Abstract
The biological recognition of human milk glycans (HMGs) is poorly understood. Because HMGs are rich in galactose we explored whether they might interact with human galectins, which bind galactose-containing glycans and are highly expressed in epithelial cells and other cell types. We screened a number of human galectins for their binding to HMGs on a shotgun glycan microarray consisting of 247 HMGs derived from human milk, as well as to a defined HMG microarray. Recombinant human galectins (hGal)-1, -3, -4, -7, -8 and -9 bound selectively to glycans, with each galectin recognizing a relatively unique binding motif; by contrast hGal-2 did not recognize HMGs, but did bind to the human blood group A Type 2 determinants on other microarrays. Unlike other galectins, hGal-7 preferentially bound to glycans expressing a terminal Type 1 (Galβ1-3GlcNAc) sequence, a motif that had eluded detection on non-HMG glycan microarrays. Interactions with HMGs were confirmed in a solution setting by isothermal titration microcalorimetry and hapten inhibition experiments. These results demonstrate that galectins selectively bind to HMGs and suggest the possibility that galectin–HMG interactions may play a role in infant immunity.
Keywords: galectins, glycan microarrays, glycan recognition, human milk glycans, shotgun glycomics
Introduction
Human milk provides infants with all essential nutrients, including proteins, lipids and the digestible carbohydrate lactose (WHO 2009). Human milk glycans (HMGs), which contain lactose at their reducing end and are further modified to contain N-acetylglucosamine (GlcNAc), galactose (Gal), fucose (Fuc) and/or sialic acid (as N-acetylneuraminic acid; Neu5Ac), are a major component of human milk (Kunz et al. 2000; Bode 2012). HMGs function as prebiotics that help shape the infant's gut microflora, glycan receptor decoys against pathogenic microbes, regulators of immune responses and even regulators of gene expression in intestinal epithelial cell cultures as well as other cell types (Newburg et al. 2005; Bhargava et al. 2012; Bode 2012; Bienenstock et al. 2013; He et al. 2014). These regulatory functions of HMGs may contribute to the infant health benefits associated with breast-feeding for the first 6 months of life (WHO 2009). Despite the known roles of HMGs in infants, the mechanism(s) by which HMGs regulate immune responses and intestinal epithelial cell gene expression are unknown.
Unlike lactose, HMGs are not appreciably digested in the infant gastrointestinal (GI) tract based on in vitro studies (Engfer et al. 2000; Gnoth et al. 2000), although the gut microflora (ex-certain Bifidobacteria species (Ward et al. 2006; Asakuma et al. 2011)) catabolizes HMGs to some degree. This lack of digestion may allow HMGs to act as physiological and/or immunological regulators in the GI tract. A key family of glycan-binding proteins implicated in immune regulation are the galectins, which are expressed by gut epithelial cells and are known for binding to Gal-rich glycans (Barondes et al. 1994; Cummings and Liu 2009). Thus, we explored whether HMGs may interact in a selective manner with specific galectins.
The Human Protein Atlas project (http://www.proteinatlas.org) (Uhlen et al. 2015) and other studies (Magnaldo et al. 1998; Huflejt and Leffler 2004; Saal et al. 2005) have shown that hGal-2, -3, -4, -7, -8 and -9, but interestingly not hGal-1, are all expressed in epithelial cells of the esophagus, stomach, duodenum, small intestine and/or large intestine under normal conditions, with some extracellular localization. Recent studies also show that a small percentage (∼1%) of HMGs enters the infant's circulation and urine (Rudloff et al. 1996, 2012; Goehring et al. 2014). Thus, HMGs in either the GI tract or blood have the potential to contact galectins in vivo, which may modulate their activity and functions in breast-fed infants.
To explore these interactions of galectins with HMGs, we have exploited the availability of a human milk shotgun glycan microarray containing natural glycans purified from human milk, termed the HM-SGM-v2 array (Ashline et al. 2014; Yu et al. 2014), as well as an array containing defined, simple HMG structures, and the extensive non-HMG glycan microarray from the Consortium for Functional Glycomics (CFG). Studies of galectin binding to glycans both on microarrays and free in solution demonstrate that human galectins, except for hGal-2, bind a unique subset of HMGs. This is the first systematic study of the binding of a lectin family to a specific metaglycome (Cummings and Pierce 2014). The results of this study suggest that galectin–HMG interactions might be relevant to infant health.
Results
Binding of human galectins to the HM-SGM-v2 glycan microarray and the array from the CFG
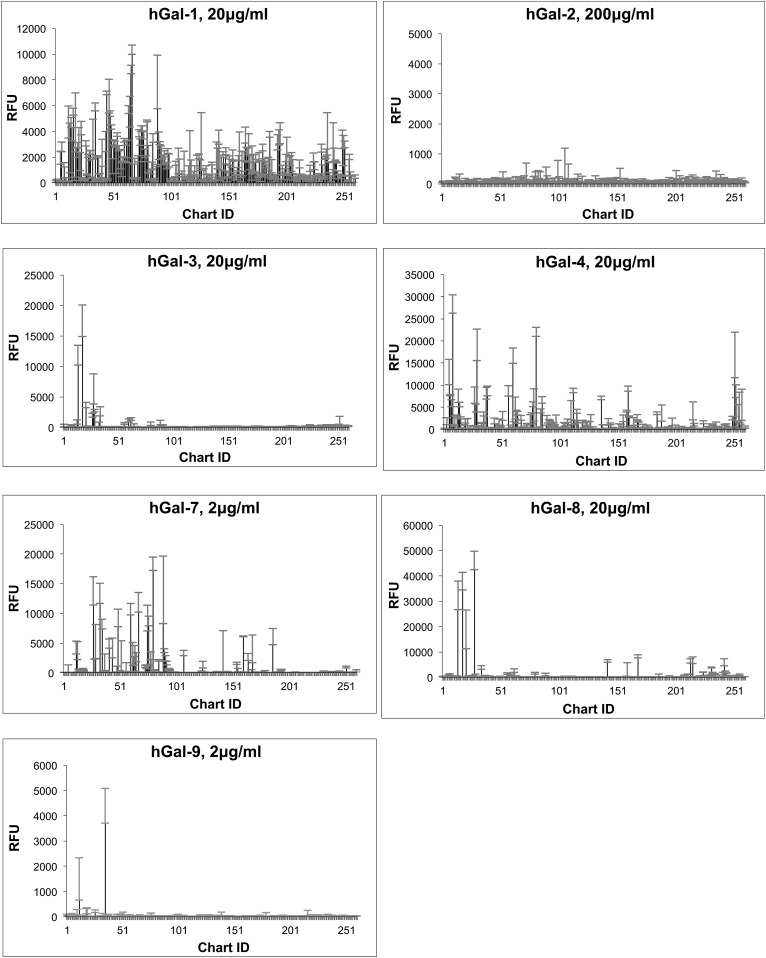
Recombinant hGal-1 (C2S mutant, refer to “Materials and methods” for more information), hGal-2, hGal-3, hGal-4, hGal-7, hGal-8 and hGal-9 were screened on the HM-SGM-v2 array at three concentrations: 2, 20 and 200 μg/mL. The results for one concentration of each galectin are shown in Figure 1 (refer to Supplementary data, File 1 for the results for all three concentrations of each galectin as well as the measured values). The results showed that all the galectins tested, with the exception of hGal-2, bound to glycans on the HM-SGM-v2 array. Most glycans that were strongly bound by galectins demonstrated binding in a dose-dependent manner (Supplementary data, File 1). Non-specific binding was minimal since binding of the galectins to the defined structure 3-fucosyllactose (3-FL), a known non-binder of most galectins (Sparrow et al. 1987), was only at background levels. Moreover, each galectin had a more or less unique binding profile on the HM-SGM-v2 array, suggesting that each galectin appeared to recognize a structural motif within the collection of HMGs. However, there were some general similarities among galectins in binding. For example, only neutral HMG samples were bound, while the sialylated HMGs were typically not bound; the few that were bound likely have a nonsialylated branch.
Fig. 1.
Summary of HM-SGM-v2 microarray binding by galectins. Data are examples of one concentration of each biotinylated galectin screened on the HM-SGM-v2 shotgun microarray, with Streptavidin-Cy5 used for detection. The concentrations fell within the approximate linear range of binding to highlight the strongest bound samples. Error bars represent the standard deviation of binding to four technical replicates printed on the array. Refer to Supplementary data, File 1 for the total data from these screenings at all concentrations of all galectins.
