Abstract
The Polycomb Repressive Complexes PRC1 and PRC2 maintain embryonic stem cell (ESC) pluripotency by silencing lineage-specifying developmental regulator genes1. Emerging evidence suggests that Polycomb complexes act through controlling spatial genome organisation2–9. We show that PRC1 functions as a master regulator of ESC genome architecture by organizing genes in three-dimensional interaction networks. The strongest spatial network is composed of the four Hox clusters and early developmental transcription factor genes, the majority of which contact poised enhancers. Removal of Polycomb repression leads to disruption of promoter-promoter contacts in the Hox network. In contrast, promoter-enhancer contacts are maintained, accompanied by widespread acquisition of active chromatin signatures at network enhancers and pronounced transcriptional up-regulation of network genes. Thus, PRC1 physically constrains developmental transcription factor genes and their enhancers in a silenced but poised spatial network. We propose that selective release of genes from this spatial network underlies cell fate specification during early embryonic development.
To understand the potential role of spatial genome organization in maintaining ESC self-renewal potential, we analyzed mouse ESC Promoter Capture Hi-C (CHi-C) data using GOTHiC (Genome Organization Through Hi-C) (B.M. et al., manuscript in preparation). Spatial genome architecture can be interrogated at high resolution by combining sequence capture with 3C10 or Hi-C11. Promoter CHi-C specifically enriches Hi-C12 libraries for promoters and their spatially contacting DNA elements13,14, providing genome wide, restriction fragment resolution chromosomal contact data for 22,225 annotated mouse promoters14. Our unbiased approach detected a strong enrichment for long-range contacts between promoters bound by Polycomb group proteins, especially the PRC1 component RING1B (Supplementary Fig. 1a). In particular, amongst the spatially interacting Polycomb-bound genes, analysis of promoter-promoter contacts revealed an unusually strong intra- and inter-chromosomal spatial network between gene promoters of the four Hox clusters (HoxA, HoxB, HoxC and HoxD). We detected extensive contacts between gene promoters across all four Hox clusters with particularly strong connections between HoxB-HoxC and HoxB-HoxA (Supplementary Fig. 1b). In addition, after filtering promoters < 10 Mb from the Hox clusters to exclude contacts based merely on genomic proximity, we found 66 other gene promoters with strong direct intra- and inter-chromosomal contacts to the Hox clusters in a central position within the global promoter network (Fig. 1a, b, Supplementary Fig. 1c, and Supplementary Table 1).
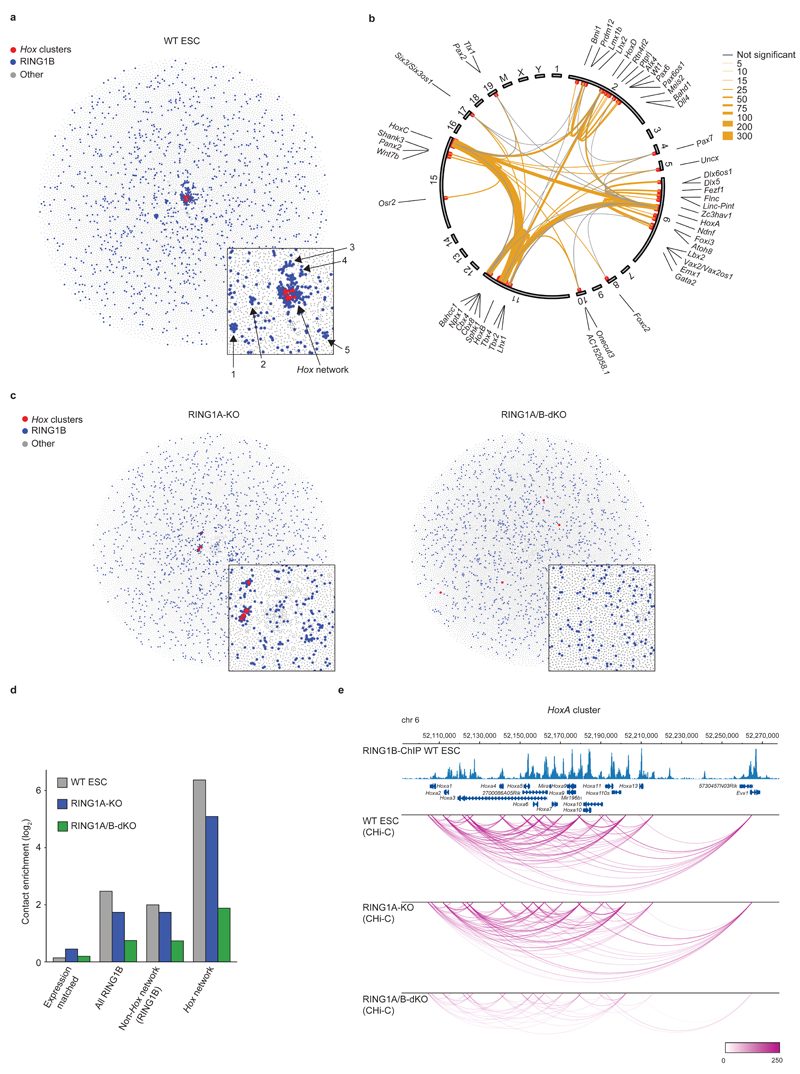
Figure 1 |. PRC1 organizes 3D promoter-promoter contact networks in ESCs.
a, Network map of spatial promoter connectivity in WT mouse ESCs. Red nodes: Hox clusters; blue nodes: RING1B-occupied promoters (non-Hox cluster); grey nodes: non RING1B-occupied promoters. Promoter nodes are connected by edges (not drawn) representing long-range cis (> 10 Mb) or trans Promoter CHi-C contacts. Promoter nodes are positioned using a force-directed layout such that connected nodes are drawn closer together. Inset; zoom in on the Hox network and the five smaller networks. b, Hox network circos contact map in WT ESCs. Inner circle: chromosomes; outer circle: genes names; red dots: significantly interacting RING1B-bound promoters; edges: significant promoter contacts (orange, weighted by read count) and non-significant contacts (grey, minimum of 3 reads). c, Network analysis in RING1A-KO and RING1A/B-dKO cells as in (a). d, Contact enrichment for Hox network promoters, RING1B non-Hox network promoters, all RING1B-bound promoters, and expression-matched non RING1B-bound promoters. e, Significant promoter-promoter CHi-C contacts (arcs) within the HoxA cluster for WT, RING1A-KO and RING1A/B-dKO cells (WashU Epigenome Browser30). Upper tracks: Genomic coordinates, RefSeq genes, and WT RING1B ChIP-Seq profile17.
Gene Ontology and protein domain analyses show that this Hox network is enriched for promoters of developmental transcription factor genes encoding homeobox and paired box domain proteins, which control body plan specification, morphogenesis and organogenesis (Supplementary Fig. 1d, e). Polycomb group proteins epigenetically repress Hox and developmental transcription factor genes which are enriched for bivalent chromatin marks in ESCs15,16. Analysis of PRC1 occupancy via RING1B ChIP-Seq data17 demonstrates that approximately 80% of the promoters in the Hox network are bound by PRC1 (Fig. 1a, b and Supplementary Table 1), 85.9% of which also harbor the active chromatin mark H3K4me3. We also identified five smaller independent spatial networks, mainly formed by long-range intra-chromosomal contacts between PRC1-bound promoters (Fig. 1a and Supplementary Table 1). The Hox network promoters are distinguished from other PRC1-regulated genes by significantly more enriched RING1B ChIP-Seq peaks (Supplementary Fig. 1f), suggesting they are high affinity, high occupancy PRC1 sites. These data suggest that the Hox clusters act as central 3D nucleation points for high affinity PRC1 bound genes, and that this spatial network is a major constraint on ESC genome organization.
