Abstract
Neutral sphingomyelinase-2 (nSMase2) is a key ceramide-producing enzyme in cellular stress responses. While many posttranslational regulators of nSMase2 are known, emerging evidence suggests a more protracted regulation of nSMase2 at the transcriptional level. Previously, we reported that nSMase2 is induced by all-trans retinoic acid (ATRA) in MCF7 cells and implicated nSMase2 in ATRA-induced growth arrest. Here, we further investigated how ATRA regulates nSMase2. We find that ATRA regulates nSMase2 transcriptionally through the retinoic acid receptor-α, but this is independent of previously identified transcriptional regulators of nSMase2 (Sp1, Sp3, Runx2) and is not through increased promoter activity. Epigenetically, the nSMase2 gene is not repressively methylated in MCF7 cells. However, inhibition of histone deacetylases (HDACs) with trichostatin A (TSA) induced nSMase2 comparably to ATRA; furthermore, combined ATRA and TSA treatment was not additive, suggesting ATRA regulates nSMase2 through direct modulation of histone acetylation. Confirming this, the histone acetyltransferases CREB-binding protein and p300 were required for ATRA induction of nSMase2. Finally, use of class-specific HDAC inhibitors suggested that HDAC4 and/or HDAC5 are negative regulators of nSMase2 expression. Collectively, these results identify a novel pathway of nSMase2 regulation and suggest that physiological or pharmacological modulation of histone acetylation can directly affect nSMase2 levels.
Keywords: sphingolipids, retinoids, nuclear receptors, transcription, signal transduction, histone deacetylase, histone acetyltransferase, epigenetic, neutral sphingomyelinase-2, all-trans retinoic acid
Sphingolipids such as ceramide (Cer) are bioactive lipids involved in many cellular processes including apoptosis, proliferation, and differentiation (1). Cer is produced through multiple pathways, and it functions as a central hub in the sphingolipid network, an interlinked system of metabolic enzymes that tightly control cellular sphingolipid levels; of these pathways, the hydrolysis of SM by the SMases is a major pathway for stress-induced Cer generation. At present, three main classes of SMase are known and classified according to the pH optima of their activity (acid, neutral, and alkaline, respectively), of which the acid and neutral SMases (nSMase) are thought to play primary roles in stress and cytokine-induced Cer production (2, 3). Currently, four mammalian nSMases have been cloned and characterized, nSMase1 (SMPD2), nSMase2 (SMPD3), nSMase3 (SMPD4), and mitochondria-associated nSMase (SMPD5). Of these, nSMase2 is by far the most studied and has been implicated in the cellular response to cytokines such as TNF and interleukin-1β, chemotherapeutic drugs, and oxidative stress (2). Physiologically, nSMase2 has been implicated in apoptosis (4), growth arrest (5, 6), inflammation (7), mitogenesis (8), aging (9), and mineralization of bone and teeth (10, 11).
Considerable research has focused on understanding the acute transient activation of nSMases, and many regulators of nSMase2 in this process have been identified, including p38 MAPK (7, 12), protein kinase C-δ (PKC-δ) (13), matrix metalloproteinase-2 and integrins (8), calcineurin (14), and the proteins FAN and EED1 (15). More recent studies have suggested that phosphorylation of nSMase2 can regulate both its activity and stability (14, 16), although the specific upstream kinases involved have yet to be determined. Posttranslational regulation of nSMase2 in response to reactive oxygen species (ROS) and glutathione depletion has also been reported (9). Clearly, nSMase2 regulation is complex, likely depending on both the cell type and the stimulus. Indeed, emerging evidence has begun to point to a more protracted regulation of at the transcriptional level. Increases in nSMase2 expression have been reported in response to bone morphogenetic protein-2 (BMP2) (17, 18), daunorubicin (19), cigarette smoke (20), confluence (6), the hedgehog signaling mediator cyclopamine (21), and all-trans retinoic acid (ATRA) (5, 22, 23). Notably, in the latter two cases, increased expression of nSMase2 was required for ATRA-induced growth arrest (23) and cyclopamine-induced apoptosis, respectively (21). Additionally, expression of nSMase2 was increased in mature osteoblasts compared with mesenchymal precursors, consistent with a role for nSMase2 in bone homeostasis (10). However, despite multiple known inducers of nSMase2, to date only three transcription factors have been implicated in regulating nSMase2 expression; Sp1 and Sp3 were suggested to be important for daunorubicin and ATRA responses (19, 22) while Runx2 was implicated in the BMP2 response in both osteoblasts and chondrocytes (17, 18). In addition, cyclopamine induction of nSMase2 required generation of reactive nitrogen species (RNS) and was sensitive to treatment with N-acetylcysteine (NAC) (21). Finally, PKC-δ was implicated in ATRA-mediated nSMase2 induction through mediating Sp1 phosphorylation (22). However, this study focused on ATRA concentrations (10 μM) far in excess of those achieved pharmacologically (<1 μM) or observed physiologically (<0.1 μM). Thus, the biological relevance of these relationships is unclear.
In our recent study, we identified nSMase2 as an early ATRA-induced gene and implicated nSMase2 in growth arrest induced by a pharmacological ATRA dose (7). In this study, we have further investigated mechanisms underlying nSMase2 induction by ATRA. We report that ATRA induces nSMase2 at the transcriptional level through the retinoic acid receptor (RAR)-α. Moreover, the data suggest that ATRA induction of nSMase2 does not require any of the previously reported regulators of nSMase2 transcription (Runx2, Sp1, Sp3, RNS) and does not appear to be through activation of the putative Smpd3 promoter. Instead, ATRA appears to regulate nSMase2 through epigenetic effects on histone acetylation and requires CBP/p300 histone acetyltransferase (HAT).
MATERIALS AND METHODS
Materials
MCF7 and MDA-MB-231 breast carcinoma cells were obtained from ATCC (Manassas, VA). RPMI and DMEM culture medium, FBS, blasticidin S HCl, and superscript reverse transcriptase were obtained from Life Technologies (Carlsbad, CA). Antibodies for nSMase2, Runx2, Sp1, and Sp3 were from Santa Cruz Biotechnology (Santa Cruz, CA). Trichostatin A (TSA) and antibodies for acetyl-histone H3 (K9) and histone H3 were from Cell Signaling (Danvers, MA). Anti-actin was from Sigma (St. Louis, MO). Enhanced chemiluminescence kit was from Pierce. Histone deacetylase (HDAC) inhibitors LMK and mocestinostat were from Selleck Biochem (Houston, TX). Retinoic acid, Am580, vorinostat, cycloheximide (CHX), and all other chemicals, unless indicated otherwise, were obtained from Sigma.
Cell culture and siRNA
MCF7 cells were maintained in RPMI media containing 10% FBS; MDA-MB-231 cells were maintained in DMEM containing 10% FBS. Cells were maintained at 37°C, 5% CO2 in a humidified atmosphere. For experiments, cells were subcultured in 60 mm (200K) and 100 mm (500K) dishes with media being changed 1–2 h prior to the start of experiments. For siRNA, cells were transfected using both forward and reverse transfection methods. For forward transections, cells were seeded in 60 mm (150K), and after 24 h, cells were transfected with 20 nM negative control (AllStar, Qiagen) or siRNA using oligofectamine or lipofectamine RNAimax (Life Technologies) according to the manufacturer’s protocol (Sp1, Sp3, Sox9, HoxA5, ID1, IRF1 siRNA from Qiagen; Runx2, CBP, p300 siRNA from Life Technologies). For reverse transfection, cells were seeded directly into media containing siRNA complexes using lipofectamine RNAimax according to the manufacturer’s protocol. In both cases, after 48 h, cells were incubated in fresh media for 1–2 h prior to stimulation as indicated.