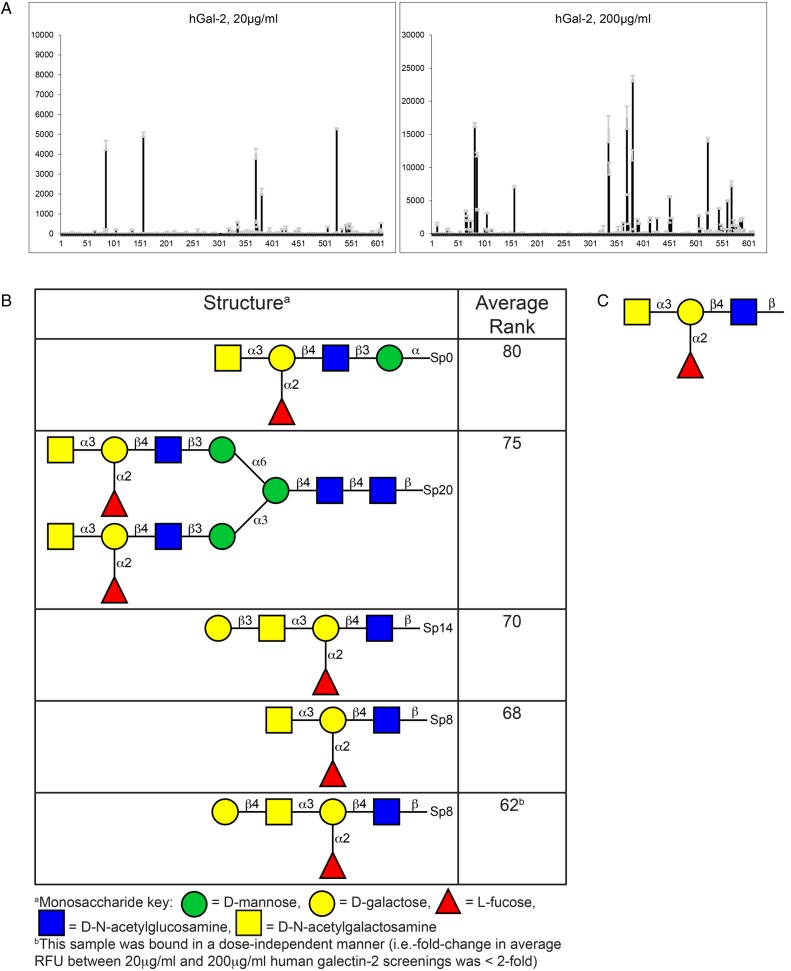
The unexpected lack of hGal-2 binding to HMGs was not due to inactivity of the hGal-2 preparation; we concurrently screened the hGal-2 on the glycan microarray from the CFG, which contains 610 defined glycan structures, and found that it bound to several glycans (Figure 2 and Supplementary data, File 2). On the CFG array, hGal-2 at both 20 and 200 μg/mL bound strongly to five glycans (Figure 2A and B); no significant binding was detected at 2 μg/mL hGal-2. Manual inspection and glycopattern analysis (Agravat et al. 2014) showed that the binding motif for hGal-2 on the CFG array was the Blood Group A Type 2 determinant (GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ-, Figure 2C), a determinant that is mainly restricted to expression on erythrocytes and epithelial tissues and not known to occur on HMGs. Indeed, we did not observe any binding of a Blood Group A-specific antibody on the HM-SGM-v2 array, further confirming the absence of Blood Group A determinants on HMGs. Therefore, the lack of hGal-2 binding to the HM-SGM-v2 was due to the absence of high affinity hGal-2 determinants on HMGs.
Fig. 2.
CFG glycan microarray Version 5.1 results for galectin-2 binding. (A) Galectin-2 binding to the CFG microarray at 20 and 200 μg/mL. Galectin-2 was also screened at 2 μg/mL (not shown here; Supplementary data, File 2), but showed no binding. (B) A list of the top five structures bound by galectin-2. These five structures were bound at both 20 and 200 μg/mL. The fifth structure shown in this table was bound in a dose-independent manner, suggesting this was a nonspecific binder. Note that additional structures were bound at 200 μg/mL galectin-2 only (Supplementary data, File 2), but are not shown here. (C) Proposed glycan-binding motif of galectin-2 based on manual inspection of the structures in (B) and glycopattern analysis. This structure represents the Histo-Blood Group Antigen A Type 2 (i.e. Blood Group A2) determinant. This figure is available in black and white in print and in color at Glycobiology online.
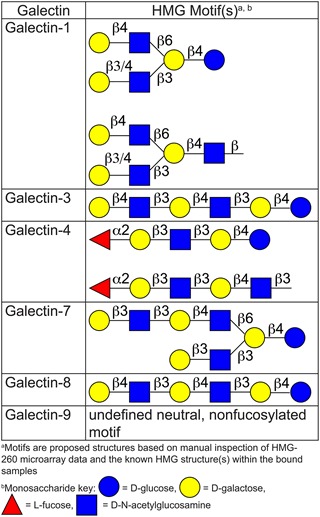
For the remaining galectins, the glycans bound on the HM-SGM-v2 array were manually examined. Using previous glycan sequencing data for these samples (Ashline et al. 2014; Yu et al. 2014), binding motifs were defined for each galectin and found to be relatively unique (Table I). hGal-1 binding to the HM-SGM-v2 array was weak and broad, with a slight preference for branched glycans terminating in Type 1 LacNAc (Galβ1-3GlcNAc) or Type 2 LacNAc (Galβ1-4GlcNAc), although linear structures were also bound. hGal-3 only bound glycans containing at least three repeating Type 2 LacNAc/lactose structures that lacked branched features, which is consistent with previous studies (Hirabayashi et al. 2002). hGal-8 had a similar preference for structures containing at least three linear repeating LacNAc structures without branching, similarly to hGal-3, although the actual specificity of hGal-8 was somewhat different as can be seen in Figure 1, including the weak binding of hGal-8 to sialylated glycans. hGal-9 bound only a relatively restricted panel of glycans unlike the other galectins. The structure of the major glycan (HMO-35) bound by hGal-9 is predicted to be a nonfucosylated, biantennary, neutral HMG structure containing terminating Type 1 LacNAc and Type 2 LacNAc epitopes based on lectin and antibody screening profiles (Yu et al. 2014). Due to limitations in sample material, the actual structures of HMO-35 and the sialylated glycans bound by hGal-8 were not determined by MSn at this point in time. hGal-4 gave a broad pattern of binding, with LNFPI (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and samples containing LNFPI-like determinants being the major structures bound, although nonfucosylated HMGs including LNT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc) and LNnT (Galβ1-4GlcNAcβ1-3Galβ1-4Glc) were also recognized.
Table I.
Major HM-SGM-v2 array motifs recognized by galectins. This table is available in black and white in print and in color at Glycobiology online.
 |
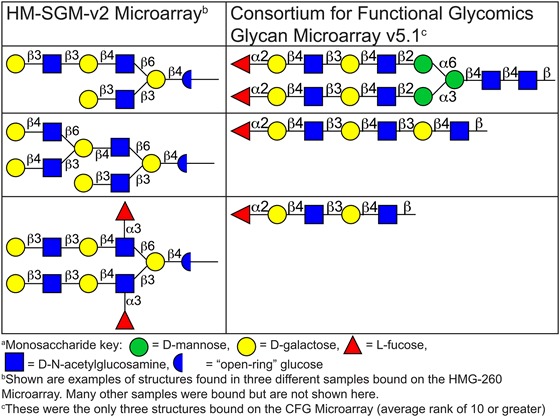
The binding pattern of hGal-7 was interesting as this galectin bound many glycans containing at least one terminal Type 1 LacNAc determinant. Branched glycans containing one or more terminal Type 1 LacNAc determinants were generally slightly preferred over non-branched structures, similarly to hGal-1. The presence of α1-2 fucosylation did not seem to increase or decrease binding to this determinant. Glycans that have a non-reducing Type 2 LacNAc-terminating sequence were typically bound much more weakly, especially linear Type 2 LacNAc-terminating glycans. This binding motif was interesting when compared with hGal-7 binding motif on the CFG array; a comparison of glycans bound on the HM-SGM-v2 and CFG arrays by hGal-7 is shown in Table II. Screening on the CFG array showed that hGal-7 was relatively specific for glycans containing Blood Group H Type 2 (Blood Group H2) determinants and also expressing two Type 2 LacNAc units (Fucα1-2Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-) (Supplementary data, File 2). Type 1 LacNAc-terminating structures are very limited on the CFG array, unlike the HM-SGM-v2 array where this determinant is very abundant based on screenings with an antibody specific for terminal Type 1 LacNAc (Yu et al. 2014). Therefore, the HM-SGM-v2 array helped to further refine the glycan specificity for hGal-7, because such branched Type 1 LacNAc determinants are not present on the CFG microarray.
Table II.
