Abstract
Reactive oxygen species (ROS) oxidize guanosines in DNA to form 8-oxo-7,8-dihydro-2-deoxyguanosine (8-oxodG), a biomarker for oxidative stress. Herein we describe a novel 64-microwell electrochemiluminescent (ECL) array enabling sensitive multiplexed detection of 8-oxodG in ds-DNA without hydrolysis. Films of Nafion and reduced graphene oxide containing ECL dye [Os(bpy)2(phen-benz-COOH)]2+ (OsNG, {bpy= 2,2′-bipyridine and phen-benz-COOH = (4-(1,10-phenanthrolin-6-yl) benzoic acid)}) were assembled into microwells on a pyrolytic graphite wafer to detect 8-oxodG in oligonucleotides by electrochemiluminescence (ECL). DNA oxidation by Fenton’s reagent or by ROS formation during redox cycles involving NADPH, CuII, and model metabolites was monitored. UPLC-MS/MS of oxidized DNA samples were used for calibration. Detection limit for the fluidic arrays was one 8-oxodG per 670 intact nucleobases, or 0.15%. The method is sensitive enough to evaluate DNA oxidation from biologically relevant ROS-generating reactions of CuII, NADPH, and model metabolites.
Keywords: electrochemiluminescence, oxidative stress, DNA oxidation sensor, microwell array, [Os(bpy)2(phen-benz-COOH)]2+
Oxidative stress in living organisms results from imbalanced reactive oxygen species (ROS) and antioxidant defense pathways. It is implicated in cancer, neurological disorders, and heart disease.1,2 DNA, lipids, and proteins in living organisms are constantly exposed to ROS that include hydroxyl radicals, singlet oxygen, and superoxide.3–5 ROS are formed from ionizing radiation, chemical reactions, and free radicals.6,7 ROS can also form when metabolites of drugs or pollutants undergo redox cycling involving metal ions and NADPH.8 Oxidized DNA is a biomarker for biomolecule damage from ROS and oxidative stress.9–11
Guanine has the lowest oxidation potential of DNA nucleobases, and is a major target for ROS.12 Oxidation of guanine leads to several products,13 but initially formed 8-oxodG causes G-to-T transversions and A-to-C substitutions in DNA.14 8-OxodG can be detected by gas chromatography/ mass spectroscopy (GC/MS),15 liquid chromatography-electrochemical detection (HPLC-ECD),16–18 and LC-MS/MS,19–21 using protocols requiring DNA hydrolysis. We recently developed an 8-electrode voltammetric array using [Os-(bpy)2(PVP)10Cl]+ {(PVP = poly(4-vinylpyridine)}22 to detect 8-oxodG lesions on intact ds-DNA. While this system provides a sensitive, accurate approach to measure DNA oxidation without hydrolysis, its ability to be multiplexed is limited.
Electrochemiluminescence (ECL)23 is useful for larger sensor arrays since individually addressable electrodes are unnecessary.24 Selective catalytic oxidation of 8-oxodG can be achieved electrochemically using Os bipyridyl complexes25 that have oxidation potentials low enough to oxidize only 8-oxodG, but not native nucleobases. We previously demonstrated that 8-oxodG in ds-DNA can serve as coreactant to metallopolymer [Os(bpy)2(PVP)10]2+ in thin films to produce ECL26 via the suggested pathway in Scheme 1.
Scheme 1.
Pathway for ECL Emission with Os-Complexes (8G = 8-Oxoguanine Site)
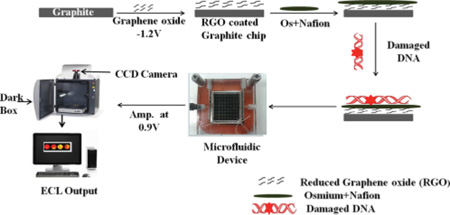
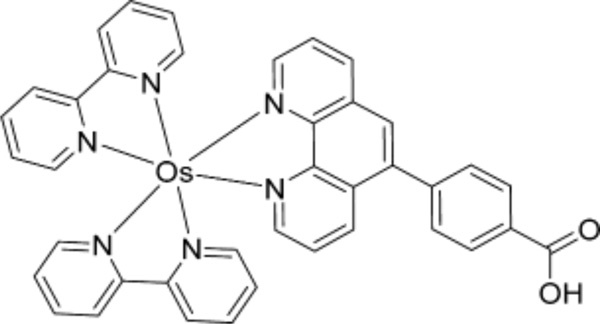
Herein we describe an ECL array capable of measuring metabolite-related DNA oxidation for multiple samples simultaneously. We used a wafer of pyrolytic graphite (PG) with computer-printed microwells as a single electrode enabling analysis of 64 samples.24 Attempts to employ [Os-(bpy)2(PVP)10]2+ in this array were unsuccessful due to weak ECL emission and a short-lived excited state. Thus, we investigated bis(2,2′-bipyridine)-(4-(1,10-phenanthrolin-6-yl)-benzoic acid) Os(II) [Os(bpy)2(phen-benz-COOH)]2+ (Scheme 2), which has a larger quantum yield than [Os-(bpy)2(PVP)10]2+,27 but should also oxidize 8-oxodG to produce ECL.
Scheme 2.
Structure of [Os(bpy)2(phen-benz-COOH)]2+
[Os(bpy)2(phen-benz-COOH)]2+ was immobilized in Nafion-graphene films inspired by an approach used earlier for Ruthenium complexes.28 [Os(bpy)2(phen-benz-COOH)]2+ in a Nafion dispersion was drop cast onto PG surfaces coated with electrochemically reduced graphene oxide (ERGO).29,30 These stable Os/ Nafion/ reduced graphene oxide (OsNG) films were used in the 64 microwell array to detect oxidized DNA induced by Fenton’s reagent31,32 or by catechol33 and N-hydroxyaminofluorene (N–OHAF)34 activated to form ROS by Cu2+ and NADPH. The high-throughput ECL array integrated into a fluidic detector measured unhydrolyzed oxidized DNA in multiple samples simultaneously.
EXPERIMENTAL SECTION
Chemicals and Materials
Polyguanylic acid (polyG), deoxyguanosine (dG), 8-oxoguanosine (8-oxodG), and salmon testes (ST) ds-DNA (2000 bp avg, 41.2% G/ C), CuCl2, MgCl2, catechol, d-glucose 6-phosphate sodium salt (G6P), β-nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP+), glucose-6-phosphate dehydrogenase (G6PDH), Nafion (20 wt %), diethylenetriaminepentaacetic acid (DTPA), deoxyribonuclease I (DNase I), phosphodiesterase I, alkaline phosphatase, and sodium oxalate were from Sigma-Aldrich. N-Hydroxyaminofluorene (N-OHAF, > 90%) was from MRI Chemical Carcinogen Repository. Pyrolytic graphite (PG, 2.5 × 2.5 × 0.3 cm3) wafers were from GraphiteStore.com (http://www.graphitestore.com), and basal plane PG was from Advanced Ceramics. [Os(bpy)2(phen-benz-COOH)]2+ was synthesized by refluxing Os(bpy)2Cl2 with 4-(1,10-phenanthrolin-6-yl) benzoic acid in ethanol/ water (1:1) for 16 h (Scheme S1, Supporting Information (SI)),35 and characterized by 1H NMR, HPLC, and spectroscopy (Figures S1 and S2 of the SI).
