Abstract
Several bacterial pathogens utilize conjugation machines to export effector molecules during infection. Such systems are members of the type IV or ‘adapted conjugation’ secretion family. The prototypical type IV system is the Agrobacterium tumefaciens T-DNA transfer machine, which delivers oncogenic nucleoprotein particles to plant cells. Other pathogens, including Bordetella pertussis, Legionella pneumophila, Brucella spp. and Helicobacter pylori, use type IV machines to export effector proteins to the extracellular milieu or the mammalian cell cytosol.
Gram-negative bacteria have adapted at least two cell-surface organelles for use in the delivery of macromolecules across kingdom boundaries by a cell-contact-dependent mechanism. The type III secretion systems are assembled from core components of the flagellar machine1. The type IV systems, the subject of this review, are built from core components of conjugation machines. This is a promiscuous secretion family both in terms of the translocated substrates – large nucleoprotein conjugation intermediates, an A/B toxin and monomeric proteins – and the phylogenetic diversity of cells targeted for substrate delivery – bacteria, fungi, plants and animals. In this review, we will summarize recent structure–function studies of the type IV systems, with an emphasis on the Agrobacterium tumefaciens T-DNA transfer machine.
Type IV family members
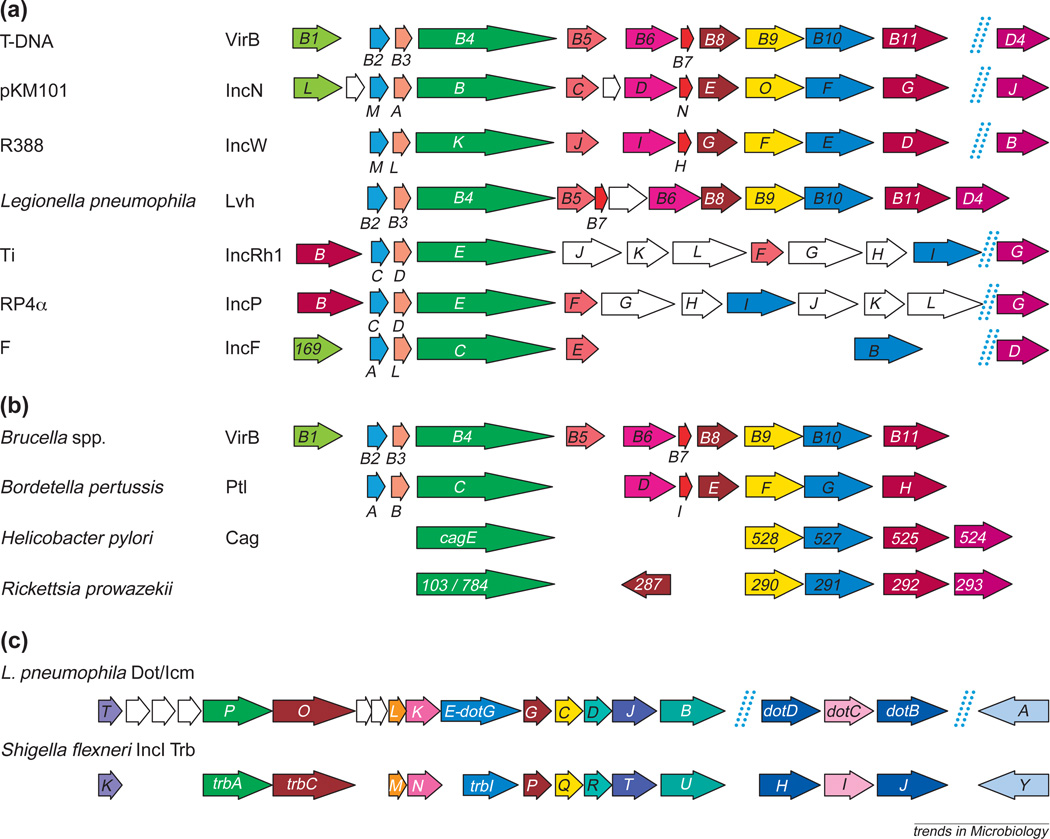
The type IV systems were initially defined on the basis of homologies between components of three different macromolecular complexes: the A. tumefaciens T-DNA transfer system required for exporting oncogenic T-DNA to susceptible plant cells; the conjugal transfer (Tra) system of the conjugative IncN plasmid pKM101; and the Bordetella pertussis pertussis toxin exporter, Ptl (Refs 2,3). As shown in Fig. 1, the list of type IV systems has recently been greatly expanded, with the identification of additional systems involved in DNA and protein translocation. Some of these systems are composed of a complete set of proteins homologous to the Agrobacterium VirB proteins; the corresponding genes are often colinearly arranged in the respective operons, highly suggestive of a common ancestral origin. Other transfer systems, such as the RP4 plasmid and the Agrobacterium pTi Tra systems, seem to be chimeras of VirB protein homologs and Tra proteins of an unrelated ancestry.
