Abstract
Heparanase is a β-d-glucuronidase which cleaves heparan sulfate chains in the extracellular matrix and on cellular membranes. A dysregulated heparanase activity is intimately associated with cell invasion, tumor metastasis and angiogenesis, making heparanase an attractive target for the development of anticancer therapies. SST0001 (roneparstat; Sigma-Tau Research Switzerland S.A.) is a non-anticoagulant 100% N-acetylated and glycol-split heparin acting as a potent heparanase inhibitor, currently in phase I in advanced multiple myeloma. Herein, the kinetics of heparanase inhibition by roneparstat is reported. The analysis of dose-inhibition curves confirmed the high potency of roneparstat (IC50 ≈ 3 nM) and showed, at higher concentrations, a Hill coefficient consistent with the engagement of two molecules of inhibitor. A homology model of human heparanase GS3 construct was built and used for docking experiments with inhibitor fragments. The model has high structural similarity with the recently reported crystal structure of human heparanase. Different interaction schemes are proposed, which support the hypothesis of a complex binding mechanism involving the recruitment of one or multiple roneparstat chains, depending on its concentration. In particular, docking solutions were obtained in which (i) a single roneparstat molecule interacts with both heparin-binding domains (HBDs) of heparanase or (ii) two fragments of roneparstat interact with either HBD-1 or HBD-2, consistent with the possibility of different inhibitor:enzyme binding stoichiometries. This study provides unique insights into the mode of action of roneparstat as well as clues of its interaction with heparanase at a molecular level, which could be exploited to design novel potential inhibitor molecules.
Keywords: heparanase, homology modeling, kinetic inhibition analysis, roneparstat, SST0001
Introduction
Mammalian heparanase is an endo-β-d-glucuronidase that hydrolyzes heparan sulfate (HS) chains from HS proteoglycans (HSPGs), which represent fundamental constituents of the cell membrane as well as of the extracellular matrix (ECM). In physiological conditions, the cleavage of HS polysaccharides by heparanase triggers a variety of functional processes intimately associated with tissue remodeling, ranging from the degradation of the ECM to favor the penetration of immune cells at sites of inflammation, to the release of HS-bound growth factors, cytokines and chemokines that stimulate migration and proliferation of endothelial cells and fibroblasts. Not surprisingly, an abnormal, upregulated ECM-disassembling activity driven by this specific glycoside hydrolase (GH) has been related to tumor cell invasion, enhanced metastatic progression and sustained tumor angiogenesis, making heparanase an attractive target for the development of novel antitumor agents (Pisano et al. 2014). Furthermore, given its role in a plethora of biological processes, heparanase was also found to be involved in the development of other, non-cancer related, pathologies and heparanase inhibitors were thus proposed as a novel potential therapeutic opportunity, especially for those diseases induced by an aberrant enzyme expression and activity (Ferro 2013; Simeonovic et al. 2013).
The mammalian genome encodes two heparanase enzymes, i.e., heparanases 1 and 2, with only the former able to cleave HS glycosaminoglycans from HSPGs. The cDNA of human heparanase 1 was first cloned in the late 1990s (Vlodavsky et al. 1999) and was found to code for a retaining endo-β-d-glucuronidase of 543 amino acids, which preferably cleaves the glycosidic bond between a glucuronic acid (GlcA) and an N-sulfated-glucosamine (GlcNS) at the non-reducing end of highly sulfated HS segments (Pikas et al. 1998). The enzyme is initially expressed as a pre-pro-form, which is subsequently processed into a catalytically inactive pro-form through the excision of the initial Met1-Ala35 peptide. The resulting pro-form is then subjected to a proteolytic cleavage by cathepsin L (Abboud-Jarrous et al. 2008), which removes the fragment Ser110-Gln157 to yield the active mature form of the enzyme, composed by a heterodimer of a 8 kDa peptide (Gln36–Glu109) noncovalently associated to a 50 kDa subunit (Lys158–Ile543). Sequence-based analyses and mutagenesis studies were applied to infer the overall three-dimensional (3D) architecture of the enzyme as well as the location and organization of the ligand-binding site. Indeed, given its high sequence similarity with other clan A GHs (Hulett et al. 2000), it was postulated that human heparanase might bear the prototypical (β/α)8 TIM-barrel domain characteristic of this clan, which should host the catalytic site. The location of the active cleft was elucidated by extensive site-directed mutagenesis studies, which helped in identifying not only the two catalytic amino acids but also the protein segments involved in ligand recognition. In particular, Glu225 and Glu343 were recognized as the catalytic proton donor and acceptor, respectively (Hulett et al. 2000), and two amino acid sequences, named heparin-binding domain (HBD) 1 (Lys158–Asp171) and 2 (Gln270–Lys280), proved to be fundamental for ligand stabilization (Levy-Adam et al. 2005). Very recently, the crystal structure of human heparanase confirmed the predicted 3D architecture of the protein dimer and the location and composition of the binding site (Wu et al. 2015).
Besides its implication in cancer growth and development, a growing body of evidence indicates a key role of heparanase in a variety of other pathological conditions, including inflammation, diabetes, diabetic nephropathy, atherosclerosis, and hereditary multiple exostoses that lead to skeletal dysfunctions (Vlodavsky et al. 2013; Huegel et al. 2015). For this reason, heparanase inhibitors represent promising therapeutic agents for a wide range of diseases. Thus far, drug development campaigns aimed at identifying heparanase inhibitors yielded two main classes of non-peptide and non-nucleic acid compounds, i.e., small molecules and HS mimetics (Ferro et al. 2004; McKenzie 2007). The former group comprises natural substances, such as RK-682 and siastatin-B, as well as compounds identified from high-throughput screening and subsequent structural optimization. However, despite the remarkable inhibitory activity exhibited by some of these derivatives, none of them progressed to clinical trials. The class of HS-mimetic inhibitors, which structurally resemble the polysaccharide chain of HS, is highly heterogeneous spanning from modified heparins and other sulfated polysaccharides to derivatives characterized by a lower molecular weight, such as sulfated oligosaccharides and cyclitol-based pseudo-sugars. Among them, PI-88, PG545, M402 and SST0001 (roneparstat) showed promising antitumorigenic activities (Basche et al. 2006; Dredge et al. 2011; Ritchie et al. 2011; Zhou et al. 2011) and are currently evaluated in clinical trials for the treatment of a variety of cancer types, including myeloma, metastatic pancreatic cancer and hepatocellular carcinoma.
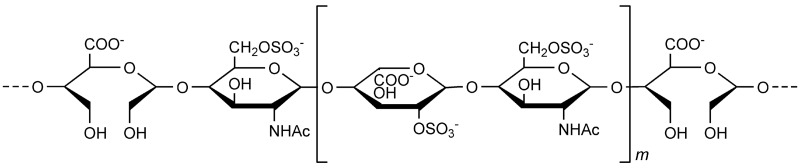
Roneparstat (SST0001, 100NA, RO-H, Figure 1) is a high molecular-weight heparin derivative obtained by glycol splitting of the nonsulfated hexuronic acids (GlcA and IdoA) present in a fully N-acetylated heparin. Glycol-split portions and 2-O-sulfated iduronic acid (IdoA2S) residues were estimated in a 1:3 ratio (Naggi et al. 2005). Roneparstat is considered one of the most potent heparanase inhibitors developed to date, with IC50 in the single-digit nM range, currently being tested in Phase I clinical trial for the treatment of multiple myeloma (Ritchie et al. 2011). While comprehensive studies on the biological activity of roneparstat were conducted in vitro and in vivo, the molecular mechanism of action of roneparstat was not described. Herein, we report the experimental evaluation of the kinetics of roneparstat inhibition of human heparanase based on the fondaparinux assay. The results thus obtained were rationalized by means of molecular modeling simulations. In particular, a 3D structure of human heparanase was built and docking experiments conducted with roneparstat led us to propose a putative binding mode consistent with experimental data.
Fig. 1.
Chemical structure of heparanase inhibitor roneparstat. Given the heterogeneous saccharide composition, only a representative sequence is depicted, with m ranging from 1 to 5. Adapted from Casu et al. (2008).
Results and discussion
Inhibition of human heparanase by roneparstat
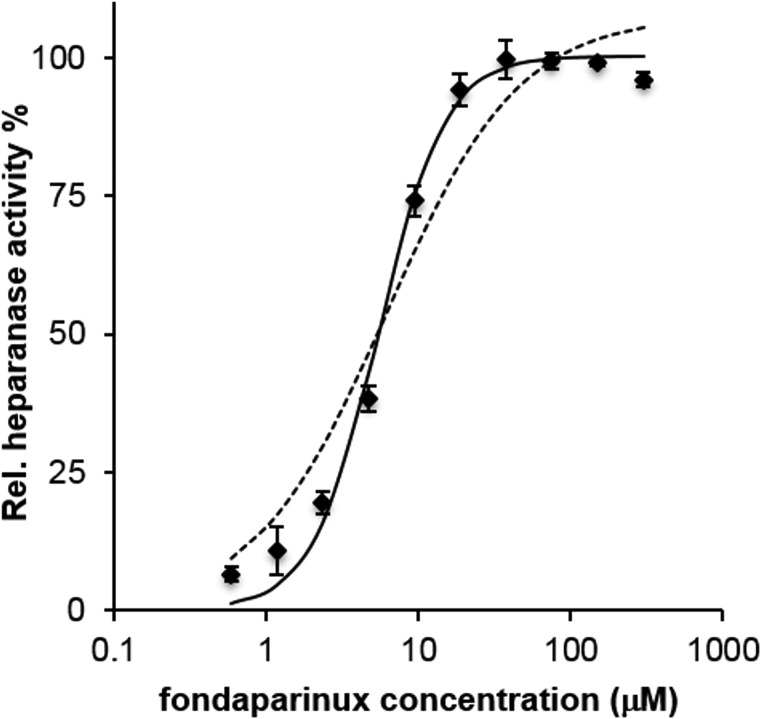
Heparanase cleaved the substrate fondaparinux with a sigmoidal dependence of velocity on the logarithm of substrate concentration. Fitting relative velocity to a classical Michaelis–Menten model:
gave a Km value of 6.18 µM with R2 = 0.96. Fitting to a Hill-like model:
with Hill coefficient fixed at the value of 2, gave a relevant improvement of R2, which resulted of 0.99, with a Km value of 5.45 µM (Figure 2). The steeper dependence of hydrolysis velocity on substrate concentration is a first sign that heparanase catalytic mechanism is probably different from a simple classical model, involving formation of an 1:1 enzyme-substrate complex.