Protein extraction and immunoblot analysis
To extract cellular protein, cells were scraped in RIPA buffer and lysed by sonication. Protein concentration was estimated by the Bradford assay, and aliquots of lysates were mixed with equal volumes of ×2 Laemmli buffer (Biorad), vortexed, and boiled for 5–10 min. Protein was separated by SDS-PAGE and immunoblotted as described previously (5, 7)
Real-time PCR
Following stimulation, mRNA from MCF7 cells was extracted using the RNEasy kit (Qiagen). RNA (0.5–1 μg) was used to synthesize cDNA with either the Superscript II Kit for first-strand synthesis (Invitrogen) or Quanta cDNA mastermix. Following cDNA conversion, samples were diluted to 300 µl with molecular-biology-grade dH2O. Real-time RT-PCR with SYBR green (Biorad) was performed on a Biorad iCycler detection system. Standard reaction volume was 25 µl containing 12.5 µl supermix, 6.5 µl dH2O (Sigma), 100–500 nM oligonucleotide primers (IDT), and 5 µl cDNA template (diluted 12× in molecular-biology-grade dH2O). Initial steps of quantitative RT-PCR (qRT-PCR) were 2 min at 50°C for UNG erase activation, followed by a 3 min hold at 95°C for enzyme activation. For all primers, cycles (n = 40) consisted of a 10 s melt at 98°C, followed by a 45 s annealing at 55°C and 45 s extension at 68°C. The final step was 55°C incubation for 1 min. All reactions were performed in triplicate, and threshold for cycle of threshold (Ct) analysis of all samples was set at 0.15 relative fluorescence units. Data were normalized to an internal control gene, actin, to control for RNA preparation. Analysis of a single PCR product was confirmed by melt-curve analysis. Real-time PCR with Taqman assays was performed on the ABI 7500 real-time system using iTaq mastermix (Biorad). Standard reaction volume was 15 µl containing 7.5 µl supermix, 3.25 µl dH2O (Sigma), 0.25 µl 60× Taqman assay, and 3.75 µl cDNA template (diluted 12× in molecular-biology-grade dH2O). The quantitative PCR protocol consisted of 2 min enzyme activation at 95°C followed by 40 cycles consisting of a 10 s melt at 98°C, followed by a 60 s anneal and extension at 60°C. All reactions were performed in triplicate. Actin was used as a reference gene in all cases, and Ct values were converted to mean normalized expression using the ΔΔCt method. All Taqman assays were purchased from Life Technologies.
Analysis of heteronuclear RNA
RNA was extracted and treated with DNase (Ambion) for two rounds according to the manufacturer’s protocol. Treated RNA was processed for cDNA and real-time RT-PCR as described above. For detection of nSMase2 heteronuclear RNA (hnRNA), Taqman assays that span intron-exon junctions were utilized. Actin was used as reference gene, and analysis of no template controls was performed to confirm no contamination of genomic DNA.
Promoter activity by luciferase assay
MCF7 cells were seeded in 60 mm dishes and, 24 h later, were transfected with 1 μg nSMase2 promoter-luciferase and 0.5 μg pEF6-LacZ using Xtremegene 9 (Roche) according to the manufacturer’s protocol. After 6 h, media was changed for 1 h prior to stimulation with vehicle (DMSO), ATRA (1 μM), or doxorubicin (200 nM) for 24 h. Luciferase and galactosidase activities were extracted and assayed using the luciferase and high-sensitivity galactosidase assay kits (Stratagene) according to the manufacturer’s protocol. Measured luciferase activity was normalized to galactosidase activity.
Transcription factor array
To facilitate analysis of transcription factors in the early ATRA response, a transcription factor array (SA Biosciences/Qiagen) was utilized (PAHS-075). To run the array, a mastermix of 1,275 µl SYBR-green, 102 µl cDNA, and 1,173 µl dH2O was prepared for each cDNA template, and 25 µl of the mastermix was loaded per well. The RT-PCR protocol consisted of 10 min at 95°C for polymerase activation followed by 40 cycles of a 15 s melt at 95°C and 60 s at 60°C for annealing and extension. After the cycles were run, a melt curve was performed to confirm a single product for each primer pair. Data were analyzed utilizing the PCR Array Data analysis portal (http://www.sabiosciences.com).
Chromatin immunoprecipitation assay
For analysis of acetyl-histone H3 (Ac-H3) enrichment at the nSMase2 and nSMase3 genes, the Magna ChIP A kit (EMD Millipore) was utilized according to the manufacturer’s instructions. For these experiments, ∼1 × 107 cells was used for immunoprecipitation with the Ac-H3 (K9) antibody (C5B11; Cell Signaling) using normal rabbit IgG (#2729; Cell Signaling) as negative control. Following chromatin immunoprecipitation (ChIP) and purification of DNA, levels of nSMase2 and nSMase3 were analyzed by qRT-PCR with the following primers: N2-F, CAGGGTCCGAGGCAGAAAT; N2-R, AGACCACATCCCCCAGAGAC; N3-F, GCGTCATTGAAGCCCAACAG; and N3-R, CGGGCGGACCAGAAAATTG. For all PCR analyses, melt curves were performed to ensure single products. Relative DNA in immunoprecipitates was normalized to total input, and enrichment is calculated as DNA in Ac-H3 over IgG negative control.
Statistical analysis
Data are represented as mean ± SEM, unless otherwise indicated. Statistical analysis was by unpaired Student’s t-test, one-way ANOVA or two-way ANOVA as indicated with P < 0.05 being considered statistically significant and n representing the number of experiments as indicated.
RESULTS
ATRA regulates nSMase2 through RAR-α receptors
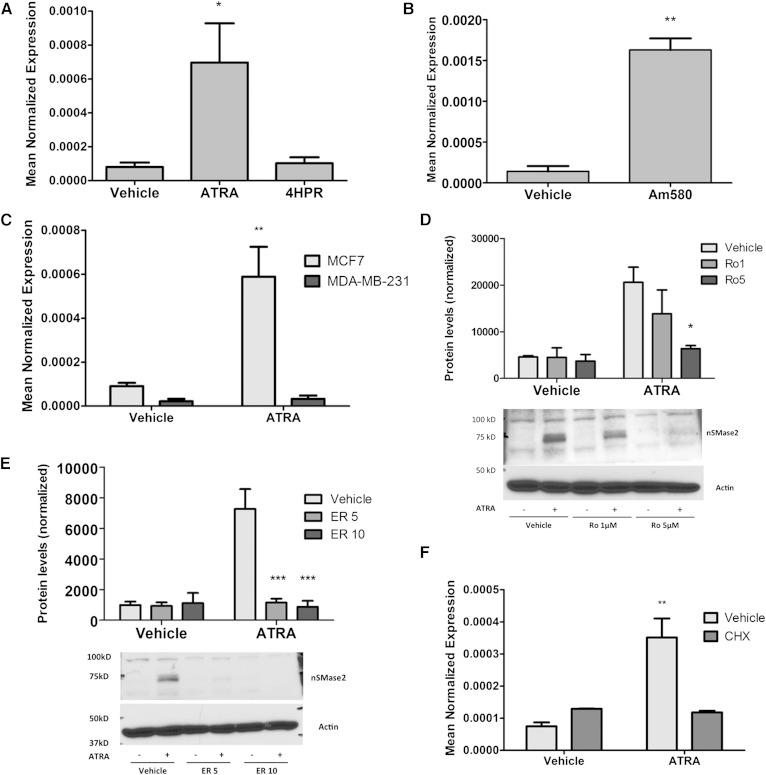
Previously, we reported that nSMase2 is an early induced gene in the ATRA response and is regulated in a time- and dose-dependent manner (7). Here, the mechanism by which ATRA regulates nSMase2 was explored. ATRA effects are primarily mediated by nuclear receptors, whereas the synthetic ATRA analog fenretinide (4HPR) has acute functions that are independent of RARs (24). Accordingly, to confirm that ATRA effects on nSMase2 were receptor mediated, the effects of 4HPR and ATRA were compared (Fig. 1A). As can be seen, treatment with 4HPR had no significant effect on nSMase2 expression compared with the strong effect seen with ATRA suggesting that nSMase2 regulation is directly through nuclear receptors. There are currently three subtypes of ATRA receptor (RAR-α, RAR-β2, and RAR-γ), and MCF7 cells express all three subtypes, whereas MDA-MB-231 cells possess primarily the RAR-γ subtype (25). To determine which receptor is important for nSMase2 induction, the effects of the RAR-α agonist Am580 on nSMase2 mRNA were investigated (Fig. 1B). As can be seen, Am580 treatment (100 nM, 12 h) significantly increased nSMase2 expression, comparable to the ATRA response. In contrast, ATRA stimulation of MDA-MB-231 cells had no effect on nSMase2 induction (Fig. 1C). To consolidate these results, two structurally distinct RAR-α antagonists, Ro-415253 and ER-50891, were used. As can be seen, preincubation with 1 and 5 μM Ro-415253 (Fig. 1D) or 5 and 10 μM ER-50891 (Fig. 1E) significantly inhibited ATRA induction of nSMase2. Taken together, this confirms ATRA regulates nSMase2 through the RAR-α subtype.
Fig. 1.
ATRA effects on nSMase2 are through the RAR-α. MCF7 and MDA-MB-231 cells shown were seeded in 60 mm dishes. A: MCF7 cells were stimulated with vehicle (DMSO), ATRA (1 μM), or 4HPR (1 μM) for 12 h (* P < 0.05 vs. vehicle, n = 4). B: MCF7 cells were stimulated with vehicle (DMSO) or Am580 (100 nM) for 12 h (RNA) (** P < 0.01 vs. vehicle, n = 3 RNA). C: MCF7 and MDA-MB-231 cells were stimulated with vehicle (DMSO) or ATRA (1 μM) for 24 h (** P < 0.01 vs. vehicle, n = 3). D: MCF7 cells were pretreated with Ro-415253 (1 or 5 μM) for 20 min prior to stimulation with DMSO or ATRA (1 μM) for 24 h (* P < 0.05 vs. vehicle, n = 4). E: MCF7 cells were pretreated with ER-50891(5 or 10 μM) for 15 min prior to stimulation with DMSO or ATRA (1 μM) for 24 h (*** P < 0.001 vs. vehicle, n = 4). F: MCF7 cells were pretreated with CHX (50 μg/ml) for 15 min prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 6 h (** P < 0.01 vs. vehicle, n = 3). As shown, RNA was extracted and converted to cDNA, and nSMase2 expression analyzed by qRT-PCR. As shown, protein was extracted in RIPA buffer, and nSMase2 levels analyzed by immunoblot using actin as loading control.