Fig. 1.
Type IV systems demonstrated or postulated to direct DNA and/or protein translocation. The Agrobacterium tumefaciens T-DNA transfer machine is composed of products of the virB operon and the virD4 gene. Genes encoding VirB and VirD4 homologs are color-coded for the other type IV systems. Dashed lines indicate the genes are physically unlinked. (a) Representative conjugation systems. (b) Transfer systems thought to function as exporters of effector proteins during infection. (c) The Legionella pneumophila Dot/Icm system encodes icmE/dotG and dotB, weak homologs of VirB10 and VirB11, but is clearly ancestrally related to the transfer region of the Shigella flexneri ColIb IncI plasmid. Additional homologies not shown exist between L. pneumophila IcmW, IcmX, IcmV and DotK and the IncI plasmid TraV, TraW, TraX and TraL proteins, respectively. Sources: A. tumefaciens virB (see Ref. 3), pKM101 (Refs 2,47), R388 (Ref. 42), RP4 (Ref. 48), F (Ref. 7), L. pneumophila lvh (Ref. 6), Brucella virB (Ref. 49), Bordetella pertussis ptl (Refs 2,18), Helicobacter pylori cag (Ref. 21), Rickettsia prowazekii (Ref. 50), L. pneumophila dot/icm (Refs 23,24).
The Legionella pneumophila Dot/Icm system, an important macromolecular transporter associated with virulence, was originally proposed to be related to the A. tumefaciens T-DNA transfer system. Very interestingly, recent work has shown that the Dot/Icm system is in fact very closely related to the Tra region of the IncI ColIb-P9 plasmid of Shigella flexneri (Fig. 1). At least 16 dot/icm genes encode plasmid ColIb Tra homologs and gene organization is generally conserved in these two transfer systems4. The Dot/Icm system is unrelated to the VirB-based systems and yet is postulated to export novel effector proteins during the infection process5. Based on these observations, we propose the type IV classification be expanded to include functional secretion systems assembled from Tra proteins of any ancestral origin. We further propose the assignment of type IV subgroups, such that machines assembled from VirB homologs are type IVA and those from IncI Tra homologs are type IVB, allowing for future classification of other type IV systems assembled from other Tra systems.
A survey of the unfinished bacterial genomes database (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) reveals additional candidate type IV systems among bacterial pathogens including Bordetella bronchiseptica, other Brucella spp., Bartonella spp. and Actinobacillus actinomycetemcomitans. Intriguingly, putative type IV systems are also present in a number of non-pathogenic bacteria, including Caulobacter crescentus, Sinorhizobium meliloti, Rhizobium etli and Thiobacillus ferroxidans, although their functions remain unknown. Finally, L. pneumophila contains a type IVA system (the Lvh system), which appears to play no role in virulence but is able to transfer a mobilizable IncQ plasmid6.