Fig. 2.
Heparanase relative activity, measured as amount of substrate cleaved at 3 h, vs. fondaparinux initial concentration (log scale). Hashed line: fitted Michaelis–Menten function. Continuous line: fitted Hill function with nH = 2 (see text). Vertical bars represent standard deviation of single measurements (n = 4).
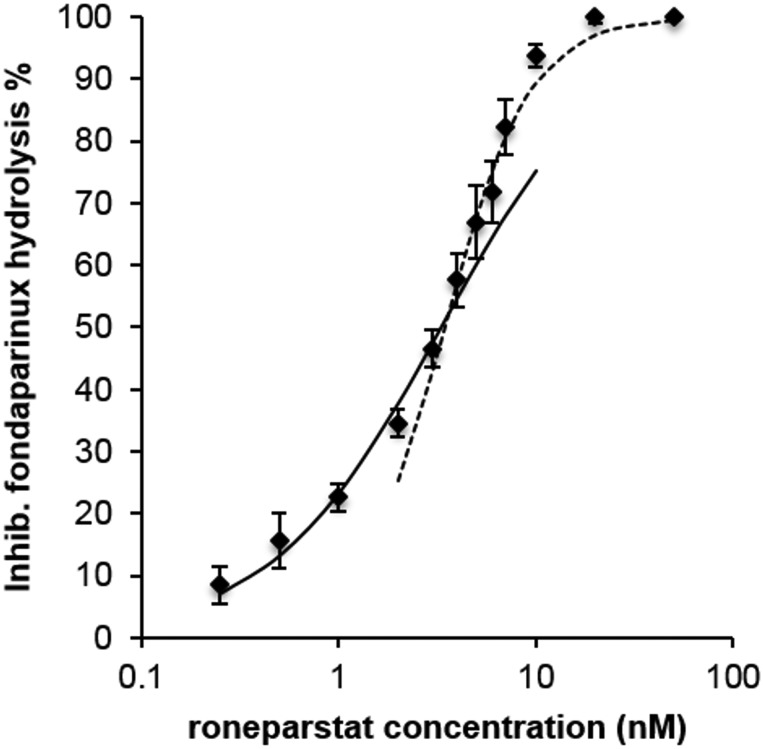
Starting from a fixed concentration of fondaparinux, increasing concentrations of the inhibitor roneparstat inhibited substrate hydrolysis with a curve that, when fitted to a single curve with equation:
gave non-optimal fit, with R2= 0.95 and a fitted Ki value of 2.42 nM. The plotted points could be better fitted by two separate curves (Figure 3). A first one, with the abovementioned equation, well fitted the data corresponding to concentrations of roneparstat ≤4 nM (R2> 0.99, Ki = 3.30 nM). For data corresponding to roneparstat concentrations ≥4 nM, a Hill-like function:
with nH = 2 was necessary to have good fit (R2> 0.99, Ki = 3.45 nM). Although this complex behavior could be interpreted in different ways (Copeland 2005), a possible explanation is that roneparstat acts as a pure competitive inhibitor at lower concentrations, while at higher ones a new mechanism occurs, which involves in some way the interference of more than one (maybe two) molecules of inhibitor. Apart from this complex dose-inhibition curve, roneparstat was confirmed as a potent heparanase inhibitor, giving 50% inhibition of fondaparinux hydrolysis at a concentration close to 3 nM (IC50 ≈3 nM).
Fig. 3.
Inhibition of fondaparinux (75 µM) hydrolysis, expressed as % of maximum inhibition, vs inhibitor concentration (log scale). Continuous line: Hill function with nH = 1, fitted to data with roneparstat concentrations ≤4 nM. Hashed line: Hill function with nH = 2, fitted to data with roneparstat concentrations ≥4 nM. Vertical bars represent standard deviation of single measurements (n = 4).
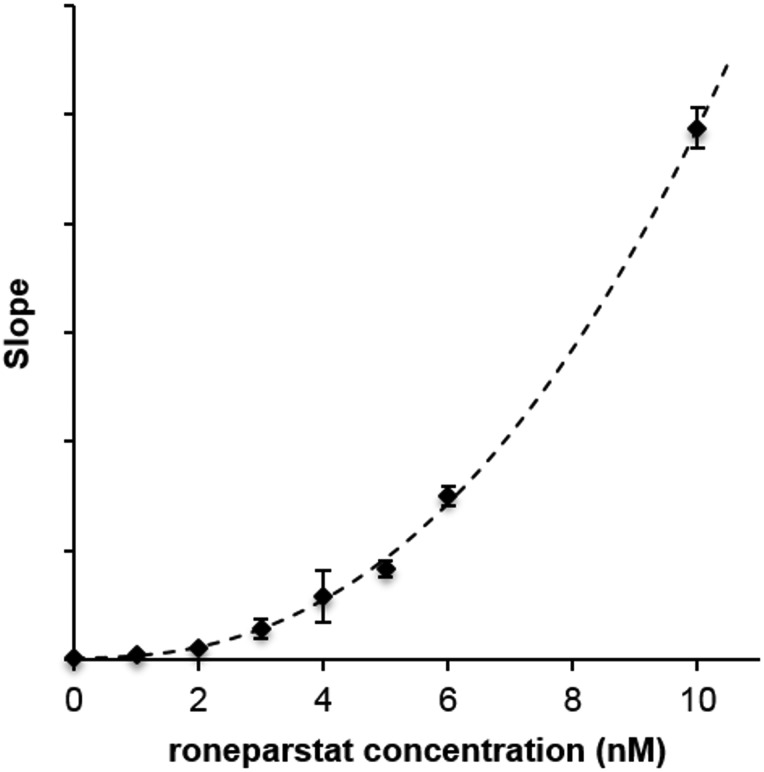
To consider different substrate and inhibitor concentrations at the same time, the slopes of double-reciprocal plots were fitted to a Hill-like model similar to that described by Cao et al. (2010) and employed by Hammond for the analysis of the heparanase inhibitor PG545 (Hammond et al. 2013). The equation used to fit the slope data (Figure 4, hashed line R2= 0.999) was slightly modified to use a single fitting parameter, A, to replace the ratio Km/Vmax.
Fig. 4.
Inhibition of fondaparinux hydrolysis by roneparstat. The slopes of double-reciprocal plots (fondaparinux starting concentrations in the range 9.375–600 µM) vs. inhibitor concentrations. Hashed line: fitted Hill function. Vertical bars represent standard deviation of single measurements (n = 4).
The Ki and Hill coefficient (nH) determined from the curve fit were 1.12 nM and 2.41, respectively.
Thus, both the analysis of the inhibition curve at fixed concentration of substrate (Figure 3) and of the slopes of double-reciprocal plots reveal that, at higher concentrations of inhibitor, monomolecular interaction between heparanase and roneparstat is not consistent with the observed increase of inhibition slope. On the other hand, in this condition a bimolecular reaction, involving two molecules of roneparstat and an undefined number of protein molecules, is consistent with the Hill coefficient around 2, employed to fit the higher concentration data in Figure 3 and obtained as a fitting parameter from double-reciprocal plots.
Molecular modeling
Heparanase homology model
Homology models of human heparanase reported so far have been built from template structures belonging to GH families different from that of heparanase (Hulett et al. 2000; Zhou et al. 2006; Gandhi et al. 2012; Sapay et al. 2012), which do not possess a recognition site for the carboxylic group of GlcA within the cleavage site. Recently, the X-ray structure of a family 79 GH, β-glucuronidase from Acidobacterium capsulatum, has become available (Michikawa et al. 2012) and was used to build a model of the catalytically inactive pre-pro form of heparanase (Vinader et al. 2013).
Starting from this β-glucuronidase as the template structure, we built a new homology model of the catalytically active GS3 construct (Nardella et al. 2004) of human heparanase in which the 8 and the 50 kDa subunits are linked through the interposition of a GSGSGS (GS3) peptide. The model was exploited (i) to investigate the recognition mechanism of substrates and (ii) to provide a possible interaction mode for the inhibitor roneparstat consistent with experimental kinetic data. In particular, the GS3-heparanase model was built using the crystal structure of β-glucuronidase co-crystallized with GlcA (PDB code 3VNZ) as the template, to take advantage of the proper arrangement of the side chains of active site residues around GlcA, which was maintained in the homology model. The alignment between heparanase and β-glucuronidase sequences is reported in Supplementary data, Figure S1. The overall identity and similarity percentages between the template and the GS3 construct sequences were 22 and 36%, respectively, which increased to 24 and 46% considering only the eight α-helix and the eight β-strand segments constituting the TIM-barrel region of the template.
The 3D structure of heparanase GS3 construct was built with Prime (Schrodinger 2013a) and energy-minimized to relax residual steric clashes, to adjust residue geometries and to optimize the hydrogen bond network stabilizing GlcA within the active site (see Materials and methods). The Ramachandran plot of the protein (Supplementary data, Figure S2) showed 78.3, 18.3 and 2% of non-proline and non-glycine residues located within the most favorable, additionally allowed and generously allowed regions, respectively, with only six amino acids (1.4%) in disallowed regions. In particular, two of them (Ser88 and Lys107) were located within two distinct loop segments of the 8 kDa domain of heparanase, whereas the remaining four amino acids (Gln198, Lys214, Val289 and Leu483) were situated in loop regions belonging to the 50 kDa subunit. All six residues were located >20 Å far from the catalytic site and their influence on ligand placement and stabilization should be negligible. Residues forming HBD-1 and HBD-2 domains, as well as the two catalytic amino acids Glu225 and Glu343 resided in either most favorable or additionally allowed regions of the Ramachandran plot.
