered by ligand binding on a membrane-bound RHK.
AgrA phosphorylation and transcription activation
Acting as the phospho-receiver in the TCS as well as a transcription activator, AgrA consists of two domains, each assuming one of its two functions. The N-terminal receiver domain is shared across all RR proteins and dimerizes upon phosphorylation at its conserved Asp residue (Gao and Stock, 2009). The C-terminal DNA binding domain (DBD) belongs to the LytTR protein family and binds to the consensus DNA elements located in the P2 and P3 promoter region (Nikolskaya and Galperin, 2002). In the crystal structure of the AgrA DBD in complex with a cognate, 16-nucleotide (nt) DNA fragment, the DBD is enriched in β-strands and harnesses residues on its inter-strand loops for the interaction with DNA (Sidote et al., 2008). This unique binding interaction causes bending of the DNA double helix by 38°. Oxidative stress inactivates DNA binding by inducing disulfide-bond formation within the DBD between two cysteine residues at positions 199 and 228 (Sun et al., 2012).
Autoinduction of agr depends on a different temporal pattern of RNAII and RNAIII transcription. Tight repression of the P3 promoter is needed to avoid premature RNAIII-mediated mRNA degradation prior to autoinduction. At the same time, a reasonable expression level of RNAII is required to prime the autoinduction circuit. In congruence with this idea, experiments have confirmed that the baseline level of RNAII transcription is higher, while its activation is less dramatic, compared to that of RNAIII (Reynolds and Wigneshweraraj, 2011). How could the same pool of AgrA possibly exercise differential regulation over two operons? The answer lies within their promoter sequences. Both P2 and P3 promoter regions contain, in the orientation of transcription, two AgrA-binding elements followed by the -35 and -10 boxes required for RNA polymerase (RNAP) recognition. In spite of the similar architecture, the P2 recognition sequence provide stronger affinity to AgrA than that in P3 (Koenig et al., 2004). Thus, the promoter occupancy of P2 would be higher than that of P3 in the case where there is limited availability of phosphorylated AgrA, i.e. prior to full autoinduction. Moreover, the spacer between the -35 and -10 boxes measures 18 nts in the P2 promoter and 20 nts in P3, both deviating from the optimal 17-nt length for RNAP binding (Reynolds and Wigneshweraraj, 2011). Strikingly, shortening the spacer in P3 to the optimal length dramatically enhances the baseline transcription activity both in vitro and in vivo (Morfeldt et al., 1995; Reynolds and Wigneshweraraj, 2011). In light of this observation, the DNA-bending effect of AgrA binding, as well as the dimerization induced by AgrA phosphorylation, has been postulated to rearrange the -35 and -10 boxes back to the optimal conformation for engagement of RNAP (Reynolds and Wigneshweraraj, 2011). This model provides an attractive explanation for the more substantial up-regulation of RNAIII production during autoinduction.
Reagent development for the manipulation of agr
S. aureus requires agr not for survival but for virulence. Interfering with the autoinduction, or quorum quenching (QQ), should therefore be effective in combating infection whilst, in principle, reducing the likelihood of resistance development versus classic, bactericidal or bacteristatic, antibiotics (Cegelski et al., 2008). It has long been known that QQ agents are effective in containing the spread of S. aureus in mouse models of infection (Mayville et al., 1999). However, the utility of this therapeutic strategy in treating an existing infection is still very much an open question (Otto, 2004). It is noteworthy that silencing of the agr system is known to strengthen the S. aureus biofilm and the QQ strategy might therefore contribute to the maintenance of chronic infection (Kong et al., 2006). By extension, activation of the agr system might represent a more attractive strategy in tackling chronic S. aureus infections: not only does it disperse the biofilm, the constant induction of agr-regulated genes also takes a toll on the fitness of the bacterium, perhaps rendering them more susceptible to classic antibiotics. Practical application of this strategy would entail a global activator with the ability to turn on the autoinduction of all four S. aureus subgroups. This reagent could potentially be a cocktail of “clean” activators each of which activates one or more agr variants without substantially affecting the rest. Unfortunately, the efficacy of agr activators in animal models has not been tested to date, nor has any clean agr activator ever been developed. Indeed, nearly all the medicinal chemistry efforts in this area to date have focused on the development of QQ agents, although based on the above discussion, the identification of global agr activators clearly merits investigation.