Type IV systems as protein secretion machines
Currently, type IV systems are known to export three types of substrates: DNA conjugation intermediates, the multisubunit pertussis toxin (PT), and monomeric proteins including primase, RecA, the A. tumefaciens VirE2 and VirF proteins, and the Helicobacter pylori CagA protein (Table 1).
Table 1.
Substrates of type IV systemsa
| System | Substrate(s) | Consequence(s) of transfer |
|---|---|---|
| Plasmid conjugation | Transfer intermediate (ss DNA) covalently bound at 59 end to transesterase; primase; SSB; RecA |
Dissemination of virulence and antibiotic-resistance genes; transferred proteins contribute to establishment of plasmid in recipient |
|
Agrobacterium tumefaciens T-DNA transfer |
T-DNA (ss T-DNA/VirD2); VirE2; VirF |
Oncogenesis (crown gall disease); T-DNA delivery to plant nucleus; broadened host range |
| Bordetella pertussis Ptl | Pertussis toxin | ADP-ribosylation of GTP-binding proteins |
| Helicobacter pylori Cag | 145-kDa CagA protein | Tyrosine phosphorylation of CagA; formation of CagA cylinders; host cell cytoskeletal rearrangements |
|
Legionella pneumophila Dot/Icm |
Unknown effector | Allows survival and growth in macrophages; prevent phagosome/lysosome fusion? |
| IncQ plasmid | Plasmid acquisition by L. pneumophila recipients | |
| Lvh | IncQ plasmid | Plasmid acquisition by L. pneumophila recipients |
|
Brucella, Bartonella, Rickettsia |
Unknown | Aid in intracellular survival? |
Abbreviation: SSB, single-stranded-DNA-binding protein.
Many type IV systems transfer DNA, but it is important to note that the conjugation intermediate is not naked DNA but rather single-stranded (ss) DNA associated with one or more proteins. The co-transported protein(s) are involved in the processing of DNA at the origin of transfer (oriT) to form the conjugation intermediate. A transesterase, with contributions from one or more auxiliary proteins, initiates the processing reaction by binding and generating a strand-specific nick at the plasmid oriT or the functionally equivalent A. tumefaciens T-DNA borders. Upon nicking, the transesterase remains covalently bound via a tyrosine to the 5′ end of the single-stranded (ss) DNA. Given that conjugal DNA transfer is a polar 5′–3′ reaction, the transesterase probably mediates the interaction between the ss DNA transfer intermediate and the transfer machine7. In the case of A. tumefaciens, the VirD2 transesterase is exported with T-DNA into the plant host cell, where nuclear localization sequences present on VirD2 mediate delivery of the T-DNA to the plant nucleus3. Thus, the type IV DNA-transfer systems are more aptly viewed as protein secretion machines, with the transesterase supplying the substrate recognition signal(s) and a ‘piloting’ function for directing its cargo, covalently bound ss DNA, through the translocation channel.
Recent work has also added to a body of early evidence that conjugation systems can transfer proteins independently of DNA to recipient cells. A clever genetic assay based on monitoring phage λ release from lysogenic recipients demonstrated transfer of RecA during the transfer of the RP4 plasmid to Escherichia coli recipients8. Similarly, there is strong genetic evidence that the A. tumefaciens T-DNA transfer machine exports VirE2, a ss-DNA-binding protein (SSB) and VirF, another virulence factor. Genetic work further suggests the protein substrates and conjugation intermediates compete for available T-DNA transfer machines3,9.
What is the nature of the interaction between conjugation machines and their DNA and protein substrates? For plasmid mobilization, a coupling protein [e.g. TraG (RP4 and Ti plasmids), TrwB (R388), TraD (F) and VirD4 (A. tumefaciens T-DNA transfer system)] is thought to link the DNA processing and transfer reactions both spatially and temporally. Studies of chimeric transfer systems composed of a translocation apparatus and a heterologous coupling protein suggest that the coupling protein provides the basis for recognition and export of specific plasmid substrates10– 12. For DNA-independent protein transfer, a chaperone-like protein is now known to be involved in mediating the transfer of the A. tumefaciens VirE2 SSB protein via the T-DNA transporter to plant cells. VirE1 is a small (7.5 kDa), acidic protein that interacts directly with an internal domain of VirE2 and is required for export of the SSB. There is also evidence that VirE1 stabilizes VirE2 in vivo and prevents VirE2 aggregation in vitro13–15. These characteristics, as well as the physical properties of VirE1, are reminiscent of the Syc chaperone or ‘bodyguard’ proteins involved in type III secretion16. Thus, by analogy to the type III systems, DNA-independent export of protein substrates via the type IV machines might generally require initial complex formation between the substrate and a cognate chaperone-like protein.