The structure of the heparanase model was characterized by two subdomains, i.e., the (β/α)8-barrel domain and a noncatalytic portion arranged in a β-sandwich fold (Figure 5A). The 8 kDa subunit of heparanase protruded within the TIM-barrel, where it formed β-strands 1 and 2, and α-helix 1. The two subunits were linked through the GS3 peptide, which is positioned on the same side of the protein surface where the binding cleft is located (Figure 5A). The 50 kDa chain formed the remaining six β-strands and seven α-helices of the TIM-barrel domain, as well as eight of the nine β-strands of the noncatalytic β-sandwich portion of the enzyme.
Fig. 5.
(A) Overview of the heparanase–GlcA complex structure. The 8 and 50 kDa subunits are depicted with yellow and purple carbons, respectively, while the arrows indicate the location of the GS3 peptide. The TIM-barrel and β-sandwich domains of heparanase are contoured with red and green lines, respectively. GlcA is represented as spheres with white carbons. (B) Close-up view of the catalytic site of heparanase (white cartoons and carbons). The two catalytic residues are colored in yellow and GlcA in green.
The catalytic residues Glu225 and Glu343 resided in two highly conserved sequences among clan A GHs and were positioned at the opposite sides of the channel branching from the catalytic pocket. In particular, Glu225 laid at the N-terminus of the loop connecting β-strand 4 and α-helix 4, while Glu343 was inside β-strand 7. A number of neighboring residues stabilized the orientation of the catalytic residues within the active cleft (Figure 5B). The carboxylate group of Glu343 was involved in two hydrogen bond interactions with the side chains of Arg93 (not shown in Figure 5B) and Tyr298, while the side chain of Glu225 formed a polar interaction with the imidazole ring of His296, which was modeled in its protonated imidazolium form (see Materials and methods) due to the peculiar environment surrounding its imidazole ring, pointing towards the carboxylate groups of Glu225 and Asp267 with its δ and ϵ nitrogen atoms, respectively. It is worth mentioning that experimental studies on another hydrolase belonging to the same GH clan as heparanase showed that the histidine residue corresponding to His296 of human heparanase was likely to be protonated in both free and inhibitor-bound forms of the enzyme (Schubert et al. 2007).
The narrow channel containing the catalytic site branched towards HBD-1 and HBD-2 segments, widening at its terminal ends in a funnel-like shape. The calculated electrostatic potential of the region encompassing the main binding channel and its neighboring residues was highly electropositive, due to the presence of several basic amino acids, including Arg70, Lys79, Lys98, Lys158, Lys159, Lys161, Lys231, Lys232, Arg272, Arg273, Lys274 and Arg303 (Supplementary data, Figure S3). This is consistent with the substrate specificity exhibited by heparanase, which preferably recognizes and cleaves glycosidic bonds located within segments of HS chains rich in sulfate groups (Pikas et al. 1998, Okada et al. 2002; Mao et al. 2014). Most of the basic residues delimiting the binding groove were located in the two terminal portions of the channel, within HBD-1 and HBD-2 domains. In the heparanase model, the HBD-2 domain (Gln270–Lys280) extended from the C-terminus of the loop connecting β-strand 5 with α-helix 5 to the first half of α-helix 5, retaining an overall structural arrangement similar to that observed for the corresponding sequence of the template β-glucuronidase (Tyr219–Lys230). Residues Gln270 and Pro271 were in close contact with α-helix 6 and β-strand 6 of the TIM-barrel, while Arg272, Arg273 and Lys274 were mainly solvent-exposed and defined one edge of the heparanase binding groove. The remaining amino acids (Thr275–Lys280) laid within α-helix 5, protruding either towards the inner portion of the (b/a)8-barrel domain or towards the solvent. HBD-1 (Lys158–Asp171) was located within the protein portion exhibiting the most remarkable differences with the template structure, i.e., the loop linking β-strand 2 with α-helix 2 of the TIM-barrel (Supplementary data, Figure S4). Indeed, in the template β-glucuronidase, this long loop (41 residues, Gly79-Glu119) occludes the active site in the proximity of the non-reducing end of the saccharide molecule. The corresponding loop sequence of the GS3-heparanase construct is shorter, being constituted by 32 residues (Supplementary data, Figure S1). In our model, this portion extended above the plane of the catalytic residues, forming a portion of a wall delimiting the binding groove. The side chains of the residues in the N-terminal region of HBD-1 (Lys158–Lys161) pointed towards the binding channel, in accordance with the experimentally verified role of these amino acids in ligand stabilization (Levy-Adam et al. 2005). The different length and architecture of the loop portion connecting β-strand 2 with α-helix 2 of the (β/α)8-barrel is consistent with the different substrate specificity of template β-glucuronidase and heparanase. Indeed, while the template β-glucuronidase is an exo-acting GH and thus cleaves the glycosidic bond located at one end of the polysaccharide chain, heparanase is an endo-acting GH (Nakajima et al. 1984) and therefore acts on a glycosidic bond located within the long HS chain. Consequently, the loop connecting β-strand 2 with α-helix 2 leads to the formation of a small and buried binding pocket in the template β-glucuronidase, whereas it shaped an extended and solvent-exposed binding groove in heparanase, which accommodates the non-reducing portion of the substrate.
The GlcA molecule laid in a sub-pocket located in the middle of the main binding channel, close to the catalytic residues, where it formed an extended network of hydrogen bonds with neighboring amino acids (Figure 5B), similar to those present in the template structure, being the active site residues highly conserved between the two proteins. The carboxylate group of GlcA formed a bidentate interaction with Gly349 and Gly350, and an additional hydrogen bond with the side chain of Tyr391. The hydroxyl group located at position 1 of the tetrahydropyran ring of GlcA was hydrogen bonded to the carboxylate group of Glu225, while the hydroxyl group at position 2 acted as hydrogen bond acceptor and donor for the side chains of Asn224 and Glu343, respectively. The hydroxyl group at position 3 interacted with the backbone of Thr97 through its oxygen atom, while its hydrogen was located close to Asp62 side chain (not shown in Figure 5B). Inspection of the modeled complex also revealed the presence of different hydrophobic interactions between the ligand and adjacent amino acids. In particular, the side chains of Gln383 (not shown in Figure 5B) and Tyr298, which delimited the floor of the sub-pocket where GlcA is accommodated, stabilized the tetrahydropyran ring, while the side chains of Thr97 and Tyr348 were found in close proximity of the C2 and the carboxylate carbon of the ligand, respectively.
The crystal structure of a heparanase from the bacterium Burkholderia pseudomallei was recently reported (Bohlmann et al. 2015). This enzyme is constituted by a single chain with a (β/α)8 TIM-barrel domain and a β-sandwich C-terminal portion. Its primary sequence has low homology with that of human heparanase, with ∼24% sequence identity. Sequence identity of the GS3 construct of human heparanase with this protein, and with the β-glucuronidase from Acidobacterium capsulatum used as the template for our model, are similar (18 vs. 22%, respectively). Superposition of Burkholderia pseudomallei heparanase with our model of human heparanase highlights an overall good resemblance of the 3D structure of the protein regions involved in substrate recognition (Supplementary data, Figure S5A). In particular, the two acidic amino acids involved in the catalytic process and the loop region where two NH groups have been observed to interact with the carboxylate of GlcA in the template structure are strictly superposed (Supplementary data, Figure S5B). Also the backbones of regions surrounding the putative glycan-sulfate binding sites are remarkably similar, with minor differences emerging in the regions interacting with the non-reducing end of the substrate. In our model, the HBD-1 loop is longer and closer to the catalytic site, compared with a corresponding loop in Burkholderia pseudomallei heparanase, due to the presence of the GS3 peptide (Supplementary data, Figure S5C).
Substrate docking studies
The heparanase GS3 model was first submitted to docking studies with substrate molecules, to validate the modeled structure and reshape the HBD regions. In fact, the enzyme model was built retaining the small GlcA within its catalytic site, and adaptation of the protein 3D structure to the bulky structure of its substrates was necessary. We docked a prototypic heptasaccharide (1, Table I) and the reference substrate fondaparinux (Table I) (Choay et al. 1983), used to experimentally assess the kinetics of roneparstat inhibitory activity, evaluating the consistency of their accommodation within the enzyme structure with available experimental observations.
Table I.
Structures of heparanase substrates used in docking simulations
| Substrate | Saccharide units |
|---|---|
| 1 | OMe-GlcNS-IdoA2S-GlcNAc6S-GlcA-GlcNS-IdoA2S-GlcNS-OMe |
| Fondaparinux | GlcNS6S-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S-OMe |
GlcA, β-d-glucuronic acid; GlcN, α-d-glucosamine; IdoA, iduronic acid; NAc, N-acetyl; NS, N-sulfate; 2S, 2-O-sulfate; 3S, 3-O-sulfate, 6S, 6-O-sulfate; OMe, O-methyl.
The saccharide composition of substrate 1 was devised from a recent study, which investigated the substrate specificity of heparanase through the identification of its most abundant cleavage products by mass spectrometry analysis (Mao et al. 2014). This heptasaccharide has a central GlcA residue bound to a GlcNS saccharide, which represents the preferred cleavage site of heparanase. These two units are flanked by repeating IdoA2S-glucosamine disaccharides, abundant in HS chains. As standard docking simulations performed on both substrates did not provide ligand poses within the binding groove of the enzyme, compounds 1 and fondaparinux were docked within the heparanase model structure applying a customized induced-fit docking (IFD) protocol (Sherman et al. 2006), which takes into account both protein and ligand flexibility (see Materials and methods). In particular, the GlcA residue of the substrates was first placed within the catalytic site by superposition with the GlcA molecule derived from the crystallized template structure. Then, an in-site conformational analysis of the ligand molecule was performed, selecting those conformations able to occupy the binding groove and performing mutual optimization of the geometries of the substrate and of the surrounding amino acid side chains.