Instrumentation
A CH 660A electrochemical analyzer was used for cyclic voltammetry (CV) and the arrays. The CV cell had a saturated calomel reference electrode (SCE), Pt-wire counter and PG disc working electrode (A = 0.2 cm2). ACCD camera (SynGene) was used for ECL data acquisition.24 Digital Instruments (NanoScope IV) atomic force microscope (AFM) was used. Thermo Scientific (DIONEX UltiMate 3000) ultrahigh performance liquid chromatography (UHPLC) and a 4000 QTRAP triple quadrupole linear ion trap mass spectrometer (AB Sciex, Foster City, CA) were used for UPLC-MS/ MS.
Preparation of Os-Nafion-Reduced Graphene Oxide (OsNG) Films
Graphene oxide was electrochemically reduced and deposited onto PG electrodes by a previously described electrophoretic method.29,30 Briefly, the electrode is placed in a graphene oxide dispersion (0.2 mg mL−1 in 0.1 M LiClO4) and −1.2 V vs SCE applied for 60 s. Reduced graphene oxide (rGO) is deposited onto the surface as monitored by increased surface conductivity. The Os-complex-Nafion catalytic ink was prepared by mixing 1 mL of 1.5 mg mL−1 Os complex in acetonitrile with 0.2 mL of aqueous 5 wt % Nafion. After ultrasonication 15–30 min, 15 µL of the “ink” mixture was drop-cast onto the rGO-PG electrodes and dried overnight.
ECL Sensor Array
The ECL microarray was fabricated by first heat transferring a laserjet-printed 64-microwell pattern on glossy paper onto a pyrolytic graphite (PG) wafer (2.5 × 2.5 × 0.3 cm3).36 Each microwell is 2 mm diameter (SI Scheme S2 and Figure S4).
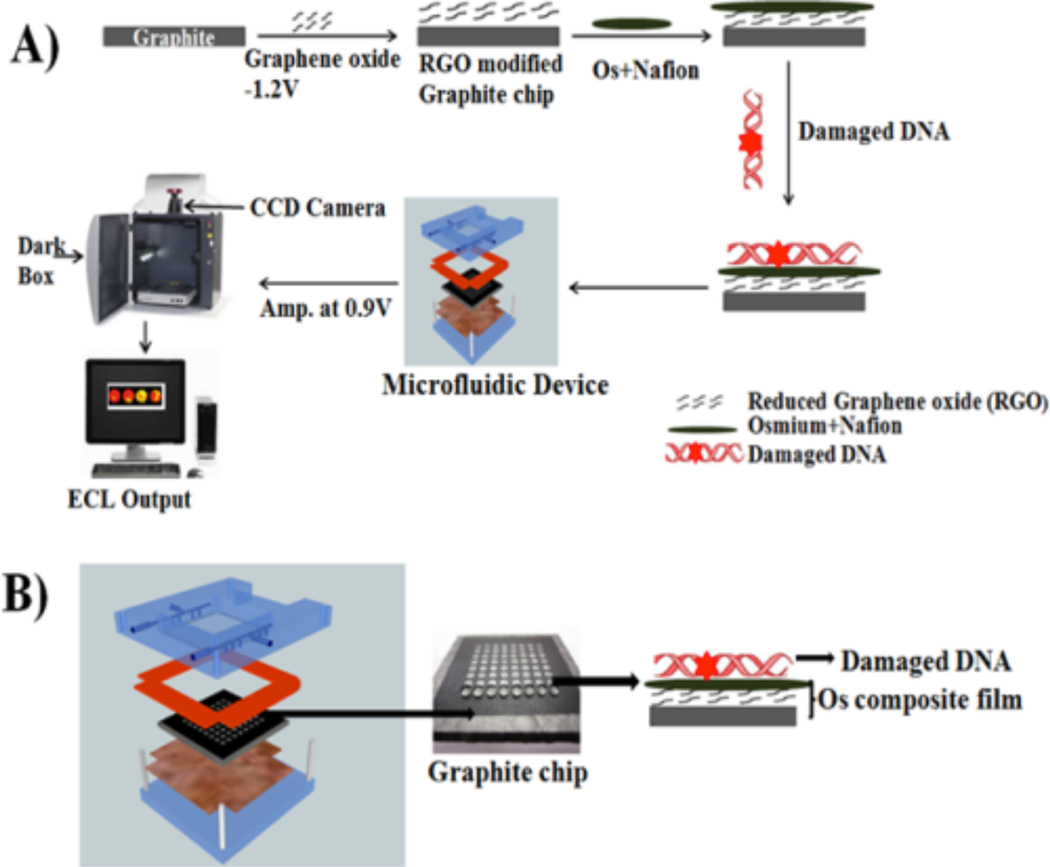
OsNG films were deposited in each microwell as described above for disc electrodes (Scheme 3A). A volume of 5.0 µL of osmium-Nafion ink was placed into each microwell on the PG chip that already contained reduced graphene oxide and dried overnight. And 5.0 µL of oxidized DNA solution was deposited into OsNG wells and dried. These arrays are reusable after removing OsNG films and hydrophobic toner with acetone, abrading on 600 grit SiC paper, and ultrasonicating in water for 2 min. Then, a new microwell pattern is heat transferred onto the surface.
Scheme 3.
Array and Film Fabricationa
a(A) Addition of OsNG and oxidized DNA layers to microwells. Chip is placed in the microfluidic device, then oxidized DNA samples are added to the wells and detected by ECL, intensity of which is proportional to the amount of 8-oxodG in the DNA. (B) Graphite chip and fluidic holder that is connected to syringe pump. Image of microfluidic device: Reproduced in part from ref 24 with permission of The Royal Society of Chemistry.
For ECL detection, the array was fitted in a fluidic device with top and bottom poly(methyl methacrylate) (PMMA) plates and an optical glass window (Scheme 3B). Pt counter and Ag/ AgCl reference electrode wires were embedded on the underside of the top plate (Figure S4 of the SI). The microfluidic device was then filled with pH 7.2, 10 mM phosphate buffer + 150 mM NaCl by using a syringe pump (Harvard Apparatus) at 500 µL min−1. Next, 0.9 V vs Ag/ AgCl was applied for 5 min to develop ECL detected by the CCD camera (Scheme 3A).24
Oxidation of Deoxyguanosine and DNA
PolyG (0.04 mg mL−1) or DNA (0.2 mg mL−1) in 10 mM phosphate buffer + 150 mM NaCl, pH 7.2 was oxidized by Fenton’s reagent containing 0.1 mM FeSO4 and 40 mM H2O2 at room temperature with constant stirring. Samples of 1 mL were taken every 5 min and frozen immediately in liquid nitrogen, followed by freeze-drying. For ECL analysis, samples were reconstituted in 1 mL 10 mM phosphate buffer, pH 7.2 + 150 mM NaCl, and 5 µL added to each microwell.