Structural-activity relationship (SAR) analysis of AIP-AgrC interactions
Native AIPs provide a rich source of information for the design of QQ agents: once the structural elements required for binding are dissected from those needed for receptor activation, selective perturbation of the later should yield competitive inhibitors of the AgrC receptor. To this end, extensive SAR studies have been performed on AIP-I, -II and –III and have reached consensus on several important points. First of all, the 5-aa, 16-membered macrocycle is of utmost importance for binding (Figure 6A) (Mayville et al., 1999; McDowell et al., 2001). To date, no linear peptide has ever shown any activity on a native AgrC. Expanding or contracting the size of the macrocycle is also deleterious to AIP activity (Johnson et al., 2015). Secondly, hydrophobic residues at the C-terminal end of the peptide, two in AIP-II and three in each of the other AIPs, are necessary (but not sufficient, see below) for tight binding to AgrC (Figure 6A). Alanine point mutations at these positions cause severe loss of potency (Mayville et al., 1999; McDowell et al., 2001; Tal-Gan et al., 2013b). Last, but not least, the agonist activity of an AIP is highly sensitive to structural modification. Hotspots required for AgrC activation locate to the exocyclic tail and the second residue within the macrocycle (Figure 6A). Modification at these sites can convert an AIP into a global agr inhibitor (Johnson et al., 2015; Lyon et al., 2002; McDowell et al., 2001; Tal-Gan et al., 2013b), although in some cases a more complex pharmacology can result (Johnson et al., 2015).
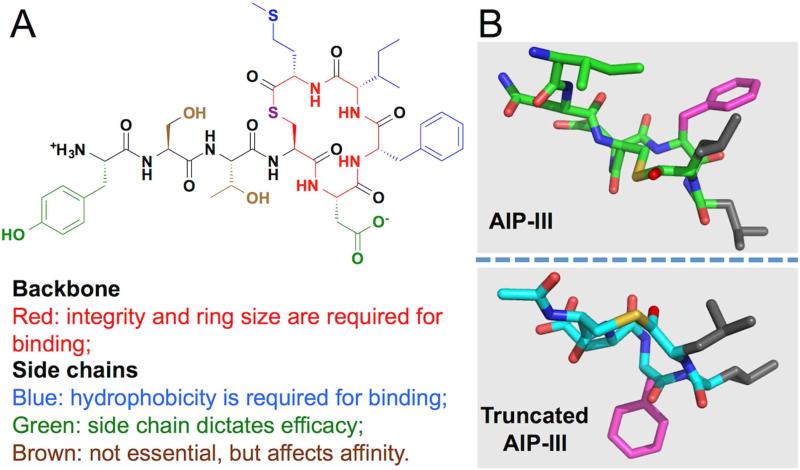
Figure 6. SAR analysis of AIPs.
(A) Summary of SAR studies of S. aureus AIPs, exemplified by AIP-I. (B) Comparison of the AIP-III solution structure to a less potent ligand, the truncated AIP-III. Side chain of Phe5 (in AIP-III numbering) is highlighted in magenta, and Leu6 and Leu7 in gray.
Recently, solution structures of all four wild-type AIPs, as well as a series of well-characterized analogs, have become available (Tal-Gan et al., 2015; Tal-Gan et al., 2013a). These structures have revealed some interesting correlations with the activities of these peptides. For instance, in all tight-binding AIP-I, -III and –IV analogs, the three C-terminal hydrophobic side chains form a distinct hydrophobic surface (Figure 6B, top panel). In contrast, truncated versions of AIP-I and AIP-III lacking the exocyclic tail, no longer maintain this surface due to their dramatically different macrocycle conformations (Figure 6B, bottom panel). Perhaps not coincidentally, both truncated peptides suffer a severe loss of potency despite the fact that their hydrophobic triad remains intact.
AgrC-targeting agents
Target-specific AgrC inhibitor design has been predominantly limited to engineering of the parent AIP scaffold. To date, AIP-I, -II and -III have all been successfully converted into QQ reagents capable of inhibiting the autoinduction of all four S. aureus subgroups (Figure 7A) (Lyon et al., 2000; Lyon et al., 2002; Tal-Gan et al., 2013b). Despite this success, the AIP backbone remains peptidic in nature and hence suffers such drawbacks as high immunogenicity and lack of stability in vivo. In an attempt to address this problem, modification of single residues in AIP-III through the substitution of amino acids with the corresponding peptoid or N-methyl mimics has generated a few global QQs (George et al., 2008; Tal-Gan et al., 2014). Full conversion of an AIP global inhibitor into a peptido-mimetic has, however, yet to be achieved.