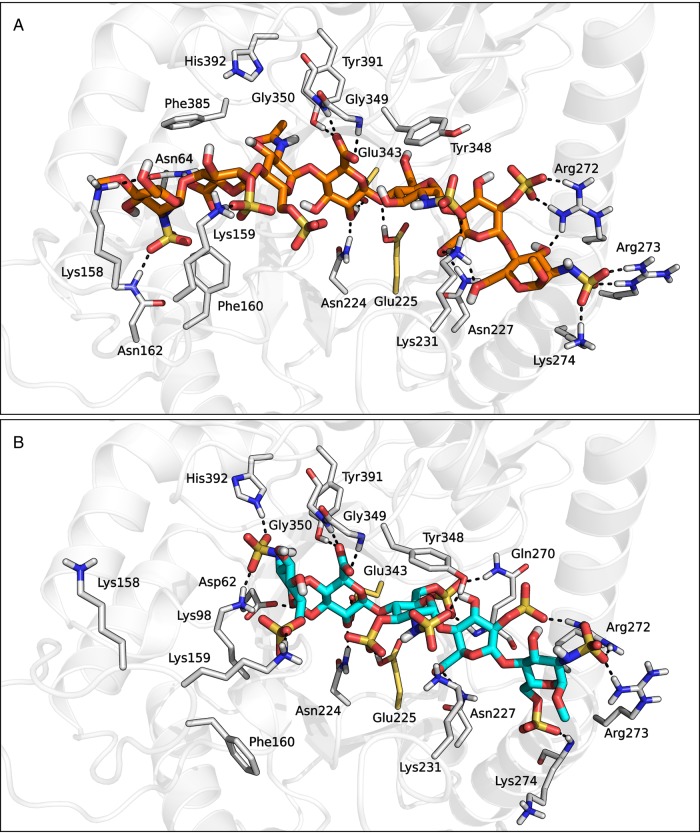
Substrate molecules were accommodated in the middle portion of the main binding groove spanning from HBD-1 to HBD-2, where they formed an extended network of hydrogen bonds and hydrophobic interactions with neighboring residues (Figure 6). In both complex structures, the central GlcA−1 [saccharides are labeled from −n to +n, with minus and plus signs indicating the non-reducing and the reducing end of the chain, respectively, and with the cleavage site being placed between positions −1 and +1 (Davies et al. 1997)] was located within the catalytic pocket of heparanase and formed the same pattern of interactions observed for the GlcA saccharide in the initial homology model structure. In particular, the hydroxyl group at position 2 of GlcA−1 was hydrogen bonded to the carboxylate group of Glu343, while the protonated carboxylic acid of Glu225 interacted with the oxygen atom linking GlcA−1 to the subsequent GlcNS(6S)+1. Furthermore, the carboxylate function of the 2-O-sulfated iduronic acid located at position +2 (IdoA2S+2) was involved in two hydrogen bond interactions with the side chains of Asn227 and Lys231, while the tetrahydropyran ring of the glucosamine at position +1 interacted with the phenyl ring of Tyr348 through hydrophobic interactions. Moreover, in accordance with site-directed mutagenesis, the non-reducing and reducing ends of both substrate ligands formed a wide network of polar interactions with HBD-1 and HBD-2 domains, respectively. For example, the terminal GlcNS6S−2 of fondaparinux formed a salt-bridge interaction with the protonated side chain of Lys159 via its 6-O-sulfate group (Figure 6B), while compound 1 simultaneously interacted with both Lys158 and Lys159 side chains through the 2-O-sulfate group of IdoA2S−3 and the 6-hydroxyl group of GlcNS−4, respectively (Figure 6A). In addition, the terminal GlcNS+3 and GlcNS6S+3 saccharides of substrate 1 and fondaparinux, respectively, formed strong electrostatic interactions with Arg272, Arg273 and Lys274 belonging to the HBD-2 domain.
Fig. 6.
Heparanase-compound 1 (A) and heparanase-fondaparinux (B) complex structures obtained from the IFD procedure. This figure is available in black and white in print and in color at Glycobiology online.
To evaluate the stability of the docked pose of substrate 1, a 150 ns-long molecular dynamics (MD) simulation was conducted on the heparanase-substrate 1 complex structure within an explicit water box. The time-evolution of the root-mean-square deviation (RMSD) calculated on the Cα of α-helices and β-strands reached a plateau after 110 ns around 3.3 Å (Supplementary data, Figure S6), which is a reasonable value given the poor sequence identity between heparanase and the template protein (Fan and Mark 2004). The average root-mean-square fluctuations (RMSF) calculated on Cα of residues constituting HBD-1 and HBD-2 were 1.45 ± 0.84 and 1.45 ± 0.65 Å, respectively, indicating that these portions did not undergo remarkable structural rearrangements and retained a stable conformation throughout the simulation. The calculated RMSF value decreased when measured on Cα of amino acids located within 5 Å from the central GlcA−1 saccharide (0.99 ± 0.44 Å), further proving the structural stability of the catalytic domain of the enzyme. RMSD value calculated on ligand heavy atoms clearly showed that the substrate molecule not only had a faster equilibration process compared with the protein structure, but also experienced smaller fluctuations, as it reached a stable conformation having an RMSD value around 2 Å after 15 ns (Supplementary data, Figure S6). The ligand molecule undertook slight movements within the enzyme binding groove and the overall network of hydrogen bond interactions was conserved throughout the simulation. During MD simulation, the polar groups of substrate 1 formed both stable and transient interactions with different hydrogen-bonding partners within the enzyme active cleft (Supplementary data, Figure S7). The most remarkable structural rearrangement detected during MD simulation was the reorganization of the loop segment connecting β-strand 6 with α-helix 6 of the TIM-barrel domain and, in particular, of the arginine residue located at position 303. Indeed, Arg303, which was initially located >15 Å far from the 2-O-sulfate group of IdoA2S+2, underwent a shift from its initial position towards the tip of α-helix 6, leading to the formation of a network of salt-bridge interactions with the exposed sulfate groups of the substrate. In addition to the 2-O-sulfate function of IdoA2S+2, Arg303 could form a transient ionic interaction with the N-sulfate group of GlcNS+1. The results of the MD simulation conducted on the heparanase-substrate 1 complex structure supported the overall geometrical quality and dynamic stability of the homology model and the reliability of the predicted binding modes of substrates, suggesting that the model structure could effectively be used to rationalize available experimental data for known heparanase ligands. Therefore, the heparanase model optimized around substrate 1 by means of the IFD procedure was used to investigate the interaction mode of roneparstat at a molecular level.
Comparison between heparanase model and crystal structure
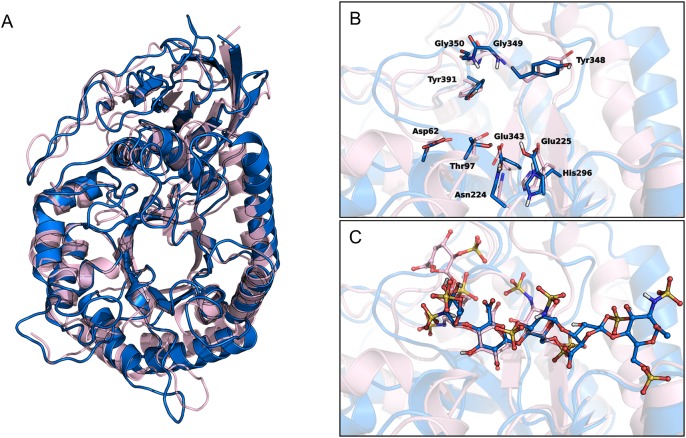
While this manuscript was under review, the crystal structure of human heparanase was reported, showing the predicted (β/α)8 TIM-barrel and β-sandwich domains (Wu et al. 2015). Superposition of our model with the crystal coordinates showed a remarkable similarity in the overall 3D structure (Figure 7A), with an RMSD value calculated for Cα atoms of α-helices and β-strands of 5.08 Å, which decreased to 3.23 Å when considering only the TIM-barrel domain. The main differences between crystal and model lied in the HBD-1 region, as the modeled structure is a monomeric GS3 construct, while the crystal structure is a heterodimer of 8 and 50 kDa chains. Modeled HBD-2 was superposable to the crystallized one. Focusing on the ligand-binding site, the RMSD calculated for Cα atoms of residues within 4 Å from catalytic Glu225 and Glu343 was 1.76 Å. The arrangement of Glu225 and Glu343 and their interaction pattern with the surrounding amino acids were the same in the two structures (Figure 7B). Comparison of the arrangement of fondaparinux saccharides at positions −2, −1 and +1 with the corresponding ones in the crystallized ligands showed a remarkable similarity, with an RMSD value of 0.52 Å calculated for heavy atoms of the three pyran rings (Figure 7C). Moreover, the common functional groups of the modeled fondaparinux and the crystallized ligands showed the same pattern of interactions with neighboring protein residues. This also applied to Lys159 of HBD-1 that undertook a hydrogen bond with the 6-O-sulfate group of the glucosamine at position −2 in both modeled and crystal structure. This strict similarity supports the reliability of our model structure and improves the confidence in the accuracy of the results obtained for the inhibitor roneparstat.
Fig. 7.
(A) Superposition of human heparanase crystal structure (pink ribbons, PDB code: 5E9C) and the homology model of GS3 construct (blue ribbons). (B and C) Close-up views of the ligand-binding site of heparanase crystal in complex with ligand dp4 (ΔHexA2S-GlcNS6S-IdoA-GlcNS6S, pink carbons) and heparanase GS3 construct in complex with fondaparinux (blue carbons). (B) depicts the residues delimiting the catalytic sites, (C) shows the ligand molecules dp4 and roneparstat.