Alternatively, DNA (0.2 mg mL−1) was incubated with 3 mM catechol or 5 mM N-hydroxyaminofluorene in a solution of 5 mM CuCl2, 1 µM DTPA, and NADPH regenerating system of 1 unit/ mL glucose-6-phosphate dehydrogenase (G6PDH), 2.5 mM glucose-6-phosphate (G6P), 0.5 mM NADP+, 1 mM MgCl2, in 50 mM pH 7.2 Tris buffer at 37 °C. DNA was oxidized, and then freeze-dried and reconstituted in buffer as above. A volume of 5 µL of oxidized DNA solution was added to each microwell and air-dried.
For measurements, the sample-loaded PG chip inside the fluidic chamber was washed and filled with 10 mM PBS, pH 7.2, and ECL measured at 0.9 V with the CCD camera for 5 min.
UPLC-MS/MS
Oxidized DNA was purified by ethanol precipitation, then reconstituted in 1 mL Tris buffer, pH 7.2. Hydrolysis of oxidized DNA was done with DNase I (400 unit mg−1 of DNA) for 3 h, followed by addition of phosphodiesterase I (0.2 unit mg−1 of DNA) and alkaline phosphatase (1.2 unit mg−1 of DNA).22 DNA samples were then incubated overnight in a rotor at 37 °C, then vacuum filtered using a 96-well filtration plate (3 kDa MW cutoff), and analyzed via UPLC-MS/MS. A Hypersil Gold C18 (100 mm × 0.3 mm i.d., 3 µm) reversed phase column was used. Mobile phase was 10 mM ammonium acetate, pH 4.0 (solvent A), and methanol +0.1% formic acid (solvent B). The gradient (Table S1, SI) was at 6 µL min−1. MS was in the positive ion mode in multiple reaction monitoring (MRM) mode.
RESULTS
AFM
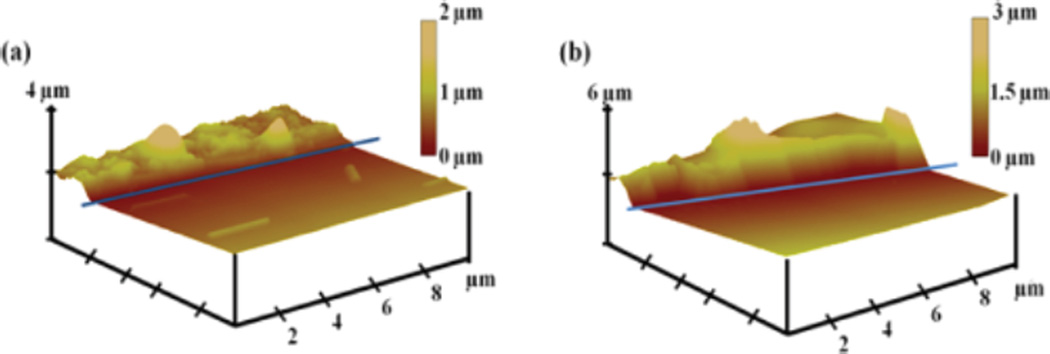
Os complex-Nafion and Os complex-Nafion/ DNA films were drop cast on half-masked (with tape) HOPG, then stripping away the mask under the films to form steps.37 The Os-Nafion film on HOPG gave hill-like images 0.7 ± 0.2 µm thick, and mean surface roughness 0.12 ± 0.03 µm (Figure 1a). For oxidatively damaged DNA cast onto the osmium-Nafion film, thickness of 1.1 ± 0.6 µm and mean surface roughness 0.09 ± 0.02 µm were found (Figure 1b).
Figure 1.
Tapping mode AFM images of films on highly ordered pyrolytic graphite (HOPG): (a) Os-complex-Nafion film deposited on half-masked HOPG, after stripping away the mask. Blue line shows edge of mask before stripping. The darker, front parts of images are stripped regions, and the brighter, upper left regions are where film remains. Analysis gave thickness 0.7 ± 0.2 µm. (b) Os-complex-Nafion/ oxidized DNA film on half-mask stripped HOPG, giving thickness 1.1 ± 0.6 µm.
Voltammetry of OsNG Films
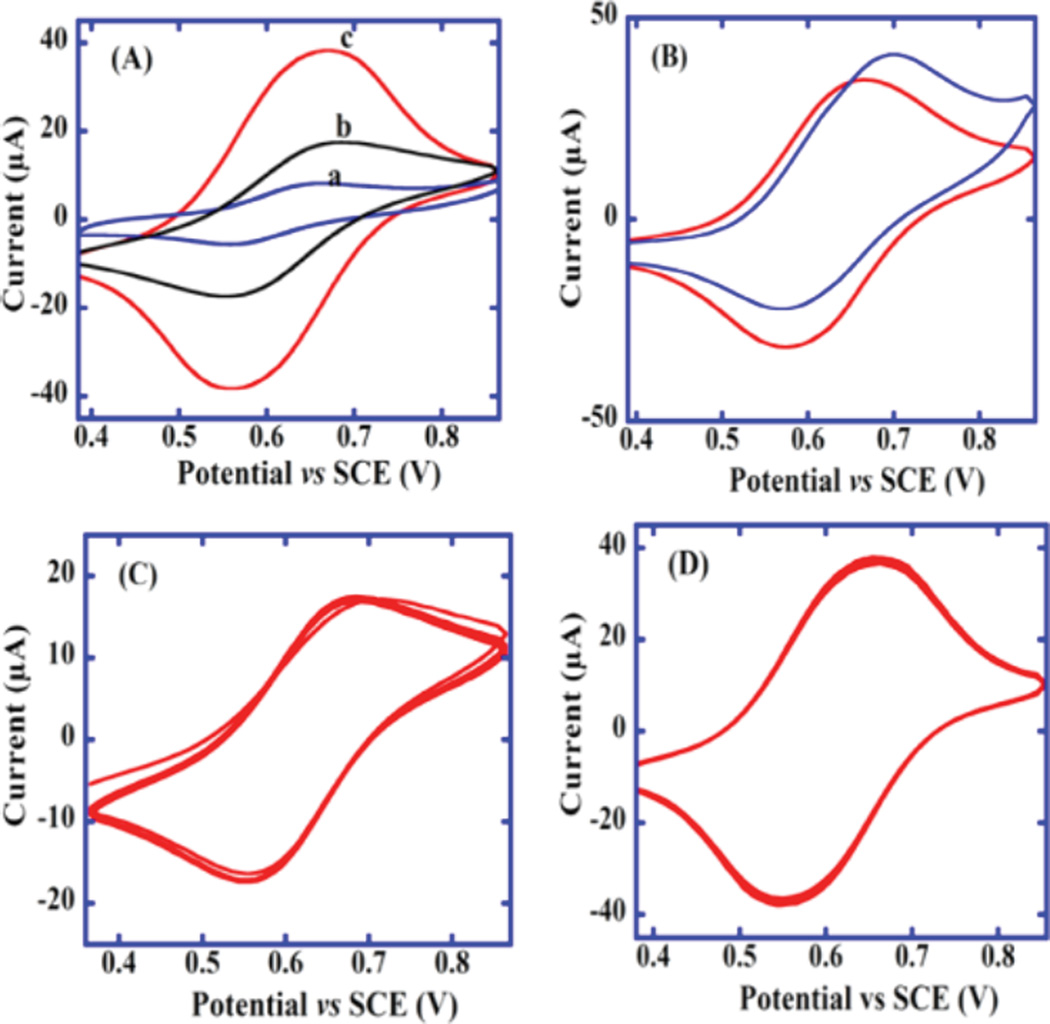
Figure 2A shows quasireversible CVs of [Os(bpy)2(phen-benz-COOH)]2+ (a) dissolved in pH 7.2 buffer on bare PG, (b) after depositing [Os(bpy)2(phen-benz-COOH)]2+ -Nafion ink on PG, and (c) after immobilizing RGO and depositing osmium-Nafion catalytic ink [as in (b)]. Oxidation peak current of [Os-(bpy)2(phen-benz-COOH)]2+ increased from (b) to (c), suggesting improved charge transport and/ or more incorporation of Os-complex when using RGO. Figure 2B shows the OsNG CV with a small catalytic oxidation peak increase when sodium oxalate was added, confirming catalytic activity of the film. Figure 2C shows 20 repetitive-scan CVs of the Os-Nafion catalytic ink drop-cast on bare PG, and Figure 2D shows 20 repetitive CVs after PG was coated with graphene oxide.