Effector proteins implicated in virulence of mammalian hosts
The protein substrates of type IV systems important for the virulence of bacterial pathogens of mammals are quite distinct from those associated with conjugation systems (Table 1). For example, the B. pertussis Ptl system secretes pertussis toxin (PT), which is a multisubunit A/B toxin composed of five subunits, S1–S5. The S1 subunit, or A domain, shares active-site ADP-ribosylating activity and structure with diphtheria toxin (DT), cholera toxin (CT) and other A/B toxins17. The B domain is a pentamer of the remaining S2–S5 subunits in a ratio of 1:1:2:1, respectively. Interestingly, despite the overall similarity in subunit composition between the T-DNA transfer and Ptl secretion systems, the TDNA and PT export routes differ significantly. Whereas T-DNA and other conjugation intermediates are thought to be delivered across the cell envelope in a single step via a transenvelope mating channel, PT export is a two-step translocation reaction. PT subunits are first secreted across the cytoplasmic membrane via the general secretion pathway, then the PT holotoxin, assembled in the periplasm, is exported via the Ptl system across the outer membrane. Upon export, the B domain interacts with host cell glycoprotein receptors and mediates translocation of the A domain across the host cell membrane. In the host cell cytosol, PT ADP-ribosylates the α subunits of Gi,o proteins, thus interfering with receptor-mediated activation of the G protein and associated signaling pathways18.
Very recently, H. pylori was shown to export the 145-kDa CagA protein via a type IV secretion system encoded by the cag pathogenicity island19,20. H. pylori is the causative agent of peptic ulcer disease and a risk factor for the development of gastric adenocarcinoma. H. pylori attaches to human gastric epithelial cells and induces a number of changes in host cell physiology, including effacement of microvilli, cup/pedestal formation, cytoskeletal rearrangements, and interleukin (IL)-8 production21. Type I strains of H. pylori carrying the cag pathogenicity island were shown to induce signal transduction pathways, resulting in tyrosine phosphorylation of proteins adjacent to the site of bacterial adherence22. Two studies have now reported that CagA is one of these tyrosine-phosphorylated proteins. Intriguingly, a proportion of H. pylori cells found attached to the host cell surface were associated with a high concentration of CagA, with the appearance of a cylindrical structure associated with regions of active actin reorganization. These findings suggest that CagA cylinders might induce host cytoskeletal rearrangements19,20.
Several type IV systems of mammalian pathogens have been identified on the basis of a requirement for virulence. However, the effector(s) exported by these systems remains unknown. The L. pneumophila Dot/Icm system was identified by screening for mutants defective in multiplication in host macrophages23,24. Loss of dot/icm function results in bacteria mistargeting to a late endocytic or lysosomal compartment. Presumably, the dot/icm system exports a virulence factor(s) that is required to prevent phagosome–lysosome fusion. As noted above, the dot/icm system is unrelated to the VirB systems. Yet, this system conjugally transfers plasmid RSF1010 to L. pneumophila recipients23,24. RSF1010 transfer is not thought to be relevant to dot/icm gene function during the infection process. Nevertheless, conjugation proficiency does provide a convenient assay for monitoring functionality of this, and possibly other, type IV systems associated with virulence.