To address the role of PRC1 in this spatial network, we generated Promoter CHi-C libraries from cells lacking one or both genes encoding the catalytic core E3 ubiquitin ligase subunits of all known PRC1 complexes1: RING1A (constitutive RING1A knock out: RING1A-KO) and RING1B (inducible RING1B knock out in a RING1A-KO background: RING1A/B-dKO). RING1A/B-dKO results in undetectable levels of RING1B and H2AK119ub1, whilst cells retain key features of pluripotency (Supplementary Fig. 2a and Supplementary Table 2).
In RING1A-KO cells the Hox network shows reduced connectivity compared to WT ESCs, as do the five smaller networks which are no longer discernible (Fig. 1c, d and Supplementary Fig. 2b). In RING1A/B-dKO cells, the Hox spatial network is completely disrupted (Fig. 1c, d and Supplementary Fig. 2c), demonstrating that PRC1 is essential to maintain the Hox spatial network. We also found that intra-chromosomal contacts within all four Hox clusters are substantially reduced, indicating that PRC1 plays a critical role in maintaining the Hox clusters in compact repressive domain structures (Fig. 1e and Supplementary Fig. 2d-f)18–21.
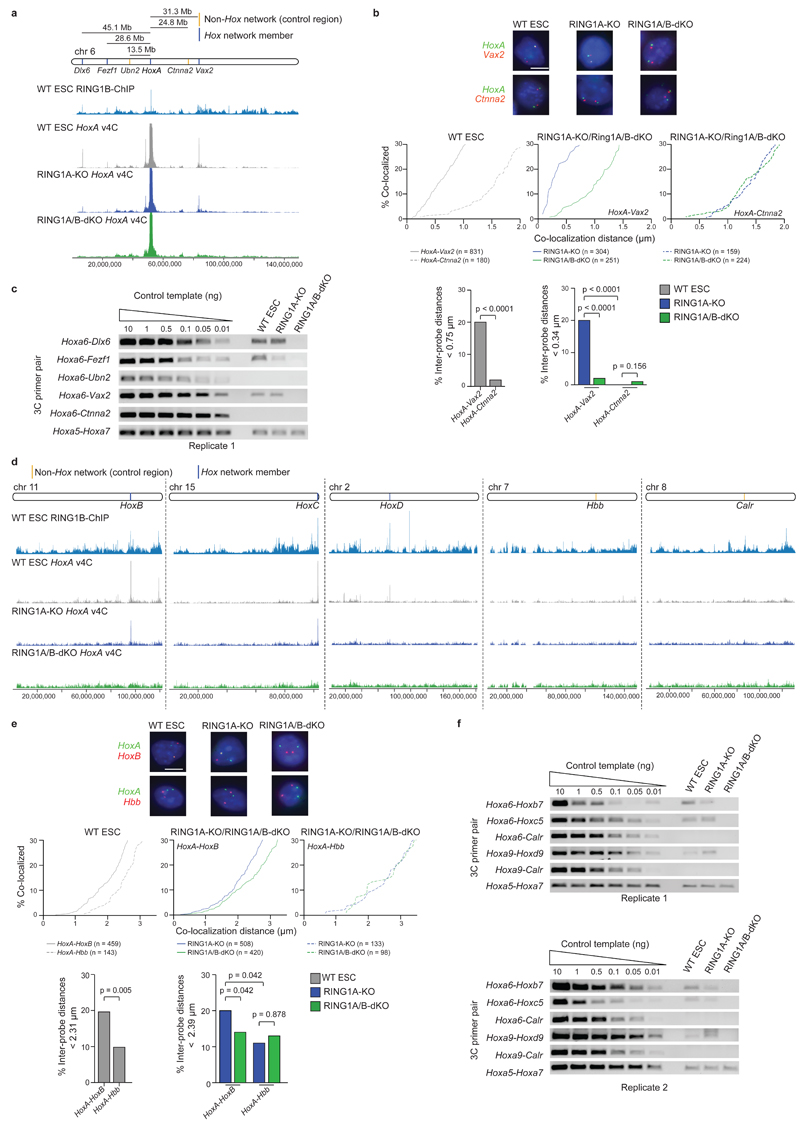
We validated selected PRC1-dependent intra- and inter-chromosomal contacts by 3C and 3D DNA FISH. 3D DNA FISH shows that for the Hox network, genes on the same chromosome are significantly closer than intervening control regions in both WT and RING1A-KO, but not in RING1A/B-dKO cells (Fig. 2a, b and Supplementary Fig. 3). Similarly, inter-chromosomal associations between Hox network members are reduced in RING1A/B-dKO cells (Fig. 2d, e). 3C analyses extend and confirm these results (Fig. 2c, f and Supplementary Fig. 4, 5). Collectively, these data validate the Promoter CHi-C results identifying a complex, PRC1-dependent spatial network centered on the four Hox clusters and key developmental regulator genes.
Figure 2 |. Validation of a PRC1-dependent 3D promoter-promoter contact network in ESCs.
Validation of selected HoxA cis (a-c) and trans (d-f) contacts by double-label 3D DNA FISH and 3C-PCR. a, d, Genomic locations of tested loci, RING1B ChIP-Seq signal from WT ESCs17 and virtual 4C (v4C) contacts, generated from Promoter CHi-C data, using the HoxA cluster as a viewpoint. b, e, Representative double-label 3D DNA FISH images showing probe signals for HoxA, Ctnna2, and Vax2 (b), and HoxA, HoxB and Hbb (e). Scale bars, 5 µm. Cumulative frequency distribution plots show the percentage of alleles co-localizing at increasing distance cut-offs. Bar plots show percentage of inter-probe distances below the specified cut-off, set at the distance which includes the lowest quintile of measurements for Hox network member alleles in WT or RING1A-KO ESCs. P values: Mann-Whitney test (comparison of inter-probe distances within the same cell) and Kruskall-Wallis/Dunn’s multiple comparisons test (comparison of inter-probe distances between RING1A-KO and RING1A/B-dKO cells). c, f, 3C-PCR validation of HoxA cluster long-range contacts for regions identified by Promoter CHi-C as making PRC1-dependent contacts with HoxA in cis (c) (Dlx6, Fezf1, and Vax2) and trans (f) (HoxB, HoxC, and HoxD); and control regions identified as non-HoxA contacting regions in cis (c) (Ubn2, Ctnna2) and trans (f) (Calr). Full-length gels are presented in Supplementary Data Set 1.