The binding of ATRA to RARs induces a conformational change leading to recruitment of transcriptional coactivators and direct upregulation of target genes. To determine whether nSMase2 is a direct target of RAR-α, MCF7 cells were preincubated with or without CHX (50 μg/ml). If ATRA is still able to induce nSMase2 mRNA in the absence of protein synthesis, this would imply that RAR-α directly interacts with the nSMase2 gene (Fig. 1F). As seen before, ATRA strongly induced nSMase2 mRNA in the absence of CHX; however, in the presence of CHX, ATRA was unable to increase nSMase2 mRNA suggesting that ATRA requires synthesis of an intermediate regulatory factor to induce nSMase2.
ATRA induces nSMase2 at the transcriptional level
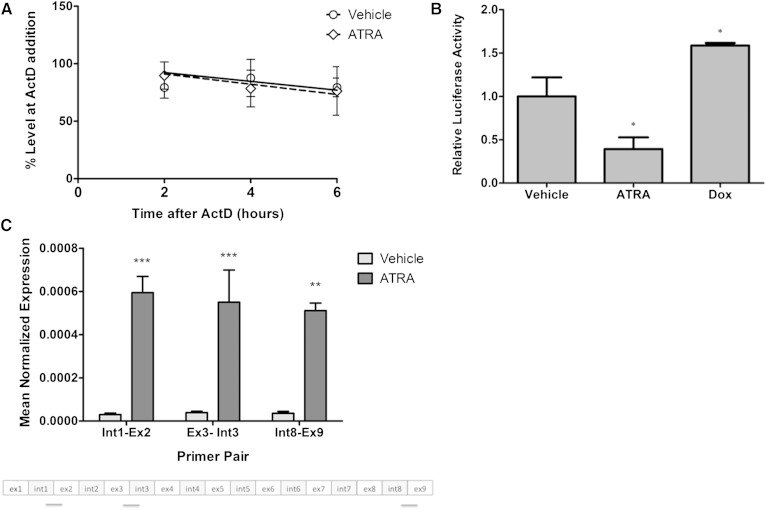
The requirement of an intermediate factor for ATRA regulation of nSMase2 raised the possibility that it may increase mRNA stability rather than affecting transcription. To rule this out, we assessed nSMase2 mRNA stability in the presence and absence of ATRA. For this, cells were stimulated with vehicle or ATRA for 12 h, and transcription was interrupted with actinomycin D. Subsequently, nSMase2 mRNA levels were assessed at 0, 3, 6, and 9 h post actinomycin treatment (Fig. 2A). Overall, there were no significant differences in mRNA half-life between vehicle-treated (7.13 h) and ATRA-treated (7.35 h) cells consistent with transcriptional regulation.
Fig. 2.
ATRA induction of nSMase2 is at the transcriptional level. A: MCF7 cells were seeded in 60 mm dishes and stimulated with vehicle (DMSO) or ATRA (1 μM) for 12 h. At this time, transcription was interrupted with actinomycin D (ActD; 1 μg/ml) for 0, 3, 6, and 9 h as shown. At these times, RNA was extracted and nSMase2 expression analyzed by qRT-PCR (P > 0.1; n = 6). B: MCF7 cells were seeded in 60 mm dishes and, 24 h later, were cotransfected with 1 μg nSMase2 promoter-luciferase and 0.5 μg pEF6-LacZ for 6 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Cells were lysed, and luciferase and galactosidase activity were determined as described in Materials and Methods (* P < 0.05 vs. vehicle, n ≥ 3). C: MCF7 cells were seeded in 100 mm dishes and stimulated with vehicle (DMSO) or ATRA (1 μM) for 24 h. hnRNA levels were analyzed as described in Materials and Methods (** P < 0.01, *** P < 0.001 vs. vehicle, n = 3).
To develop these data and further evaluate transcriptional regulation, the effect of ATRA on activity of the Smpd3 promoter was determined. For this, we cloned the region spanning from −1,100 bp to +25 bp upstream of exon 1 as described previously (17–19, 22), and reporter assays were performed as described above. For these experiments, we utilized doxorubicin as a positive control as its analog daunorubicin was previously reported to increase nSMase2 promoter activity (19). Indeed, as seen with daunorubicin, low-dose doxorubicin increased the activity of the nSMase2 promoter compared with vehicle control (Fig. 2B). Surprisingly, and in contrast to doxorubicin, ATRA did not increase promoter activity. This was also the case when a longer (−2,000 bp to +25 bp) promoter construct was utilized (data not shown).
Given the lack of effect of ATRA on the nSMase2 promoter, it was important to confirm transcriptional induction of nSMase2 by other means. Accordingly, the effects of ATRA on nSMase2 hnRNA were determined. The hnRNA represents the immediate product of gene transcription that contains both transcribed introns and exons prior to mRNA splicing; thus, increased levels of transcription result in increased hnRNA levels. For this purpose, extracted RNA was treated with DNase to remove any possible contaminating genomic DNA, and qRT-PCR was performed with primers designed to span intron-exon junctions (Fig. 2C). Results indicated that ATRA strongly increased levels of hnRNA as detected by both primer sets confirming that induction of nSMase2 primarily occurs at the transcriptional level.
ATRA induction of nSMase2 is independent of previously reported transcriptional regulators
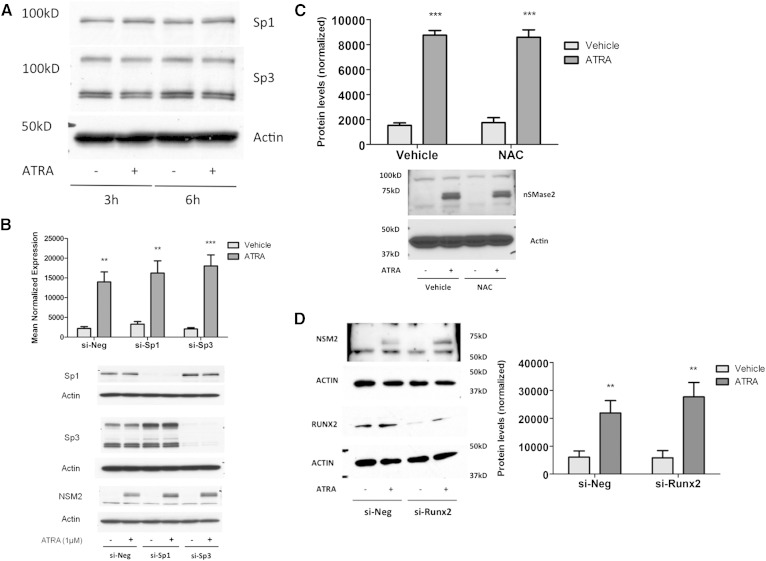
Previous studies have identified Runx2, Sp1, Sp3, and RNS as transcriptional regulators of nSMase2 in response to various stimuli (17–19, 21). Indeed, Sp1 and Sp3 were reported to be upstream of nSMase2 in the ATRA response, although this was inferred by use of pharmacological inhibition (22). Consequently, we elected to confirm their role in ATRA regulation of nSMase2 by utilizing an siRNA approach to more specifically downregulate Sp1 and Sp3. Notably, ATRA stimulation had no effects on levels of Sp1 or Sp3 alone (Fig. 3A), and thus we were able to achieve >80% knockdown of both at the protein level. Strikingly, loss of Sp1, Sp3, or both together had no inhibitory effect on nSMase2 induction by ATRA at the protein level; indeed, levels appeared somewhat increased (Fig. 3B). This was also seen when we analyzed ATRA effects on nSMase activity following Sp1 and Sp3 knockdown (data not shown). Thus, Sp1 and Sp3 are dispensable for ATRA induction of nSMase2 in MCF7 cells.
Fig. 3.
Sp1, Sp3, Runx2, and RNS are not required for ATRA effects on nSMase2. A: MCF7 cells were stimulated with vehicle (DMSO) or ATRA (1 μM) as shown. Protein was extracted and analyzed for Sp1 and Sp3 levels by immunoblot using actin as loading control. Immunoblot is representative of n = 3. B: MCF7 cells were seeded in 60 mm dishes and transfected with AStar, Sp1, or Sp3 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted and analyzed for nSMase2, Sp1, and Sp3 levels by immunoblot using actin as loading control. Immunoblot is representative of n = 4. C: MCF7 cells were pretreated with NAC (500 μM) for 3 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted, and nSMase2 levels analyzed by immunoblot (n = 4). D: MCF7 cells were seeded in 60 mm dishes and transfected with AStar or Runx2 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted and analyzed for nSMase2 and Runx2 levels by immunoblot using actin as loading control. Immunoblot is representative of n = 3.