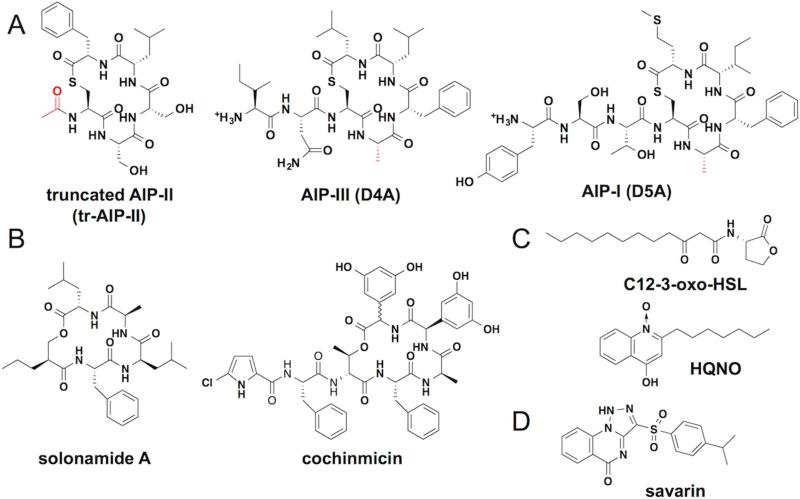
Figure 7. Synthetic molecules and natural products that target agr.
(A) Global AgrC inhibitors derived from native AIPs. Groups that differ from the wild-type AIP are highlighted in red. (B) AIP-mimicking natural products. (C) The Pseudomonas autoinducers 3-oxo-C12-HSL and HQNO. (D) The AgrA-targeting lead compound, savarin.
Aside from synthetic peptides derived from the native AIP scaffold, a few secondary metabolites from other microbes have been shown to inhibit S. aureus autoinduction. It should be pointed out that the mechanism of action of these compounds is yet to be rigorously determined. Nonetheless, some of these inhibitors share astonishing structural similarity to the native AIP architecture, despite their disparate origin. For instance, solonamides, cochinmicin and avellanin, identified from marine bacteria, actinomycetes and sponges, respectively, each possesses a 16-membered macrocycle and are therefore believed to function through competitive inhibition of AgrC (Figure 7B) (Desouky et al., 2015; Igarashi et al., 2015; Mansson et al., 2011). Two other natural products, 3-oxo-C12-HSL and 4-hydroxy-2-heptylquinoline N-oxide (HQNO) originating from Pseudomonas aeruginosa, are capable of quenching S. aureus autoinduction with a low-micromolar IC50 (Figure 7C) (Gordon et al., 2013). In particular, the HSL may act as an allosteric inhibitor of AgrC at lower concentrations as was inferred from its functional interaction with AIPs in cell-based assays (Murray et al., 2014). Given the similar, amphiphilic structure of these two compounds and the sensitivity of AgrC activation to lipid composition (Wang et al., 2014), it is tempting to speculate that they function through interacting with the cell membrane.
The recently available nanodisc-reconstitution system of AgrC and the solution NMR structures of AIPs potentially open new avenues for the screening or design of nonpeptidic AgrC ligands. Nanodiscs are particularly amenable to small-molecule library screening employing affinity-based approaches, which have given rise to potent ligands of several G-protein coupled receptors (Annis et al., 2007). Solution structures of AIPs and the configurations of the structural determinants for receptor binding observed therein, conceivably could be harnessed as the template to search for small-molecule AgrC ligands employing chemoinformatic strategies (Kolb et al., 2009).
