Abstract
A major challenge in cancer research is the use of appropriate models with which to study a specific biological question. Cell lines have long been used to study cellular processes and the effects of individual molecules because they are easy to use, grow rapidly, produce reproducible results and have a strong track record in research. In uveal melanoma in particular, the absence of animal models that faithfully replicate the behavior of the human disease has propagated the generation and use of numerous cell lines by individual research groups. This in itself, however, can be viewed as a problem due to the lack of standardization when characterizing these entities to determine how closely they reflect the genetic and phenotypic characteristics of this disease. The alternative is to use in vitro primary cultures of cells obtained directly from uveal melanoma patient samples, but this too has its difficulties. Primary cell cultures are difficult to use, hard to obtain and can show considerable heterogeneity. In this article, we review the following: (1) the uveal melanoma cell lines that are currently available, discussing the importance of establishing a bank of those that represent the molecular heterogeneity of uveal melanoma; (2) the methods used to isolate and perform short-term cultures of primary uveal melanoma cells, and (3) the establishment of 3D tissue culture models that bridge the gap between 2D in vitro systems and in vivo models with which to dissect cancer biology and perform therapeutic screens.
Key Words: Uveal melanoma, Primary cell cultures, Culture models
Uveal Melanoma Cell Lines
Stratified medicine, a priority area in cancer research, highlights the importance of patient heterogeneity when determining which patients will have clinical benefit from a particular therapeutic agent. Because each patient's tumor responds to drugs differently, the use of cell lines is often criticized as being a poor cancer model. However, for hypothesis-driven basic research, it should be argued that they are still a crucial resource provided that they are well matched to the cancer of interest and reflect the spectrum of tumor genetics as closely as possible. This is already being achieved for ovarian cancer such that data from approximately 50 ovarian cancer cell lines has been matched with The Cancer Genome Atlas (TCGA) data from more than 300 patients with the disease [1]. The cell lines are then rated based on their genetic similarity to the TCGA tumor data.
In uveal melanoma research, cell lines have been widely used in vitro to dissect the basic biology and identify the therapeutic efficacy of drugs due to an absence of well-characterized and representative animal model systems. The current status of animal model systems for uveal melanoma is discussed in subsequent papers in this issue. Perhaps because of the lack of good animal model systems, there are more than 20 uveal melanoma cell lines reported and used in in vitro studies variably throughout the literature. Unlike other areas of cancer research, most of the uveal melanoma cell lines are not available from central repositories such as the American Type Culture Collection (ATCC; www.atcc.org) or the European Collection of Cell Cultures (ECACC; http://www.phe-culturecollections.org.uk/collections/ecacc.aspx), which perhaps makes it more difficult to ensure that rigorous quality control procedures have been employed by each supplying laboratory or to avoid cross-contamination and/or misidentification of the cell lines amongst other factors. Indeed, cell line misidentification within the ocular oncology community was highlighted in 2008 by Folberg et al. [2] when the MUM2B and M619 cell lines in their laboratory did not match the description of their origin in the literature. The short tandem repeat (STR) analysis conducted inferred that MUM2B and M619 were both derived from the same primary tumor as another uveal melanoma cell line, C918. The authors also made a number of important recommendations which, just over 5 years later, are generally still lacking within the ocular oncology research community: minimally, all uveal melanoma cell lines should be validated according to (1) species, (2) whether they are mycoplasma free, and (3) STR analysis against a benchmark standard.
Although STR profiling is useful for cell line authentication, other data are also essential for researchers to select the most appropriate cell lines for their study. Genomic and molecular profiles as well as phenotypical and immunohistochemical characteristics can provide important information regarding subtle differences between and within the cell lines over time. In 2012, Griewank et al. [3] published a comprehensive overview of 19 cell lines providing information determined either directly in their laboratory or from information previously reported in the literature on the following factors: (1) the presence or absence of GNAQ/GNA11 and BRAF mutations; (2) tumor site of origin, i.e. primary or metastatic; (3) the expression of the melanocyte lineage markers Melan-A⁄MART-1, gp100, tyrosinase, tyrosinase-related protein-1, dopachrome tautomerase, S100, HMB45 and MCSP1, and (4) STR analysis. Despite this, the information for each cell line was incomplete and also identified some concerns as to cell line origin/authenticity. For example, the OCM1, OCM3, OCM8, SP6.5 and MUM2C cell lines all harbored BRAF mutations, a genotype that requires further validation regarding its role in uveal melanoma pathogenesis [4,5]. In addition, the Mel285 and Mel290 cell lines, which represent approximately 15% of uveal melanomas without GNAQ/GNA11 mutations, also lack expression of any melanocyte-associated lineage markers. Whether this phenotype is observed in tumors without detectable GNAQ/GNA11 mutations is unclear. Importantly, however, researchers are now using this molecular information when designing their experiments and have shown that the GNAQ/GNA11 genotype of the uveal melanoma cell lines drives differential drug sensitivity [6,7,8].