Roneparstat docking studies
Given the intrinsic structural complexity of the inhibitor molecule (Figure 1), which is constituted by a heterogeneous assembly of uronic acids and glucosamines with different degrees of sulfation (Naggi et al. 2005), a representative subunit of the polysaccharide chain was identified and selected for docking simulations (compound 2, Table II).
Table II.
Structures of roneparstat fragments used in docking simulations
| Compound | Saccharide units |
|---|---|
| 2 | OMe-GlcNAc-IdoA2S-GlcNAc6S-Gsplit-GlcNAc-IdoA2S-GlcNAc-OMe |
| 3 | OiPr-GlcNAc-IdoA2S-GlcNAc-OiPr |
| 4 | MeO-GlcNAc6S-Gsplit-GlcNAc-OMe |
Gsplit, glycol-split; GlcN, α-d-glucosamine; IdoA, iduronic acid; NAc, N-acetyl; 2S, 2-O-sulfate; 6S, 6-O-sulfate; OMe, O-methyl, OiPr, O-isopropyl.
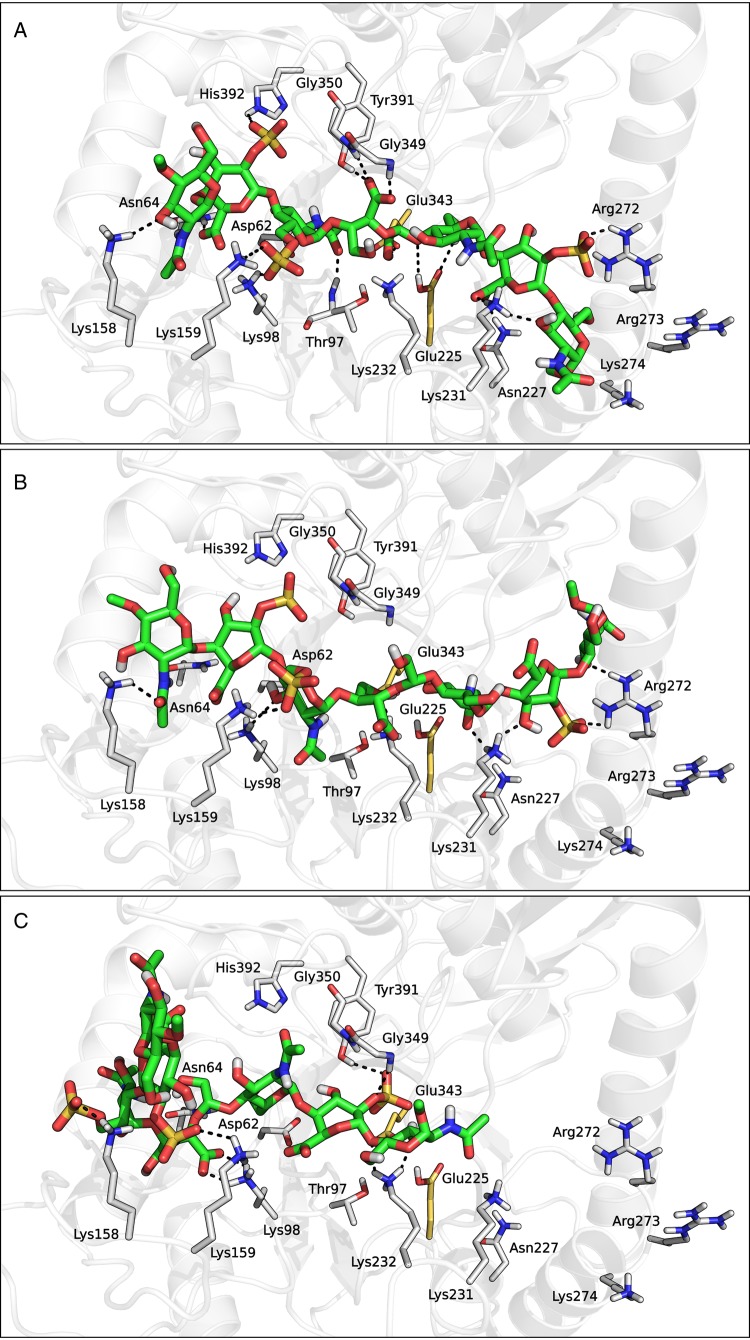
Repeated docking calculations on compound 2 returned three different clusters of poses (Figure 8). In the first cluster, the ligand molecule was accommodated in the middle of the binding groove, with the glycol-split fragment placed within the catalytic pocket (Figure 8A). In these poses, the glycol-split fragment formed an extended network of hydrogen bond contacts with the surrounding residues, including a bidentate interaction between its carboxylate group and the NH backbone hydrogens of Gly349 and Gly350, and between its two hydroxyl groups and the side chains of Glu225 and Glu343. Moreover, the terminal saccharide units of 2 formed a number of polar and hydrophobic interactions with the residues lining HBD-1 and HBD-2 segments, including Lys158, Lys159, Phe160, Arg272, Lys274 and Thr275. A similar pattern of contacts with HBD-1 and HBD-2 was also detected in the docking solutions belonging to cluster 2 (Figure 8B). However, in these poses the glycol-split fragment was not accommodated within the catalytic pocket of heparanase, but laid above the plane defined by the two catalytic residues. Finally, the poses belonging to cluster 3 greatly differed from those of clusters 1 and 2. These poses did not span the central portion of the binding groove and were thus not able to simultaneously interact with both HBDs. As an example, the pose depicted in Figure 8C was bent around the glycol-split unit, which acted as a pivot point, and was accommodated within the main binding channel with only one half of the molecule, while the remaining portion pointed towards the bulk solvent. The ligand segment within the binding groove extensively interacted with residues from HBD-1, while the catalytic cavity of the enzyme was occupied by one IdoA2S saccharide. The best docking solutions belonging to the three clusters had their non-reducing end towards HBD-1 and reducing end towards HBD-2, as seen for the substrates. We also obtained solutions with opposite orientation, but these were characterized by lower stereo-electronic complementarity with the binding site, as apparent from their lower mean Docking Score (approx. −7.5 vs approx. −8.9 kcal/mol for the best ones).
Fig. 8.
Representative docking solutions of compound 2 taken from clusters 1 (A), 2 (B) and 3 (C). This figure is available in black and white in print and in color at Glycobiology online.
To assess the validity of the binding accommodations obtained for compound 2, 150 ns-long MD simulations were conducted for the three docking solutions reported in Figure 8. During the simulations, the three poses retained their starting interactions with the protein residues involved in the recognition of the trisaccharide portions of compound 2 (Supplementary data, Figure S8). In the case of pose 1, the glycol-split fragment remained within the catalytic site, while the interaction between its carboxylate group and the two glycines 349 and 350 was lost. Pose 2 slightly increased its bent arrangement, strongly interacting with Arg303, which stabilized the neighboring negatively charged groups through salt-bridge interactions. Pose 3, characterized by a disaccharide terminus pointing towards the bulk solvent, conserved the position of the portion interacting with the protein structure, while the solvent-exposed disaccharide terminus rearranged into a new conformation after ∼100 ns.
The results of MD simulations thus confirmed that roneparstat can interact with heparanase with different binding modes. In some cases, one inhibitor molecule might simultaneously bind both HBDs of the same protein, as seen in poses 1 and 2 of compound 2. In such cases, the stoichiometry of interaction would be 1:1, which is consistent with a dose-inhibition curve showing a Hill coefficient close to 1. Alternatively, one polysaccharide chain might bind only one HBD segment on the heparanase structure (e.g., in pose 3). Given the length of the molecule, in such a case roneparstat could also interact with one HBD segment of another heparanase molecule. Particularly at increasing concentrations of the inhibitor, one protein entity could thus host two fragments belonging to different molecules of roneparstat. This model could provide a possible explanation for the remarkable improvement in fitting, for data points describing the percentage of inhibition of fondaparinux hydrolysis as a function of roneparstat concentration, obtained with the use of two distinct curves for low (≤4 nM, Hill coefficient = 1) and high (≥4 nM, Hill coefficient = 2) inhibitor concentrations. In fact, a Hill coefficient larger than 1 could result from binding of different molecules of inhibitor to different patches on enzyme surface (Prinz and Schönichen 2008), and steep Hill slopes are often observed for aggregating compounds (Acker and Auld 2014).