Figure 2.
Cyclic voltammograms at 50 mV/ s in 10 mM PBS, pH 7.2: (A) (a) 2 mM [Os(bpy)2(phen-benz-COOH)]2+ at bare PG, (b) [Os(bpy)2(phen-benz-COOH)]2+-Nafion catalytic ink on PG, and (c) [Os(bpy)2(phen-benz-COOH)]2+-Nafion catalytic ink and reduced graphene oxide on PG. (B) [Os(bpy)2(phen-benz-COOH)]2+-Nafion catalytic ink on reduced graphene oxide-PG electrode in the absence (red) and presence (blue) of 1 mM sodium oxalate. (C) 20 repetitive scans of [Os(bpy)2(phen-benz-COOH)]2+-Nafion catalytic ink on a PG electrode. (D) 20 repetitive scans of [Os(bpy)2(phen-benz-COOH)]2+-Nafion catalytic ink with reduced graphene oxide on PG electrode modified. Initial CVs were first recorded after 10 cycles, when electrodes had reached reproducible steady-state scans.
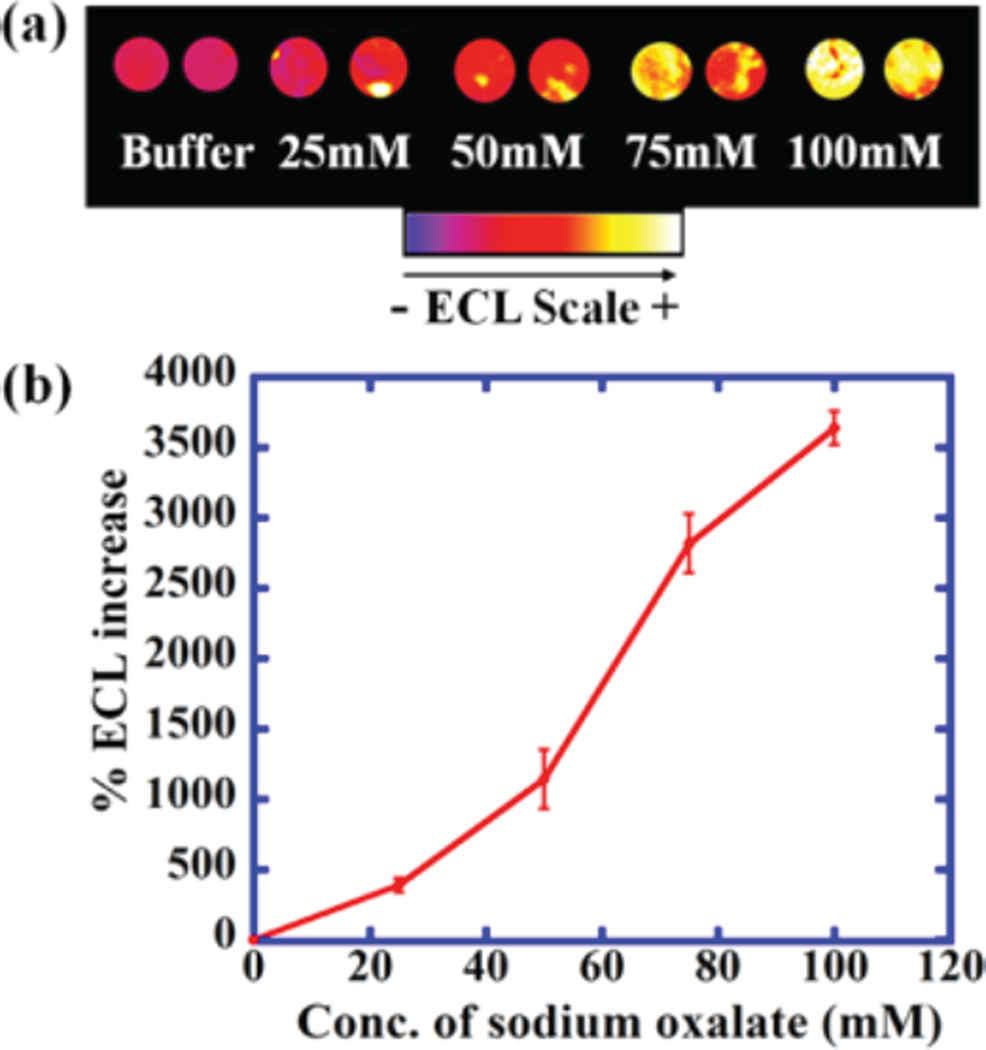
The 64-well OsNG array was evaluated for ECL emission using sodium oxalate, a known ECL coreactant.38 An increase in ECL intensity was found with increasing concentration of sodium oxalate (Figure 3). Relative spot-to-spot standard deviation was ±6.8% and between arrays was ±7.6% (SI Figure S5). Figure 3 shows ECL output related to CVs in Figure 2B, and Figure S7 represents the ECL related to CVs in Figure 2C, D.
Figure 3.
Reconstructed, recolorized ECL array images and calibration curve for sodium oxalate in 10 mM PBS, pH 7.2. (a) ECL at different concentrations of sodium oxalate in wells containing OsNG. (b) Influence of sodium oxalate concentration on %ECL increase.