Comparison of the major galectin-7 binders on HM-SGM-v2 and CFG microarraysa. This table is available in black and white in print and in color at Glycobiology online.
 |
Binding of galectins to a defined HMG microarray
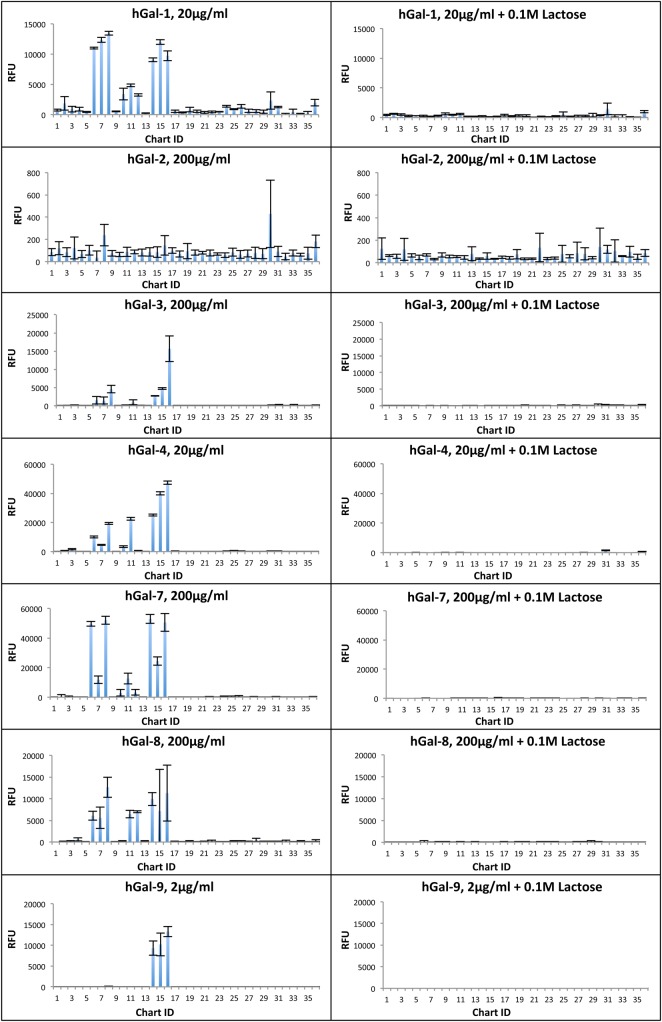
To complement the HM-SGM-v2 shotgun microarray, a second microarray, termed the “defined HMG microarray”, was generated that contained chemically defined HMG structures (as opposed to HMG fractions purified from human milk) that are commercially available. Additionally, galactooligosaccharides (GOS), a prebiotic oligosaccharide mixture that has been proposed as an HMG alternative for infant formula supplementation (Oozeer et al. 2013), were also included on the defined HMG microarray both before and after fractionation to have semi-purified GOS fractions. Galectins were then screened on this array at three concentrations in the presence or absence of lactose, a specific inhibitor of galectins. Figure 3 shows the results of the screening of galectins on the defined HMG array at one concentration with and without lactose (see Supplementary data, File 3 for data from all screenings). As seen on the HM-SGM-v2 array, all galectins except hGal-2 bound glycans on the defined HMG microarray. Binding was typically observed with LNT, LNnT, LNFPI and 2′-FL but not 6′-sialyllactose (6′-SL), 3-FL or any of the GOS samples. Lactose was poorly bound, and 3′-sialyllactose (3′-SL) was only bound (albeit weakly) by hGal-1 and hGal-8. hGal-4, -7 and -9 bound well at relatively low concentrations to glycans on the defined HMG array, while hGal-1, -3 and -8 required much higher protein concentrations for detectable binding. Binding of hGal-8 binding was weak. The HMGs bound by the galectins were largely bound in a dose-dependent manner (Supplementary data, File 3). Co-incubation of galectins with 0.1 M lactose during screening greatly reduced galectin binding (Figure 3 and Supplementary data, File 3), indicating that binding required carbohydrate recognition.
Fig. 3.
Summary of defined HMG microarray binding by galectins. One concentration of each biotinylated galectin screened on the HM-SGM-v2 shotgun microarray (left panels) along with the same concentration of galectin screened in the presence of 0.1 M lactose (right panels). Streptavidin-Cy5 was used for detection. Error bars represent the standard deviation of binding to four technical replicates printed on the array after removing the highest and lowest RFU value of six total technical replicates. Refer to Supplementary data, File 3 for the total data from these screenings at all concentrations of all galectins in the presence and absence of 0.1 M lactose. This figure is available in black and white in print and in color at Glycobiology online.
Influence of the reducing end of HMGs on binding to galectins
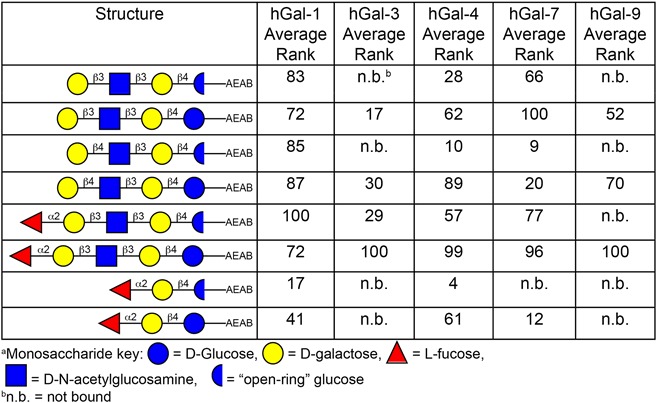
Due to the relatively small mass of the HMGs on the defined HMG array (mostly two to five monosaccharides), we hypothesized that the reducing end glucose of these small glycans might contribute to binding by galectins. The glycans on the HM-SGM-v2 were derivatized with the bifunctional linker 2-amino-N-(2-aminoethyl)-benzamide (AEAB) (Song, Xia, et al. 2009) by reductive amination, which converts the reducing end glucose into a sugar alcohol, an “open-ring” structure. We considered that glucose might possibly be part of the binding motif, in which case the “open-ring” glucose may reduce or even eliminate binding. Thus, the HMGs on the defined HMG array were also derivatized with AEAB in a manner that maintained the reducing end glucose in a cyclic (“closed-ring”) conformation (Song, Lasanajak, et al. 2009) (α/β mixture) and simultaneously printed. Comparison of the corresponding “open-ring” and “closed-ring” glycan derivatives is highlighted in Table III, which shows a clear preference for most galectins to bind to the “closed-ring” conformation of HMGs. In fact, this “closed-ring” reducing end was required for hGal-9 binding on the defined HMG array; hGal-9 bound LNT, LNnT and LNFPI, but only when the reducing end glucose ring was intact. Another dramatic example is hGal-4 and hGal-7 binding to 2′-FL, which was almost completely dependent on the reducing end glucose ring being intact. Therefore, these results suggest that the HM-SGM-v2 average rank data should be interpreted with caution because the “open-ring” glucose structures may bias the results to longer HMG structures, where the reducing end glucose is no longer a part of the galectin binding motif. A caveat to this interpretation is that the derivatization method maintaining the reducing end glucose in a “closed-ring” conformation also introduces an additional glycyl group not found in the reductively aminated structure, and reducing end linkers have been found to directly participate in some galectin binding (Carlsson et al. 2007). However, while this linker may partially increase affinity, the loss of part of the binding motif should produce a much more dramatic effect. Additionally, it has been noted by others that reductively aminated lactose greatly reduces affinity (Hirabayashi et al. 2002). Thus, the major explanation for the preference for “closed-ring” vs. “open-ring” HMG binding is most likely the reducing end glucose conformation, not the longer linker in the “closed-ring” glucose.
Table III.
Comparison of galectin binding to “open-ring” vs. corresponding “closed-ring” HMG structures on defined HMG array. This table is available in black and white in print and in color at Glycobiology online.
 |
Galectin binding to free, underivatized HMGs
While the glycan microarray results demonstrate galectin binding to HMGs, this solid-phase presentation of HMGs is not the “natural” form of HMGs regardless of the linker strategy or reducing status. HMGs are unique because they naturally exist as free, reducing glycans in solution. To ensure that the results seen for galectin–HMG interactions also applied to the more natural solution-based setting, two experimental approaches were taken. In one approach, we examined the ability of free, underivatized HMGs to inhibit galectin binding to the defined HMG array, and in the second approach we measured the binding of galectins to free, underivatized HMGs by isothermal titration microcalorimetry (ITC). hGal-7 and hGal-4 were used for the hapten inhibition experiments as the model prototypical and tandem-repeat galectins, respectively, due to their relatively unique glycan specificities vs. other galectins and robust binding to the HMG microarrays. hGal-7 was used as the model galectin for the ITC studies because hGal-7 only contains a single carbohydrate-binding site and no ITC data exist for hGal-7 with HMGs.