Inactivating mutations of the tumor suppressor gene BAP1 (BRCA1-associated protein-1) have also recently been associated with an increased metastatic risk in uveal melanoma patients [9]. To address the effects of this genotype, studies have used short hairpin RNA (shRNA) knockdown of BAP1 in the OCM1A, 92.1 and Mel290 uveal melanoma cell lines [10]. Loss of BAP1 expression, however, did not promote in vitro tumorigenicity but rather caused downregulation of canonical genes of the melanocyte differentiation program (MITF, TRPM1, TYR and DCT) and consistent upregulation of the stem cell factor NANOG, inducing a stem-like cellular phenotype. The fact that uveal melanoma cell lines contain a population of stem-like cells that self-renew, produce differentiated progeny and survive chemotherapy had been previously reported for the Mel270 and OMM2.5 uveal melanoma cell lines [11]. Most recently, uveal melanoma cell lines have been established that exhibit mutant BAP1 and lack BAP1 protein expression. These cell lines recapitulate many aspects of the diverse molecular landscape seen in uveal melanoma patients and will be fundamental to enhancing our understanding of the disease [12].
In uveal melanoma patients, one of the strongest indicators of a poor prognosis is the loss of one copy of chromosome 3 (monosomy 3) [13]. Despite this, there have been few reports of the existence of long-term monosomy 3 uveal melanoma cell lines in the research arena. Nareyeck et al. [14] reported heterozygosity at four or more chromosome 3 loci by microsatellite analysis in OMM1, Mel290, Mel285, OCM1, OCM3, OCM8 and 92.1 uveal melanoma cells. Interestingly, they also demonstrated a disomy 3 status by cytogenetic analysis for Mel202 cells, but loss of heterozygosity was indicated by microsatellite analysis, suggesting that both cytogenetically identified chromosome 3 alleles were identical (isodisomy). In this article, they then went on to describe the establishment and characterization of two novel cell lines, UPMM-1 and UPMM-2, originating from primary uveal melanoma tissue samples with monosomy 3. The UPMM-1 cell line retained the monosomy 3 status of the primary tumor, whilst in UPMM-2 cells, chromosome 3 had undergone duplication (isodisomy). The UPMM cell lines have since been used only in a handful of publications, and thus their behavior in established assays as compared with the widely available disomy/isodisomy 3 uveal melanoma cell lines is still unclear [10,15].
Burgess et al. [16] have been systematically culturing cells obtained from transscleral fine-needle aspiration biopsy of eyes harboring uveal melanoma and undergoing brachytherapy or enucleation. All biopsies were evaluated by fluorescence in situ hybridization for the centromere of chromosome 3, by 250K Mapping Array for chromosomal copy number variation, by U133 plus 2.0 Arrays for gene expression and plated out as adherent cultures in standard medium [Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS)]. Out of 101 cultures, only three cell lines could be established (MEL20-06-039, MEL20-06-045 and MEL20-07-070). These were defined as cultures that propagated successfully for at least six passages over a period of 2.5 years, showed continuous proliferation following cryo-freezing and demonstrated no evidence of contact inhibition. All three cell lines showed epithelioid morphology as well as chromosome 3 loss and were derived from patients who developed metastatic disease within 1.5 years from diagnosis of the primary tumor. Again, these cell lines have not since been reported in the experimental uveal melanoma literature.
Short-Term Primary Cultures of Uveal Melanoma Cells: Practical Aspects
Some investigators have preferred to work with short-term primary cultures to circumvent the limitations of homogeneity and the genetic or phenotypic drift often noted for long-term cell lines. Although their unadulterated nature translates to the preservation of a molecular phenotype more representative of that found in vivo, short-term cultures cannot be reliably shared between research groups, making it difficult to obtain independent validation of results. Additionally, primary cells are heterogeneous, require constant monitoring, are sensitive to passaging, often have slow proliferation rates and may undergo early senescence after only a few expansions. All of these factors require the need for research staff with advanced tissue culture training.
Over the years, a number of research groups have reported the successful culture and research use of primary uveal melanoma cells in vitro. There is, however, no standard procedure that will guarantee the successful development of these cells in culture, and so different groups have developed different approaches. A detailed description of the exact methodologies is beyond the scope of this article, which rather aims to provide an overview of the procedure; thus, the reader is invited to refer to the original articles cited in the text for further details.
