Abstract
Introduction
Banking of high-quality placental tissue specimens will enable biomarker discovery and molecular studies on diseases involving placental dysfunction. Systematic studies aimed at developing feasible standardized methodology for placental collection in a typical clinical setting are lacking.
Methods
To determine the acceptable timeframe for placental collection, we collected multiple samples from first and third trimester placentas at serial timepoints in a 2-h window after delivery, simultaneously comparing the traditional snap-freeze technique to commercial solutions designed to preserve RNA (RNAlater™), and DNA (DNAgard®). The performance of RNAlater for preserving DNA was also tested. Nucleic acid quality was assessed by determining the RNA integrity number (RIN) and genome-wide microarray profiling for gene expression and DNA methylation.
Results
We found that samples collected in RNAlater had higher and more consistent RINs compared to snap-frozen tissue. Similar RINs were obtained for tissue collected in RNAlater as large (1 cm3) and small (∼0.1 cm3) pieces. RNAlater appeared to better stabilize the time zero gene expression profile compared to snap-freezing for first trimester placenta. DNA methylation profiles remained quite stable over a 2 h time period after removal of the placenta from the uterus, with DNAgard being superior to other treatments.
Discussion and conclusion
The collection of placental samples in RNAlater and DNAgard is simple, and eliminates the need for liquid nitrogen or a freezer on-site. Moreover, the quality of the nucleic acids and the resulting data from samples collected in these preservation solutions is higher than samples collected using the snap-freeze method and easier to implement in busy clinical environments.
Keywords: Placenta, RNA integrity, Gene expression, DNA methylation, Snap-freezing, RNAlater, DNAgard, Microarray
1. Introduction
The placenta is a transient organ. It is responsible for establishing the maternal–fetal interface, and allows for proper growth and development of the fetus. A well-functioning placenta is essential to a healthy pregnancy. Adverse pregnancy outcomes, such as fetal growth restriction (FGR) and preeclampsia, are often attributed to poor placental implantation, development and/or function [1]. Placental abnormalities associated with these conditions can lead to significant maternal and fetal morbidity and mortality, and contribute to the need for iatrogenic preterm delivery [2–8].
Banking of high-quality placental tissue specimens will enable biomarker discovery and molecular studies on diseases involving placental dysfunction. Microarray-based gene expression profiling of placental tissue has been used for discovery of disease-specific biomarkers [9,10], including the anti-angiogenic molecules, sFlt and sEng [11], which are associated with the development of pre-eclampsia. Altered DNA methylation of certain genes has also been associated with IUGR and preeclampsia [12,13]. In earlier studies, placental tissue samples have been collected by snap-freezing in liquid nitrogen. Placental collections using liquid nitrogen can be logistically challenging, with the quality of RNA isolated from snap-frozen tissue often variable and overall quite poor. We sought to answer whether nucleic acids could be used as reliable biomarkers of placental function.
We concluded that systematic studies needed to be conducted to determine the best collection method for placental tissue in the clinical setting for downstream genomic and epigenomic analysis. Collection of high quality placental samples is dependent on many parameters, including timing of collection in busy Labor and Delivery units, as well as the protection of nucleic acids, particularly RNA, in samples of this RNase-rich tissue [14]. In obstetrics, the time of delivery is unpredictable, making immediate tissue collection difficult. It has generally been presumed that immediate collection of placental tissue within the first hour after removal of the placenta is critical for adequate preservation of nucleic acid integrity. However, there are no prior studies to support this time cut-off. In addition, while the traditional method for placental collection is snap-freezing in liquid nitrogen, commercially available preservation solutions, which are easier to use and designed to allow for storage of tissues from days to weeks at ambient temperature, have also not been systematically tested.
In this study, we sought to identify the optimal timing and mode of placental collection for nucleic acids of sufficient quality to perform genome-wide RNA gene expression and DNA methylation studies for downstream molecular and functional enrichment analysis. To address this, we evaluated three different tissue collection methods: snap-freezing in liquid nitrogen, RNAlater, and DNAgard, over a 2-h window upon removal of the placenta or placental tissues from the uterus, to determine: 1) the optimal collection method(s) for evaluation of mRNA expression and DNA methylation; and 2) the time period after delivery during which such optimal samples should be collected.
2. Materials and methods
This study was conducted under approval from the Institutional Review Board at the University of California, San Diego Human Research Protection Program. Clinical characteristics of placental samples (where available) are provided in Supplementary Table 1. All microarray data can be accessed at the Gene Expression Omnibus database (GSE55440).