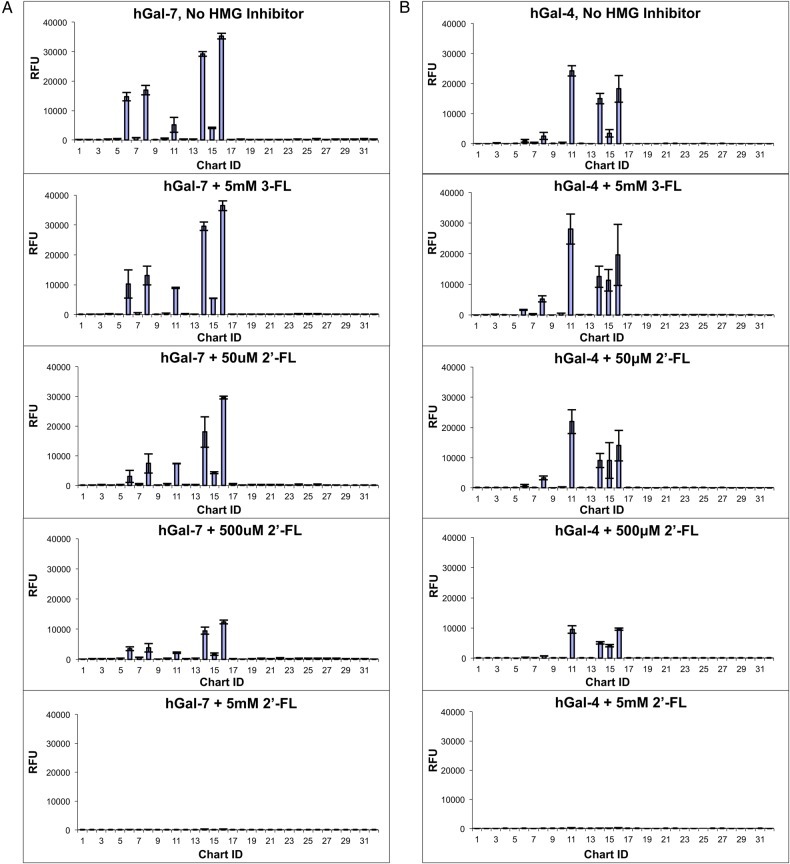
For the free HMG inhibition studies, 20 μg/mL of hGal-4 or hGal-7 were used, which was determined to be the approximate apparent Kd for hGal-4 and hGal-7 binding to most of the major bound glycans on the defined HMG array by screening multiple galectin concentrations on the defined HMG microarray. hGal-4 or hGal-7 was preincubated with 0.05, 0.5 or 5 mM of free HMGs prior to screening on the defined HMG array. An example of these results is shown in Figure 4 (see Supplementary data, File 4 for the total results for hGal-4 and hGal-7). The results show that LNT, LNnT, 2′-FL and LNFPI, but not 3-FL, inhibited hGal-4 and hGal-7 binding to the defined HMG array in a dose-dependent manner; these results are consistent with the binding of these galectins to the defined HMG array where LNT, LNnT, LNFPI and 2′-FL but not 3-FL were bound. For both hGal-4 and hGal-7, the free HMGs caused little or no inhibition at 50 μM, about 50% inhibition at 500 μM, and >95% inhibition at 5 mM. These percent inhibition values mirror the measured Kd of hGal-7 for these HMGs by ITC (see below) and those reported by surface plasmon resonance for hGal-4 (Ideo et al. 2002). Additionally, these results show that the free HMGs not only inhibited binding to the same HMG structure printed on the array but also different structures; all bound HMGs on the defined HMG array were inhibited by a single free HMG. Therefore, these experiments indicate that hGal-4 and hGal-7 bind these HMG structures in solution and that binding to galectins is specific (i.e. via the lactose-binding carbohydrate recognition domain) since the inhibition profiles looked similar to galectins screened on the defined HMG array in the presence of the specific inhibitor 0.1 M lactose. These results can likely be extended to other galectins as well.
Fig. 4.
Summary of inhibition of galectin binding to the defined HMG microarray by free HMGs. Biotinylated galectin-7 (A) and biotinylated galectin-4 (B) were screened on the defined HMG microarray in the presence or absence of 50 μM 2′-FL, 500 μM 2′-FL, 5 mM 2′-FL or 5 mM 3-FL. Streptavidin-Cy5 was used for detection. Error bars represent the standard deviation of binding to four technical replicates printed on the array after removing the highest and lowest RFU value of six total technical replicates. The y-axis is set to the same scale for all graphs. Similar results were seen for 50 μM, 500 μM and 5 mM LNT, LNnT and LNFPI as 2′-FL but are not shown here (refer to Supplementary data, Figure S4, for the total inhibition data for hGal-4 and hGal-7). This figure is available in black and white in print and in color at Glycobiology online.
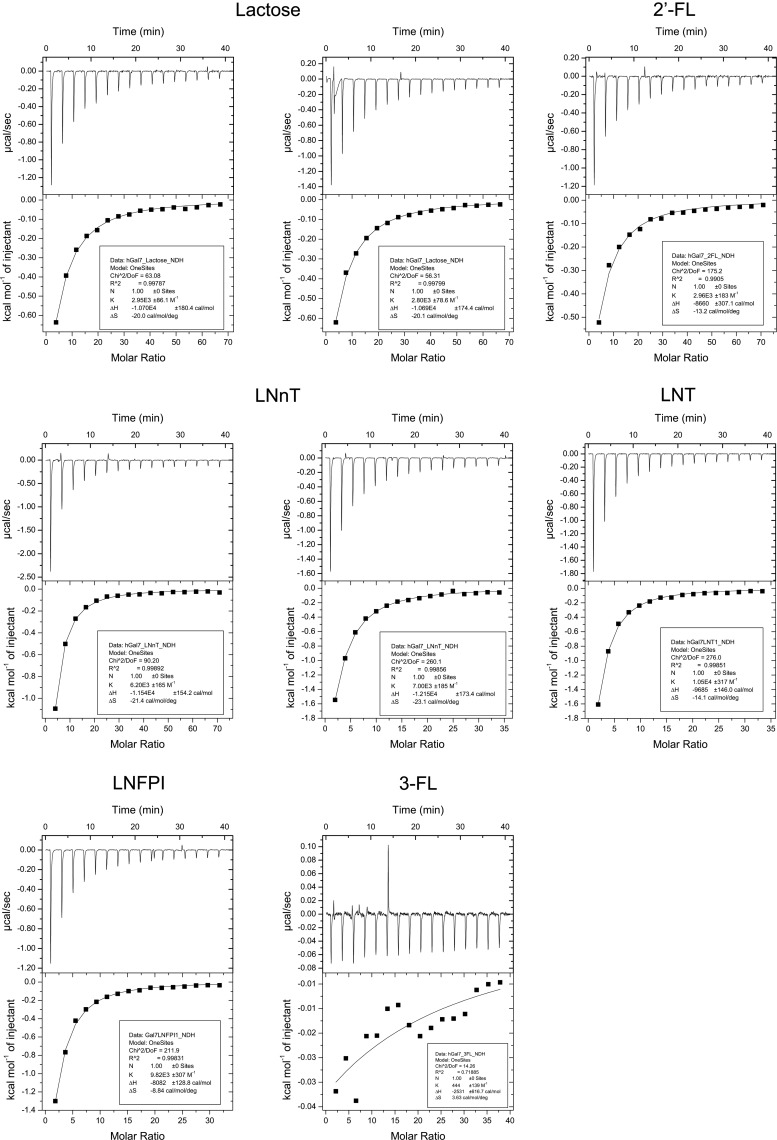
Thermodynamics of galectin binding to free, underivatized HMGs by ITC
To further corroborate solution binding of galectins to HMGs, hGal-7 was tested with the HMGs LNT, LNnT, LNFPI, 2′-FL and 3-FL by ITC. Lactose was also included in this experiment as a reference because ITC data for human hGal-7 with lactose have been published (Ahmad et al. 2002). Due to the relatively low affinity of hGal-7 for these HMGs (>50 μM) in pilot ITC experiments, the “low c-value” method (Turnbull and Daranas 2003) was used to perform ITC, with n fixed to 1.00 based on previous knowledge (Ahmad et al. 2002). The ITC results are shown in Figure 5, and the measured thermodynamic parameters (with associated uncertainties) are presented in Table IV. The results show that all tested glycans except 3-FL showed measurable binding to hGal-7, as seen in the microarrays and free HMG inhibition experiments. The hGal-7-lactose parameters measured at 298 K (Ka = 2.880 × 103 M1 and ΔH = −10.7 kcal/mol) were highly similar to the previous ITC data that did not use the low c-value method (Ka = 2.2 × 103 M−1 and ΔH = −10.6 kcal/mol at 300 K). This finding was important because it not only validated the use of “low c-value” ITC in our hands, but also demonstrated that the Glutathione S-transferase (GST) fusion tag on the recombinant hGal-7 protein used in this study did not significantly affect the solution-based binding studies. An upward trend for 3-FL heat generation was seen in the thermogram and curve, suggesting that hGal-7 may still bind 3-FL but only at very high concentrations (>1 mM), which is significantly higher than the Kd for lactose and 2′-FL. Thus, fucosylation of the 3 –OH group of glucose is disruptive to hGal-7 binding, a feature common to most galectins due to the requirement of a free 3 –OH group on Glc/GlcNAc for binding to most galectins (Lobsanov et al. 1993; Hirabayashi et al. 2002). α1–2 fucosylation of Type 1 LacNAc or Type 2 LacNAc, however, did not affect binding (comparing the results of lactose to 2′-FL and LNT to LNFPI), although a trend toward decreased enthalpic favorability and increased entropic favorability was seen for the α1-2 fucosylated HMGs. Additionally, only a 1.5-fold difference was seen in binding to the Type 1 LacNAc-terminating LNT vs. Type 2 LacNAc-terminating LNnT structures, which is most likely insignificant from a receptor–ligand interaction standpoint, in contrast to the results seen by glycan microarray studies (discussed below). Therefore, the ITC results further confirmed solution binding of galectins to HMGs and also provided previously untested thermodynamic data for hGal-7 binding to glycans, which has helped to better define the glycan specificity of hGal-7.