The first step in culture establishment is the harvest of fresh tumor tissue from the eyes of consenting patients affected by uveal melanoma who are undergoing surgery for therapeutic purposes. Traditionally, the fragment would come from a wider surgical excision specimen, such as enucleation or local resection, but successful cultures have also been established from fine-needle aspiration biopsies [16]. The tumor fragments should be processed for culture as soon as possible after removal as storing the tissue in transport medium in the fridge overnight can reduce viability [H. Kalirai, pers. commun.]. The sample is minced with a surgical blade and either transferred directly to a culture dish for tumor cell outgrowth from the explants or a single-cell suspension obtained so that the number of cells seeded in the flask is kept constant between experiments. A single-cell suspension can be achieved mechanically or by enzymatic digestion. Mechanically, the tissue fragments can be strained through a metal mesh or a linen cloth by continuous irrigation with the culture medium [17]. Whatever the method, a detailed record of cell yield per wet tissue weight should be kept. The choice of cell culture medium is yet another variable with little information in the literature to support a particular medium choice. Traditionally, cells have been grown in a basal medium, such as DMEM or RPMI, supplemented with 10% heat-inactivated FBS, glutamine, antibiotics (penicillin, streptomycin or amphotericin-B) and a variety of other factors, such as epithelial growth factor [18] or insulin [16]. Albert et al. [19] have also reported the use of a human fibroblast feeder layer. Alternatively, the use of media originally formulated for the culture of amniotic fluid cells, such as AmnioChrome Pro (Lonza), Quantum 3-21 (PAA Laboratories; fig. 1) or Chang D (Irvine Scientific), appears to improve the plating efficiency and the growth rate of the primary uveal melanoma cells [20]. More recently, Suesskind et al. [21] have described the successful propagation of uveal melanoma cells in suspension using a serum-free neuronal medium (Neurobasal A; Invitrogen) supplemented with B27.
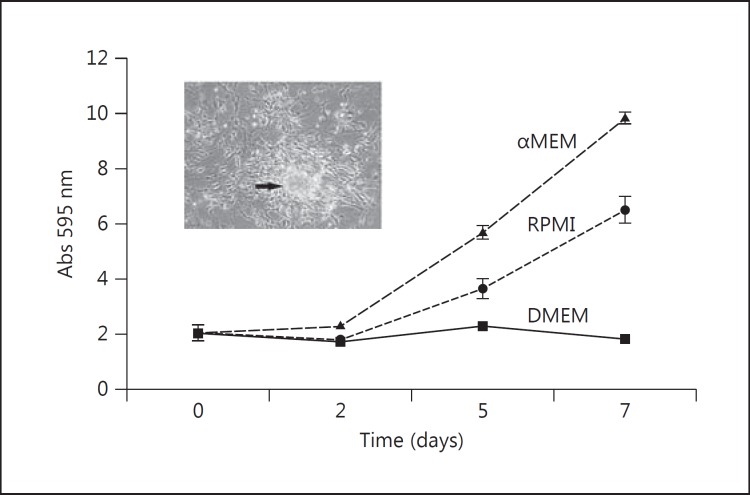
Fig. 1.
Growth rate of isolated primary uveal melanoma cells from a monosomy 3 patient in different media. The cells were grown in either RPMI + 10% FCS, DMEM + 10% FCS, or 1:1 αMEM: Quantum 3-21 + 10% FCS. The cells grew more efficiently in the latter medium and demonstrated the formation of small clumps of cells that could be detached into the medium.
Primary uveal melanoma cells can grow as a monolayer of spindle cells creating a neural-like network or as a carpet of more epithelioid-like cells. Occasionally, clumps/small spheres of cells form that are then spontaneously shed into the culture medium (fig. 1; cell image). This has previously been documented for human skin melanoma cells [22] and sarcoma cells [23] in monolayer culture. These different phenotypes are not mutually exclusive as they reflect the variety of cells in the primary tissue. Over time, it is common to observe morphological changes of the cells in culture in comparison to their tumors of origin [24]. This is most likely due to spontaneous selection for the fittest subpopulation in the in vitro environment, resulting in clonal expansion and eventually in an immortalized cell line.
It is therefore important to meticulously characterize short-term primary cultures both initially and at later stages of culture, both immunophenotypically (for the expression of melanocyte lineage markers) and genetically (in particular for chromosome 1, 3, 6 and 8 status, GNAQ/11 and other relevant mutations) so that alterations from the original tumor phenotype/genotype can be identified.