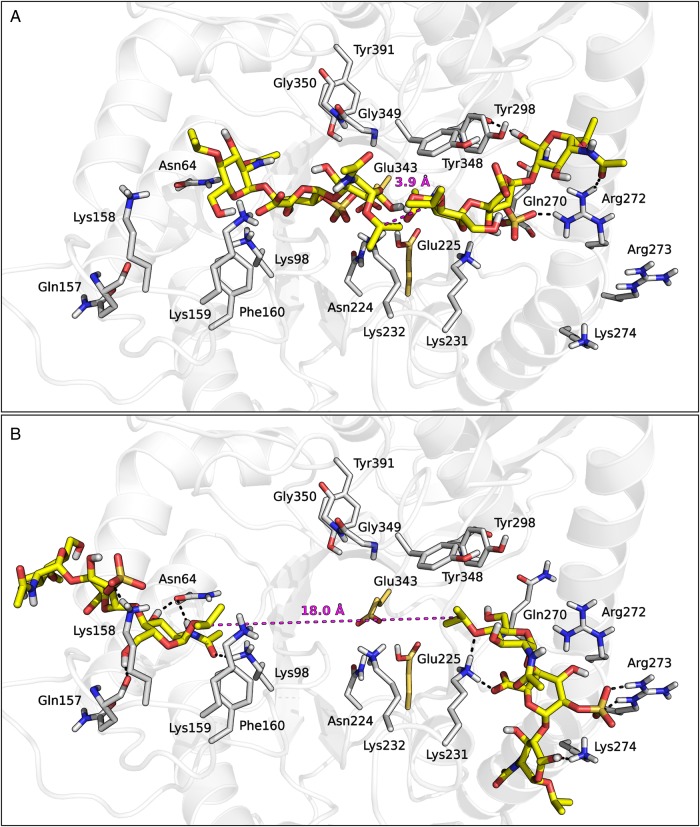
To check whether simultaneous accommodation of multiple inhibitor molecules within the heparanase structure was feasible, we set up a series of docking experiments with the trisaccharide 3 (Table II). This ligand was selected as the shortest representative unit of roneparstat fragments flanking the glycol-split, to evaluate its ability to interact with HBD-1 and HBD-2 regions. The terminal O-isopropyl substituents were inserted to mimic the branched structure of the glycol-split fragment. One hundred docking poses were collected for 3 within both HBD-1 and HBD-2 and clustered according to their pattern of interaction with surrounding residues (see Supplementary data). Cluster analysis identified five and four clusters of docking poses for HBD-1 and HBD-2, respectively (Supplementary data, Figures S9 and S10). The nine clusters had comparable average Docking Scores (Supplementary data, Table SI), but they spanned highly dissimilar protein regions (Supplementary data, Figures S9 and S10). Due to the small size of compound 3 and the flexibility of the docking protocol, poses with both orientations of their reducing and non-reducing ends were obtained. Given the docking results obtained for the roneparstat fragment 2 (vide supra), in the following part of the analysis, we only considered those consistent with its preferred docking mode, i.e., with the reducing end towards HBD-2 and the non-reducing end towards HBD-1.
We wondered if there were couples of solutions consistent with the hypothesis of either 1:1 heparanase:roneparstat interaction or accommodation of multiple inhibitor molecules within the heparanase structure. The inter-pose distances between the central carbon atoms of the closest isopropyl groups of two compounds 3 docked in the two HBDs were measured. These distances were compared with the intra-ligand distances measured between the terminal carbon atoms of the glycol-split fragment of a prototypic trisaccharide ligand 4 (MeO-GlcNAc6S-Gsplit-GlcNAc-OMe) during a 500 ns-long MD simulation (Supplementary data, Figure S11). In only a small fraction of pose pairs of compound 3 (<1%), the isopropyl groups were found within a distance compatible with interposition of a glycol-split fragment (<3.95 Å) (Figure 9A). Conversely, most of the pose pairs interacted with the outer region of both HBDs through their sulfate groups, often orienting their isopropyl termini towards the solvent (Figure 9B). The isopropyl termini of these couples of poses were separated by distances not compatible with the presence of a glycol-split linker (>>4 Å), suggesting that while simultaneous accommodation of two directly-linked fragments to HBD-1 and HBD-2 of a heparanase molecule, with the glycol-split portion covering the catalytic pocket, is possible, this is probably not the unique binding mode of roneparstat. Particularly when inhibitor concentration increases, the two HBDs of a protein could interact with fragments of different roneparstat molecules, causing an aggregation process which increases Hill slope.
Fig. 9.
Representative pose pairs of compound 3 compatible (A) and not compatible (B) with the presence of a linking glycol-split fragment. The inter-pose distance is represented with dashes. This figure is available in black and white in print and in color at Glycobiology online.
Conclusions
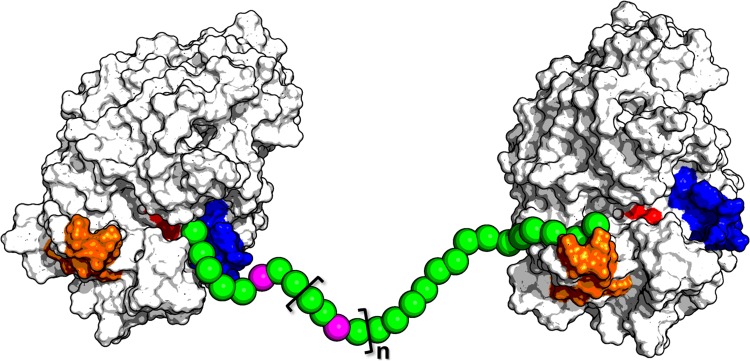
In the present study, we investigated the kinetics and mechanism of heparanase inhibition by roneparstat, a fully N-acetylated high molecular-weight glycol-split heparin currently in clinical trials for the treatment of myeloma. Best fitting of experimental data was obtained using two different curves for low and high inhibitor concentrations, suggesting the existence of multiple protein–ligand interaction modes, with one of them prevailing over the others depending upon the inhibitor concentration. A homology model of the catalytically active GS3 construct of human heparanase was built to investigate the putative inhibitor–protein interactions at a molecular level. Docking results for representative fragments of roneparstat indicated that heparanase HBD-1 and HBD-2 could interact with one inhibitor molecule or with two fragments belonging to different inhibitor molecules. These results also suggested that the glycol-split fragment might act as a flexible spacer within the inhibitor structure, rather than being a key anchor point for ligand stabilization (Naggi et al. 2005), which could favor the harboring of the roneparstat polysaccharide chain within the HBDs of two adjacent protein molecules (Figure 10). This is also consistent with the hypothesis that high concentrations of roneparstat would promote oligomerization of heparanase, with multiple protein molecules kept in proximity by the interaction with different fragments of the polysaccharide chain of roneparstat, which could also be related to the cooperative component of roneparstat binding.
Fig. 10.
Schematic representation of the hypothetic interaction between a single roneparstat chain (spheres) and two heparanase molecules (light gray). The spheres corresponding to glycol-split fragments are colored in magenta, while HBD-1, HBD-2 and the two catalytic residues are highlighted in orange, blue and red, respectively.
Besides our hypothesis, alternative explanations may be invoked for the steeper concentration–inhibition curves. In particular, it cannot be ruled out that the cooperative behavior seen for roneparstat at high concentrations might be triggered by the engagement of a different protein region. In fact, a putative third HBD had been previously described as important for stabilization of the heparanase-heterodimer quaternary structure (Levy-Adam et al. 2005). While this positively charged domain (Lys411–Arg432, see Supplementary data, Figure S3) might trigger an allosteric mechanism, it would be too speculative to devise an interaction model with this region, given its distance from the catalytic site. On the other hand, modeling the HBD regions close to the conserved catalytic site allowed us to present the first report on a possible mechanism, consistent with experimental data, of human heparanase inhibition by the nanomolar inhibitor roneparstat.
Materials and methods
The heparanase inhibitor roneparstat (SST0001) was provided by Sigma-Tau Research Switzerland S.A. (Mendrisio, CH). Preparation and characterization of roneparstat (100NA, RO-H) were previously described in detail (Naggi et al. 2005). Molecular weight of roneparstat is batch-to-batch slightly different and, in any case, it is an average value. Average molecular weight of the batch used for this experiment was 20.3 kDa.
Recombinant human heparanase GS3 was kindly provided by Prof. Israel Vlodavsky (Rappaport Faculty of Medicine & Research Institute, Technion, Haifa, Israel).
Fondaparinux sodium (Arixtra) was purchased from Glaxo Smith Kline.
The tetrazolium salt WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) was purchased from Santa Cruz Biotechnology.
Fondaparinux heparanase assay
Assays were essentially performed as described by Hammond et al. (2010), with minor modifications. Briefly, NUNC ELISA 96-well plates were pre-treated with a solution of 4% BSA in phosphate-buffered saline containing 0.05% Tween 20 (PBST), for 2 h at 37°C, then washed three times with PBST, dried by tapping on paper towel and stored at −20°C. Before use, once again, the plates were washed with PBST.
Assays were carried out with 100 µl/well of assay solution containing 40 mM sodium acetate buffer (pH 5.0), varying concentrations of fondaparinux, 2.5 nM heparansase (GS3) and serial dilutions of roneparstat (tested in quadruplicate). Plates were sealed with adhesive tape and incubated, in the dark, for 3 h at 37°C, followed by development with 1.69 mM WST-1 solution in 0.1 M NaOH, for 1 h at 60°C. Then, the absorbance at 560 nm was measured through a microplate reader (Victor 3, Perkin Elmer). Finally, the measurements were corrected by subtracting both the reagent background and the inner absorbance value of test compound.
Results of the assays were expressed as inhibition of substrate hydrolysis (in percentage) in the presence of roneparstat with respect to control assays containing no study compound.
Data analysis
Kinetic constants of the assay (Vmax, Km, Ki) and Hill slope (nH) were determined by fitting the relative velocity of fondaparinux hydrolysis, or percent inhibition by roneparstat, versus substrate or inhibitor concentrations, respectively. Curve fitting was carried out minimizing the sum of squared residuals from values calculated with the equations reported in the Results section, employing an Excel spreadsheet. R2 was used to assess the goodness of fit. Double-reciprocal analysis was also performed and the slopes of curves replotted as a function of inhibitor concentration and fitted to a Hill-like model.