We next turned our attention to additional pathways implicated in nSMase2 regulation, namely, ROS/RNS generation and Runx2 (17, 18, 21). To determine whether ROS are required for ATRA induction of nSMase2, the ROS/RNS scavenger NAC, previously shown to inhibit nSMase2 induction by cyclopamine (21), was used. Cells were pretreated for 3 h with 500 μM NAC prior to stimulation with ATRA for 24 h, with previous studies demonstrating this is sufficient time for NAC to exert its antioxidant effects (26, 27). Results indicated that ATRA induction of nSMase2 was comparable in the presence and absence of NAC (Fig. 3D) suggesting that ROS/RNS are not required for this process.
Finally, siRNA was utilized to determine whether Runx2 was important for ATRA induction of nSMase2 (Fig. 3D). Results showed a strong Runx2 knockdown in the presence of Runx2 siRNA compared with negative control siRNA. Importantly, loss of Runx2 had no effect on ATRA induction of nSMase2. Taken together, these results demonstrate that ATRA induces nSMase2 independently of previously identified transcriptional regulators of nSMase2.
Identification of transcription factors acutely induced by ATRA
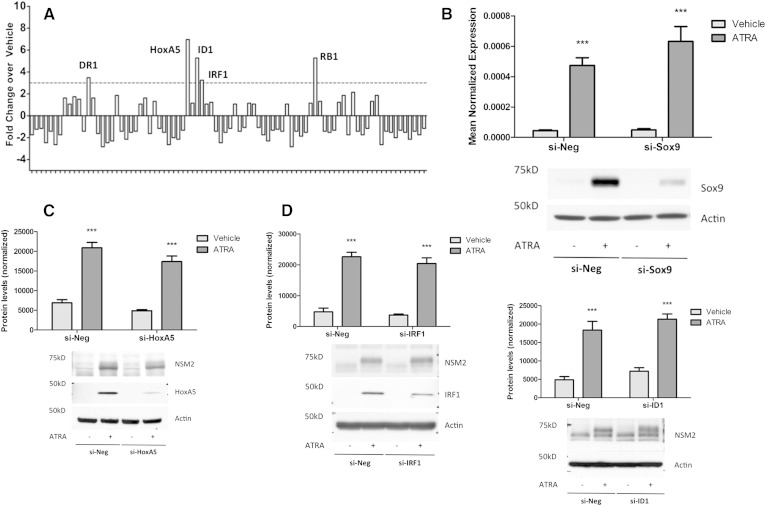
Having ruled out known transcriptional regulators, it was important to identify other candidate transcription factors induced by ATRA that may be upstream of nSMase2. To this end, a PCR array that measures expression of 84 transcription factors was utilized (Qiagen/SA Biosciences). For this, cells were stimulated with ATRA for 3 h, chosen as this was prior to nSMase2 induction (7), and mRNA was extracted (Fig. 4A). Following array analysis, transcription factors that were induced above 3-fold were chosen. As can be seen, ATRA induced DR1, HoxA5, ID1, IRF1, and RB1 compared with vehicle controls. Validation of these with independently designed qRT-PCR primers confirmed increases in ID1 and trends to increases in IRF1 and HoxA5 (data not shown). Additional analysis of the literature suggested ATRA could also induce Sox9 at early time points (28), and this was confirmed by immunoblot (Fig. 4B).
Fig. 4.
Early ATRA-induced transcription factors are not upstream of nSMase2. A: MCF7 cells were seeded in 60 mm dishes and stimulated with vehicle (DMSO) or ATRA (1 μM) for 3 h. RNA was extracted, converted to cDNA, and used as template for the Transcription Factor qRT-PCR array. Data are shown as fold change in ATRA stimulated cells compared with vehicle, n = 1. B: MCF7 cells were transfected with AStar or Sox9 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted and analyzed for nSMase2 and Sox9 levels by immunoblot using actin as loading control; RNA was extracted and analyzed for nSMase2 by qRT-PCR. Immunoblot is representative of n = 3 (* P < 0.05, n ≥ 3). C: MCF7 cells were transfected with AStar or HoxA5 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted and analyzed for nSMase2 and HoxA5 levels by immunoblot using actin as loading control. Immunoblot is representative of n = 3. D: MCF7 cells were transfected with AStar or IRF1 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. Protein was extracted and analyzed for nSMase2 and Sox9 levels by immunoblot using actin as loading control. Immunoblot is representative of n = 3. E: MCF7 cells were transfected with AStar or ID1 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. RNA and protein were extracted and analyzed for ID1 (RNA) and nSMase2 (protein) levels by qRT-PCR and immunoblot respectively. Actin was used as reference gene and loading control. Immunoblot is representative of n = 3 (* P < 0.05, n = 3).
In order to determine whether any of these candidates were upstream of nSMase2, a systematic siRNA approach was taken. Cells were treated with negative control or target siRNA for 48 h prior to stimulation with ATRA for 24 h, and the effects on both nSMase2 and transcription factor levels were determined. In cases in which an antibody for the transcription factor of interest was not available, siRNA efficiency was determined by qRT-PCR. As can be seen, ATRA strongly induced Sox9 (Fig. 4B), HoxA5 (Fig. 4C), IRF1 (Fig. 4D), and ID1 (Fig. 4E), consistent with previous results and the transcription factor array. Importantly, siRNA for all transcription factors reduced ATRA induction of the transcription factor by >60%. However, in all cases, the effects of ATRA on nSMase2 induction were not significantly inhibited (Fig. 4B–E). Thus, none of the identified transcription factors appear to act upstream of nSMase2 in the ATRA response.
The Smpd3 gene is not hypermethylated in MCF7 cells
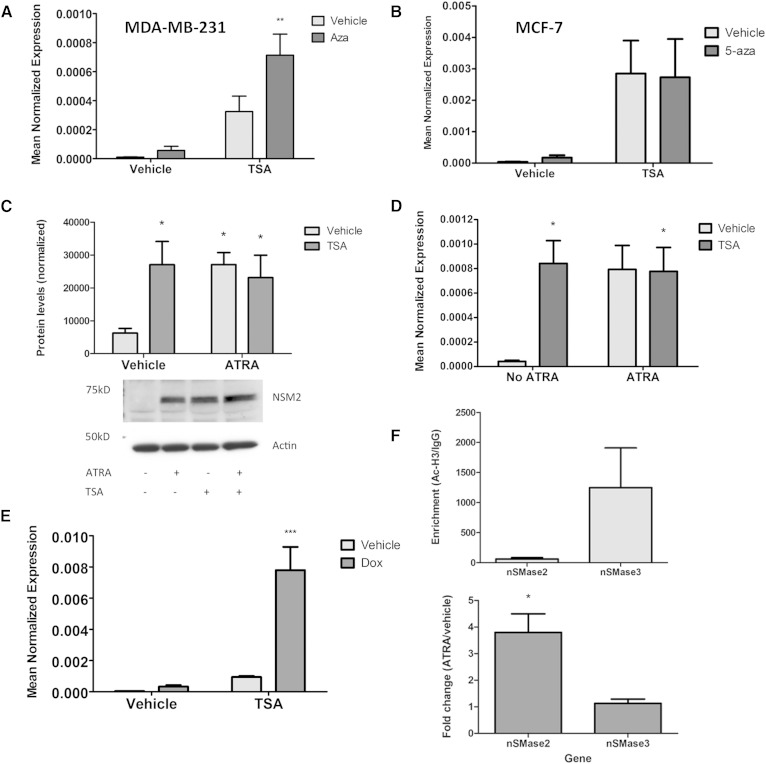
Having ruled out a number of candidate transcription factors upstream of nSMase2, other possibilities by which ATRA might affect nSMase2 expression were considered. Previous studies have reported that loss of ATRA signaling can lead to hypermethylation of downstream genes in breast epithelial cells (29, 30). Moreover, the Smpd3 gene (nSMase2) is reported to be hypermethylated in some breast cancer and hepatocellular carcinoma (HCC) cells (31, 32). Thus, we speculated that ATRA might induce nSMase2 by modulating methylation. To begin to probe this, it was first necessary to establish that the nSMase2 gene is repressively methylated in MCF7 cells. For this, epigenetic unmasking experiments were performed in which cells are treated in tandem with the DNA methyltransferase inhibitor 5-azacytidine (5-aza; 1 μM) for 48 h followed by TSA for 24 h with subsequent analysis of nSMase2 mRNA by qRT-PCR (Fig. 5A, B). To be able to distinguish repressive methylation from effects of single agents alone, dual treatments of 5-aza/TSA were compared with individual treatments of 5-aza/vehicle and vehicle/TSA, as well as with vehicle only. For these experiments, MDA-MB-231 cells were utilized as positive controls because the Smpd3 gene was shown to be hypermethylated in this cell line (32). In MDA-MB-231 cells, 5-aza/TSA treatment significantly increased nSMase2 expression compared with vehicle control to an extent that was significantly higher than either single agent (Fig. 5A), consistent with hypermethylation of the nSMase2 gene as reported previously (32). In contrast, while 5-aza/TSA treatment of MCF7 cells significantly increased nSMase2 expression over vehicle and 5-aza-treated cells, they were not significantly different from the effects seen in cells treated with TSA alone (Fig. 5B). Thus, we conclude that the Smpd3 gene is not repressively hypermethylated in MCF7 cells, and consequently, ATRA is not likely to be regulating nSMase2 through effects on methylation. Strikingly, TSA treatment alone was sufficient to significantly enhance nSMase2 expression over vehicle in both MCF7 and MDA-MB-231 cells. This suggests that histones associated with the nSMase2 gene may be hypoacetylated in these cells, and consequently, direct modulation of histone acetylation might be an alternative mechanism to increase nSMase2 expression.