Architecture of the T-DNA transfer machine
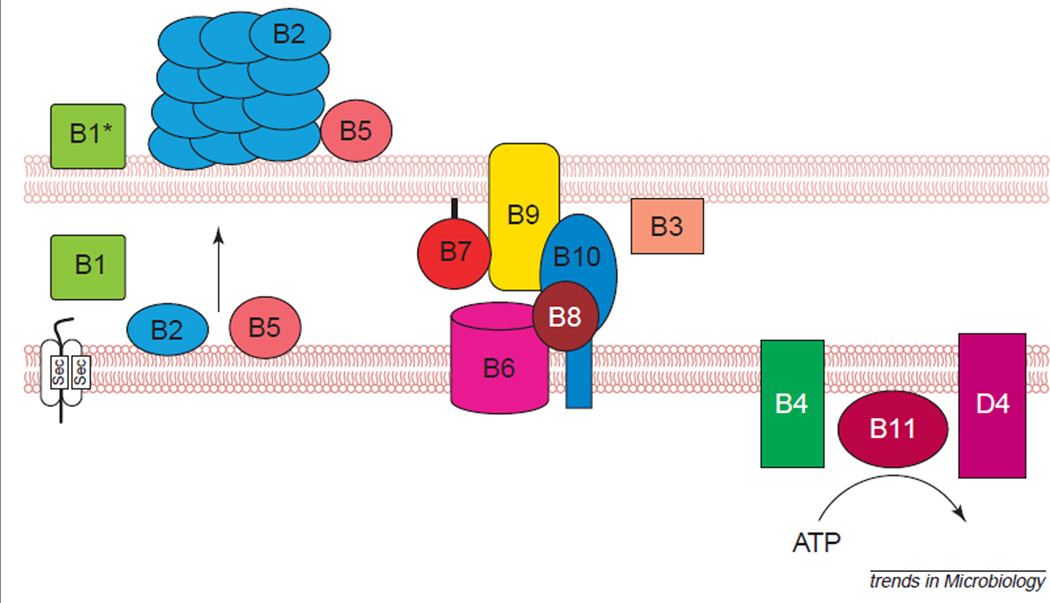
Conjugation machines of Gram-negative bacteria consist of two surface structures, the mating channel through which the DNA transfer intermediate and proteins are translocated and the conjugal pilus for contacting recipient cells2,7. Various conjugative pili have been visualized, but to date there is no ultrastructural information about the mating channel. Recent work on the A. tumefaciens T-DNA transfer system has focused on identifying interactions among the VirB protein subunits and defining steps in the transporter assembly pathway. As depicted in Fig. 2, there are three functional groups of VirB proteins: (1) proteins localized exocellularly forming the T-pilus or other adhesive structures; (2) mating-channel components; and (3) cytoplasmic membrane ATPases. Although all of these proteins probably assemble as a supramolecular complex, as yet there is no direct evidence for a physical association between the conjugative pilus and the mating channel.
Fig. 2.
Locations of VirB components of the T-DNA transfer system. The VirB and VirD4 proteins are grouped according to probable functions: exocellular proteins mediating attachment (VirB1*, VirB2 pilin and VirB5), channel proteins (VirB3, VirB6, VirB7, VirB8, VirB9 and VirB10), and ATPases (VirB4, VirB11 and VirD4).
Exocellular proteins
In an accompanying review in this issue of Trends in Microbiology (pp. 361–369) E-M. Lai and C.I. Kado present a comprehensive update on the structure and function of the A. tumefaciens T-pilus. VirB2, the major subunit of the T-pilus, is processed from a 12.5-kDa proprotein to a 7-kDa mature peptide that partitions with the cytoplasmic membrane during cell fractionation. Interestingly, a recent study demonstrated that VirB2 and its homolog, TrbC of plasmid RP4, undergo novel cyclization reactions following processing such that the amino and carboxy termini are joined via an intramolecular covalent head-to-tail peptide bond25. Upon an unknown signal, VirB2 polymerizes to form the T-pilus, a thin (10 nm) filament found in abundance on cells grown at 18°C but rarely seen on cells grown above 28°C (Ref. 26; Lai and Kado, this issue). Recent work has shown that VirB5 co-purifies in low amounts with the T-pilus and thus is a probable minor pilus subunit27. Similar findings were reported for the VirB5 homolog, TraC of plasmid pKM101 (Ref. 28).