Future Directions
There is increasing acceptance that the use of cell lines and primary cells must be improved for in vitro culture to contribute more effectively to our understanding of disease biology and therapeutic efficacy. The recent move from cell monolayers to 3D cultures has been motivated by the need to work with cellular models that mimic the functions of living tissues. In 1999, Maniotis et al. [25] reported the ability of several uveal melanoma cell lines (M619, C918 and MUM-2B) to form loops and networks in vitro when placed on gels containing matrigel or diluted type I collagen. As in patient tissue samples, these in vitro tumor cell-generated vascular channels were periodic acid-Schiff positive.
Tumor spheroids are perhaps one of the simpler and more cost-effective 3D systems described in the literature [reviewed in [26,27,28]]. Indeed, spheroids have been produced from either a single-cell type or as complex multicellular spheroids, reflecting the in vivo cells found in the tumor of interest. The multicellular tumor spheroids resemble avascular tumor nodules/micrometastases with respect to growth kinetics and some histomorphological features. With increasing size of either the mono- or multicellular spheroids, proliferation and oxygen/nutrient gradients are observed – features that affect not only cellular RNA and protein expression but also the penetration and bioactivity of therapeutic drug candidates [27,29]. Tumor spheroids can be produced by a number of methods, including hanging drop, liquid overlay or by using rotating wall vessel cultures. Establishing 3D cultures as a mainstream approach, however, is not without a number of challenges, including minimally the development of standard protocols for experimental analysis and imaging. Moreover, although tumor spheroids take advantage of the natural tendency of many cell types to aggregate without the requirement for external scaffolds, neither all primary tumor cells nor all established cell lines have this capacity. Some cells require the addition of extracellular matrix compounds, methylcellulose or agarose.
Although a 3D organotypic approach is more applicable as a human skin melanoma model [30], the development of 3D tumor spheroids has relevance for both primary and secondary uveal melanoma disease phenotypes. The possibility of a tissue-engineered model of uveal melanoma that recreates as far as possible the 3D structure, stromal environment and signalling milieu present in vivo could dramatically improve preclinical drug screening for this rare disease.
Conclusion
Exploitation of in vitro cell culture systems will continue to prove to be a valuable tool in uveal melanoma cell biology only by systematically collecting molecular data on a select panel of cell lines that phenotypically and genotypically reflect patient tumors. This will then enable researchers to match either uveal melanoma cell lines or short-term cell cultures with the most appropriate experimental system in which to address the physiological question.
References
- 1.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folberg R, Kadkol SS, Frenkel S, Valyi-Nagy K, Jager MJ, Pe'er J, Maniotis AJ. Authenticating cell lines in ophthalmic research laboratories. Invest Ophthalmol Vis Sci. 2008;49:4697–4701. doi: 10.1167/iovs.08-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griewank KG, Yu X, Khalili J, Sozen MM, Stempke-Hale K, Bernatchez C, Wardell S, Bastian BC, Woodman SE. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012;25:182–187. doi: 10.1111/j.1755-148X.2012.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen CS, Sibbett R, Henriquez FL, McKay IC, Kemp EG, Roberts F. The T1799A point mutation is present in posterior uveal melanoma. Br J Cancer. 2008;99:1673–1677. doi: 10.1038/sj.bjc.6604731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maat W, Kilic E, Luyten GP, de Klein A, Jager MJ, Gruis NA, Van der Velden PA. Pyrophosphorolysis detects B-RAF mutations in primary uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49:23–27. doi: 10.1167/iovs.07-0722. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Wu Q, Tan L, Porter D, Jager MJ, Emery C, Bastian BC. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2013;33:4724–4734. doi: 10.1038/onc.2013.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho AL, Musi E, Ambrosini G, Nair JS, Deraje Vasudeva S, de Stanchina E, Schwartz GK. Impact of combined mTOR and MEK inhibition in uveal melanoma is driven by tumor genotype. PLoS One. 2012;7:e40439. doi: 10.1371/journal.pone.0040439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitsiades N, Chew SA, He B, Riechardt AI, Karadedou T, Kotoula V, Poulaki V. Genotype-dependent sensitivity of uveal melanoma cell lines to inhibition of B-RaF, MEK, and Akt kinases: rationale for personalized therapy. Invest Ophthalmol Vis Sci. 2011;52:7248–7255. doi: 10.1167/iovs.11-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013;13:371. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalirai H, Damato BE, Coupland SE. Uveal melanoma cell lines contain stem-like cells that self-renew, produce differentiated progeny, and survive chemotherapy. Invest Ophthalmol Vis Sci. 2011;52:8458–8466. doi: 10.1167/iovs.11-7379. [DOI] [PubMed] [Google Scholar]