Fig. 5.
ATRA regulates nSMase2 epigenetically through histone acetylation but not gene methylation. MDA-MB-231 (A) or MCF7 (B) cells were seeded in 60 mm dishes and treated with vehicle or 5-aza (1 μM) for 48 h prior to treatment with vehicle (DMSO) or TSA (300 nM) for 24 h. RNA was extracted and nSMase2 levels analyzed by qRT-PCR using actin as reference gene (** P < 0.01, n = 5). C: MCF7 cells were treated with vehicle (DMSO), ATRA (1 μM), TSA (300 nM), or both for 24 h. Protein lysates were prepared and nSMase2 levels analyzed by immunoblot using actin as loading control (* P < 0.05, n = 3). D: MCF7 cells were treated with vehicle (DMSO), ATRA (1 μM), TSA (300 nM), or both for 24 h. RNA was extracted, and nSMase2 levels were analyzed by qRT-PCR using actin as reference gene (* P < 0.05, n = 3). E: MCF7 cells were treated with vehicle (DMSO), doxorubicin (200 nM), TSA (300 nM), or both for 24 h. RNA was extracted, and nSMase2 levels were analyzed by qRT-PCR using actin as reference gene (** P < 0.01, n = 4). F: MCF7 cells were treated with vehicle (DMSO) or ATRA (1 μM, 24 h), and acetyl-H3 levels at the nSMase2 and nSMase3 genes were probed by ChIP analysis (* P < 0.05, n = 4).
ATRA regulates nSMase2 through effects on histone acetylation
On binding to RAR receptors, ATRA induces conformational changes, releasing corepressors and allowing recruitment of coactivators that enable remodeling of chromatin (33, 34). The marked effects of TSA on nSMase2 expression led us to speculate that ATRA is primarily inducing nSMase2 epigenetically. If this were the case, we reasoned that TSA and ATRA would have comparable effects on nSMase2 expression and, more importantly, that combined treatment would not be additive. Accordingly, MCF7 cells were treated for 24 h with TSA (400 nM), ATRA (1 μM), or both, and the effects on nSMase2 expression were determined (Fig. 5C). As seen above, both ATRA and TSA increased nSMase2 expression to a very comparable level. Furthermore, combined treatment of the two did not increase nSMase2 mRNA further, suggesting that ATRA and TSA are acting within the same pathway. In contrast to this, combined treatment with doxorubicin and TSA led to synergistic induction of nSMase2 (Fig. 5D), consistent with doxorubicin and TSA acting in distinct pathways [i.e., doxorubicin increasing nSMase2 promoter activity as seen above (Fig. 2B), and TSA inducing histone acetylation].
To further confirm that effects of ATRA on nSMase2 are through histone acetylation, we performed ChIP analysis to determine enrichment of acetyl-histone H3 (K9) at the proximal nSMase2 promoter region and whether ATRA affected this (Fig. 5E). As a control for the effects of ATRA, we designed primers for the proximal region of nSMase3 (Smpd4), which we had previously observed to be unresponsive to ATRA stimulation (7). In the absence of ATRA stimulation, we observed that Ac-H3 was basally enriched at the nSMase2 proximal promoter region (62- ± 22-fold over IgG). This was also true for the nSMase3 proximal promoter region (1,248- ± 662-fold over IgG) with the higher enrichment of Ac-H3 observed here consistent with the considerably higher expression of nSMase3 compared with nSMase2 in MCF7 cells (7). Importantly, ATRA stimulation for 24 h significantly increased Ac-H3 levels at the nSMase2 gene (3.80- ± 0.70-fold increase over vehicle) but not at the nSMase3 gene (1.15- ± 0.16-fold increase over vehicle). Thus, ATRA significantly increases acetyl-histone H3 at the nSMase2 gene.
ATRA induction of nSMase2 requires CBP/p300
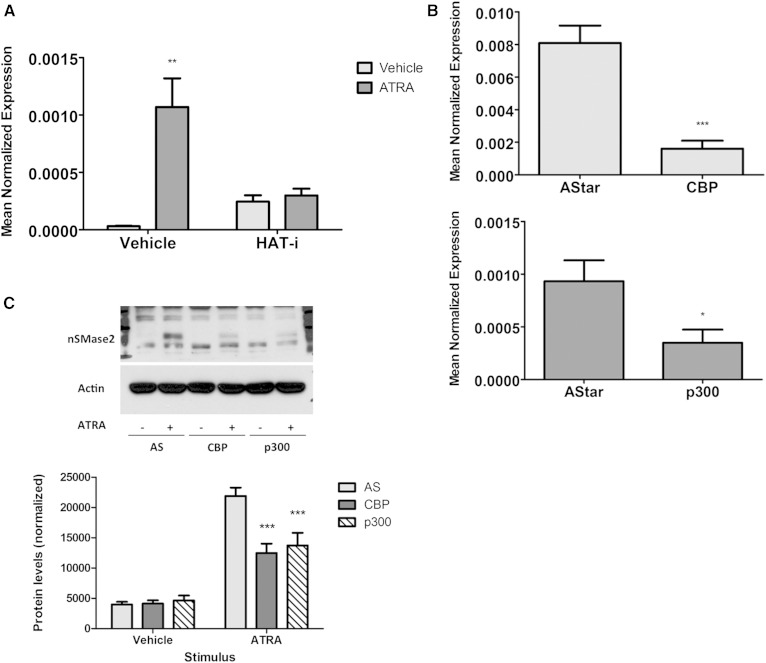
If ATRA primarily induces nSMase2 through effects on histone acetylation, it was reasoned that blocking the transfer of acetyl groups onto histones should prevent this. MCF7 cells were preincubated with an inhibitor of HATs (HAT-i; 10 μM) for 15 min prior to ATRA stimulation for 24 h (Fig. 6A). Interestingly, the results showed that HAT-i treatment alone was able to induce nSMase2 mRNA over vehicle control. However, more importantly, pretreatment with HAT-i completely inhibited ATRA induction of nSMase2. Taken together, these results suggest that ATRA effects on nSMase2 are through regulating acetylation of histones.
Fig. 6.
Knockdown of CBP and p300 blunts ATRA induction of nSMase2. A: MCF7 cells were pretreated with HAT-i (10 μM) for 15 min prior to treatment with vehicle (DMSO) or ATRA (1 μM) for 24 h. RNA was extracted, and nSMase2 levels were analyzed by qRT-PCR using actin as reference gene (* P < 0.05, n = 4). B, C: MCF7 cells were seeded in 60 mm dishes and reverse-transfected with AStar (AS), CBP, or p300 siRNA (20 nM) for 48 h prior to stimulation with vehicle (DMSO) or ATRA (1 μM) for 24 h. B: RNA was extracted with CBP and p300 levels analyzed by qRT-PCR using actin as reference gene (* P < 0.05, *** P < 0.001 vs. AS, n = 4). C: Protein was extracted and analyzed for nSMase2 levels by immunoblot using actin as loading control (*** P < 0.001 vs. AS, n = 4).