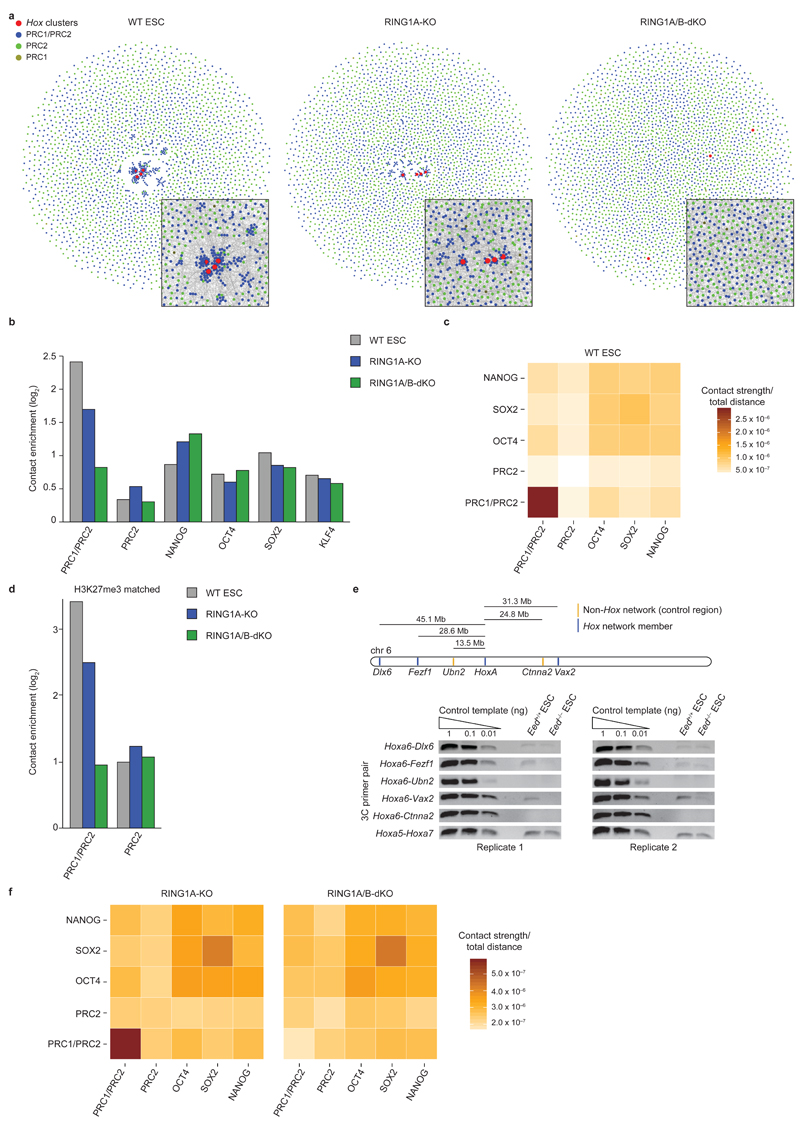
Recent work has implicated EED, a PRC2 complex component, in mediating contacts between Hox clusters5. However the relative contributions of PRC1 and PRC2 to spatial chromosome organization are unknown. We used ChIP-Seq data (Supplementary Table 3) to classify polycomb-bound promoters as ‘PRC2’ or ‘PRC1/PRC2’ bound (note that 98% of PRC1-bound promoters are also occupied by PRC2) (Supplementary Fig. 6a, b), and compared the spatial network connectivity of these promoters. We found the Hox network again to be highly centralized and composed almost exclusively of PRC1/PRC2 promoters with extremely high connectivity (Fig. 3a). In general we found that PRC1/PRC2 promoters have substantially higher connectivity than PRC2 promoters (Fig. 3a-c). PRC1/PRC2 promoters generally have higher occupancy of EZH2 and SUZ12 (two PRC2 complex components) (Supplementary Fig. 6b), and more often overlap with H3K27me3 peaks than PRC2 promoters (Supplementary Fig. 6a). However even when comparing promoter subsets with similar H3K27me3 or EZH2/SUZ12 occupancy, connectivity between PRC1/PRC2 promoters is higher than between PRC2 promoters (Fig. 3d and data not shown). We next interrogated selected Hox cluster long-range interactions in EED-KO ESCs15 (Supplementary Fig. 6c) by 3C. We found that Hox cluster interactions are still detectable in EED-KO cells, although in some cases at reduced ligation product frequencies compared to WT ESCs (Fig. 3e and Supplementary Fig. 6d-g).
Figure 3 |. PRC1 is a key regulator of 3D genome architecture in ESCs.
a, Network maps of spatial connectivity between PRC-bound gene promoters (display as in Fig. 1a). Red nodes: Hox gene promoters; blue nodes: PRC1/PRC2-bound promoters; green nodes: PRC2-bound promoters; gold nodes: PRC1-bound promoters. Insets: zoom-in of network center, but showing edges representing significant promoter contacts. b, Contact enrichment for PRC1/PRC2, PRC2, and pluripotency factor occupied promoters. c, Cumulative contact strengths between polycomb and pluripotency factor bound promoters in WT ESCs, normalized with the sum of all possible contact-distances within that group. d, Contact enrichment for subsets of PRC1/PRC2- and PRC2-bound promoters with matched H3K27me3 occupancy. e, 3C-PCR analyses of HoxA contacts in WT (Eed+/+) and EED-KO (Eed-/-) ESCs. Full-length gels are presented in Supplementary Data Set 1. f, Cumulative contact strengths between polycomb and pluripotency factor bound promoters in RING1A-KO and RING1A/B-dKO cells, normalized with the sum of all possible contact-distances within that group.
We observed that PRC1/PRC2 promoters are spatially segregated from, and have significantly stronger connectivity than the previously identified 3D pluripotency networks14,22 which are formed through contacts between promoters bound by OCT4, SOX2 and NANOG (Fig. 3b, c). The pluripotency networks are largely unaltered in RING1A/B-dKO ESCs (Fig. 3b, f and Supplementary Fig. 7a). In contrast, genes in linear genomic proximity to PRC1-bound genes show higher co-localization than control genes, and this co-localization is reduced in RING1A/B-dKO cells (Supplementary Fig. 7b), indicative of ‘bystander’ effects as a result of spatial genome rearrangement upon loss of RING1B. Collectively, our data identify PRC1 as a major regulator of 3D genome architecture in ESCs.