Detection of Oxidized PolyG and DNA
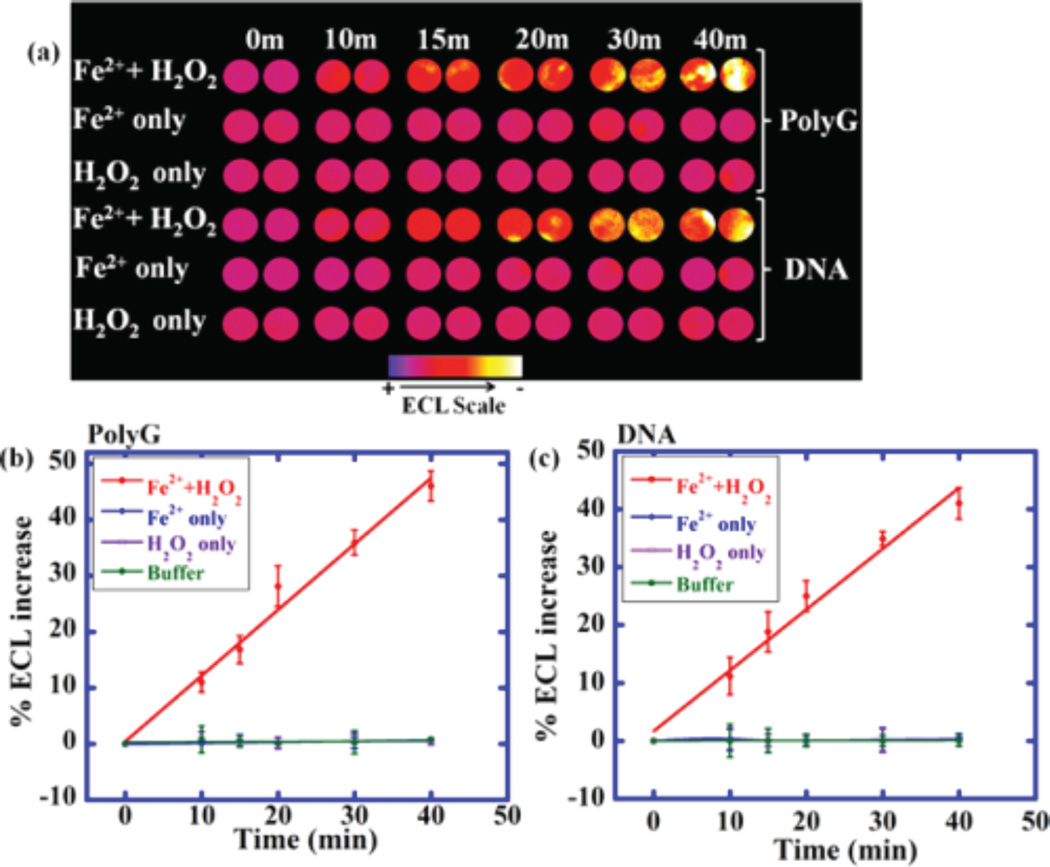
Fenton’s reagent was used to generate 8-oxodG on polynucleotides. Oxidized polyG of DNA solution was dropped into the OsNG microwells in the array and dried. Phosphate buffer was delivered to the detection chamber, flow was stopped, and ECL was measured at 0.9 V versus Ag/ AgCl. Increase in ECL over controls was found due to selective catalytic oxidation of 8-oxodG by [Os(bpy)2(phen-benz-COOH)]2+ which in turn generates electronically excited [Os(bpy)2(phen-benz-COOH)]2+* that decays to ground state by emitting ECL at 746 nm. Here, 8-oxodG serves as sacrificial reductant. Longer periods of oxidation of polyG or DNA produced more ECL, suggesting increasing quantities of 8-oxodG (Figure 4). Controls exposing polyG or DNA to buffer, FeSO4 alone, or H2O2 alone gave negligible ECL.
Figure 4.
OsNG array results on polynucleotides oxidized by Fenton’s reagent: (a) Reconstructed, recolorized ECL image showing ECL for polyG and DNA treated with Fenton’s reagent in pH 7.2 PBS for time in min (top image). Controls are polyG and DNA treated, only with buffer, with only FeSO4, or with only H2O2. Graphs show relative %ECL increase for (b) polyG and (c) DNA. Error bars represent standard deviation for three independent trials.
UPLC-MS/MS Analysis
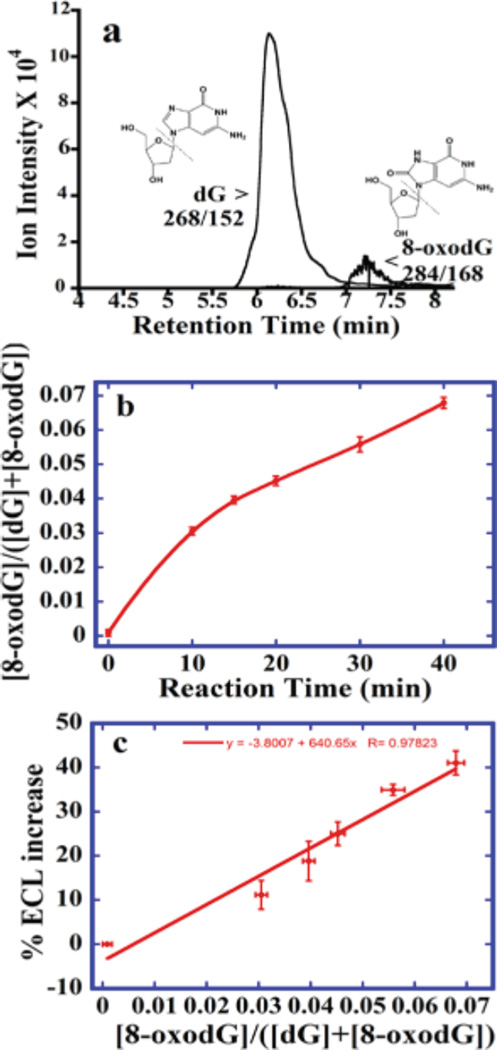
To enable calibration, we measured relative amounts of 8-oxodG directly by UPLC-MS/ MS in DNA oxidized by Fenton’s reagent. Figure 5a depicts an MRM chromatogram with m/ z 268 → 152 for dG eluting at 6.5 min and m/ z 284 → 168 for 8-oxodG at 7.2 min. Figure 5b shows ratio of 8-oxodG to total dG vs reaction time. These data were used to construct a calibration curve for the sensor, that is, % ECL increase data vs ratio of 8-oxodG to the total amount of dG (Figure 5c). This linear calibration curve enabled quantitation of relative amounts of 8-oxodG in oxidized DNA from array results. The LC-MS/ MS limit of detection (LOD) for 8-oxodG was 0.039 pmol 8-oxodG per 15.5 pmol of DNA (one 8-oxodG per 106 nucleobases). LOD for unreacted DNA was 218 8-oxodG per 106 nucleobases (nine 8-oxodG per 10 000 intact guanosines). ECL array LOD was 1500 8-oxodG per 106 nucleobases, or one 8-oxodG per 670 nucleobases.
Figure 5.
UPLC-MS/MS for ECL arrray calibration. (a) MRM chromatogram with mass transition m/ z 268− 152 indicating dG and m/ z 284− 168 measuring 8-oxodG for DNA reacted 15 min with Fenton’s reagent. (b) Ratio [8-oxo dG]/ ([dG]+[8-oxodG]) vs reaction time with Fenton’s reagent. (c) Calibration as % ECL increase vs ratio [8-oxo dG]/ ([dG]+[8-oxodG]).