Notably, the HAT inhibitor utilized above is reported to have somewhat preferential activity for CBP/p300, two proteins with known HAT activity that have previously been implicated in some ATRA responses (35–37). Thus, it became important to determine whether they were required for ATRA induction of nSMase2, and for this an RNAi approach was utilized (Fig. 6B, C). Knockdown of CBP and p300 with siRNA induced a marked downregulation of each protein with minimal effects on the other (Fig. 6B). Importantly, the effects of ATRA on nSMase2 protein were significantly blunted following loss of either CBP or p300 (Fig. 6C). This was also true at the mRNA level, although observed effects were somewhat stronger with p300 knockdown than seen with CBP knockdown (supplementary Fig. 1). Nonetheless, taken together with the inhibitor data above, this confirms that both CBP and p300 are necessary for ATRA induction of nSMase2.
nSMase2 expression is regulated by class II HDACs
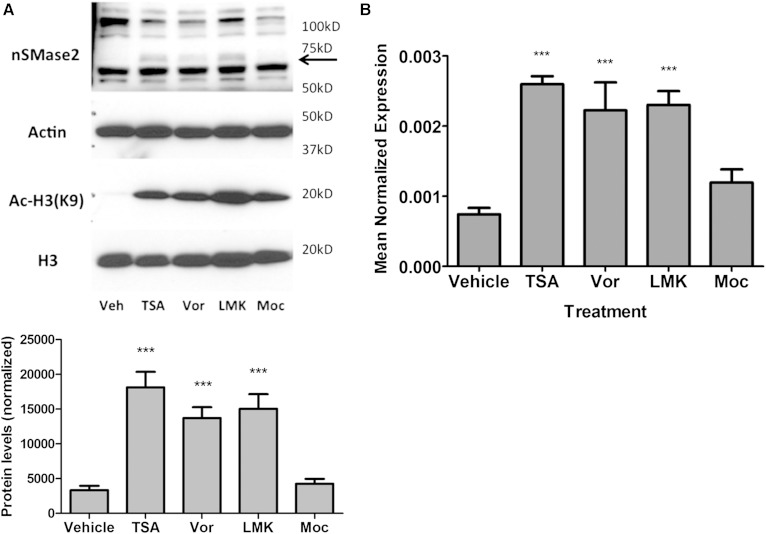
The levels of histone acetylation are a reflection of the balance of activities of both HAT and HDAC enzymes (38, 39). Identification of CBP and p300 as positive regulators of nSMase2 through histone acetylation led us to speculate if specific HDACs could negatively regulate nSMase2 expression. Having already observed the effects of TSA, a broad-specificity HDAC inhibitor, on nSMase2 expression (Fig. 5), the availability of more class-specific HDAC inhibitors was utilized. Accordingly, the inhibitors vorinostat (class I/II), LMK (HDAC4 and 5), and mocestinostat (class I) were utilized with TSA as a positive control. Analysis of acetyl-H3 (K9) levels demonstrated that all inhibitors were effective at the concentrations used and increased acetyl-H3(K9) levels to a somewhat comparable level (Fig. 7A). Strikingly, both vorinostat (10 μM) and LMK (5 μM) were effective at increasing nSMase2 to levels comparable to TSA, both at the protein (Fig. 7A) and mRNA (Fig. 7B) level. In contrast, mocestinostat (5 μM) had no effect on nSMase2 expression, despite its effects on H3 acetylation. Collectively, this suggests that class II HDACs, most likely HDAC4 and/or HDAC5, suppress nSMase2 expression and that regulation of histone acetylation is a primary mechanism by which nSMase2 levels are determined.
Fig. 7.
Class II HDACs negatively regulate nSMase2 expression. MCF7 cells were seeded into 60 mm dishes and stimulated with vehicle (DMSO), TSA (300 nM), vorinostat (10 μM), LMK (5 μM), and mocestinostat (5 μM) for 24 h. A: Proteins were extracted in RIPA buffer and analyzed for Ac-H3 (K9), H3, and nSMase2 levels by immunoblot. Actin was used as loading control (*** P < 0.001, n = 4). B: RNA was extracted and converted to cDNA, and nSMase2 expression analyzed by qRT-PCR using actin as reference gene (*** P < 0.001, n = 5).
DISCUSSION
In this study, we have explored the regulation of nSMase2 by ATRA in MCF7 cells. We report that ATRA regulation of nSMase2 occurs transcriptionally through RAR-α and is independent of both previously reported transcriptional regulators of nSMase2 (Runx2, Sp1 and Sp3, and RNS) and other candidate transcription factors induced by ATRA. Instead, ATRA was found to induce nSMase2 epigenetically, increasing histone acetylation at the nSMase2 gene, most likely through CBP/p300. Additionally, a class II HDAC, most likely HDAC4 and/or HDAC5, negatively regulates nSMase2 expression. This identifies a novel pathway that regulates nSMase2 expression, placing nSMase2 in the subset of ATRA-induced genes regulated by CBP and p300. Moreover, these data also suggest that physiological or pharmacological modulation of histone acetylation can directly affect nSMase2 levels.
To date, multiple studies have identified posttranslational regulators of nSMase2 in acute responses including p38 MAPK (7, 12), EEA1 (15), calcineurin (14), matrix metalloproteinases and integrins (9), as well as suggesting a role for phosphorylation in nSMase2 regulation (14, 16). However, recent emerging evidence has pointed to a more protracted regulation of nSMase2 occurring at the level of expression. Indeed, increased expression of nSMase2 has been reported in response to chemotherapies (19, 21), BMP2 (17, 18), confluence (6), and ATRA (5, 22, 23). Previously, we characterized nSMase2 as an early ATRA response gene (<6 h of ATRA treatment) that was important for ATRA-induced growth arrest of breast epithelial cells (5). Here, we have extended this work to define the mechanism by which ATRA regulates nSMase2 expression.
Cellular effects of ATRA are primarily mediated by transcriptional upregulation of genes through binding to nuclear RAR and retinoid X receptor. Results here demonstrate that nSMase2 is transcriptionally upregulated by ATRA, as shown by increased hnRNA, and place the Smpd3 gene downstream of RAR-α, as shown by the use of specific agonists, antagonists, and the lack of response of MDA-MB-231 cells to ATRA. In ChIP experiments, we also observed enrichment of RAR-α at the nSMase2 gene (data not shown), as was seen before (22). Collectively, these data agree with previous studies in MCF7 and T47D cells (9, 22, 23); moreover, the role that nSMase2 plays in ATRA-induced growth arrest (5) is consistent with the well-established antiproliferative role of RAR-α (23, 29, 40, 41). Additionally, as numerous studies have found that loss of RAR-α signaling leads to alterations in mammary acini formation both in vivo and in vitro (27, 42–44), this suggests that the role of nSMase2 in ATRA-induced growth arrest may be of particular relevance in mammary physiology. However, it should also be noted that the ATRA doses of 1 μM used in the current study are more relevant pharmacologically. Thus, further study utilizing physiological doses of ATRA (<100 nM) would be required to determine whether nSMase2 is of importance in these contexts. Nonetheless, we had previously observed upregulation of nSMase2 at physiological doses, albeit not to the same extent as the pharmacological concentrations used here (7).
Currently, only a handful of studies have investigated transcriptional regulation of nSMase2 in response to BMP2 (17, 18), ATRA (22), daunorubicin (19), and cyclopamine (21) demonstrating both increased nSMase2 mRNA and promoter activity. This research placed Runx 2 (17, 18), Sp1 and Sp3 (19, 22), and RNS (21) upstream of nSMase2 transcription. Here, although we confirmed that ATRA induced nSMase2 transcriptionally, this did not translate to increased activity of the Smpd3 promoter. The lack of effect of ATRA on an exogenous promoter is consistent with its regulation being epigenetic, as transfected plasmids do not have the same chromatin structure as endogenous genomic DNA, although why this would result in a decrease of promoter activity by ATRA is unclear. Indeed, this was not due to the promoter constructs used as the chemotherapeutic agent doxorobucin increased promoter activity, similar to its analog daunorubicin as reported previously (19). However, while we cannot completely rule out effects of ATRA on the nSMase2 promoter, we contend that the significant epigenetic effects of ATRA on nSMase2 would outweigh any such contribution. That being said, the identity of the newly synthesized intermediate factor revealed by CHX treatment (Fig. 1) and its relation to the epigenetic regulation of nSMase2 remain unclear. Our data do not reveal a specific transcription factor; thus, it is possible that recruitment or activation of CBP or p300 at the RAR-α receptor (as discussed below) requires a newly synthesized intermediary protein. Beyond this, importantly, our results indicated that Sp1 and Sp3 are not required for ATRA effects on nSMase2 expression; indeed, nSMase2 was clearly induced despite >90% knockdown of either Sp1 or Sp3. Moreover, the induction of nSMase2 in the presence of a double Sp1/Sp3 knockdown ruled out possible compensatory effects (data not shown). Notably, this contrasts with previous work on ATRA regulation of nSMase2 (22), which reported that ATRA enhances Smpd3 promoter activity and placed Sp1 and Sp3 upstream of nSMase2 induction. The reasons for these discrepancies are not wholly clear as both studies utilize MCF7 cells. One possible explanation relates to dose of ATRA used; here we have used a pharmacological 1 μM dose compared with 10 μM used previously. Thus, Sp1 and Sp3 may become more relevant for nSMase2 induction as dose increases. However, it should also be noted that our study utilized specific siRNA for Sp1 and Sp3, whereas previous work used the pharmacological inhibitor mithramycin A. Thus, the possibility of off-target effects cannot be ruled out. Beyond Sp1 and Sp3, none of the reported regulators of nSMase2 were required for the ATRA response in our hands as demonstrated by siRNA (Runx2) and inhibitors (NAC). Taken together, this points to a distinct pathway of nSMase2 regulation in ATRA-stimulated MCF7 cells.