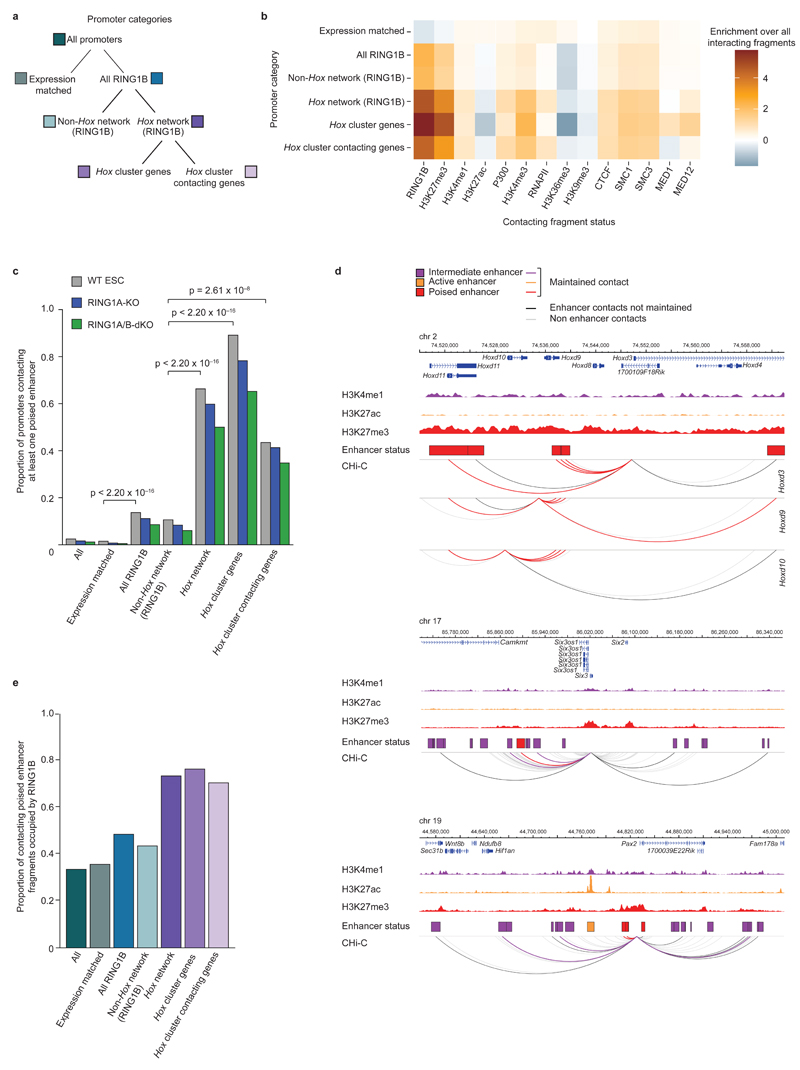
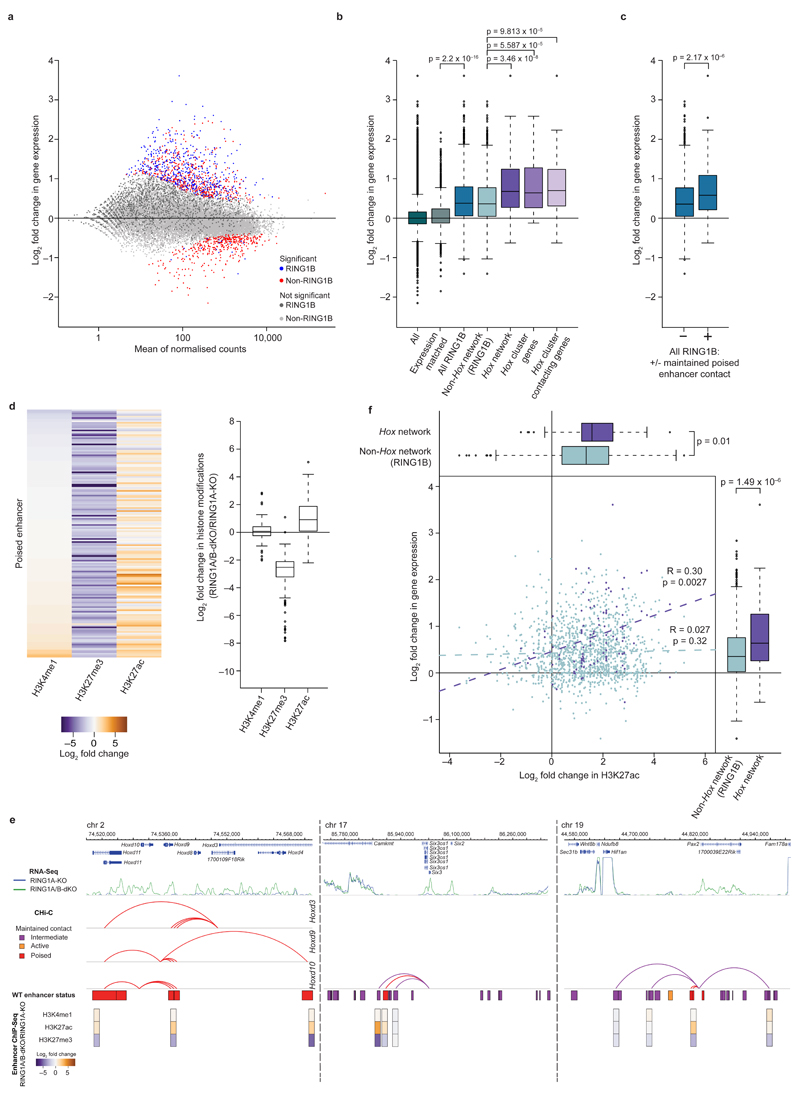
Following on from our analysis of promoter-promoter contacts, we next focused on promoter contacts with putative regulatory (non-promoter) elements (Supplementary Table 2), by integrating ChIP-Seq data (Supplementary Table 3) with Promoter CHi-C data. We found that Hox network promoters contact genomic regions enriched for RING1B, H3K4me1, H3K4me3, H3K27me3, P300, CTCF, and cohesin, but depleted for H3K27ac, H3K36me3 and H3K9me3 (Fig. 4a, b). Combinations of histone marks have been used to distinguish functional classes of enhancers23–25. To characterize RING1B-bound promoter contacts with enhancers, we classified enhancers as previously described23–25: active (H3K4me1 and H3K27ac), intermediate (H3K4me1) or poised (H3K4me1 and H3K27me3) (Supplementary Table 4). We found that Hox network promoters and (to a lesser but still highly significant extent) RING1B-bound promoters are highly enriched in contacts with poised enhancers compared to expression-matched promoters (Fig. 4c and Supplementary Fig. 8a, b). The majority of these Hox network promoters maintain contacts with at least one poised enhancer in RING1A/B-dKO cells (Fig. 4c, d), demonstrating that RING1B is not necessary to maintain these contacts, despite a substantial fraction of these enhancers being occupied by RING1B in WT ESCs (Fig. 4e). Thus, RING1B is necessary to maintain promoter-promoter contacts, but not promoter-enhancer contacts in the Hox network.
Figure 4 |. PRC1-bound promoters preferentially contact poised enhancers.
a, Schematic of promoter categories used for subsequent analyses. b, Heat map showing the enrichment/depletion of histone modifications and chromatin proteins at promoter-contacting regions. c, Proportion of promoters contacting one or more poised enhancers in WT ESCs (grey), and maintaining one or more poised enhancer contacts in RING1A-KO (blue) and RING1A/B-dKO (green) cells. P values: Fisher’s exact test (WT ESC proportions). d, WT ESC CHi-C contact profiles of Hoxd9/Hoxd10/Hoxd13, Six3 and Pax2 promoters. Upper track within each panel: genomic coordinates and RefSeq genes. Middle tracks: H3K4me1, H3K27ac, and H3K27me3 ChIP-Seq signals; boxes represent HindIII fragments overlapping an enhancer (purple: intermediate; orange: active; red: poised). Lower track: Promoter CHi-C contacts for viewpoints are displayed as arcs (WashU Epigenome Browser30). Purple, orange and red arcs: promoter-enhancer contacts maintained in RING1A/B-dKO cells. Black arcs: promoter-enhancer contacts lost in RING1A/B-dKO cells. Grey arcs: promoter contacts with non-enhancer regions. e, WT RING1B occupancy at poised enhancers (for enhancers with maintained promoter contacts).