Oxidized DNA Induced by N-OHAF and Catechol
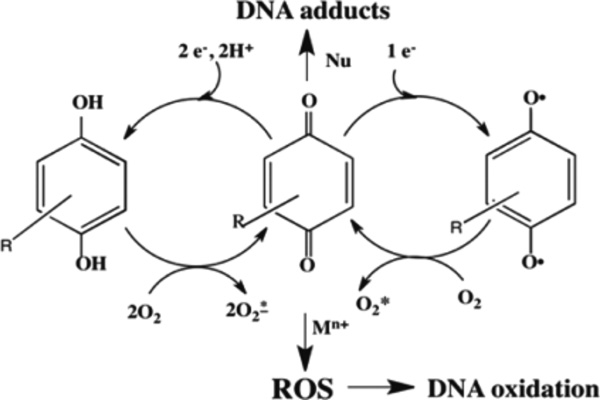
N-OHAF is a metabolite of nitrofluorene (NF) found in emissions from fuel combustion39 that can cause oxidative DNA damage.40 Catechol is an industrial chemical and constituent of tobacco smoke.41 Redox cycling between catechol, semiquinone radical, and ortho-benzoquinone in the presence of Cu2+ and NADPH lead to ROS that can oxidize DNA (Scheme 4).42
Scheme 4.
General Pathway for Oxidative DNA Damage Induced by Metabolites in the Presence of Cu2+ and NADPH
Cu2+ in cell nuclei may play a role along with NADPH in oxidative DNA damage42,43 mediated by redox active compounds.44 Typical pathways may produce ROS and DNA nucleobase adducts (Scheme 4). Thus, we explored Cu2+-NADPH assisted DNA oxidation by catechol and N-OHAF as proof-of-concept for measuring biological relevant DNA oxidations using the OsNG array.
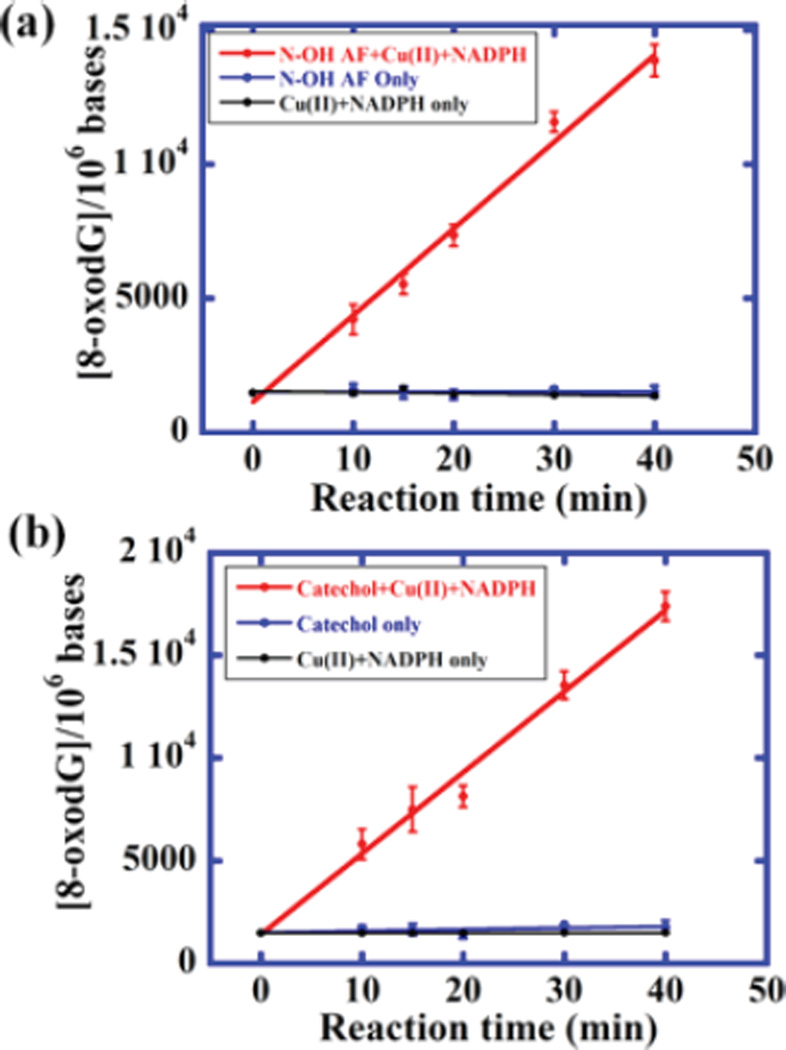
ROS were generated with catechol and N-OHAF in the presence of NADPH and Cu2+ via redox cycling pathways.39,40 ECL intensity increased with reaction time with DNA (Figure S6). Data in Figure 5c was used to convert %ECL to amount of 8-oxodG formed (Figure 6). No increases in ECL were found in controls with catechol or N-OHAF but no CuCl2 or NADPH (Figures 6 and S6) or with CuCl2 and NADPH but no catechol or N-OHAF. In each microwell, 1 µg of DNA (0.78 pmol) was analyzed for DNA oxidation. Thus, 64 µg of DNA provided 64 experiments in one array run. DNA used is significantly less than that in our microfluidic voltammetric array that required 2.5 µg.22
Figure 6.
Oxidations of DNA mediated by N-OHAF and catechol at 37 °C. (a) Dependence of 8-oxodG per 106 nucleobases on reaction time for 0.2 mg mL−1 DNA incubated with 5 mM N-OHAF, 5 mM CuCl2, and NADPH regeneration system. (b) Dependence of 8-oxodG per 106 nucleobases on reaction time using 3 mM catechol, CuCl2, and NADPH regenerating system.
DISCUSSION
Results above show that stable 1 µm thick OsNG composite films capable of detecting oxidized DNA by ECL (Figure 4) can be made from graphene, Nafion, and [Os(bpy)2(phen-benz-COOH)]2+. The 64 microwell array provides a new screening platform capable of multiplexed 8-oxodG detection in intact, slightly oxidized ds-DNA. The new array was used to analyze DNA oxidized in biologically relevant ROS-generating reactions of Cu2+, NADPH, and model metabolites. Detection can be achieved in 1 µg of DNA, with detection limit 1500 8-oxodG per 106 nucleobases. While this is not as good as the value of 150 8-oxoG’s per 106 nucleobases for our 6-sensor voltammetric array,27 it is nevertheless sufficiently sensitive to examine the chemistry of metabolite-mediated ROS production for multiple samples (Figures 6 and S6), which was our original goal.
The OsNG films incorporate the catalytic hydrophobic cation [Os(bpy)2(phen-benz-COOH)]2+ into the ionomer Nafion. Figure 2 demonstrates voltammetric and catalytic behavior and stability of OsNG films, and the importance of graphene to enhance the oxidation current and presumably ECL yields. Incorporation of [Os(bpy)2(phen-benz-COOH)]2+ into Nafion helps achieve stability which is likely related to the very large ion-exchange selectivity coefficients (106–107) of Nafion for hydrophobic cations.45
The OsNG films effectively detected oxidative damage to polyG and DNA induced by Fenton’s reagent (Figure 4), demonstrating that these films containing [Os(bpy)2(phenbenz-COOH)]2+ react with 8-oxodG coreactant to generate ECL. Given our experience with low ECL output of [Os(bpy)2(PVP)10]2+32 thin films with oxidized DNA, the OsNG films appear to provide superior performance.