Fig. 5.
ITC thermograms for galectin-7 with HMGs and curve-fitting results after subtraction of buffer-HMG titration data. Thermograms are 28 μM hGal-7 with 8.6 mM lactose (top left and top center), 9.1 mM 2′-FL (top right), 9.1 mM LNnT (middle left), 4.6 mM LNnT (center), 4.28 mM LNT (middle right), 4.11 mM LNFPI (bottom left) and 4.89 mM 3-FL (bottom center). One-Site Model curve-fitting results are shown in the box below each thermogram.
Table IV.
ITC measurements of human galectin-7 with HMGs
| HMG | Ka (M−1 × 10−3) | Kd (mM) | ΔH (kcal/mol) | ΔS (cal/mol/K) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| Lactose | 2.88 (0.075)a | 0.348 | −10.695 (0.0050) | −20.1 | −4.72 |
| LNnT | 6.6 (0.40) | 0.15 | −11.8 (0.31) | −22.3 | −5.2 |
| LNT | 10.5 (0.32) | 0.0952 | −9.7 (0.15) | −14.1 | −5.48 |
| LNFPI | 9.8 (0.31) | 0.10 | −8.1 (0.13) | −8.8 | −5.4 |
| 2′-FL | 3.0 (0.18) | 0.34 | −8.7 (0.31) | −13.2 | −4.7 |
| 3-FL | n/ab | >1 | n/a | n/a | n/a |
aNumbers in parentheses represent the uncertainty. For lactose and LNnT, the uncertainty is the standard error of the mean (SEM) of two independent experiments. For all other HMGs, the uncertainties are the Origin 7-calculated curve-fitting uncertainties from a single experiment.
bn/a = data not available due to lack of measurable binding.
Absence of detectable galectins in human milk
We also tested whether human milk itself might contain galectins. For this, we utilized dialyzed, defatted human milk, recombinant galectins as standards and defined rabbit anti-sera to the galectins. In western blot analyses, we did not detect any galectins in human milk (Supplementary data, Figure S1), although standard galectins were easily detectable. Using recombinant galectins as standards at different amounts we established that ∼5 ng per 300 μg milk protein loaded onto the gels was the limit of sensitivity by this approach.
Discussion
A major finding in our study is that all but one of the human galectins tested interact with specific HMGs at their physiologically relevant concentrations. While such interactions have been predicted to occur (Bode 2006), this represents the first systematic study to directly test interactions of human galectins with a large variety of HMGs other than a few relatively simple glycans, e.g. lactose, LNnT, 2′-FL (Sparrow et al. 1987; Bachhawat-Sikder et al. 2001; Hirabayashi et al. 2002; Ideo et al. 2002; Carlsson et al. 2007). The results of our study extend these earlier observations and also identify more complex HMGs as additional targets of specific galectins.
While lactose is present at sufficiently high concentrations in human milk (∼0.2 M) to inhibit galectin activity (Saarela et al. 2005), lactose is utilized as a carbohydrate source by the infant and is thus metabolized in the proximal small intestine of the infant by lactase. With the possible exception of newborns and some preterm infants who may not quantitatively digest lactose (Kien et al. 1996; Commare and Tappenden 2007), it is predicted that lactose would only be an efficient galectin binder in the upper GI tract prior to reaching the small intestine. In contrast, in vitro studies suggest that HMGs are not significantly digested by the conditions and human digestive enzymes of the GI tract (Engfer et al. 2000; Gnoth et al. 2000), although digestion can occur by the colonic microflora. Therefore, HMGs may be relatively intact within most of the infant GI tract.
Previous studies have demonstrated galectin expression by human GI tract epithelial cells (Magnaldo et al. 1998; Huflejt and Leffler 2004; Saal et al. 2005), indicating that galectins are expressed in anatomical regions that may come in contact with HMGs. It is unknown, however, whether galectins are properly positioned for contact with HMGs in all cases. For example, some galectins may lack extracellular localization or may only be expressed in deeper levels of the tissue. In addition, whether or not the galectin expression and localization in newborn, infant and toddler tissues mimics that of adult human tissues is unclear. Nonetheless, the current literature on GI tract cell/tissue expression along with membrane localization and secretion of galectins suggest that galectins are likely to be exposed to HMGs and could directly interact physiologically. For example, the Kd of hGal-7 (and other galectins) for simple HMGs is in the high micromolar range, which is below or near the concentration of ∼0.5–5 mM of these simple HMGs (2′-FL, LNT, LNnT and LNFPI) in human milk (Urashima et al. 2012). On the other hand, the ∼1% of HMGs that enters the circulation is likely not at a sufficiently high enough concentration to bind HMGs (Rudloff et al. 1996, 2012; Goehring et al. 2014). Clearly, more studies are needed in the future to explore the positioning and exposure of specific human galectins in the infant gut and the potential of physiological interactions there between HMGs and galectins. As certain galectin–glycan interactions have been found to promote beneficial health effects such as gut homeostasis and oral tolerance (Shan et al. 2013), the importance of galectin–HMG interactions in GI tract physiology will be addressed in future studies.
In addition to HMG binding by galectins, a striking result of our study was the unique binding signature of each galectin on the HMG arrays. This result is consistent with the data observed with galectins screened on the CFG glycan microarray (microarray data available from http://www.functionalglycomics.org/fg/; see also Stowell, Arthur, Mehta, et al. 2008; Stowell et al. 2010) and other shotgun glycan microarrays (Song, Xia, et al. 2009). However, this finding was important because this was the first study to reveal differential glycan specificity of galectins for a human metaglycome (Cummings and Pierce 2014), in this case the human milk metaglycome. These and previous results demonstrate that human galectins have relatively unique glycan specificities, including in the context of a natural metaglycome, despite the ability of all of these galectins to bind, albeit typically with lower affinity, to lactose and LacNAc. This glycan specificity may relate to the fact that each galectin has more or less unique physiological activities (Matsushita et al. 2000; Paclik et al. 2008; Stowell, Qian, et al. 2008; Cerliani et al. 2011; Di Lella et al. 2011). In other words, most human galectins do not appear to be redundant in their activities, which may be at least partially explained by their non-redundant glycan specificities. HMGs might be superior ligands for galectins compared with other oligosaccharides such as GOS and fructooligosaccharides (FOS), which are proposed infant formula additives as an “HMG substitute” particularly because of their prebiotic properties (Oozeer et al. 2013). Interestingly, we found no binding of galectins to GOS on the defined HMG array and thus such components are unlikely to modulate galectin activities.
For hGal-2, no binding was seen on the HM-SGM-v2 or defined HMG microarrays, even though binding to the CFG Glycan Microarray was observed. Although the lack of binding to the HM-SGM-v2 may be due to the absence of high affinity ligands such as the Type 2 Blood Group A determinant, this result was still surprising because rat galectin-2 was previously shown to bind the simple HMG structures including LNnT, LNT and LNFPI, with micromolar affinity (Kd's of 130, 68 and 23 μM, respectively) by frontal affinity chromatography with reductively aminated (and thus “open-ring”) glycans (Hirabayashi et al. 2002). Thus, human and rat galectin-2 may differ in glycan-binding specificity, but in any case these results raise the question as to the physiological relevance of hGal-2 interactions with HMGs in vivo.
Compared with previous studies on HMG–GBP interactions, our study used a rather unique high-throughput approach to explore binding to glycans on multiple microarrays. A key new technology to make this study possible was the development of the HMG shotgun microarray (Yu et al. 2012, 2014). We could identify a novel branched Type 1 LacNAc binding motif for hGal-7 that was not seen on the CFG microarray because, with the exception of simple HMGs, such HMG structures are not present on the CFG glycan microarray. On the other hand, the Type 2 Blood Group H (H2) antigen with at least two LacNAc motifs (i.e. -Fucα1-2Galβ1-4GlcNAcGalβ1-4GlcNAcβ-) was the only motif recognized on the CFG glycan microarray, although binding to other glycan structures, including non-HMG structures with terminal Type 1 LacNAc, could be detected when hGal-7 was screened at a much higher concentration of 200 μg/mL (Supplementary data, File 3). This was an interesting result because, while α1-2 fucosylation of Type 2 LacNAc-terminating glycans greatly improved binding, α1-2 fucosylation of Type 1 LacNAc-terminating glycans and glycans with only one lactose or Type 2 LacNAc repeat had no effect on binding by ITC (Table IV). The mechanism of how α1-2 fucosylation only improves binding to a specific subset of glycans is currently unclear, but this will be addressed in future studies examining the thermodynamics of binding along with co-crystallization of hGal-7 with defined glycan structures with or without α1-2 fucosylation.