To correlate 3D promoter-promoter and promoter-enhancer circuitry with transcriptional output, we generated nuclear RNA-Seq libraries from RING1A-KO and RING1A/B-dKO ESCs. In agreement with previous microarray steady-state expression analysis26, RING1B-occupied promoters show significant de-repression in RING1A/B-dKO cells (Fig. 5a, b). RING1B target genes preferentially form new contacts with active genes in RING1A/B-dKO cells (Supplementary Fig. 8c), concomitant with their loss of contacts with other RING1B-regulated genes (Fig. 1d). Strikingly, Hox network genes, most of which maintain contacts with poised enhancers (Fig. 4c), show the most significant transcriptional up-regulation, correlating with the disruption of promoter-promoter contacts within the network (Fig. 5b). We further find that the subset of RING1B-regulated genes which maintain contacts with poised enhancers also show significant up-regulation (Fig. 5c). In contrast, only a small proportion of RING1B-occupied promoters (25.9%) gain contacts with active enhancers, and this correlates poorly with changes in gene expression (Supplementary Fig. 8d, e). This shows that gaining contacts with active enhancers is not the major mechanism of transcriptional up-regulation at PRC1 target genes. Thus, our data suggest that silencing is maintained through PRC1-mediated promoter-promoter contacts, and that pre-formed contacts between promoters and poised enhancers may play a role in transcriptional up-regulation upon PRC1 removal.
Figure 5 |. The Hox spatial network is poised for transcription.
a, Expression in RING1A/B-dKO relative to RING1A-KO cells (nuclear RNA-Seq). Blue, red: significant changes (FDR < 0.05); light and dark grey: non-significant changes. Dark grey, blue: RING1B-occupied promoters. b, Expression in RING1A/B-dKO relative to RING1A-KO cells, grouped by promoter categories. Boxplots: median, interquartile range, range and outliers (> 1.5 times the interquartile range away from the box). P values: Wilcoxon rank-sum test. c, Expression in RING1A/B-dKO relative to RING1A-KO cells for RING1B-occupied promoters with (+) or without (−) a maintained poised enhancer contact. P value: Wilcoxon rank-sum test. d, Histone modification changes in RING1A/B-dKO relative to RING1A-KO cells at poised enhancers with maintained RING1B promoter contacts. Left: heat map representing individual enhancers. Right: boxplot summaries. e, Transcription, Promoter CHi-C contacts and contacting enhancer status for Hoxd9/Hoxd10/Hoxd13, Six3 and Pax2. Upper tracks: genomic coordinates, RefSeq genes and RNA-Seq signal (RING1A-KO and RING1A/B-dKO cells). Middle tracks: arcs30 represent promoter-enhancer contacts maintained in RING1A/B-dKO cells, boxes show HindIII fragments overlapping enhancers defined in WT cells. Lower tracks: histone modification changes at enhancers in RING1A/B-dKO relative to RING1A-KO cells (boxes centered on the precise enhancer location). f, Scatter plot: Enhancer H3K27ac and gene expression changes in RING1A/B-dKO relative to RING1A-KO cells, for RING1B Hox network promoters (purple) and RING1B non-Hox network promoters (light blue) with maintained contacts to poised or intermediate enhancers. P and R values: Pearson's product-moment correlation. Boxplots: maximal H3K27ac change at enhancers (top) and expression changes (right) for RING1B Hox network and non-Hox network genes. P values: Wilcoxon rank-sum test.
To investigate potential epigenetic changes at enhancers in RING1A-KO and RING1A/B-dKO cells, we generated ChIP-Seq profiles for H3K4me1 and H3K27ac, and used published H3K27me3 data27 (Supplementary Fig. 8f and Supplementary Table 3). We find that poised enhancers which maintain contacts with Hox network and RING1B-bound promoters show a transition towards an active state (loss of H3K27me3 and gain of H3K27ac) in RING1A/B-dKO cells (Fig. 5d). The status of active and intermediate enhancers contacting RING1B-bound promoters remains largely unchanged (Supplementary Fig. 8g), whereas a subset of intermediate enhancers which maintain contacts with Hox network promoters undergo a transition towards an active state, although this is less pronounced than observed for poised enhancers (Supplementary Fig. 8h). Thus the gain in acetylation is most prominent at enhancers with pre-existing Hox network promoter contacts, and this correlates with the most pronounced transcriptional up-regulation (Fig. 5e, f).
Here we identify an unusually strong PRC1-dependent spatial ESC network, composed of the four Hox clusters, key developmental genes, and their associated poised enhancers. We observe a complete dissociation of the promoter-promoter contacts in this network upon PRC1 knockout. However pre-existing contacts between Hox network genes and poised enhancers are largely maintained. These enhancers transition to an active chromatin state in the knockout, which correlates with significant up-regulation of the genes they contact. Thus we speculate that this higher-order genome organization mediated by PRC1 is a key determinant in maintaining network genes in a silent state, poised for activation during early development. Similar silencing mechanisms involving 3D genome organization may be evolutionarily conserved, as contacts between Polycomb target genes exist in distantly related species2,3,28, and pre-formed contacts between developmental genes and regulatory sequences have been observed in Drosophila development29. Thus, PRC1 physically constrains developmental genes in a repressive 3D spatial network in pluripotent stem cells, and we propose that selective release of genes from this network results in transcriptional up-regulation and underlies key cell fate decisions associated with organogenesis and body plan specification in early development.
URLs
ArrayExpress data repository, www.ebi.ac.uk/arrayexpress; DiffBind BioConductor package, http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf; HiCUP pipeline, http://www.bioinformatics.babraham.ac.uk/projects/hicup/; GOTHiC BioConductor package, http://www.bioconductor.org/packages/release/bioc/html/GOTHiC.html; Seqmonk, http://www.bioinformatics.babraham.ac.uk/projects/seqmonk
Accession codes
Nuclear RNA-Seq (E-MTAB-3125), ChIP-Seq (E-MTAB-3156), Promoter CHi-C raw data and lists of significant contacts (E-MTAB-3109) are available from the ArrayExpress data repository under indicated accession numbers. WT CHi-C raw data14 is available under accession number E-MTAB-2414.
Online Methods
Cell culture
J1 (129S4/SvJae) and 129SvJae/Cast WT mouse ESCs were grown under standard ESC culture conditions7. Ring1A–/–;Ring1Bfl/fl;Rosa26::CreERT2 mouse ESCs (RING1A-KO) were cultured as described previously7. Eed+/+ G2.1 and Eed-/- G8.1 mESCs15 were cultured as described previously17. For WT, RING1A-KO, Eed+/+ G2.1 and Eed-/- G8.1 mouse ESCs, dishes were coated with 0.1% gelatin and irradiated mouse embryonic fibroblasts (MEFs). Conditional deletion of Ring1B in RING1A-KO cells was carried out by addition of 800 nM tamoxifen for 48 hrs as previously described7. For harvesting, ESCs were trypsinized and pre-plated twice for 30 min to remove contaminating feeder cells.