One of our goals was to develop an array protocol capable of measuring DNA oxidation resulting from metabolite redox chemistry. Catechol is related to quinoid metabolites of polyaromatic hydrocarbons and N-OHAF is a metabolite of an aromatic amine. These compounds cause oxidation of DNA via a redox cycling pathway with Cu2+ and NADPH (cf. Scheme 4) that was readily detected on the array (Figure S6). Results are consistent with previous reports of ROS generation by these compounds.39,40
In summary, this paper describes a novel microwell array for detection of oxidized ds-DNA without the need for DNA hydrolysis. The array can accommodate up to 64 different experiments in one run as opposed to our previous voltammetric array capable of only one experiment per run. To our knowledge, this is the first multiplexed array that measures oxidation in intact ds-DNA. In the future, we hope to incorporate this approach into arrays that include other types of DNA damage from enzyme-generated metabolites.46
Supplementary Material
Acknowledgments
The authors thank National Institute of Environmental Health Sciences (NIEHS), NIH, Grant No. ES03154 (JFR) and Science Foundation Ireland Grant No. [12/ TIDA/ B2382] for financial support of this work.
Footnotes
ASSOCIATED CONTENT
* Supporting Information
- Additional experimental methods, NMR spectra, UV spectra, CV of [Os(bpy)2(phen-benz-COOH)]2+, fabrication of array, fluidic device setup, ECL reproducibility studies, UPLC-MS/MS method, and ECL results of DNA oxidation induced by metabolites (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 2.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Kehrer JP. Free Radicals as Mediators of Tissue Injury and Disease. Crit. Rev. Toxicol. 1993;23:21–48. doi: 10.3109/10408449309104073. [DOI] [PubMed] [Google Scholar]
- 4.Durackova Z. Some current insights into oxidative stress. Physiol. Res. 2010;59:459–469. doi: 10.33549/physiolres.931844. [DOI] [PubMed] [Google Scholar]
- 5.Kawanishi S, Hiraku Y, Oikawa S. Mechanism of guanine-specific DNA damage by oxidative stress and its role in carcinogenesis and aging. Mutat. Res., Rev. Mutat. Res. 2001;488:65–76. doi: 10.1016/s1383-5742(00)00059-4. [DOI] [PubMed] [Google Scholar]
- 6.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free. Free Radical Biol. Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw WHR. Cation Toxicity and the Stability of Transition-Metal Complexes. Nature. 1961;192:754–755. doi: 10.1038/192754a0. [DOI] [PubMed] [Google Scholar]
- 8.(a) Spencer WA, Vadhanam MV, Jeyabalan J, Gupta RC. Oxidative DNA damage following microsome/ Cu(II)-mediated activation of the estrogens, 17β-estradiol, equilenin, and equilin: Role of ROS. Chem. Res. Toxicol. 2012;25:305–314. doi: 10.1021/tx200356v. [DOI] [PubMed] [Google Scholar]; (b) Lin PH, Pan WC, Kang YW, Chen YL, Lin CH, Lee MC, Chou YH, Nakamura J. Effects of naphthalene quinonoids on the induction of oxidative DNA damage and cytotoxicity in calf thymus DNA and in human cultured cells. Chem. Res. Toxicol. 2005;18:1262–1270. doi: 10.1021/tx050018t. [DOI] [PubMed] [Google Scholar]
- 9.Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A Critical Biomarker of Oxidative Stress and Carcinogenesis. J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 11.Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2′-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res., Rev. Mutat. Res. 1997;387:147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 12.Steenken S, Jovanovic SV. Howeasily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 13.(a) Hickerson RP, Prat F, Foote CS, Burrows CJ. Sequence and Stacking Dependence of 8-Oxoguanine Oxidation: Comparison of One-Electron vs Singlet Oxygen Mechanisms. J. Am. Chem. Soc. 1999;121:9423–9428. [Google Scholar]; (b) White B, Smyth MR, Stuart JD, Rusling JF. Oscillating formation of 8-oxoguanine during DNA oxidation. J. Am. Chem. Soc. 2003;125:6604–6605. doi: 10.1021/ja0343252. [DOI] [PubMed] [Google Scholar]; (c) White B, Tarun MC, Gathergood N, Rusling JF, Smyth MR. Oxidised guanidinohydantoin (Ghox) and spiroiminodihydantoin (Sp) are major products of iron- and copper-mediated 8-oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2′-deoxyguanosine oxidation. Mol. BioSyst. 2005;1:373–381. doi: 10.1039/b511756a. [DOI] [PubMed] [Google Scholar]
- 14.(a) David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 15.Cadet J, Douki T, Ravanat JL. Measurement of oxidatively generated base damage in cellular DNA. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2011;711:3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc. Natl. Acad. Sci. Proc. Natl. Acad. Sci. U. S. A. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagadu S, Pottier I, Sichel F, Laurent C, Lefaix JL, Prevost V. Detection of extracellular 8-oxo-7,8-dihydro-2′-deoxyguanosine as a biomarker of oxidative damage in X-irradiated fibroblast cultures: optimization of analytical procedure. Biomarkers. 2010;15:707–714. doi: 10.3109/1354750X.2010.511269. [DOI] [PubMed] [Google Scholar]
- 18.Kasai H. A new automated method to analyze urinary 8-hydroxydeoxyguanosine by a high-performance liquid chromatography-electrochemical detector system. J. Radiat. Res. 2003;44:185–189. doi: 10.1269/jrr.44.185. [DOI] [PubMed] [Google Scholar]
- 19.Serrano J, Palmeira CM, Wallace KB, Kuehl DW. Determination of 8-Hydroxydeoxyguanosine in Biological Tissue by Liquid Chromatography/ Electrospray Ionization-Mass Spectrometry/ Mass Spectrometry. Rapid Commun. Mass Spectrom. 1996;10:1789–1791. doi: 10.1002/(SICI)1097-0231(199611)10:14<1789::AID-RCM752>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 2008;3:1287–1298. doi: 10.1038/nprot.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu CW, Wu MT, Chao MR, Pan CH, Wang CJ, Swenberg JA, Wu KY. Comparison of analyses of urinary 8-hydroxy-2′-deoxyguanosine by isotope-dilution liquid chromatography with electrospray tandem mass spectrometry and by enzyme-linked immunosorbent assay. Rapid Commun. Mass Spectrom. 2004;18:505–510. doi: 10.1002/rcm.1367. [DOI] [PubMed] [Google Scholar]
- 22.Song B, Pan S, Tang C, Li D, Rusling JF. Voltammetric microwell array for oxidized guanosine in intact ds-DNA. Anal. Chem. 2013;85:11061–11067. doi: 10.1021/ac402736q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forster RJ, Bertoncello P, Keyes TE. Electrogenerated Chemiluminescence. Annu. Rev. Anal. Chem. 2009;2:359–385. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]