AgrA-targeting reagents
In contrast to AgrC, which has four variants in S. aureus, AgrA has uniform sequence in all four subgroups, potentially making it a better therapeutic target. Analysis of the crystal structure of AgrA LytTR domain in the absence of bound DNA suggests that targeting a small molecule to an exposed hydrophobic cleft might disrupt the AgrA-DNA interaction (Leonard et al., 2012). As a proof of principle, Leonard et al. screened a focused small molecule library and identified a few hits with low-millimolar affinity for AgrA (Leonard et al., 2012). Screening of a much larger library led to the discovery of Staphylococcus aureus virulence inhibitor (savirin), which blocks S. aureus autoinduction in the mid to low micromolar range (Figure 7D) (Sully et al., 2014). Biochemical studies indicate that the small molecule blocks the AgrA association with DNA. This compound robustly inhibits autoinduction phenotypes of S. aureus cultures and attenuates the lesion size in the classic murine abscess model. Importantly, resistance did not emerge upon extensive passage of S. aureus in the presence of savirin. Thus, savirin is viewed as a promising lead compound for further medicinal chemistry studies.
Conclusions and Outlook
The agr locus plays a key role in the onset of S. aureus pathogenicity. This regulon has been the focus of intense study for well over two decades, making it one of the best-understood QS circuits in any bacterium. Indeed, all biochemical events directly involving agr-encoded proteins have now been reconstituted in vitro, allowing the associated processes to be carefully scrutinized. These investigations have revealed much about the inner workings of the QS circuit, but at the same time have exposed several hitherto unknown features of the process that await further biochemical characterization. First and foremost, it has become increasingly evident that agr-encoded proteins cannot account for all the steps in the core autoinduction circuit. Key protein participants remain to be identified in the translocation of the AgrD-thiolactone intermediate, the AIP-releasing proteolysis (at least in some subgroups), and the degradation of the AgrD-C fragment. Encoded apart from the agr locus, these proteins may well carry out other essential functions. Therefore, their identification and characterization will shed light on the driving forces, and restraints, governing the evolution of a hyper-variable system on a predominantly uniform genetic background. In addition, the biochemical properties of the AgrCA TCS suggest it may be responsive to spurious changes in cellular redox potential, ATP levels or lipid composition (Sun et al., 2012; Wang et al., 2014). These regulatory connections still await further examination in vivo. Once confirmed, they may be applicable to a wide range of Gram-positive QS systems as the underlying biochemical properties are dependent upon conserved sequence features.
The past decade has seen a rapid growth in the number of reagents that modulate agr. The vast majority of these tools are agr inhibitors and, perhaps as a consequence, all pharmacological studies performed on animal models involve, to our best knowledge, the administration of QQ agents. By contrast, the development of clean and/or global agr activators remains a persistent challenge for the field. Only when these reagents become available can the therapeutic value of S. aureus biofilm dispersion be explored in the context of chronic infections. Critically, medicinal chemistry efforts in this area are hampered by the absence of high-resolution structural information on how the AIP is recognized by the AgrC sensor domain. The challenges here can hardly be overstated – to date, there is no high-resolution structure available for any membrane-embedded RHK sensor domain. Nonetheless, advances in membrane protein X-ray crystallography and in single particle cryo-EM approaches offer some hope for the future, especially given the availability of numerous AgrC orthologs, including those from thermophilic species, for screening purposes (Wang et al., 2014).
As a result of the knowledge gaps listed above, we believe that the potential of targeting the agr circuit as an anti-infective modality is far from being fully explored. We imagine that interdisciplinary efforts will be needed to address these outstanding problems. Given the widespread occurrence of agr-like TCS systems in gram-positive bacteria, the concepts and tools that emerge form such efforts are likely to have wide-spread implications for our understanding of the complex behavior of bacterial pathogens.