Extracts, western blot analysis and antibodies
For whole cell extracts, cells were harvested, washed once with PBS, and twice with cold TB buffer (20 mM Hepes, pH 7.3, 110 mM KOAc, 5 mM NaOAc, 2 mM MgOAc, 1 mM EGTA, 2 mM DTT, protease inhibitors (Roche)). Extracts were then incubated for 30 min at 37°C in TB buffer with 0.1% NP-40, 10 mM MnCl2, and 20 μg/ml DNaseI (Life Technologies). For acid extraction of histones, cells were trypsinized, washed with room temperature PBS, and incubated in ice-cold PBS containing protease inhibitors for 10 min. Cells were spun for 4 min at 235×g. The pellet was resuspended in 1 ml of ice cold 0.2 M H2SO4 and incubated on ice for 30 min. Samples were then spun for 2 min at 20,000×g at 4°C. Histones were precipitated with 25% TCA for 30 min at 4°C. Precipitated histones were pelleted by spinning for 10 min at 20,000×g at 4°C and washed twice with 1 ml of ice cold acetone for 10 min on ice. Histones were centrifuged for 10 min at 20,000×g at 4°C and dissolved in 100 mM Tris (pH 7.6).
Western blot blocking, antibody incubations and washes were in PBS pH8, 5% milk powder, 0.1% Tween-20. Primary antibodies (H2AK119ub1 (05-678, Millipore) 1:500; H3K27me3 (07-449, Millipore), EED (09-774, Millipore), EZH2 (pAB-039-050, Diagenode), RING1B31 1:1,000; H3 (ab1791, Abcam) 1:10,000; OCT4 (ab19857, Abcam) 1:200; and NANOG (RCAB0002, Reprocell) 1:500) were incubated overnight. Secondary antibodies (either sheep anti-mouse IgG HPR linked (GE Healthcare) 1:2,000; donkey anti-rabbit IgG HRP linked (GE Healthcare) 1:2,000; or anti-mouse IgM HRP linked (DAKO) 1:1,500) were incubated for 1 hr.
Immunofluorescence
Immunofluorescence analysis was carried out essentially as described previously32,33. ESCs were split onto poly-L-lysine slides coated with PBS 0.1% gelatin for 3 hrs. Antibodies used were RING1B31 1:50 and OCT4 (ab19857, Abcam) 1:200. Secondary antibodies used were Alexa Fluor 488 goat anti-rabbit IgG (H+L) (A11008, Molecular Probes) 1:400, and Alexa Fluor 568 goat anti-mouse IgG (H+L) (A11031, Molecular Probes) 1:400. Images were captured using an Olympus BX61 multicolor fluorescence microscope.
Promoter Capture Hi-C
3–4 x 107 ESCs (RING1A-KO or RING1A/B-dKO) were fixed in 2% formaldehyde for 10 min, and Promoter Capture Hi-C was performed essentially as described previously14. Hi-C DNA was amplified with 9 pre-capture PCR amplification cycles using the PE PCR 1.0 and PE PCR 2.0 primers (Illumina). Hi-C DNA was hybridized to a custom-designed capture bait system consisting of biotinylated RNAs targeting the HindIII restriction fragment ends of 22,225 mouse gene promoters14 (Agilent Technologies). Biotin pull-down (MyOne Streptavidin T1 Dynabeads (Life Technologies)) and washes were performed following the Sure Select Target enrichment protocol (Agilent Technologies), and a post-capture PCR (4 amplification cycles using Illumina PE PCR 1.0 and PE PCR 2.0 primers) was performed on DNA bound to the beads via biotinylated RNA. Promoter Capture Hi-C libraries were sequenced (50 bp paired end) on the HiSeq1000 platform (Illumina).
3C-PCR validation
Cells were fixed in 2% formaldehyde for 10 min, and 3C was performed essentially as previously described34. 3C DNA was purified using an Amicon Ultracel 0.5 ml column. For Promoter CHi-C validation, long-range 3C-PCR amplicons were designed by combining a ‘bait’ primer (located within a captured promoter HindIII fragment) with primers (Supplementary Table 5) in contacting or non-contacting HindIII fragments, as determined by Promoter CHi-C. To generate a standard curve for PCR, the corresponding ligation products were generated from a template library by digestion and ligation of the corresponding BAC DNA (Life Technologies) (Supplementary Table 5). In order to control for crosslinking and ligation efficiency within individual 3C libraries, short-range 3C-PCR amplicons were designed for each of the Hox clusters (Hoxa5-Hoxa7, Hoxb7-Hoxb9, Hoxc10 5’-Hoxc10 3’, and Hoxd12-Hoxd13) and the Hist1 cluster (Hist1h2ae 1-2). In the case of inter-chromosomal contacts, this control was performed by analyzing the contact frequency between the corresponding Hox cluster and Calr. For each of the three cell types (J1 WT ESCs, RING1A-KO cells and RING1A/B-dKO cells), two independent biological replicates were analyzed. The identity of 3C ligation products was verified by DNA sequencing.
3D DNA FISH
BAC (Life Technologies) DNA (Supplementary Table 5) was purified and chemically coupled with Alexa Fluor 488 or Alexa Fluor 555 reactive dyes (Life Technologies) according to manufacturer's instructions, as described previously14. 3D DNA FISH was performed as described previously35. DNA FISH signals were imaged and analyzed with the MetaCyte automated imaging system (MetaSystems). 3D distances between the specified genomic loci were calculated (Supplementary Table 6). For the comparison of inter-probe distances within the same cell, the Mann-Whitney test was applied. For the comparison of inter-probe distances between RING1A-KO and RING1A/B-dKO cells, the Kruskall-Wallis/Dunn’s multiple comparisons test was used.
Nuclear strand-specific RNA-Seq
Mouse ESCs (WT 129SvJae/Cast, RING1A-KO and RING1A/B-dKO cells) were washed in PBS and approximately 30–50 x 106 ESCs were lysed for 5 min in 0.5 ml cold buffer RLN (50 mM Tris-HCl pH 7.5, 140 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.4% Igepal). Nuclei were pelleted by spinning at 300×g for 10 min at 4°C. Nuclear RNA was isolated using TRIsure (Bioline), treated with DNaseI (Roche) and re-purified using an RNeasy Mini Kit (Qiagen). Strand-specific RNA-Seq libraries were prepared as described previously by marking the second strand with dUTP36,37 but with some modifications. 250 ng nuclear RNA was fragmented using a Covaris E220 instrument at standard RNA settings for 60 sec. Fragmented RNA was precipitated and first strand synthesis was carried out using SuperScript III (Invitrogen) with 4 µg of actinomycin D (Sigma). Nucleotides were removed with mini quick spin DNA columns (Roche) and second strand synthesis was performed using E.coli DNA ligase (NEB), DNA Polymerase I (NEB) and RNase H (Fermentas), replacing dTTP with dUTP (Fermentas). Following purification on QIAquick columns (Qiagen), TruSeq Illumina adapters were ligated with T4 DNA Ligase (Enzymatics). Libraries were purified on QIAquick columns, treated with USER (NEB) to destroy the second strand and size selected using AMPure XP beads. Libraries were amplified with 9–11 PCR cycles and sequenced (50 bp paired end) on a HiSeq1000 platform (Illumina).