- 24.Wasalathanthri DP, Malla S, Bist I, Tang CK, Faria RC, Rusling JF. High-throughput metabolic genotoxicity screening with a fluidic microwell chip and electrochemiluminescence. Lab Chip. 2013;13:4554–4562. doi: 10.1039/c3lc50698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ropp PA, Thorp HH. Site-selective electron transfer from purines to electrocatalysts: voltammetric detection of a biologically relevant deletion in hybridized DNA duplexes. Chem. Biol. 1999;6:599–605. doi: 10.1016/s1074-5521(99)80111-2. [DOI] [PubMed] [Google Scholar]
- 26.Dennany L, Forster RJ, White B, Smyth M, Rusling JF. Direct electrochemiluminescence detection of oxidized DNA in ultrathin films containing [Os(bpy)2(PVP)10]2+ J. Am. Chem. Soc. 2004;126:8835–8841. doi: 10.1021/ja048615+. [DOI] [PubMed] [Google Scholar]
- 27.Forster RJ, Vos JG. Synthesis, characterization, and properties of a series of osmium-and ruthenium-containing metallopolymers. Macromolecules. 1990;23:4372–4377. [Google Scholar]
- 28.Li H, Chen J, Han S, Niu W, Liu X, Xu G. Electrochemiluminescence from tris (2, 2′-bipyridyl) ruthenium (II)–graphene–Nafion modified electrode. Talanta. 2009;79:165–170. doi: 10.1016/j.talanta.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Hilder M, Winther-Jensen B, Li D, Forsyth M, MacFarlane DR. Direct electro-deposition of graphene from aqueous suspensions. Phys. Chem. Chem. Phys. 2011;13:9187–9193. doi: 10.1039/c1cp20173e. [DOI] [PubMed] [Google Scholar]
- 30.Sheng K, Sun Y, Li C, Yuan W, Shi G. Ultrahigh-rate supercapacitors based on eletrochemically reduced graphene oxide for ac line-filtering. Sci. Rep. 2012;2:247. doi: 10.1038/srep00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Walling C. Fenton’s reagent revisited. Acc. Chem. Res. 1975;8:125–131. [Google Scholar]; (b) Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases in DNA induced by hydrogen peroxide and ferric ion chelates. J. Biol. Chem. 1989;264:20509–20512. [PubMed] [Google Scholar]
- 32.Luo YC, Han ZX, Chin SM, Linn S. Three chemically distinct types of oxidants formed by iron-mediated Fenton reactions in the presence of DNA. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12438–12442. doi: 10.1073/pnas.91.26.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oikawa S, Hirosawa I, Hirakawa K, Kawanishi S. Site specificity and mechanism of oxidative DNA damage induced by carcinogenic catechol. Carcinogenesis. 2001;22:1239–1245. doi: 10.1093/carcin/22.8.1239. [DOI] [PubMed] [Google Scholar]
- 34.Murata Y, Yoshiki Y, Tada M, Kawanishi S. Oxidative DNA damage by a common metabolite of carcinogenic nitrofluorene and Nacetylaminofluorene. Int. J. Cancer. 2002;102:311–317. doi: 10.1002/ijc.10717. [DOI] [PubMed] [Google Scholar]
- 35.(a) Martin A, Byrne A, Burke CS, Forster RJ, Keyes TE. Peptide-Bridged Dinuclear Ru(II) Complex for Mitochondrial Targeted Monitoring of Dynamic Changes to Oxygen Concentration and ROS Generation in Live Mammalian Cells. J. Am. Chem. Soc. 2014;136:15300–15309. doi: 10.1021/ja508043q. [DOI] [PubMed] [Google Scholar]; (b) Byrne A, Moriarty R, Dolan C, Martin A, Neugebauer U, Forster RJ, Davies A, Volkov Y, Keyes TE. An Osmium (II) Polypyridyl Polyarginine Conjugate as a Probe for Live Cell Imaging; A Comparison of Uptake, Localization and Cytotoxicity with its Ru(II) Analogue. Dalton Trans. 2015;44:14323–14332. doi: 10.1039/c5dt01833a. [DOI] [PubMed] [Google Scholar]
- 36.Tang CK, Vaze A, Rusling JF. Fabrication of immunosensor microwell arrays from gold compact discs for detection of cancer biomarker proteins. Lab Chip. 2012;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munge B, Das SK, Ilagan R, Pendon Z, Yang J, Frank HA, Rusling JF. Electron transfer reactions of redox cofactors in spinach photosystem I reaction center protein in lipid films on electrodes. J. Am. Chem. Soc. 2003;125:12457–12463. doi: 10.1021/ja036671p. [DOI] [PubMed] [Google Scholar]
- 38.Chang MM, Saji T, Bard AJ. Electrochemical oxidation of oxalate ion in the presence of luminescers in acetonitrile solutions. J. Am. Chem. Soc. 1977;99:5399–5403. [Google Scholar]
- 39.Kriek E. Fifty years of research on N-acetyl-2-aminofluorene, one of the most versatile compounds in experimental cancer research. J. Cancer Res. Clin. Oncol. 1992;118:481–489. doi: 10.1007/BF01225261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.(a) Burger MS, Torino JL, Swaminathan S. DNA damage in human transitional cell carcinoma cells after exposure to the proximate metabolite of the bladder carcinogen 4-aminobiphenyl. Environ. Mol. Mutagen. 2001;38:1–11. doi: 10.1002/em.1044. [DOI] [PubMed] [Google Scholar]; (b) Ohnishi S, Murata M, Oikawa S, Totsuka Y, Takamura T, Wakabayashi K, Kawanishi S. Oxidative DNA damage by an N-hydroxy metabolite of the mutagenic compound formed from norharman and aniline. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2001;494:63–72. doi: 10.1016/s1383-5718(01)00177-2. [DOI] [PubMed] [Google Scholar]
- 41.Stone K, BermuÂdez E, Zang LY, Carter KM, Queenan KE, Pryor WA. The ESR properties, DNA nicking, and DNA association of aged solutions of catechol versus aqueous extracts of tar from cigarette smoke. Arch. Biochem. Biophys. 1995;319:196–203. doi: 10.1006/abbi.1995.1282. [DOI] [PubMed] [Google Scholar]
- 42.Malaisse WJ, Hutton JC, Kawazu S, Herchuelz A, Valverde I, Sener A. The stimulus-secretion coupling of glucose-induced insulin release. XXXV. The links between metabolic and cationic events. Diabetologia. 1979;16:331–341. doi: 10.1007/BF01223623. [DOI] [PubMed] [Google Scholar]
- 43.Bryan SE, Vizard DL, Beary DA, LaBiche R, Hardy KJ. Partitioning of zinc and copper within subnuclear nucleoprotein particles. Nucleic Acids Res. 1981;9:5811–5823. doi: 10.1093/nar/9.21.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of Quinones in Toxicology. Chem. Res. Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 45.Szentirmay MN, Martin CR. Ion-exchange selectivity of Nafion films on electrode surfaces. Anal. Chem. 1984;56:1898–1902. [Google Scholar]
- 46.Wasalathanthri DP, Li D, Song D, Zheng Z, Choudhary D, Jansson I, Lu X, Schenkman JB, Rusling JF. Elucidating organ-specific metabolic toxicity chemistry from electrochemiluminescent enzyme/DNA arrays and bioreactor bead-LC-MS/MS. Chem. Sci. 2015;6:2457–2468. doi: 10.1039/c4sc03401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.