Homology modeling
The X-ray structure of β-glucuronidase from Acidobacterium capsulatum co-crystallized with GlcA (PDB code 3VNZ) (Michikawa et al. 2012) was selected as the template to build the model of heparanase. The sequence of the full-length human heparanase pre-pro-form (amino acids 1–543) was retrieved from the Universal Protein Resource (UniProt ID Q9Y251) (UniProt Consortium 2015) and modified to create the catalytically active GS3 construct of the enzyme (Nardella et al. 2004), in which the 8 kDa (amino acids Gln36–Glu109) and 50 kDa (amino acids Gln157–Ile543) subunits of the mature protein are linked through a GSGSGS peptide. Sequence alignment (Supplementary data, Figure S1) and homology modeling were performed with Prime 3.4 (Schrodinger 2013a). To optimize the orientation of residues within the catalytic site of the enzyme, which are highly conserved between the two proteins, the molecule of GlcA co-crystallized within the template structure was maintained in the 3D structure of the heparanase model during the homology modeling procedure. The resulting crude heparanase model was processed with the Protein Preparation Wizard (Schrodinger 2013b), which was used to cap the C- and N-termini with methylamino and acetyl groups, respectively, and to optimize the overall hydrogen-bonding network by sampling the orientations of the side chains of polar amino acids. In the optimized structure, all glutamate and aspartate residues were deprotonated, all lysines and arginines were protonated and all histidines were in their neutral form. The only exception was His296, which was protonated to allow the formation of two hydrogen bonds with the adjacent carboxylate groups of Glu225 and Asp267. To relax the overall system geometry, the heparanase–GlcA complex was submitted to two stages of energy minimization with the OPLS 2005 force field (Banks et al. 2005). In the first minimization, all heavy atoms were kept fixed and only hydrogen atoms were free to move; the second minimization was performed on all atoms, applying a harmonic potential of 25 kcal/mol Å2 on protein heavy atoms and setting a heavy atom RMSD value of 0.5 Å as the convergence threshold. According to experimental evidences supporting the role of Glu225 as the proton donor responsible for the cleavage activity of heparanase, the residue Glu225 was protonated before docking calculations.
Docking simulations of substrates
The structures of substrate 1 and fondaparinux were built with Maestro 9.6 (Schrodinger 2013c), arranging the tetrahydropyran rings of GlcA and glucosamine in a chair conformation and those of iduronic acids in a 2S0 skew-boat conformation (Mulloy et al. 1993). Initial docking attempts for substrate 1 and fondaparinux, applying both a rigid docking or an induced-fit protocol were unsuccessful, likely due to the bulkiness of the ligands docked into a binding site modeled in its empty form (with the exception of the small GlcA bound to the catalytic residues).
Therefore, to induce a structural rearrangement of the enzyme binding region that could favor the accommodation of the bulky substrates, a modified version of the IFD protocol (Sherman et al. 2006) was applied, which was essentially based on a conformational search of the substrate inside the binding cavity. Each substrate was initially placed in an arbitrary conformation within the heparanase catalytic site, superposing its GlcA unit on the GlcA molecule coming from the crystallized template complex (see Homology modeling paragraph). Then, a conformational search was conducted on the ligand molecules, applying MacroModel 10.2 (Schrodinger 2013d) and the OPLS 2005 force field. 1000 Steps of mixed-torsional/large scale-low mode search (MCMM/LLMOD) (Kolossvàry and Guida 1999) were used to sample the rotatable bonds of the substrates, fixing the heavy atom positions of their glucuronic acid and applying torsional restraints of 500 kJ/mol on the internal dihedral angles of tetrahydropyran rings to keep their conformation unaltered. During this conformational search vdW radii of all atoms were scaled by a factor of 0.5 and the side chains of amino acids potentially hampering the accommodation of the substrate molecule, i.e. Asp62, Thr97, Lys98, Lys158, Lys159, Phe160, Asn227, Leu230, Lys231, Lys232, Gln270, Arg272, Lys274, Tyr298, Gln383, Phe385 and His392, were temporarily removed. To avoid redundant conformations, only those ligand conformations showing a pairwise RMSD value calculated on heavy atoms >1 Å were retained. At the end of this stage, 20 and 9 different complexes of heparanase with compound 1 and fondaparinux, respectively, were collected. Each complex was subjected to a Prime structural refinement stage, which (i) reintroduced the side chains previously removed, (ii) performed three cycles of conformational search on all the side chains of residues located within 5 Å from the substrate molecule (comprising all the side chains deleted during the MCMM/LLMOD search) and (iii) performed an energy minimization on the refined complexes. To retain the network of hydrogen bond interactions between the central GlcA of the substrates and the residues delimiting the catalytic site of the enzyme, the amino acids Glu225, His296, Glu343, Gly349, Gly350 and Tyr391 were kept fixed during the Prime 3.9 (Schrodinger 2015a) structural optimization. Each ligand molecule was then optimized in the field of the induced-fit protein structure applying default Glide 6.6 (Schrodinger 2015b) settings, maintaining the tetrahydropyran rings in their initial chair or skew-boat conformations. A combined score, called IFDScore, was calculated for each protein–ligand complex on the basis of both the protein–ligand interaction energy (GScore) and the total energy of the system (Prime Energy) and was used to rank the heparanase–substrate complexes.
A recent study highlighted the importance of HBD-1 and, in particular, of the sequence Lys158–Lys161 for substrate binding (Levy-Adam et al. 2005). Accordingly, the best-scored heparanase–substrate complexes in which substrates 1 and fondaparinux interacted with Lys158–Lys161 sequence were selected and then energy-minimized applying the OPLS 2005 force field and the GB/SA water solvation treatment (Still et al. 1990) to an energy gradient of 2 kJ/mol Å. The energy-minimized heparanase-substrate 1 complex was selected as the starting structure for docking studies of compounds 2 and 3.
Docking simulations of compounds 2 and 3
Compound 2 was built from the structure of compound 1 by deleting the bond connecting C2 and C3 atoms of GlcA−1 and by N-acetylating all glucosamine units. Compound 3 was built with Maestro 9.6, fixing the tetrahydropyran rings of glucosamines and iduronic acid to chair and 2S0 skew-boat conformations, respectively. Two isopropyloxy groups were introduced to cap the C1 and C4 atoms of the two terminal glucosamines to mimic the branched structure of the glycol-split fragment. The two structures were energy-minimized with MacroModel, applying the OPLS 2005 force field and the GB/SA water solvation treatment to an energy gradient of 0.05 kJ/mol Å.
Glide 6.6 was used to create three docking grids. Two grids encompassed the whole protein region surrounding either HBD-1 or HBD-2 domains and were used for docking of compound 3. The third grid was built to embrace the whole channel spanning from HBD-1 to HBD-2 and was applied for docking of compound 2. The first two grids were centered on a residue located in close proximity of HBD-1 or HBD-2, i.e., Asn64 or Gly269, whereas the third grid was centered on the energy-minimized pose of compound 1 obtained at the end of the IFD procedure. Enclosing and bounding box dimensions were set to 40 and 20 Å, respectively, for the two grids used for compound 3, and to 41.6 and 20 Å, respectively, for the grid used for compound 2, applying no scaling factor on vdW radii of non-polar protein atoms. During docking simulations, the conformations of tetrahydropyran rings were kept fixed and a vdW scaling factor of 0.8 was applied to non-polar ligand atoms. For each docking run, 100 ligand poses were collected and ranked according to their Docking Score value.
Cluster analysis
To identify the most representative conformations of compound 3 interacting with HBD-1 or HBD-2 domain, the two sets of 100 docking poses obtained for 3 were subjected to a cluster analysis, based on structural interaction fingerprints and calculation of Tanimoto similarity indexes. Five and four clusters were identified for accommodation of compound 3 in the region surrounding HBD-1 and HBD-2, respectively. The details of fingerprints calculation and cluster analysis are reported in the Supplementary data.
MD simulations
The energy-minimized complexes of heparanase with compound 1 and 2 and the structure of compound 4 (alone) were subjected to an MD simulation in an explicit water box. Desmond 3.6 (D. E. Shaw Research 2013) was used to build the solvated systems, applying the TIP3P model for water and ensuring that the solute molecules were placed at least 20 Å far from their neighboring periodic images. The OPLS 2005 force field was applied for the solvated systems containing compound 4 and heparanase in complex with compound 2, whereas heparanase-compound 1 complex was parametrized applying the AMBER99SB-ILDN (Lindorff-Larsen et al. 2010) and GLYCAM06j (Kirschner et al. 2008) force fields. In particular, the ligand partial charges were recalculated by fitting the HF/6-31G* electrostatic potential of compound 1 calculated within the field of the protein binding site with QSite 6.1 (Schrodinger 2013e).
The equilibrated systems were subjected to MD simulations performed in the NPT ensemble at 300 K and 1 atm, applying the Langevin coupling scheme (Feller et al. 1995). Long-range electrostatic interactions were calculated applying the smooth particle mesh Ewald method (Essmann et al. 1995), while short-range electrostatic and vdW interactions were cut off at 9 Å. The M-SHAKE algorithm (Krautler et al. 2001) was used to constraint all bond lengths to hydrogens and a RESPA integrator (Tuckerman et al. 1992) was applied to schedule the calculation of short-range nonbonded interactions and long-range electrostatic interactions every 2 and 6 fs, respectively.
During the simulation of compound 4, 10,000 snapshots were collected and distances were measured between atoms 1 and 2 specified in Supplementary data, Figure S11, belonging to the glycol-split portion.
Supplementary data
Supplementary data for this article were available online at http://glycob.oxfordjournals.org/.
Funding
The Open Access publication charges for this article was provided by Sigma-Tau Research Switzerland S.A., Mendrisio, CH.
Conflict of interest statement
F.M.M. and G.G. are employees of Sigma-Tau I.F.R. SpA.
Abbreviations
ECM, extracellular matrix; GH, glycoside hydrolase; GlcA, β-d-glucuronic acid; GlcN, α-d-glucosamine; HS, heparan sulfate; HBD, heparin-binding domain; HSPGs, HS proteoglycans; IFD, induced-fit docking; IdoA, iduronic acid; MCMM/LLMOD, mixed-torsional/large-scale low mode search; MD, molecular dynamics; PBST, phosphate-buffered saline containing 0.05% Tween 20; RMSD, root-mean-square deviation; RMSF, root-mean-square fluctuation.
Supplementary Material
Acknowledgements
The authors thank Prof. Israel Vlodavsky (Technion-Israel Institute of Technology's; Rappaport Faculty of Medicine & Research Institute, Technion, Haifa, Israel) who kindly provided the recombinant human heparanase GS3; Dr. Alessandro Noseda (Sigma-Tau Research Switzerland S.A. (STRCH)) for facilitating this study and for drug roneparstat availability.