Based on the ITC data, there was a trend toward decreased enthalpic favorability and increased entropic favorability for binding to LNT and LNnT (Type 1 LacNAc- and Type 2 LacNAc-terminating structures, respectively), suggesting slightly different mechanisms for hGal-7 binding to Type 1 LacNAc- and Type 2 LacNAc-terminating structures. However, the Kd values for LNT and LNnT binding were <2-fold different. This result was in contrast to the defined HMG array studies, where LNT and LNFPI were consistently bound 3–5-fold better than LNnT. This suggests that hGal-7 may have greater avidity for Type 1 LacNAc vs. Type 2 LacNAc structures, which further suggests a different mechanism of Type 1 LacNAc vs. Type 2 LacNAc binding despite similar affinity. While likely not important from an HMG binding standpoint, this concept may be important in hGal-7 cell surface glycan interactions and warrants further study. Previous ITC studies with hGal-1 and hGal-3 (Bachhawat-Sikder et al. 2001; Ahmad et al. 2002) show that these two galectins can also bind some simple HMGs. Due to the complexity of tandem-repeat galectins (two non-identical CRDs), these were not tested by ITC with HMGs. Additionally, the contribution of the individual CRDs to HMG binding by tandem-repeat galectins was of interest but beyond the scope of this study. Current efforts are underway in the laboratory to measure the affinities of tandem-repeat galectins to HMGs as well as the glycan-binding specificity, affinity and binding mechanism of individual CRDs toward HMGs.
The binding of HMGs to galectins raises another interesting point: are galectins themselves present in human milk? Galectins are present in most human tissues, but no studies have explored their presence in human milk. Using a western blot approach, we did not detect galectins in human milk (<5 ng per 300 μg milk protein, or <0.002% of total milk protein by mass; Supplementary data, Figure S1). This also result corroborates previous proteomics studies that did not identify galectins in human milk, although hGal-7 was detected but only at trace levels in one of these studies (Coscia et al. 2012; Molinari et al. 2012). Therefore, despite their antimicrobial properties (Kohatsu et al. 2006; Stowell et al. 2010, 2014) and other activities that might otherwise be thought to be beneficial to infants, human milk lacks significant quantities of galectins. Such a result might suggest that endogenous galectins in the infant gut might be privileged to interact with HMGs in those locations. An additional issue to be considered in the future is the degree to which HMGs or endogenous galectins in the GI tract might alter the microbiota independently of the HMGs' prebiotic functions. Finally, the expression/localization of galectins and/or galectin glycan ligands in the neonatal and infant GI epithelium are also of interest to determine galectin expression and localization. In any case, our results suggest that differential interactions of HMGs with human galectins might impact infant health and immune development.
Materials and methods
Recombinant human galectin expression and purification
The recombinant human galectins used in this study were hGal-1, -2, -3, -4, -7, -8 and -9. The hGal-1 used had a C2S substitution; this substitution greatly improves stability but does not alter affinity for lactose (Hirabayashi and Kasai 1991). Recombinant hGal-3 cloning was previously described (Massa et al. 1993). Recombinant hGal-9 protein was purchased from R&D. The hGal-2, -4 and -7 CDS were polymerase chain reaction (PCR)-amplified from human genomic DNA, and hGal-8 short isoform CDS were generated by gene synthesis with OptimumGene™ Codon Optimization (Genescript) based on the NCBI reference sequences. Recombinant hGal-1 C2S was generated by PCR-based mutagenesis of wild-type hGal-1 CDS and cloned into pQE-50 at the BamHI and HindIII cut sites. The hGal-2 CDS was cloned into pET11b (Novagen) at the NdeI and BamHI cut sites, hGal-4 CDS into a modified pET29a vector (Novagen), hGal-7 CDS into pGEX-4T-1 (GE Healthcare) at the BamHI and XhoI sites and hGal-8 CDS into pET22b (Novagen) at the NdeI and HindIII cut sites. The galectins were expressed from Escherichia coli M15 (Qiagen), BL21 (DE3) (Life Technologies) or BL21 star (DE3) (Life Technologies). hGal-1, -2, -3, -4 and -8 were expressed as untagged, native proteins. hGal-7 was expressed as a GST-tagged fusion protein, and the GST tag was not removed prior to glycan microarray screening. Sanger sequencing (Genewiz) was used to confirm the nucleotide sequence of each galectin.
Galectins except hGal-9 were expressed for 4 h after induction with 0.1–1 mM isopropyl β-D-1-thiogalactopyranoside (USB) when the OD600 of cultures was 0.5–0.7 with a UV-1700 UV-Vis Spectrophotometer (Shimadzu). The cells were then pelleted by centrifugation and frozen at −20°C overnight. Cell pellets were lysed with CellLytic B Buffer (Sigma) containing 14 mM β-mercaptoethanol (Fisher Scientific), 1 mM lysozyme (Sigma), 10 U/mL Benzonase Nuclease (Novagen) and Complete, Ethylenediaminetetraacetic acid-free protease inhibitor cocktail (Roche). For hGal-2, CellLytic B was replaced with PBS (6.7 mM KH2PO4 pH 7.5, 150 mM NaCl) and sonication was performed. Cell debris was removed by centrifugation at 15,000 × g for 30 min at 4°C. The supernatants were applied to columns containing 10–25 mL lactosyl-Sepharose gel (prepared as previously described (Levi and Teichberg 1981)) that was equilibrated with PBS containing 14 mM β-mercaptoethanol. The columns were washed with PBS + 14 mM β-mercaptoethanol and then eluted with lactose elution buffer (PBS containing 14 mM β-mercaptoethanol and 0.1 M lactose (Fisher Scientific)). Elution fractions positive for protein by absorbance at 280 nm were pooled, and aliquots were stored at −80°C in the lactose elution buffer until immediately before use. SDS-PAGE, Coomassie Brilliant Blue R-250 staining and densitometry analysis using the Gel Analysis feature in ImageJ (http://imagej.nih.gov/ij/) were used to confirm that all galectin preparations were >90% pure.
Generation and printing of on human milk shotgun glycan microarray and defined HMG microarray
The Human Milk Shotgun Glycan Microarray Version 2 (HM-SGM-v2) was generated and printed as previously described (Yu et al. 2014). The HMGs and other glycans used for generating the defined HMG microarray were all purchased from V-Labs, except GOS, which was a gift from Abbott. All structures (except GOS, which is a mixture) are shown in Supplementary data, Table SI. Each HMG was derivatized with AEAB (Song, Xia, et al. 2009) by reductive amination, or with a procedure that maintains the reducing end ring structure, as previously described (Song et al. 2011). GOS was further fractionated into six fractions (F1–F6) with a Shimadzu CBM-20A HPLC system using a Zorbax NH2 normal-phase column, which were detected by absorbance at 330 nm with an SPD-20A UV detector. All the AEAB-labeled glycans shown in Supplementary data, Table SI, as well as the crude GOS mixture and six chromatography fractions, were printed on N-hydroxylsuccinamide (NHS)-activated slides (Schott) as previously described (Heimburg-Molinaro et al. 2011).
Screening of galectins on HM-SGM-v2 shotgun microarray and defined HMG microarray
Prior to screening, all galectins except hGal-9 were biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) according to manufacturer's instructions. Biotinylation was performed in lactose elution buffer as the solvent. Excess biotinylation reagent, free NHS and lactose were removed by passing the biotinylated galectins over a PD-10 desalting column (GE Healthcare) according to manufacturer's instructions, and eluted with PBS. This biotinylation procedure has been found to retain >95% galectin activity as measured by lactosyl-Sepharose binding and not compromise binding specificity when compared with antibody-based detection (refer to the CFG website, http://www.functionalglycomics.org/fg/, for galectin glycan microarray data with anti-galectin antibody detection). Galectins were quantitated by measuring the absorbance at 280 nm and comparing to the theoretical molar absorptivity of each galectin (calculated using the ExPASY Protparam tool, http://web.expasy.org/protparam/). β-mercaptoethanol at ∼14 mM final was then added to each biotinylated galectin preparation.
For glycan microarray screening, the biotinylated galectins and full-length hGal-9 were diluted in TSM binding buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% v/v Tween-20 and 1% w/v BSA) containing 14 mM β-mercaptoethanol. The galectins were screened on the HMG microarrays and, for hGal-2 and -7, the CFG glycan microarray as previously described (Heimburg-Molinaro et al. 2011; Yu et al. 2012). All biotinylated galectins were screened at 2, 20 and 200 μg/mL and detected with Cy5-labeled streptavidin (Molecular Probes) at 0.5 μg/mL on both the defined HMG and HM-SGM-v2 microarrays. Biotinylated hGal-2 was also similarly screened on the CFG Glycan Microarray Version 5.1, but was only screened at 200 μg/mL on the HM-SGM-v2 and defined HMG arrays. hGal-9 was screened at 0.2 and 2 μg/mL and detected with goat anti-human galectin-9 affinity purified polyclonal antibody (R&D) at a final concentration of 20 μg/mL, followed by incubation with Alexa Fluor 488-labeled rabbit anti-goat IgG (Molecular Probes) at 5 μg/mL. hGal-7 was screened on the CFG Glycan Microarray at 0.5, 2.0, 5.0 and 200 μg/mL; only the first three concentrations were used for rank analysis due to detector saturation by the high affinity binders at 200 μg/mL. As controls for specificity, the 200 µg/mL concentration of each galectin (and sometimes the 2 and 20 μg/mL concentrations) was screened in the presence of 0.1 M lactose in TSM binding buffer, but only on the defined HMG microarray since HM-SGM-v2 arrays were highly limited. For hGal-9, only the 2 μg/mL concentration was screened on the defined HMG microarray in the presence or absence of 0.1 M lactose.