VirB1 has a motif found among lytic transglycosylases, and therefore might cause local lysis of the peptidoglycan during transporter assembly. VirB1*, a proteolytic product corresponding to the carboxy-terminal 73 residues of VirB1, localizes exocellularly and chemically crosslinks with VirB9. VirB1* is not detected in purified T-pilus preparations, and thus might mediate close contacts between donor and recipient cells29,30.
Putative channel components
Several lines of evidence suggest that VirB6–VirB10 as well as one or more of the three ATPases (see below) are probable channel subunits3. VirB6, a highly hydrophobic protein, is thought to span the cytoplasmic membrane several times and presently is the best candidate for a channel-forming protein3. VirB7, an outer membrane lipoprotein31, interacts with itself and with VirB9 via disulfide bonds between unique reactive cysteines present in each protein32,33. The VirB7–VirB9 heterodimer localizes at the outer membrane and plays a critical role in stabilizing other VirB proteins during assembly of the transfer machine34. Recent yeast two-hybrid studies supplied evidence for pairwise interactions between VirB8, VirB9 and VirB10 (Ref. 35). VirB9 is also required for formation of chemically crosslinked VirB10 oligomers probably corresponding to homotrimers. VirB10 has a short amino-terminal cytoplasmic domain and a large carboxy-terminal periplasmic domain and thus is proposed to link cytoplasmic and outer membrane VirB subcomplexes36.
Cytoplasmic membrane ATPases
Two VirB proteins, VirB4 and VirB11, possess conserved Walker A nucleotide-binding motifs required for function, and purified forms of both proteins possess weak ATPase activities3. Recent work suggests both proteins assemble minimally as homodimers in vivo. Self-interaction is mediated by a domain in the amino-terminal third of VirB4 and by domains located in each half of VirB11 (Refs 37–39). Further studies have now established that VirB11 and its homologs, TrbB of plasmid RP4, TrwD of plasmid R388, and RP0525 of the H. pylori Cag pathogenicity island, form higher-order homomultimers39–41. Examination of complexes by electron microscopy led to the intriguing discovery that TrbB, TrwD and RP0525 assemble as homohexameric rings in solution40,41. TrbB hexamer formation is dependent on an intact Walker A motif, suggesting that ATP-induced conformational changes in TrbB might contribute to transporter morphogenesis or substrate translocation40.
As noted above, VirD4 is a member of a family of putative ATPases thought to couple DNA processing and transfer reactions and to supply the basis for plasmid substrate discrimination42,12. However, VirD4 homologs are also found among systems thought to function exclusively in protein export (see Fig. 1). For example, a VirD4 homolog is required for H. pylori CagA protein export20. Thus, these proteins are not limited to DNA translocation reactions and might instead play a more general role in substrate trafficking via type IV machines.
Perspectives and the future
The excitement surrounding the type IV secretion pathway builds with the identification and demonstration of functionality of each new member. Not only are these systems widespread in nature, they are also highly versatile as evidenced by their various uses by mammalian and plant pathogens. The A. tumefaciens T-DNA transporter seems to be the most promiscuous of these machines in its capacity to deliver DNA and proteins to an impressively wide array of cell types – numerous species of plants, bacteria, Saccharomyces, Aspergillus, Fusarium, Trichoderma, Neurospora and Agaricus3,43. The mammalian pathogens use their type IV systems for distinct purposes: B. pertussis to export PT to the extracellular milieu, H. pylori to deliver CagA to the mammalian cell cytosol and L. pneumophila and other intracellular pathogens probably to export an effector molecule(s) following uptake by macrophages. Comparative studies of these systems will certainly unveil common mechanistic principles as well as unique features appropriated for novel purposes.