Upon binding to its receptors, ATRA induces conformational changes allowing release of corepressors and recruitment of coactivators such as CBP and p300 (33, 34). Furthermore, a loss of ATRA signaling can result in repressive methylation of downstream target genes (29, 30). This, together with the lack of effect of ATRA on promoter activity, led us to explore epigenetic mechanisms of regulation, and our data clearly indicate that ATRA primarily regulates nSMase2 through directly modulating histone acetylation, which is most likely through CBP/p300. This is supported by multiple lines of evidence: 1) the nSMase2 gene is not repressively hypermethylated in MCF7 cells suggesting alterations in methylation are not relevant for the ATRA response; 2) HDAC inhibition (TSA) induces nSMase2 comparably to ATRA and does not have additive responses when combined; 3) ATRA stimulation increased enrichment of Ac-H3 at the nSMase2 gene but not at the ATRA-unresponsive nSMase3 gene; and 4) inhibition of HAT pharmacologically or with CBP/p300 siRNA blunted ATRA induction of nSMase2 both at the protein and mRNA levels. Collectively, these results identify two novel regulators of nSMase2 and place nSMase2 as part of the CBP and p300 responses. Functionally, the role of CBP and p300 in the cellular effects of ATRA in F9 cells was previously investigated (35), and interestingly, it was found that although both were important for ATRA induction of genes, each protein controlled different biologies through regulating distinct sets of genes. Thus, CBP was important for ATRA-induced growth arrest and apoptosis, while p300 seemed to also be important for ATRA-induced differentiation. Notably, as both CBP and p300 seem to be upstream of nSMase2, this would imply that nSMase2 might have roles within all of these outcomes. Thus, we speculate that nSMase2 may be a “core” gene of ATRA responses, which would be consistent with its early induction time in MCF7 cells (5). According to this hypothesis, additional targets of CBP and p300 would then determine specific cellular responses. This scenario is also consistent with the various functions of nSMase2 that depend on specific cell types even with the same stimulus. It will be particularly important in future studies to determine whether the relationships between CBP, p300, and nSMase2 hold in vivo and if these relationships are also relevant for physiological roles of ATRA such as in mammary gland development as discussed above.
Finally, in addition to identifying CBP and p300 as positive regulators of nSMase2 expression, we also explored the regulation of nSMase2 by HDACs. Using class-specific inhibitors, data suggest that class II HDACs, specifically HDAC4 and/or HDAC5, are primary candidates for inhibiting nSMase2 expression. While this suggests that physiological and pharmacological modulation of histone acetylation can induce nSMase2, it should be noted that mocestinostat increased overall Ac-H3 to comparable levels as TSA, vorinostat, and LMK, yet was unable to induce nSMase2. One possible explanation is that while mocestinostat increases Ac-H3 in general, this increase occurs at genes other than nSMase2, dependent on genomic localization of the specific HDACs targeted. Alternatively, it is equally plausible that this single acetylation event alone is insufficient for nSMase2 induction and that additional acetylation events that are unaffected by mocestinostat are required for full nSMase2 induction to occur. Future studies probing the significance of the epigenetic regulation of nSMase2 will be able to explore this further. Notably, HDAC activities are often dysregulated in cancer. For example, HDAC4 is important for maintenance of colon cancer cell proliferation (45). Similarly, HDAC5 is upregulated in HCC and can promote cell proliferation (46). Strikingly, this is consistent with the reported decrease in nSMase2 reported in HCC (31). Indeed, it would be of particular interest to determine whether a loss of nSMase2 expression is relevant in these or other pathologies where HDAC activities are increased.
In summary, this study has found that ATRA induces nSMase2 through epigenetic modulation of histones and independently of promoter activity in MCF7 cells. Moreover, we have identified CBP and p300 as novel upstream regulators of nSMase2, find that HDAC inhibition alone is sufficient to increase nSMase2 expression, and identify HDAC4/5 as candidates for negative regulators of nSMase2. Collectively, these results suggest that modulation of histone acetylation is a primary mechanism by which nSMase2 expression is regulated. Indeed, we have observed comparable effects of TSA across multiple cell lines (data not shown). Furthermore, given the increasing development of HDAC inhibitors such as vorinostat as therapeutic agents in a variety of cancers, it would be particularly interesting to investigate whether nSMase2 plays a functional or biomarker role in the therapeutic response to such agents.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Cungui Mao and Ruijuan Xu for their advice and expertise on ChIP, and Drs. Chiara Luberto and Can Senkal for their constructive comments on the manuscript.
Footnotes
Abbreviations:
- ATRA
- all-trans retinoic acid
- BMP2
- bone morphogenetic protein-2
- CBP
- CREB-binding protein
- Cer
- ceramide
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- 5-aza
- 5-azacytidine
- 4HPR
- fenretinide
- HAT
- histone acetyltransferase
- HCC
- hepatocellular carcinoma
- HDAC
- histone deacetylase
- hnRNA
- heteronuclear RNA
- NAC
- N-acetylcysteine
- nSMase
- neutral sphingomyelinase
- PKC-δ
- protein kinase C-δ qRT-PCR, quantitative RT-PCR
- RAR
- retinoic acid receptor
- RNS
- reactive nitrogen species
- ROS
- reactive oxygen species
- TSA
- trichostatin A
This work was supported by National Institutes of Health Grant GM43815 (to Y.A.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Hannun Y. A., and Obeid L. M.. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 2.Wu B. X., Clarke C. J., and Hannun Y. A.. 2010. Mammalian neutral sphingomyelinases: regulation and roles in cell signalling responses. Neuromolecular Med. 12: 320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins R. W., Canals D., and Hannun Y. A.. 2009. Roles and regulation of secretory and lysosomal acid sphingomyelianase. Cell. Signal. 21: 836–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chipuk J. E., McStay G. P., Bharti A., Kuwana T., Schafer B., Clarke C. J., Siskind L. J., Obeid L. M., and Green D. R.. 2012. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 148: 988–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke C. J., Mediwala K., Jenkins R. W., Sutton C. A., Tholanikunnel B. G., and Hannun Y. A.. 2011. Neutral sphingomyelinase-2 mediates growth arrest by retinoic acid through modulation of ribosomal S6 kinase. J. Biol. Chem. 286: 21565–21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchesini N., Osta W., Bielawski J., Luberto C., Obeid L. M., and Hannun Y. A.. 2004. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J. Biol. Chem. 279: 25101–25111. [DOI] [PubMed] [Google Scholar]
- 7.Clarke C. J., Truong T-G., and Hannun Y. A.. 2007. Role for neutral sphingomyelinase-2 in tumor necrosis factor alpha-stimulated expression of vascular cell adhesion molecule-1 (VCAM1) and intercellular adhesion molecule-1 (ICAM1) in lung epithelial cells: p38 MAPK is an upstream regulator of nSMase2. J. Biol. Chem. 282: 1384–1396. [DOI] [PubMed] [Google Scholar]
- 8.Maupas-Schwalm F., Bedel A., Augé N., Grazide M. H., Mucher E., Thiers J. C., Salvayre R., and Nègre-Salvayre A.. 2009. Integrin alpha(v) beta(3), metalloproteinases, and sphingomyelinase-2 mediate urokinase mitogenic effect. Cell. Signal. 21: 1925–1934. [DOI] [PubMed] [Google Scholar]
- 9.Rutkute K., Karakashian A. A., Giltiay N. V., Dobierzewska A., and Nikolova-Karakashian M. N.. 2007. Aging in rat causes hepatic hyperresposiveness to interleukin-1beta which is mediated by neutral sphingomyelinase-2. Hepatology. 46: 1166–1176. [DOI] [PubMed] [Google Scholar]
- 10.Khavandgar Z., Alebrahim S., Eimar H., Tamimi F., McKee M. D., and Murshed M.. 2013. Local regulation of tooth mineralization by sphingomyelinase phosphodiesterase 3. J. Dent. Res. 92: 358–364. [DOI] [PubMed] [Google Scholar]