ChIP-Seq
RING1A-KO and RING1A/B-dKO cells (2 x 108 cells) were fixed in ChIP fix buffer (1% formaldehyde, 5 μM EGTA, 10 μM EDTA, 1 mM NaCl, and 0.5 mM HEPES in PBS) for 10 min at RT. Fixation was stopped by glycine addition (final concentration 125 mM). Cells were washed with PBS, buffer A (10 mM Hepes pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100) and buffer B (10 mM Hepes pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cell extracts were lysed in lysis buffer (25 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 1% SDS, 0.5% deoxycholate, complete protease inhibitor (Roche)) for 30 min on ice. Sonication was performed using a BioRupter sonicator (Diagenode) to obtain an average DNA fragment size of 300 bp. Chromatin was diluted with ChIP dilution buffer (25 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton-X 100, 0.1% SDS, 0.5% deoxycholate, complete protease inhibitor (Roche)). Dynabeads® Protein G beads (Life Technologies) were blocked for 1 hr at 4°C with 1 mg/ml BSA and 1 mg/ml yeast tRNA (Life Technologies). For each immuno-precipitation, 150 µg of chromatin and 5 µg of antibodies recognizing H3K4me1 (ab8895, Abcam) or H3K27ac (ab4729, Abcam) were used. Chromatin was pre-cleared with blocked beads for 1 hr at 4°C. Chromatin was incubated with antibodies overnight at 4°C with rotation. Protein-antibody complexes were immuno-precipitated by addition of beads for 2 hrs. Complexes were washed twice with wash buffer A (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP40, 1 mM EDTA), once with wash buffer B (50 mM Tris pH8.0, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP40, 1 mM EDTA), once with wash buffer C (50 mM Tris pH8.0, 250 mM LiCl, 0.5% deoxycholate, 1% NP40, 1 mM EDTA) and once with TE. Samples were treated with RNase A and Proteinase K, and cross-links were reversed overnight. DNA was purified using a ChIP DNA clean and concentrator column (Zymo Research). Libraries were prepared using NEB Next Fast DNA Fragmentation and library preparation set for Ion Torrent kit (E6285S) following manufacturer’s instructions. Briefly, 40 ng of ChIP or input DNA was used for library generation. Libraries were size-selected for 250 bp fragments using 2% E-gel Size Select agarose gel (Life Technologies) and were amplified with 5 PCR cycles. Libraries were sequenced on the Ion Proton Sequencer using Ion PI™ chips v2 (Life Technologies). Templates were generated using Ion PI™ Template OT2 200 kit v3 and Ion PI™ Sequencing 200 kit v3 or using Ion PI IC 200 kit (Life Technologies).
ChIP-Seq data processing
Publically available ChIP-Seq datasets used are listed in Supplementary Table 3. ChIP-Seq peaks from the ENCODE project were directly imported without reprocessing the data. RING1B ChIP-Seq peaks were defined previously17, and EZH2 and SUZ12 ChIP-Seq peaks were called in the same way. For other publically available datasets, raw reads were mapped to the mm9 mouse genome using Bowtie238, with a seed length of 25 bp, allowing reads that had at most only one mismatched nucleotide in the seed, returning only one possible mapping and with the remaining parameters set to default values. After mapping, MACS39 was used to call peaks using default parameters.
For H3K4me1 and H3K27ac ChIP-Seq in RING1A-KO and RING1A/B-dKO cells, raw reads were aligned to the mouse mm9 genome using Bowtie238 with default alignment parameters but excluding non-unique mappings (-m 1) and removing duplicated reads. Each replicate was down-sampled to the same number of aligned reads (Supplementary Table 2).
HindIII fragments were considered to be occupied by a protein factor or histone modification if they overlapped a ChIP-Seq peak. Baited promoter fragments that overlapped a RING1B peak were considered to be bound by PRC1. Promoter fragments that overlapped EZH2 and SUZ12 peaks were considered to be bound by PRC2. Promoter fragments that overlapped any OCT4, SOX2 or NANOG peaks (but were depleted for both PRC1 and PRC2) were designated OSN.
Defining enhancers
Enhancers were defined as previously described23 in WT ESCs using ENCODE data (Supplementary Table 3): H3K4me1 peaks were filtered to remove peaks within 1000 bp (edge-to-edge) of a RefSeq promoter or an H3K4me3 peak. Remaining H3K4me1 peaks were overlapped with H3K27ac and H3K27me3 peaks. H3K4me1 peaks without either mark were designated ‘intermediate’ enhancers, those overlapping only H3K27ac were designated ‘active’ enhancers and those overlapping only H3K27me3 were designated ‘poised’ enhancers (Supplementary Table 4). HindIII fragments which overlapped only one type of enhancer were annotated with this classification (Supplementary Table 4). Differential histone modification occupancy analysis was carried out using the DiffBind BioConductor package40 (EdgeR method) at all WT H3K4me1 peaks.
RNA-Seq analysis
Reads were mapped with Tophat41 using default parameters and filtered to remove read pairs also aligning to ribosomal DNA sequences. Seqmonk was used to generate read counts for genes > 200 bp (considering the entire gene body, using read pairs separated by < 1 kb) (Supplementary Table 2). Downstream analysis was performed using the DESeq2 BioConductor package42. Differential gene expression analysis was performed using the default settings of DESeq2 but without independent filtering of the results and with an FDR of 0.05.
Mean normalized WT FPKM values were used to categorize promoter fragments into non-expressed (0 FPKM) and four quartiles of expressed promoters. To form a control set of non-RING1B-bound promoters with a similar expression profile to RING1B-bound promoters (expression matched promoters), RING1B-bound promoters were counted in each expression category and an equal number of non-RING1B-bound promoters were randomly selected from each category.
Promoter CHi-C contact calling
Raw sequencing reads from RING1A-KO and RING1A/B-dKO CHi-C libraries were processed using the HiCUP pipeline, which maps the positions of the di-tags against the mouse genome (mm9), filters experimental artefacts, such as circularized reads and re-ligations, and removes duplicate reads (Supplementary Table 2). Pre-processed reads from two replicates of WT ESC Promoter CHi-C data (E-MTAB-2414)14 were combined and a random subset of 98,842,763 reads was selected to correspond to the averaged number of reads from the RING1A-KO and RING1A/B-dKO CHi-C data sets.
Significantly contacting regions were identified using the GOTHiC BioConductor package. This assumes that biases occurring in Hi-C and CHi-C experiments are captured in the coverage (total number of reads mapping to a given fragment or larger bin), and therefore significantly contacting regions or true contacts can be separated from background noise using a cumulative binomial test followed by Benjamini-Hochberg multiple testing (FDR cut-off 0.05). Biological replicates were pooled for analysis and promoter-promoter and promoter-genome contacts were handled separately.
Promoter-promoter contacts
For promoter-promoter contacts, we calculated a modified null distribution to account for the non-multiplicative capture bias in products targeted by two baits. A random ligation CHi-C sample14 was used to build a generalized linear model. The product and the sum of the coverage values of the two ends were used as input variables, whereas the contact frequencies of random ligation events were used as dependent variables. Predicted contact frequencies for the actual samples were calculated from the model using logit regression. The GOTHiC binomial test was then applied with this modified background distribution to identify significant contacts. Short-range intra-chromosomal contacts were excluded by filtering contacts separated by < 10 Mb. Strong promoter-promoter contacts were defined as those represented by 3 or more independent reads.