References
- Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, Vlodavsky I. 2008. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem. 283:18167–18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker MG, Auld DS. 2014. Considerations for the design and reporting of enzyme assays in high-throughput screening applications. Perspect Sci. 1:56–73. [Google Scholar]
- Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR et al. 2005. Integrated modeling program, applied chemical theory (IMPACT). J Comput Chem. 26:1752–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basche M, Gustafson DL, Holden SN, O'Bryant CL, Gore L, Witta S, Schultz MK, Morrow M, Levin A, Creese BR et al. 2006. A phase I biological and pharmacologic study of the heparanase inhibitor PI-88 in patients with advanced solid tumors. Clin Cancer Res Off J Am Assoc Cancer Res. 12:5471–5480. [DOI] [PubMed] [Google Scholar]
- Bohlmann L, Tredwell GD, Yu X, Chang C-W, Haselhorst T, Winger M, Dyason JC, Thomson RJ, Tiralongo J, Beacham IR et al. 2015. Functional and structural characterization of a heparanase. Nat Chem Biol. 11:955–957. [DOI] [PubMed] [Google Scholar]
- Cao R, Zeidan AA, Rådström P, van Niel EWJ. 2010. Inhibition kinetics of catabolic dehydrogenases by elevated moieties of ATP and ADP − Implication for a new regulation mechanism in Lactococcus lactis. FEBS J. 277:1843–1852. [DOI] [PubMed] [Google Scholar]
- Casu B, Vlodavsky I, Sanderson RD. 2008. Non-anticoagulant heparins and inhibition of cancer. Pathophysiol Haemost Thromb. 36:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choay J, Petitou M, Lormeau JC, Sinaÿ P, Casu B, Gatti G. 1983. Structure-activity relationship in heparin: A synthetic pentasaccharide with high affinity for antithrombin III and eliciting high anti-factor Xa activity. Biochem Biophys Res Commun. 116:492–499. [DOI] [PubMed] [Google Scholar]
- Copeland RA. 2005. Evaluation of Enzyme Inhibitors in Drug Discovery. A Guide for Medicinal Chemists and Pharmacologists. 1st ed Hoboken: (NJ: ): John Wiley & Sons, Inc. [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 321(Pt 2):557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D. E. Shaw Research. 2013. Desmond Molecular Dynamics System, Version 3.6, Maestro-Desmond Interoperability Tools, Version 3.6. New York: (NY: ). [Google Scholar]
- Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, Brandt R, Ferro V, Bytheway I. 2011. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 104:635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J Chem Phys. 103:8577. [Google Scholar]
- Fan H, Mark AE. 2004. Refinement of homology-based protein structures by molecular dynamics simulation techniques. Protein Sci Publ Protein Soc. 13:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. 1995. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 103:4613. [Google Scholar]
- Ferro V. 2013. Heparan sulfate inhibitors and their therapeutic implications in inflammatory illnesses. Expert Opin Ther Targets. 17:965–975. [DOI] [PubMed] [Google Scholar]
- Ferro V, Hammond E, Fairweather JK. 2004. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 4:693–702. [DOI] [PubMed] [Google Scholar]
- Gandhi NS, Freeman C, Parish CR, Mancera RL. 2012. Computational analyses of the catalytic and heparin-binding sites and their interactions with glycosaminoglycans in glycoside hydrolase family 79 endo-β-D-glucuronidase (heparanase). Glycobiology. 22:35–55. [DOI] [PubMed] [Google Scholar]
- Hammond E, Handley P, Dredge K, Bytheway I. 2013. Mechanisms of heparanase inhibition by the heparan sulfate mimetic PG545 and three structural analogues. FEBS Open Bio. 3:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond E, Li CP, Ferro V. 2010. Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Anal Biochem. 396:112–116. [DOI] [PubMed] [Google Scholar]
- Huegel J, Enomoto-Iwamoto M, Sgariglia F, Koyama E, Pacifici M. 2015. Heparanase stimulates chondrogenesis and is up-regulated in human ectopic cartilage: A mechanism possibly involved in hereditary multiple exostoses. Am J Pathol. 185:1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, Parish CR. 2000. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry (Mosc). 39:15659–15667. [DOI] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, González-Outeiriño J, Daniels CR, Foley BL, Woods RJ. 2008. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 29:622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossvàry I, Guida WC. 1999. Low-mode conformational search elucidated: Application to C39H80 and flexible docking of 9-deazaguanine inhibitors into PNP. J Comput Chem. 20:1671–1684. [Google Scholar]
- Krautler V, van Gunsteren WF, Hunenberger PH. 2001. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 22:501–508. [Google Scholar]
- Levy-Adam F, Abboud-Jarrous G, Guerrini M, Beccati D, Vlodavsky I, Ilan N. 2005. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 280:20457–20466. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 78:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Huang Y, Buczek-Thomas JA, Ethen CM, Nugent MA, Wu ZL, Zaia J. 2014. A liquid chromatography-mass spectrometry-based approach to characterize the substrate specificity of mammalian heparanase. J Biol Chem. 289:34141–34151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie EA. 2007. Heparanase: A target for drug discovery in cancer and inflammation. Br J Pharmacol. 151:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa M, Ichinose H, Momma M, Biely P, Jongkees S, Yoshida M, Kotake T, Tsumuraya Y, Withers SG, Fujimoto Z et al. 2012. Structural and biochemical characterization of glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum. J Biol Chem. 287:14069–14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy B, Forster MJ, Jones C, Davies DB. 1993. N.m.r. and molecular-modelling studies of the solution conformation of heparin. Biochem J. 293(Pt 3):849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, Pisano C, Giannini G, Ishai-Michaeli R, Vlodavsky I. 2005. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J Biol Chem. 280:12103–12113. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Irimura T, Di Ferrante N, Nicolson GL. 1984. Metastatic melanoma cell heparanase. Characterization of heparan sulfate degradation fragments produced by B16 melanoma endoglucuronidase. J Biol Chem. 259:2283–2290. [PubMed] [Google Scholar]
- Nardella C, Lahm A, Pallaoro M, Brunetti M, Vannini A, Steinkühler C. 2004. Mechanism of activation of human heparanase investigated by protein engineering. Biochemistry (Mosc). 43:1862–1873. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamada S, Toyoshima M, Dong J, Nakajima M, Sugahara K. 2002. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. Hierarchical sulfate groups with different effects and the essential target disulfated trisaccharide sequence. J Biol Chem. 277:42488–42495. [DOI] [PubMed] [Google Scholar]
- Pikas DS, Li JP, Vlodavsky I, Lindahl U. 1998. Substrate specificity of heparanases from human hepatoma and platelets. J Biol Chem. 273:18770–18777. [DOI] [PubMed] [Google Scholar]
- Pisano C, Vlodavsky I, Ilan N, Zunino F. 2014. The potential of heparanase as a therapeutic target in cancer. Biochem Pharmacol. 89:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz H, Schönichen A. 2008. Transient binding patches: A plausible concept for drug binding. J Chem Biol. 1:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, Penco S, Pisano C, Carminati P, Tortoreto M et al. 2011. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res Off J Am Assoc Cancer Res. 17:1382–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapay N, Cabannes E, Petitou M, Imberty A. 2012. Molecular model of human heparanase with proposed binding mode of a heparan sulfate oligosaccharide and catalytic amino acids. Biopolymers. 97:21–34. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC. 2015a. Prime, Version 3.9. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2015b. Glide, Version 6.6. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2013a. Prime, Version 3.4. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2013b. Protein Preparation Wizard 2013-3; Epik, Version 2.4, Impact, Version 5.9, Prime, Version 3.2. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2013c. Maestro, Version 9.6. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2013d. MacroModel, Version 10.2. New York: (NY: ). [Google Scholar]
- Schrodinger LLC. 2013e. Qsite, Version 6.1. New York: (NY: ). [Google Scholar]
- Schubert M, Poon DKY, Wicki J, Tarling CA, Kwan EM, Nielsen JE, Withers SG, McIntosh LP. 2007. Probing electrostatic interactions along the reaction pathway of a glycoside hydrolase: Histidine characterization by NMR spectroscopy. Biochemistry (Mosc). 46:7383–7395. [DOI] [PubMed] [Google Scholar]
- Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 49:534–553. [DOI] [PubMed] [Google Scholar]
- Simeonovic CJ, Ziolkowski AF, Wu Z, Choong FJ, Freeman C, Parish CR. 2013. Heparanase and autoimmune diabetes. Front Immunol. 4:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still WC, Tempczyk A, Hawley RC, Hendrickson T. 1990. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 112:6127–6129. [Google Scholar]
- Tuckerman M, Berne BJ, Martyna GJ. 1992. Reversible multiple time scale molecular dynamics. J Chem Phys. 97:1990. [Google Scholar]
- UniProt Consortium. 2015. UniProt: A hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinader V, Haji-Abdullahi MH, Patterson LH, Afarinkia K. 2013. Synthesis of a pseudo-disaccharide library and its application to the characterisation of the heparanase catalytic site. PloS ONE. 8:e82111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I et al. 1999. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat Med. 5:793–802. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Iozzo RV, Sanderson RD. 2013. Heparanase: Multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol J Int Soc Matrix Biol. 32:220–222. [DOI] [PubMed] [Google Scholar]
- Wu L, Viola CM, Brzozowski AM, Davies GJ. 2015. Structural characterization of human heparanase reveals insights into substrate recognition. Nat Struct Mol Biol. 22:1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, Zhao G, Smith S, Galcheva-Gargova Z, Karlgren J et al. 2011. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PloS One. 6:e21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Bates M, Madura JD. 2006. Structure modeling, ligand binding, and binding affinity calculation (LR-MM-PBSA) of human heparanase for inhibition and drug design. Proteins. 65:580–592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.