- 12.Amirouchene-Angelozzi N, Nemati F, Gentien D, Nicolas A, Dumont A, Carita G, Camonis J, Desjardins L, Cassoux N, Piperno-Neumann S, Mariani P, Sastre X, Decaudin D, Roman-Roman S. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol. 2014;8:1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damato BE, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16:6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 14.Nareyeck G, Zeschnigk M, Prescher G, Lohmann DR, Anastassiou G. Establishment and characterization of two uveal melanoma cell lines derived from tumors with loss of one chromosome 3. Exp Eye Res. 2006;83:858–864. doi: 10.1016/j.exer.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Nareyeck G, Zeschnigk M, Bornfeld N, Anastassiou G. Novel cell lines derived by long-term culture of primary uveal melanomas. Ophthalmologica. 2009;223:196–201. doi: 10.1159/000201566. [DOI] [PubMed] [Google Scholar]
- 16.Burgess BL, Rao NP, Eskin A, Nelson SF, McCannel TA. Characterization of three cell lines derived from fine needle biopsy of choroidal melanoma with metastatic outcome. Mol Vis. 2011;17:607–615. [PMC free article] [PubMed] [Google Scholar]
- 17.Luyten GP, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, Van Drunen E, Vuzevski V, De Jong PT, Luider TM. Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer. 1996;66:380–387. doi: 10.1002/(SICI)1097-0215(19960503)66:3<380::AID-IJC19>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Pardo M, Pineiro A, de la Fuente M, Garcia A, Prabhakar S, Zitzmann N, Dwek RA, Sanchez-Salorio M, Dominguez F, Capeans C. Abnormal cell cycle regulation in primary human uveal melanoma cultures. J Cell Biochem. 2004;93:708–720. doi: 10.1002/jcb.20230. [DOI] [PubMed] [Google Scholar]
- 19.Albert DM, Ruzzo MA, McLaughlin MA, Robinson NL, Craft JL, Epstein J. Establishment of cell lines of uveal melanoma. Methodology and characteristics. Invest Ophthalmol Vis Sci. 1984;25:1284–1299. [PubMed] [Google Scholar]
- 20.Angi M, Jenkins R, Damato BE, Coupland SE, Kalirai H: Optimization of culture conditions for the analysis of the secretome of primary uveal melanoma cells (abstract 3392/A585). ARVO 2012 Annual Meeting Abstracts.
- 21.Suesskind D, Gauss S, Faust UE, Bauer P, Schrader M, Bartz-Schmidt KU, Henke-Fahle S. Characterisation of novel uveal melanoma cell lines under serum-free conditions. Graefes Arch Clin Exp Ophthalmol. 2013;251:2063–2070. doi: 10.1007/s00417-013-2292-9. [DOI] [PubMed] [Google Scholar]
- 22.Thurber AE, Douglas G, Sturm EC, Zabierowski SE, Smit DJ, Ramakrishnan SN, Hacker E, Leonard JH, Herlyn M, Sturm RA. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene. 2011;30:3036–3048. doi: 10.1038/onc.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skehan P, Thomas JE, Friedman SJ. Spontaneous cell shedding by tumor cells in monolayer culture. In Vitro Cell Dev Biol. 1986;22:632–636. doi: 10.1007/BF02623476. [DOI] [PubMed] [Google Scholar]
- 24.Aubert C, Rouge F, Reillaudou M, Metge P. Establishment and characterization of human ocular melanoma cell lines. Int J Cancer. 1993;54:784–792. doi: 10.1002/ijc.2910540513. [DOI] [PubMed] [Google Scholar]
- 25.Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe'er J, Trent JM, Meltzer PS, Hendrix MJ. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Multicellular tumor spheroids: An underestimated tool is catching up again. J Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaitan D, Dwarakanath BS. Multicellular spheroids as an in vitro model in experimental oncology: applications in translational medicine. Expert Opin Drug Discov. 2006;1:663–675. doi: 10.1517/17460441.1.7.663. [DOI] [PubMed] [Google Scholar]
- 30.Vorsmann H, Groeber F, Walles H, Busch S, Beissert S, Walczak H, Kulms D. Development of a human three-dimensional organotypic skin-melanoma spheroid model for in vitro drug testing. Cell Death Dis. 2013;4:e719. doi: 10.1038/cddis.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]