Acknowledgements
This work was supported by NIH grants AI042783 and GM095880.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annis DA, Nickbarg E, Yang X, Ziebell MR, Whitehurst CE. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Current opinion in chemical biology. 2007;11:518–526. doi: 10.1016/j.cbpa.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of Two-Component Signal Transduction Systems. Annual Review of Microbiology. Vol 66. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar EA, Geisinger E, Muir TW, Novick RP. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol. 2009;74:44–57. doi: 10.1111/j.1365-2958.2009.06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- Desouky SE, Shojima A, Singh RP, Matsufuji T, Igarashi Y, Suzuki T, Yamagaki T, Okubo K, Ohtani K, Sonomoto K, et al. Cyclodepsipeptides produced by actinomycetes inhibit cyclic-peptide-mediated quorum sensing in Gram-positive bacteria. Fems Microbiol Lett. 2015;362 doi: 10.1093/femsle/fnv109. [DOI] [PubMed] [Google Scholar]
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter P, Caldelari I, Lioliou E, Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS letters. 2014;588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological Insights from Structures of Two-Component Proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Chen J, Novick RP. Allele-Dependent Differences in Quorum-Sensing Dynamics Result in Variant Expression of Virulence Genes in Staphylococcus aureus. J Bacteriol. 2012;194:2854–2864. doi: 10.1128/JB.06685-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Muir TW, Novick RP. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. P Natl Acad Sci USA. 2009;106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- Gonzalez DJ, Corriden R, Akong-Moore K, Olson J, Dorrestein PC, Nizet V. N-Terminal ArgD Peptides from the Classical Staphylococcus aureus Agr System Have Cytotoxic and Proinflammatory Activities. Chem Biol. 2014;21:1457–1462. doi: 10.1016/j.chembiol.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CP, Williams P, Chan WC. Attenuating Staphylococcus aureus Virulence Gene Regulation: A Medicinal Chemistry Perspective. J Med Chem. 2013;56:1389–1404. doi: 10.1021/jm3014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B, Hall P, Gresham H. Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors. 2013;13:5130–5166. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Advances in Microbial Physiology. 1999;41(41):139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Igarashi Y, Gohda F, Kadoshima T, Fukuda T, Hanafusa T, Shojima A, Nakayama J, Bills GF, Peterson S. Avellanin C, an inhibitor of quorum- sensing signaling in Staphylococcus aureus, from Hamigera ingelheimensis. The Journal of antibiotics. 2015 doi: 10.1038/ja.2015.50. [DOI] [PubMed] [Google Scholar]
- Jarraud S, Lyon GJ, Figueiredo AMS, Gerard L, Vandenesch F, Etienne J, Muir TW, Novick RP. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji GY, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- Johnson JG, Wang BY, Debelouchina GT, Novick RP, Muir TW. Increasing AIP Macrocycle Size Reveals Key Features of agr Activation in Staphylococcus aureus. Chembiochem. 2015;16:1093–1100. doi: 10.1002/cbic.201500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol186. 2004:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb P, Ferreira RS, Irwin JJ, Shoichet BK. Docking and chemoinformatic screens for new ligands and targets. Curr Opin Biotech. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Leonard PG, Bezar IF, Sidote DJ, Stock AM. Identification of a Hydrophobic Cleft in the LytTR Domain of AgrA as a Locus for Small Molecule Interactions That Inhibit DNA Binding. Biochemistry-Us. 2012;51:10035–10043. doi: 10.1021/bi3011785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. P Natl Acad Sci USA. 2012;109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Jarraud S, Ji GY, Greenland T, Pedraza A, Etienne J, Novick RP, Vandenesch F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- Lowy FD. Medical progress - Staphylococcus aureus infections. New Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Lyon GJ, Mayville P, Muir TW, Novick RP. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. P Natl Acad Sci USA. 2000;97:13330–13335. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GJ, Wright JS, Muir TW, Novick RP. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry-Us. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- Mansson M, Nielsen A, Kjaerulff L, Gotfredsen CH, Wietz M, Ingmer H, Gram L, Larsen TO. Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium. Mar Drugs. 2011;9:2537–2552. doi: 10.3390/md9122537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayville P, Ji GY, Beavis R, Yang HM, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. P Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell P, Affas Z, Reynolds C, Holden MTG, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CER, Bycroft BW, et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E, Taylor D, Vongabain A, Arvidson S. Activation of Alpha-Toxin Translation in Staphylococcus-Aureus by the Trans-Encoded Antisense Rna, Rnaiii. Embo J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EJ, Crowley RC, Truman A, Clarke SR, Cottam JA, Jadhav GP, Steele VR, O'Shea P, Lindholm C, Cockayne A, et al. Targeting Staphylococcus aureus Quorum Sensing with Nonpeptidic Small Molecule Inhibitors. J Med Chem. 2014;57:2813–2819. doi: 10.1021/jm500215s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Galperin MY. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 2002;30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum Sensing in Staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The Agr P2 Operon - an Autocatalytic Sensory Transduction System in Staphylococcus-Aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of Staphylococcal Virulence Factors Is Controlled by a Regulatory Rna Molecule. Embo J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, Dunman PM, Rohde H, Cech NB, Fey PD, et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol. 2014;196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Quorum-sensing control in Staphylococci - a target for antimicrobial drug therapy? Fems Microbiol Lett. 2004;241:135–141. doi: 10.1016/j.femsle.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Qiu RD, Pei WH, Zhang LS, Lin JQ, Ji GY. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]
- Queck SY, Jameson-Lee M, Villaruz AE, Bach THL, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Wigneshweraraj S. Molecular Insights into the Control of Transcription Initiation at the Staphylococcus aureus agr operon. J Mol Biol. 2011;412:862–881. doi: 10.1016/j.jmb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Methods in Enzymology; Liposomes, Pt F. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-Homoserine Lactone Quorum Sensing: From Evolution to Application. Annual Review of Microbiology. Vol 67. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Sekedat MD, Syed AK, O'Hara B, Payne DE, Lamb A, Boles BR. The AgrD N-Terminal Leader Peptide of Staphylococcus aureus Has Cytolytic and Amyloidogenic Properties. Infect Immun. 2014;82:3837–3844. doi: 10.1128/IAI.02111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote DJ, Barbieri CM, Wu T, Stock AM. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava SK, Rajasree K, Fasim A, Arakere G, Gopal B. Influence of the AgrC-AgrA Complex on the Response Time of Staphylococcus aureus Quorum Sensing. J Bacteriol. 2014;196:2876–2888. doi: 10.1128/JB.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturme MH, Nakayama J, Molenaar D, Murakami Y, Kunugi R, Fujii T, Vaughan EE, Kleerebezem M, de Vos WM. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J Bacteriol. 2005;187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sully EK, Malachowa N, Elmore BO, Alexander SM, Femling JK, Gray BM, DeLeo FR, Otto M, Cheung AL, Edwards BS, et al. Selective Chemical Inhibition of agr Quorum Sensing in Staphylococcus aureus Promotes Host Defense with Minimal Impact on Resistance. Plos Pathog. 2014;10 doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Liang HH, Kong XQ, Xie S, Cho H, Deng X, Ji QJ, Zhang HY, Alvarez S, Hicks LM, et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. P Natl Acad Sci USA. 2012;109:9095–9100. doi: 10.1073/pnas.1200603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y, Ivancic M, Cornilescu G, Blackwell HE. Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus. Organic & biomolecular chemistry. 2015 doi: 10.1039/c5ob01735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus. J Am Chem Soc. 2013a;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y, Stacy DM, Blackwell HE. N-Methyl and peptoid scans of an autoinducing peptide reveal new structural features required for inhibition and activation of AgrC quorum sensing receptors in Staphylococcus aureus. Chem Commun. 2014;50:3000–3003. doi: 10.1039/c4cc00117f. [DOI] [PubMed] [Google Scholar]
- Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed Through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J Am Chem Soc. 2013b;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD Residues Required for Autoinducing Peptide Biosynthesis. J Biol Chem. 2009;284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide Signaling in the Staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. agr function in clinical Staphylococcus aureus isolates. Microbiol-Sgm. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran QH, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. European Journal of Biochemistry. 1998;251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhao A, Novick RP, Muir TW. Key driving forces in the biosynthesis of autoinducing peptides required for staphylococcal virulence. Proc Natl Acad Sci U S A. 2015;112:10679–10684. doi: 10.1073/pnas.1506030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Zhao AS, Novick RP, Muir TW. Activation and Inhibition of the Receptor Histidine Kinase AgrC Occurs through Opposite Helical Transduction Motions. Mol Cell. 2014;53:929–940. doi: 10.1016/j.molcel.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sang JY, Wang JW, Su MY, Downey JS, Wu QG, Wang SD, Cai YF, Xu XZ, Wu J, et al. Mechanistic Insights Revealed by the Crystal Structure of a Histidine Kinase with Signal Transducer and Sensor Domains. Plos Biol. 2013;11 doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wright JS, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. P Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LS, Gray L, Novick RP, Ji GY. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J Biol Chem. 2002;277:34736–34742. doi: 10.1074/jbc.M205367200. [DOI] [PubMed] [Google Scholar]
- Zhang LS, Lin JQ, Ji GY. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. J Biol Chem. 2004;279:19448–19456. doi: 10.1074/jbc.M311349200. [DOI] [PubMed] [Google Scholar]