Rank analysis was performed as previously described (Smith et al. 2010). Briefly, the glycan structure with the highest RFU at a given concentration was ranked 100 and all other RFU's were normalized to this value. The average rank was calculated as the average of the ranks at all concentrations screened for a given structure. Rank analysis of glycan binding was performed using all three concentrations of galectins screened, if possible. However, the lowest concentration(s) tended to show weak or no binding and thus were excluded from the rank analysis. For this reason, the 2 μg/mL hGal-3 and 2 μg/mL hGal-8 screens on the HM-SGM-v2 array were excluded from rank analysis. For the defined HMG microarray, only the 200 μg/mL concentration was used for rank analysis of hGal-3. For hGal-2 and -7 screening on the CFG glycan microarray, rank analysis and analysis of high binding structures and motifs were performed using Glycopattern (https://glycopattern.emory.edu/) (Agravat et al. 2014). For the HM-SGM-v2 results, all the known structures referred to in this article have been previously sequenced by MSn (Ashline et al. 2014; Yu et al. 2014).
Free HMG inhibition of defined HMG microarray binding by galectins
The HMGs 2′-fucosyllactose (2′-FL), 3-FL, lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT) and lacto-N-fucopentaose I (LNFPI) were purchased from V-Labs. hGal-4 and hGal-7 at 1–4 mg/mL was biotinylated as described above. The biotinylated hGal-4 or hGal-7 was then desalted on a Bio-Gel P10 column to quantitatively remove lactose; the absence of detectable lactose in the desalted galectin preparations was confirmed by phenol–sulfuric acid assay analysis. The biotinylated hGal-4 or hGal-7 at 20 μg/mL (0.55 μM or 0.49 μM, respectively) was preincubated with 50 μM, 500 μM or 5 mM of 2′-FL, 3-FL, LNT, LNnT or LNFPI (V-Labs) in TSM binding buffer containing 14 mM β-mercaptoethanol for 30 min prior to glycan microarray screening. In the case of hGal-7, 3-FL was only used at 5 mM. Then, 20 μg/mL hGal-4 or hGal-7 preincubated with TSM binding buffer containing 14 mM β-mercaptoethanol was used as the mock control. Each sample was then incubated on a separate subarray on a single slide containing 14 arrays. In the case of hGal-4, two slides had to be used; a mock control was included on both slides as the reference control. Glycan microarray screening was otherwise performed as described above, except that the slides were scanned and analyzed using a GenePix 4300A scanner and GenePix Pro 7 software (Molecular Devices).
ITC
The HMGs 2′-FL, 3-FL, LNT, LNnT, and LNFPI were purchased from V-Labs. Lactose was purchased from Fisher Scientific. All these HMGs were dissolved in PBS (6.7 mM KH2PO4 pH 7.4, 150 mM NaCl) and accurately quantitated by the phenol–sulfuric acid assay as previously described (DuBois et al. 1956) with slight modifications; a monosaccharide mixture that best represented each HMG's monosaccharide composition was used as the standard. The measured concentrations of each HMG were as follows: 8.6 mM lactose, 9.1 mM 2′-FL, 4.89 mM 3-FL, 4.28 mM LNT, 4.11 mM LNFPI and 4.6 & 9.1 mM LNnT.
Prior to beginning the ITC, hGal-7 was passed over a Bio-Gel P10 column to quantitatively remove lactose; the absence of detectable lactose in the desalted galectin preparations was confirmed by phenol–sulfuric acid assay analysis. PBS + 14 mM β-mercaptoethanol was the final buffer. The hGal-7 concentration was then measured by the absorbance at 280 nm using the theoretical molar absorptivity calculated using the ExPASY Protparam tool (http://web.expasy.org/protparam/). hGal-7 was measured to be 27.9 μM in the first experiment and 28.1 μM in the second experiment.
ITC was performed using a MicroCal auto-iTC200 instrument (GE Healthcare). An initial water–water titration was performed to ensure that background titration heats were negligible and the noise was low. Four galectin–glycan titrations were performed in a given experiment; lactose and LNnT were included in both experiments to examine inter-experimental variability, which was determined to be minimal. LNnT was also used at two different concentrations in these two experiments (4.6 and 9.1 mM); no significant differences in the curve-fitting parameters were observed. Additionally, the corresponding buffer–glycan titrations were also performed and subtracted from the corresponding galectin–glycan titration data. PBS + 14 mM β-mercaptoethanol eluted from the Bio-Gel P10 column (prior to sample application but after column washing) was used for the buffer–glycan titrations. The data were analyzed with Origin 7 software with manual adjustment of the integration ranges and baseline when deemed appropriate due to the relatively low heats generated. The data were fit to a One Site model with n fixed at 1.00; initial parameters for Ka and ΔH ranged from 1000 to 10,000 and 1000 to 20,000, respectively. One independent experiment of hGal-7 with LNT, LNFPI, 2′-FL and 3-FL was performed, with the reported uncertainties representing the Origin 7-calculated curve-fit uncertainties. Two independent experiments of hGal-7 with lactose and LNnT were performed (although LNnT was used at two different concentrations, but this did not significantly alter the results), with the reported uncertainties representing the standard error of the mean (SEM).
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the National Institutes of Health [P41GM103694 and R24GM09879 to R.D.C.] and Abbott Laboratories [to D.F.S. and R.D.C.].
Conflict of interest statement
R.D.C. and D.F.S. are consultants for Abbott Nutrition. No other authors have a potential conflict of interest.
Abbreviations
2′-FL, 2′-fucosyllactose; 3′-SL, 3′-sialyllactose; 3-FL, 3-fucosyllactose; 6′-SL, 6′-sialyllactose; AEAB, 2-ethyl-N-(aminoethyl)-benzamide; CFG, Consortium for Functional Glycomics; CRD, carbohydrate recognition domain; EDTA, ethylenediaminetetraacetic acid; FOS, fructooligosaccharides; Fuc, fucose; Gal, galactose; GalNAc, N-acetylgalactosamine; GI, gastrointestinal; GlcNAc, N-acetylglucosamine; GOS, galactooligosaccharides; GST, glutathione S-transferase; hGal, human galectin; HMG, human milk glycan; IPTG, isopropyl β-D-1-thiogalactopyranoside; ITC, isothermal titration microcalorimetry; Lac, lactose; LacNAc, N-acetyllactosamine; LNFPI, lacto-N-fucopentaose I; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; Neu5Ac, 5-N-acetylneuraminic acid; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; RFU, relative fluorescence units.
Supplementary Material
Acknowledgements
We thank Melinda Hanes for assistance with and training for the ITC experiments, Jamie Heimburg-Molinaro for critical review of the manuscript, Sandra Cummings and Hong Ju for technical assistance and Oskar Laur at the Emory Custom Cloning Core (ECCC) facility for generating the pET22b-hGal-8 construct.