Hox cluster contacts were calculated with HindIII fragments of the Hox cluster regions binned together, and normalized for the number of captured HindIII fragments within the cluster region. The following mm9 coordinates were used to define the Hox clusters: HoxA: chr 6: 52,099,000–52,277,000; HoxB: chr 11: 96,045,674–96,240,000; HoxC: chr 15: 102,740,000–102,877,000; HoxD: chr 2: 74,486,000–74,614,000.
Promoter-promoter network connectivity maps were generated using significant contacts between all 22,225 captured promoters or considering only Polycomb-bound promoters (in each case contacts from the binned Hox cluster regions were used). Networks were visualized with Cytoscape43 using a force-directed layout with the following parameters: number of iterations: 100; weight attribute: read count; minimum weight to consider: 3; no partitioning of sub-graphs before layout.
The Hox network was defined as comprising of RING1B-bound promoters within the defined Hox cluster regions (Hox cluster genes) and RING1B-bound promoters making direct inter-chromosomal or long range (> 10 Mb) intra-chromosomal contacts with the Hox clusters (Hox cluster contacting genes) (Supplementary Table 1). Contacts between Hox cluster promoters and contacts between Hox network members were visualized as circos plots using the ggbio R package. Non-Hox network promoters were defined as RING1B-bound promoters not making direct contacts to the Hox cluster regions and located > 10 Mb away from them (Supplementary Table 1).
Contact enrichment and contact strengths between promoters
To measure the enrichment of contacts within a set of promoters, 100 random promoter sets were generated with comparable pair-wise distance distributions to the experimental set. The p value by its classical definition was acquired by counting the number of random control sets with more contacts than the experimental set. Since contact counts in control sets generally followed a near-normal distribution, a T-test with 99 degrees of freedom was used to more accurately estimate low p values. Contact enrichment was derived by dividing the number of contacts in the experimental set by the average expected number of contacts in the control sets.
To compare contact strengths within and between sets of promoters (e.g. between PRC1/PRC2 and OCT4-bound promoters) a distance-normalized contact strength value for each set or pair of sets was calculated by summing the log fold observed/expected values of contacts and dividing it by the summed contact distances of all possible contacts within or between promoter sets.
Promoter-genome contacts
Promoter-genome contacts (i.e. contacts between promoters and non-promoter regions in the genome) were calculated using an unmodified null distribution; however contacts were subsequently filtered in three steps. First, contacts in which one of the fragments had extremely high coverage (among the 10 most covered bait fragments in at least two out of three samples or among 50 most covered non-bait fragment in any of the samples) were removed. A ‘neighbor filter’ was applied to control for spurious contact spikes involving a single fragment pair by only keeping contacts which were supported by at least one valid read pair involving a neighboring fragment. Finally, since log fold observed/expected values of all contacts have a bimodal distribution, contacts that are likely to be background contacts were removed by fitting a normal distribution to the lower peak and applying a cut-off at the 95th percentile of the normal distribution (~10). Contacts were considered ‘maintained’ if present in WT, RING1A-KO and RING1A/B-dKO samples. Contacts which were only present in RING1A/B-dKO samples were considered to be ‘gained’.
Enrichment at non-bait promoter-contacting fragments
To calculate enrichment for chromatin marks or transcription factors (Supplementary Table 3) at non-bait promoter-contacting HindIII fragments for each promoter category, the proportion of promoter-category-contacting fragments occupied by the factor or mark was divided by the proportion of all promoter-contacting fragments occupied by the same mark or factor. The resulting value was converted to its log2 value, so that positive values represent an enrichment compared with all promoter-contacting non-bait fragments, and a negative value represents depletion.
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust (WT085102MA) (SE), the Biotechnology and Biological Science Research Council, the Medical Research Council UK (PF), and the EU FP7 Epigenesys Network of Excellence (NML). We thank members of the Elderkin, Fraser, and Luscombe groups for discussions and Jon Houseley and Peter Rugg-Gunn for commenting on the manuscript. We thank Felix Krueger for help with data processing and formatting. We thank Rob J. Klose and Neil Brockdorff for sequencing. We thank Daniel Bolland, Joana Martins and Anne Corcoran for help and advice with 3D DNA FISH and MetaCyte data analyses.
Footnotes
Author Contributions
S.S. conceptualized, designed and performed Promoter CHi-C, and helped with data interpretation and writing the manuscript. R.S. analyzed promoter-promoter (Promoter CHi-C) data and performed network analysis. A.D. performed nuclear RNA-Seq, analyzed promoter-genome (Promoter CHi-C) data, gene expression, and ChIP-Seq data and helped write the manuscript. B-M.J. performed 3C-PCR and commented on the manuscript. H.A. performed 3D DNA FISH, analyzed MetaCyte data, and helped with CHi-C data analysis. S.S., R.S., A.D., B-M.J. and HA contributed equally to this manuscript. B.M. analyzed ChIP-Seq data, performed promoter-promoter contact enrichment analysis and commented on the manuscript. E.D. performed experiments and histone ChIP-Seq. L.M. performed 3D DNA FISH, analyzed MetaCyte data, and helped with CHi-C data analysis. Work by B.M., E.D. and L.M. represents an equal contribution. F.T-C. mapped ChIP-Seq data and analyzed promoter-genome enrichments. M.F-M. helped with Promoter CHi-C protocol development and WT ESC Promoter CHi-C. W.J. analyzed ChIP-Seq data. A.S-P. analyzed 3D DNA FISH data. S.W.W. helped with mapping and analysis of Promoter CHi-C data. K.T. carried out sequencing. B.H., E.L., C.S.O., S.A., and S.W.W. designed and provided the capture system. H.K. provided RING1A-KO ESCs and helped with manuscript preparation. P.F. helped with study design, data interpretation and manuscript preparation. N.M.L. was involved in study design, data interpretation and manuscript preparation. S.E. conceptualized and designed the study, designed and performed experiments, interpreted data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 3.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isono K, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Endoh M, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T, et al. Polycomb potentiates Meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev Cell. 2014;28:94–101. doi: 10.1016/j.devcel.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci USA. 2015;112:4672–4677. doi: 10.1073/pnas.1504783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes JR, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 11.Dryden NH, et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014;24:1854–1868. doi: 10.1101/gr.175034.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mifsud B, et al. High-resolution capture Hi-C to map long-range promoter contacts in human cells. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 14.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Noordermeer D, et al. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife. 2014;3:e02557. doi: 10.7554/eLife.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 21.Williamson I, et al. Anterior-posterior differences in HoxD chromatin topology in limb development. Development. 2012;139:3157–3167. doi: 10.1242/dev.081174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Wit E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 23.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endoh M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 27.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosa S, et al. Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev. 2013;27:1845–1850. doi: 10.1101/gad.221713.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atsuta T, et al. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma. 2001;20:43–46. doi: 10.1089/027245701300060427. [DOI] [PubMed] [Google Scholar]
- 32.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Silva J, et al. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 34.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 35.Bolland DJ, King MR, Reik W, Corcoran AE, Krueger C. Robust 3D DNA FISH using directly labeled probes. J Vis Exp. 2013 doi: 10.3791/50587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold changes and dispersion for RNA-Seq data with DESeq2. bioRxiv. 2014 doi: 10.1101/002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 47.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.