Studies of the types II–IV secretion systems have revealed remarkable examples of convergent evolution. For example, in function, the type IV systems very closely resemble the type III systems, which utilize a flagellar export machine to inject effector molecules into host cells1,16. Both systems deliver substrates by a process requiring physical contact with target cells. Both systems require coupling or chaperone- like proteins for delivery of substrates to the respective transfer machines. Both systems are generally thought to export substrates in a one-step reaction via a transenvelope channel. Finally, both systems elaborate extracellular pili or filaments that contribute in some way to substrate delivery. However, at least one fundamental difference exists between these two systems – the type IV systems can export long ss DNA polymers to recipient cells, whereas there is currently no evidence for transmission of nucleic acids via type III machines.
The type IV systems also mechanistically resemble the type II systems (reviewed in Ref. 44). Most notably, the B. pertussis PT export pathway is an exception to the one-step translocation route envisaged for type IV machines in that it involves a two-step translocation route much more reminiscent of the type II secretion systems. Indeed, cholera toxin (CT), an A/B toxin that is structurally highly related to PT (Ref. 17), is exported via a type II secretion pathway45. In addition, both the type IV and II systems permit transmission of nucleic acids across the cell envelope. A recent report showed that the same type II system (Eps) that mediates CT export also serves as the conduit for secretion of the filamentous phage CTXϕ across the outer membrane of Vibrio cholerae45. Thus, although the type IV secretion systems are configured for exporting plasmid DNA in association with conjugation proteins, a type II system has now been shown to export a phage genome in association with phage proteins.
Many fundamental questions surrounding the type IV systems await further exploration. The Tra operons encoding the type IV conjugation systems are often induced by extracellular or growth-phase-dependent signals2,3,7,46, as are the genes for the B. pertussis Ptl system2,18. What extracellular signals regulate expression of genes for type IV systems implicated in virulence of intracellular pathogens such as L. pneumophila or Brucella spp.? What is the molecular basis for dual recognition of protein and nucleoprotein substrates? What specific cell–cell interactions enable the delivery of substrates to such a wide array of target cell types? Answers to these questions will not only supply basic knowledge about this fascinating transporter family but might also pave the way for development of strategies aimed at incapacitating these systems to suppress virulence, or for manipulating bacterial pathogens as vectors for the targeted delivery of therapeutic DNA or proteins.
Questions for future research.
What is the basis for substrate recognition? Are chaperone-like proteins of general importance for type IV protein transfer?
What is the architecture of the channel and the nature of the contact with extracellular pili? Do all type IV systems elaborate extracellular pili or other appendages?
What host signals are required for elaboration of type IV transfer machines? In addition to transcriptional regulation, what translational or post-translational regulatory processes are required for transporter biogenesis?
Are type IV systems viable targets for antibiotic intervention or vectors for delivery of therapeutic molecules?
Can the principle of interkingdom transfer of effector DNA or proteins via conjugation machines be generalized to the Gram-positive bacteria?
Acknowledgments
We express special thanks to scientists working on conjugation systems for laying a foundation of knowledge about these newly termed type IV systems. We apologize for any omissions of citations to original data cited herein owing to space limitations; the original references can be found in the reviews cited. Additional information on the type IV systems can be found in the accompanying review by E-M. Lai and C.I. Kado. Work in the P.J.C. laboratory is funded by the NIH (GM48746).
Footnotes
Note added in press
Recently, Chen et al.51 identified VirE2 and VirD2 proteins in supernatant fractions independently of the A. tumefaciens VirB proteins. The authors speculate that VirE2 and VirD2 are translocated by a route that does not require the VirB proteins. Because only trace amounts (~1%) of the total cellular VirD2 and VirE2 proteins were detected in the supernatant, we suggest the alternative possibility that these proteins are associated with membrane vesicles or other cell surface structures that are released into the supernatant under laboratory growth conditions.
Contributor Information
Peter J. Christie, Dept of Microbiology and Molecular Genetics, The University of Texas-Houston Medical School, 6431 Fannin, Houston, TX 77030, USA.