- 11.Khavandgar Z., Poirier C., Clarke C. J., Li J. J., Wang N., McKee M. D., Hannun Y. A., and Murshed M.. 2011. A cell-autonomous requirement for neutral sphingomyelinase-2 in bone mineralization. J. Cell Biol. 194: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohanian J., Forman S. P., Katzenberg G., and Ohanian V.. 2012. Age-related remodeling of small arteries is accompanied by increased sphingomyelinase activity and accumulation of long-chain ceramides. J. Vasc. Res. 49: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke C. J., Guthrie J. M., and Hannun Y. A.. 2008. Regulation of neutral sphingomyelinase-2 by tumor necrosis factor-alpha involves protein kinase C-delta in lung epithelial cells. Mol. Pharmacol. 74: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filosto S., Fry W., Knowlton A. A., and Goldkorn T.. 2010. Neutral sphingomyelinase-2 (nSMase2) is a phosphoprotein regulated by calcineurin (PP2B). J. Biol. Chem. 285: 10213–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philipp S., Puchert M., Adam-Klages S., Tchikov V., Winoto-Morbach S., Mathieu S., Deerberg A., Kolker L., Marchesini N., Kabelitz D., et al. 2010. The Polycomb group protein EED couples TNF receptor 1 to neutral sphingomyelinase. Proc. Natl. Acad. Sci. USA. 107: 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filosto S., Ashfaq M., Chung S., Fry W., and Goldkorn T.. 2012. Neutral sphingomyelinase 2 activity and protein stability are modulated by phosphorylation of five conserved serines. J. Biol. Chem. 287: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakoi H., Maeda S., Shinohara N., Matsuyama K., Imamura K., Kawamura I., Nagano S., Setoguchi T., Yokouchi M., Ishidou Y., et al. 2014. Bone morphogenic protein (BMP) signaling up-regulates neutral sphingomyelinase 2 to suppress chondrocyte maturation via the Akt protein signaling pathway as a negative feedback mechanism. J. Biol. Chem. 289: 8135–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae Y. M., Heo S. H., Kim J. Y., Lee J. M., Ryoo H. M., and Cho J. Y.. 2009. Upregulation of smpd3 via BMP2 stimulation and Runx2. BMB Rep. 42: 86–90. [DOI] [PubMed] [Google Scholar]
- 19.Ito H., Murakami M., Furuhata A., Gao S., Yoshida K., Sobue S., Hagiwara K., Takagi A., Kojima T., Suzuki M., et al. 2009. Transcriptional regulation of neutral sphingomyelinase 2 gene expression of a human breast cancer cell line, MCF-7, induced by the anti-cancer drug, daunorubicin. Biochim. Biophys. Acta. 1789: 681–690. [DOI] [PubMed] [Google Scholar]
- 20.Filosto S., Castillo S., Danielson A., Franzi L., Khan E., Kenyon N., Last J., Pinkerton K., Tuder R., and Goldkorn T.. 2011. Neutral sphingomyelinase 2: a novel target in cigarette smoke-induced apoptosis and lung injury. Am. J. Respir. Cell Mol. Biol. 44: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers-Needham M., Lewis J., Gencer S., Sentelle R. D., Clarke C. J., Hannun Y. A., Norell H., Nishimura M., Khavandgar Z., Murshed M., et al. 2012. Off-target function of the Sonic hedgehog inhibitor cyclopamine in mediating apoptosis via nitric oxide-dependent neutral sphingomyelinase 2/ceramide induction. Mol. Cancer Ther. 11: 1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito H., Tanaka K., Hagiwara K., Kobayashi M., Hoshikawa A., Mizutani N., Takagi A., Kojima T., Sobue S., Ichihara M., et al. 2012. Transcriptional regulation of neutral sphingomyelinase 2 in all-trans retinoic acid-treated human breast cancer cell line, MCF-7. J. Biochem. 151: 599–610. [DOI] [PubMed] [Google Scholar]
- 23.Somenzi G., Sala G., Rossetti S., Ren M., Ghidoni R., and Sacchi N.. 2007. Disruption of retinoic acid receptor alpha reveals the growth promoter face of retinoic acid. PLoS One. 2: e836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabichi A. L., Xu H., Fischer S., Zao C., Yang X., Steele V. E., Kelloff G. J., Lotan R., and Clifford J. L.. 2003. Retinoid receptor-dependent and independent biological activities of novel fenretinide analogues and metabolites. Clin. Cancer Res. 9: 4606–4613. [PubMed] [Google Scholar]
- 25.Terao M., Fratelli M., Kurosaki M., Zanetti A., Guarnaccia V., Paroni G., Tsykin A., Lupi M., Gianni M., Goodall G. J., et al. 2011. Induction of miR-21 by retinoic acid in estrogen receptor-positive breast carcinoma cells: biological correlates and molecular targets. J. Biol. Chem. 286: 4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K. S., Choi H. W., Yoon H. E., and Kim I. Y.. 2010. Reactive oxygen species generated by NADPH Oxidase 2 and 4 are required for chondrogenic differentiation. J. Biol. Chem. 285: 40294–40302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y-B., Gao J-L., Zhong Z-F., Hoi P-M., Lee S. M., and Wang Y-T.. 2013. Bisdemethoxycurcumin suppresses MCF-7 cell proliferation by inducing ROS accumulation and modulating senescence-related pathways. Pharmacol. Rep. 65: 700–709. [DOI] [PubMed] [Google Scholar]
- 28.Afonja O., Raaka B. M., Huang A., Das S., Zhao X., Helmer E., Juste D., and Samuels H. H.. 2002. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene. 21: 7850–7860. [DOI] [PubMed] [Google Scholar]
- 29.Corlazzoli F., Rossetti S., Bistulfi G., Ren M., and Sacchi N.. 2009. Derangement of a factor upstream of RARalpha triggers the repression of a pleiotropic epigenetic network. PLoS One. 4: e4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bistulfi G., Pozzi S., Ren M., Rossetti S., and Sacchi N.. 2006. A repressive epigenetic domino effect confers susceptibility to breast epithelial cell transformation: implications for predicting breast cancer risk. Cancer Res. 66: 10308–10314. [DOI] [PubMed] [Google Scholar]
- 31.Revill K., Wang T., Lachenmayer A., Kojima K., Harrington A., Li J., Hoshida Y., Llovet J. M., and Powers S.. 2013. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 145: 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demircan B., Dyer L. M., Gerace M., Lobenhofer E. K., Robertson K. D., and Brown K. D.. 2009. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer. 48: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilworth F. J., and Chambon P.. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 20: 3047–3054. [DOI] [PubMed] [Google Scholar]
- 34.Bastien J., and Rochette-Egly C.. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 328: 1–16. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki H., Eckner R., Yao T-P., Taira K., Chiu R., Livingston D. M., and Yokoyama K. K.. 1998. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 393: 284–289. [DOI] [PubMed] [Google Scholar]
- 36.Dietze E. C., Troch M. M., Bowie M. L., Yee L., Bean G. R., and Seewaldt V. L.. 2003. CBP/p300 induction is required for retinoic acid sensitivity in human mammary cells. Biochem. Biophys. Res. Commun. 302: 841–848. [DOI] [PubMed] [Google Scholar]
- 37.Chakravarti D., LaMorte V. J., Nelson M. C., Nakajimi T., Schulman I. G., Jugulion H., Montminy M., and Evans R. M.. 1996. Role of CBP/P300 in nuclear receptor signalling. Nature. 383: 99–103. [DOI] [PubMed] [Google Scholar]
- 38.Chen H. P., Zhao Y. T., and Zhao T. C.. 2015. Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 20: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberharter A., and Becker P. B.. 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seewaldt V. L., Caldwell L. E., Johnson B. S., Swisshelm K., Collins S. J., and Tsai S.. 1997. Inhibition of retinoic acid receptor function in normal human mammary epithelial cells results in increased cellular proliferation and inhibits the formation of a polarized epithelium in vitro. Exp. Cell Res. 236: 16–28. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y., Bertran S., Samuels T-A., Mira-y-Lopez R., and Farias E.. 2010. Mechanism of inhibition of MMTV-neu and MMTV-wnt1 induced mammary oncogenesis by RARalpha agonist AM580. Oncogene. 29: 3665–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohn E., Ossowski L., Bertran S., Marzan C., and Farias E. F.. 2010. RARα1 control of mammary gland ductal morphogenesis and wnt1-tumorigenesis. Breast Cancer Res. 12: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Offterdinger M., Schneider S. M., and Grunt T. W.. 2003. Heregulin and retinoids synergistically induce branching morphogenesis of breast cancer cells cultivated in 3D collagen gels. J. Cell. Physiol. 195: 260–275. [DOI] [PubMed] [Google Scholar]
- 44.Cho K-W., Kwon H-J., Shin J-O., Lee J-M., Cho S-W., Tickle C., and Jung H-S.. 2012. Retinoic acid signaling and the initiation of mammary gland development. Dev. Biol. 365: 259–266. [DOI] [PubMed] [Google Scholar]
- 45.Wilson A. J., Byun D-S., Nasser S., Murray L. B., Ayyanar K., Arango D., Figueroa M., Melnick A., Kao G. D., Augenlicht L. H., et al. 2008. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol. Biol. Cell. 19: 4062–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan J., Lou B., Chen W., Zhang J., Lin S., Lv F. F., and Chen Y.. 2014. Down-regulation of HDAC5 inhibits growth of human hepatocellular carcinoma by induction of apoptosis and cell cycle arrest. Tumour Biol. 35: 11523–11532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.