References
- Agravat SB, Saltz JH, Cummings RD, Smith DF. 2014. GlycoPattern: A web platform for glycan array mining. Bioinformatics. 30:3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N, Gabius HJ, Kaltner H, Andre S, Kuwabara I, Liu FT, Oscarson S, Norberg T, Brewer CF. 2002. Thermodynamic binding studies of cell surface carbohydrate epitopes to galectins-1,-3, and-7: Evidence for differential binding specificities. Can J Chem. 80:1096–1104. [Google Scholar]
- Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 286:34583–34592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashline DJ, Yu Y, Lasanajak Y, Song X, Hu L, Ramani S, Prasad BV, Estes MK, Cummings RD, Smith DF et al. 2014. Structural characterization by MSn of human milk glycans recognized by human rotaviruses. Mol Cell Proteomics. 13:2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat-Sikder K, Thomas CJ, Surolia A. 2001. Thermodynamic analysis of the binding of galactose and poly-N-acetyllactosamine derivatives to human galectin-3. FEBS Lett. 500:75–79. [DOI] [PubMed] [Google Scholar]
- Barondes SH, Castronovo V, Cooper DN, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K et al. 1994. Galectins: A family of animal beta-galactoside-binding lectins. Cell. 76:597–598. [DOI] [PubMed] [Google Scholar]
- Bhargava P, Li C, Stanya KJ, Jacobi D, Dai L, Liu S, Gangl MR, Harn DA, Lee CH. 2012. Immunomodulatory glycan LNFPIII alleviates hepatosteatosis and insulin resistance through direct and indirect control of metabolic pathways. Nat Med. 18:1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienenstock J, Buck RH, Linke H, Forsythe P, Stanisz AM, Kunze WA. 2013. Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions. PLoS One. 8:e76236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. 2006. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 136:2127–2130. [DOI] [PubMed] [Google Scholar]
- Bode L. 2012. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 22:1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S, Oberg CT, Carlsson MC, Sundin A, Nilsson UJ, Smith D, Cummings RD, Almkvist J, Karlsson A, Leffler H. 2007. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology. 17:663–676. [DOI] [PubMed] [Google Scholar]
- Cerliani JP, Stowell SR, Mascanfroni ID, Arthur CM, Cummings RD, Rabinovich GA. 2011. Expanding the universe of cytokines and pattern recognition receptors: Galectins and glycans in innate immunity. J Clin Immunol. 31:10–21. [DOI] [PubMed] [Google Scholar]
- Commare CE, Tappenden KA. 2007. Development of the infant intestine: Implications for nutrition support. Nutr Clin Pract. 22:159–173. [DOI] [PubMed] [Google Scholar]
- Coscia A, Orru S, Di Nicola P, Giuliani F, Varalda A, Peila C, Fabris C, Conti A, Bertino E. 2012. Detection of cow's milk proteins and minor components in human milk using proteomics techniques. J Matern Fetal Neonatal Med. 25 Suppl 4:54–56. [DOI] [PubMed] [Google Scholar]
- Cummings RD, Liu FT. 2009. Galectins. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. Cold Spring Harbor: (NY: ): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Cummings RD, Pierce JM. 2014. The challenge and promise of glycomics. Chem Biol. 21:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lella S, Sundblad V, Cerliani JP, Guardia CM, Estrin DA, Vasta GR, Rabinovich GA. 2011. When galectins recognize glycans: From biochemistry to physiology and back again. Biochemistry. 50:7842–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem. 28:350–356. [Google Scholar]
- Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. 2000. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 71:1589–1596. [DOI] [PubMed] [Google Scholar]
- Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. 2000. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 130:3014–3020. [DOI] [PubMed] [Google Scholar]
- Goehring KC, Kennedy AD, Prieto PA, Buck RH. 2014. Direct evidence for the presence of human milk oligosaccharides in the circulation of breastfed infants. PLoS One. 9:e101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Liu S, Leone S, Newburg DS. 2014. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 7:1326–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg-Molinaro J, Song X, Smith DF, Cummings RD. 2011. Preparation and analysis of glycan microarrays. Curr Protoc Protein Sci. Chapter 12:64:12.10.11–12.10.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J, Hashidate T, Arata Y, Nishi N, Nakamura T, Hirashima M, Urashima T, Oka T, Futai M, Muller WE et al. 2002. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim Biophys Acta. 1572:232–254. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Kasai K. 1991. Effect of amino acid substitution by sited-directed mutagenesis on the carbohydrate recognition and stability of human 14-kDa beta-galactoside-binding lectin. J Biol Chem. 266:23648–23653. [PubMed] [Google Scholar]
- Huflejt ME, Leffler H. 2004. Galectin-4 in normal tissues and cancer. Glycoconj J. 20:247–255. [DOI] [PubMed] [Google Scholar]
- Ideo H, Seko A, Ohkura T, Matta KL, Yamashita K. 2002. High-affinity binding of recombinant human galectin-4 to SO(3)(−)-->3Galbeta1-->3GalNAc pyranoside. Glycobiology. 12:199–208. [DOI] [PubMed] [Google Scholar]
- Kien CL, McClead RE, Cordero L Jr. 1996. In vivo lactose digestion in preterm infants. Am J Clin Nutr. 64:700–705. [DOI] [PubMed] [Google Scholar]
- Kohatsu L, Hsu DK, Jegalian AG, Liu FT, Baum LG. 2006. Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J Immunol. 177:4718–4726. [DOI] [PubMed] [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu Rev Nutr. 20:699–722. [DOI] [PubMed] [Google Scholar]
- Levi G, Teichberg VI. 1981. Isolation and physicochemical characterization of electrolectin, a beta-d-galactoside binding lectin from the electric organ of Electrophorus electricus. J Biol Chem. 256:5735–5740. [PubMed] [Google Scholar]
- Lobsanov YD, Gitt MA, Leffler H, Barondes SH, Rini JM. 1993. X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution. J Biol Chem. 268:27034–27038. [DOI] [PubMed] [Google Scholar]
- Magnaldo T, Fowlis D, Darmon M. 1998. Galectin-7, a marker of all types of stratified epithelia. Differentiation. 63:159–168. [DOI] [PubMed] [Google Scholar]
- Massa SM, Cooper DN, Leffler H, Barondes SH. 1993. L-29, an endogenous lectin, binds to glycoconjugate ligands with positive cooperativity. Biochemistry. 32:260–267. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Nishi N, Seki M, Matsumoto R, Kuwabara I, Liu FT, Hata Y, Nakamura T, Hirashima M. 2000. Requirement of divalent galactoside-binding activity of ecalectin/galectin-9 for eosinophil chemoattraction. J Biol Chem. 275:8355–8360. [DOI] [PubMed] [Google Scholar]
- Molinari CE, Casadio YS, Hartmann BT, Livk A, Bringans S, Arthur PG, Hartmann PE. 2012. Proteome mapping of human skim milk proteins in term and preterm milk. J Proteome Res. 11:1696–1714. [DOI] [PubMed] [Google Scholar]
- Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 25:37–58. [DOI] [PubMed] [Google Scholar]
- Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, Boehm G, Knol J. 2013. Intestinal microbiology in early life: Specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr. 98:561S–571S. [DOI] [PubMed] [Google Scholar]
- Paclik D, Lohse K, Wiedenmann B, Dignass AU, Sturm A. 2008. Galectin-2 and -4, but not galectin-1, promote intestinal epithelial wound healing in vitro through a TGF-beta-independent mechanism. Inflamm Bowel Dis. 14:1366–1372. [DOI] [PubMed] [Google Scholar]
- Rudloff S, Pohlentz G, Borsch C, Lentze MJ, Kunz C. 2012. Urinary excretion of in vivo (1)(3)C-labelled milk oligosaccharides in breastfed infants. Br J Nutr. 107:957–963. [DOI] [PubMed] [Google Scholar]
- Rudloff S, Pohlentz G, Diekmann L, Egge H, Kunz C. 1996. Urinary excretion of lactose and oligosaccharides in preterm infants fed human milk or infant formula. Acta Paediatr. 85:598–603. [DOI] [PubMed] [Google Scholar]
- Saal I, Nagy N, Lensch M, Lohr M, Manning JC, Decaestecker C, Andre S, Kiss R, Salmon I, Gabius HJ. 2005. Human galectin-2: Expression profiling by RT-PCR/immunohistochemistry and its introduction as a histochemical tool for ligand localization. Histol Histopathol. 20:1191–1208. [DOI] [PubMed] [Google Scholar]
- Saarela T, Kokkonen J, Koivisto M. 2005. Macronutrient and energy contents of human milk fractions during the first six months of lactation. Acta Paediatr. 94:1176–1181. [DOI] [PubMed] [Google Scholar]
- Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M et al. 2013. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 342:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Song X, Cummings RD. 2010. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 480:417–444. [DOI] [PubMed] [Google Scholar]
- Song X, Heimburg-Molinaro J, Smith DF, Cummings RD. 2011. Derivatization of free natural glycans for incorporation onto glycan arrays: Derivatizing glycans on the microscale for microarray and other applications (ms# CP-10-0194). Curr Protoc Chem Biol. 3:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Xia B, Smith DF, Cummings RD. 2009. Fluorescent glycosylamides produced by microscale derivatization of free glycans for natural glycan microarrays. ACS Chem Biol. 4:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. 2009. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 16:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow CP, Leffler H, Barondes SH. 1987. Multiple soluble beta-galactoside-binding lectins from human lung. J Biol Chem. 262:7383–7390. [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B et al. 2010. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 16:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S et al. 2014. Microbial glycan microarrays define key features of host–microbial interactions. Nat Chem Biol. 10:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. 2008. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem. 283:10109–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Qian Y, Karmakar S, Koyama NS, Dias-Baruffi M, Leffler H, McEver RP, Cummings RD. 2008. Differential roles of galectin-1 and galectin-3 in regulating leukocyte viability and cytokine secretion. J Immunol. 180:3091–3102. [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Daranas AH. 2003. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 125:14859–14866. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A et al. 2015. Proteomics. Tissue-based map of the Human Proteome. Science. 347:1260419. [DOI] [PubMed] [Google Scholar]
- Urashima T, Asakuma S, Leo F, Fukuda K, Messer M, Oftedal OT. 2012. The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv Nutr. 3:473S–482S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 72:4497–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2009. Infant and Young Child Feeding: Model Chapter. Geneva, Switzerland: WHO Press; p. 112. [Google Scholar]
- Yu Y, Lasanajak Y, Song X, Hu L, Ramani S, Mickum ML, Ashline DJ, Prasad BV, Estes MK, Reinhold VN et al. 2014. Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Mol Cell Proteomics. 13:2944–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Mishra S, Song X, Lasanajak Y, Bradley KC, Tappert MM, Air GM, Steinhauer DA, Halder S, Cotmore S et al. 2012. Functional glycomic analysis of human milk glycans reveals presence of virus receptors and embryonic stem cell biomarkers. J Biol Chem. 287:44784–44799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.