Joseph P. Vogel, Dept of Molecular Microbiology, Washington University School of Medicine, 660 S. Euclid Ave, St Louis, MO 63110, USA
References
- 1.Macnab RM. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winans SC, et al. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie PJ. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segal G, Shuman HA. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol. Microbiol. 1999;33:669–670. doi: 10.1046/j.1365-2958.1999.01511.x. [DOI] [PubMed] [Google Scholar]
- 5.Vogel JP, Isberg RR. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 6.Segal G, et al. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 7.Frost L, et al. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemann J. Genetic evidence of protein transfer during bacterial conjugation. Plasmid. 1999;41:240–247. doi: 10.1006/plas.1999.1392. [DOI] [PubMed] [Google Scholar]
- 9.Stahl LE, et al. The conjugal intermediate of plasmid RSF1010 inhibits Agrobacterium tumefaciens virulence and VirB-dependent export of VirE2. J. Bacteriol. 1998;180:3933–3939. doi: 10.1128/jb.180.15.3933-3939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabezon E, et al. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 1994;176:4455–4458. doi: 10.1128/jb.176.14.4455-4458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sastre JI, et al. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 1998;180:6039–6042. doi: 10.1128/jb.180.22.6039-6042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton CM, et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundberg C, et al. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng W, et al. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 1999;31:795–807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X-R, Christie PJ. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng LW, Schneewind O. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 2000;8:214–220. doi: 10.1016/s0966-842x(99)01665-0. [DOI] [PubMed] [Google Scholar]
- 17.Bazan J, Koch-Nolte F. Sequence and structural links between distant ADP-ribosyltransferase families. Adv. Exp. Med. Biol. 1997;419:99–107. doi: 10.1007/978-1-4419-8632-0_12. [DOI] [PubMed] [Google Scholar]
- 18.Burns DL. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 19.Segal E, et al. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein M, et al. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covacci A, et al. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 22.Segal EE, et al. Induction of host signal transduction pathways by Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7595–7599. doi: 10.1073/pnas.94.14.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel JP, et al. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 24.Segal G, et al. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenbrandt R, et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 26.Fullner KJ, et al. ) Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1110. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt-Eisenlohr H, et al. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt-Eisenlohr H, et al. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J. Bacteriol. 1999;181:5563–5571. doi: 10.1128/jb.181.18.5563-5571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llosa M, et al. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 2000;182:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron C, et al. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez D, et al. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spudich GM, et al. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson LB, et al. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez D, et al. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beaupré CE, et al. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang TA, et al. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol. Microbiol. 1999;32:1239–1253. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X-R, Christie PJ. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J. Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rashkova S, et al. Self–assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J. Bacteriol. doi: 10.1128/jb.182.15.4137-4145.2000. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause S, et al. Sequence related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krause S, et al. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moncalian G, et al. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 1999;274:36117–36124. doi: 10.1074/jbc.274.51.36117. [DOI] [PubMed] [Google Scholar]
- 43.de Groot MJA, et al. Agrobacterium tumefaciens mediated transformation of filamentous fungi. Nat. Biotech. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 44.Pugsley A. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria – a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 45.Davis BM, et al. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXf. Science. 2000;288:333–335. doi: 10.1126/science.288.5464.333. [DOI] [PubMed] [Google Scholar]
- 46.Piper KR, Farrand SK. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J. Bacteriol. 2000;182:1080–1088. doi: 10.1128/jb.182.4.1080-1088.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paterson E, et al. Genetic analysis of the mobilization and leading regions of the IncN plasmids pKM101 and pCU1. J. Bacteriol. 1999;181:2572–2583. doi: 10.1128/jb.181.8.2572-2583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grahn AM, et al. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Callaghan D, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 50.Anderson SGE, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, et al. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7545–7550. doi: 10.1073/pnas.120156997. [DOI] [PMC free article] [PubMed] [Google Scholar]