2.1. Third trimester placentas
Samples were collected from four placentas, from women with normal pregnancies undergoing a term (38–40 weeks of gestation) scheduled cesarean section. Placentas were processed immediately after delivery as follows: Initially, samples approximately 1 cm3 in size were obtained from the placental disc, halfway between the umbilical cord insertion point and the placental margin. Following removal of the maternal and fetal surfaces, the sample was washed twice in cold PBS and some samples were cut into smaller pieces measuring approximately 0.1 cm3 (corresponding to cubes of tissue measuring ∼0.5 cm per side). Care was taken to exclude large blood vessels. The small samples were then banked as follows: 1) samples were placed into sterile DNase- and RNase-free 1.5 ml microfuge tubes and snap-frozen in liquid nitrogen and 2) samples were placed into tubes containing 1 ml of preservative solution (RNAlater™ RNA Stabilization Reagent (Qiagen) or DNAgard® Tissue (Sigma Aldrich)). Biological triplicate samples were collected at 0, 30, 60, and 120 min following placental removal from the uterus, with the placenta remaining at room temperature between samplings. Samples that were snap-frozen in liquid nitrogen were moved immediately to a −80°C freezer. Samples in DNAgard were stored at room temperature until the time of DNA extraction (within 1 month). Samples in RNAlater were immediately placed at 4°C. After a period of 24–72 h, excess RNAlater was removed from the microfuge tubes and the samples were placed in the −80°C freezer for storage until RNA isolation was performed.
Larger samples (“chunk”), 1 cm3 in size, were collected at 0 and 60 min following delivery and placed in 10 ml RNAlater. These samples were also stored at 4°C for 24–72 h, after which they were removed from the preservative, cut into smaller pieces (0.1 cm3), placed into RNase- and DNase-free microfuge tubes, and stored in the −80 °C freezer.
2.2. First trimester placentas
First trimester (10–12 weeks gestation) placental samples were collected from women undergoing abortion procedures. Sterile tubing and collection jars were used in procedures for placental collection. After extraction of products of conception, the placenta was identified and small samples ∼0.1 cm3 were collected by snap-freezing and in RNAlater and DNAgard as described for term placenta. Larger samples were not obtained for first trimester placentas, as the majority of available tissue samples were present as smaller fragments.
2.3. RNA extraction, quantification, and quality control
Tissue was lysed in mirVana (Life Technologies) lysis buffer, using a Mini-Beadbeater-16 (Biospec), with agitation for 1 min in the presence of 1 mm zirconia beads. Samples were then centrifuged at maximum speed for 1 min and the lysed solution was transferred to a fresh microfuge tube. The remainder of the extraction was per the manufacturer's protocol for the mirVana kit (Life Technologies). After extraction, RNA was quantified using the Quant-iT RNA BR Assay Kit (Life Technologies). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent). RNA Nano-chips were prepared and loaded according to the manufacturer's protocol. The RNA integrity number (RIN) was obtained using the software provided by the manufacturer.
2.4. Data analysis and statistics: gene expression
Non-parametric Wilcoxon signed-rank test was used to compare RIN values of small and large sample, and different treatments at time 0 and 60 min for all placental samples (performed in triplicates) using the R statistical environment [15]. Gene expression profiling was performed using HumanHT-12 v4 Expression Bead-Chips according to the manufacturer's instructions (Illumina). Samples were prepared using the TotalPrep RNA Amplification Kit (Life Technologies) according to the manufacturer's instructions. Probes were filtered with a detection P-value ≤0.01 using GenomeStudio, and normalized using the lumi package in R with the RSN (Robust spline normalization) method. Samples were also subjected to batch correction using ComBat R package [16] with default settings. Principal component analysis (PCA), hierarchical clustering, and differential gene expression analysis was performed using Qlucore Omics Explorer (version 2.3). Student's t-test (P ≤ 0.01) was applied where multiple testing correction did not yield any probes (q ≤ 0.01). Area proportional Venn diagrams were generated using BioVenn [17]. A total of 72 samples were used for analysis.
2.5. DNA extraction and quantification
Samples were lysed in Buffer ATL (Qiagen), using a Mini-Beadbeater-16 (Biospec), with agitation for 1 min in the presence of 1 mm zirconia beads. Samples were then centrifuged at maximum speed for 1 min and the lysed solution was transferred to a new tube. The remainder of the extraction was followed per the DNeasy handbook using the animal tissue (spin column) protocol, without the optional RNase treatment. DNA was quantified using the Quant-iT DNA Assay kit (Life Technologies).
2.6. Data analysis and statistics: DNA methylation
DNA quality was checked using the BioAnalyzer 6000 (Agilent) according to the manufacturer's protocol. DNA was bisulfite converted using the EZ DNA methylation kit (Zymo Research) according to the manufacturer's protocol. Bisulfite converted DNA was processed and hybridized to HumanMethylation450 BeadChips (Illumina), which were scanned with an Illumina iScan Bead Array Scanner per the manufacturer's protocol. DNA methylation data IDAT files for each samples were used for processing and SWAN normalization and differential analysis on M values using the minfi Bioconductor package in R [18]. A total of 48 samples were used for analysis. principal component analysis (PCA), hierarchical clustering, was performed using Qlucore Omics Explorer (version 2.3).
3. Results
The experimental study design is summarized in Table 1 and Fig. 1. Briefly, placentas were obtained from four first trimester and four term pregnancies (see Material and Methods) and were cut into large “chunks” (1 cm3, third trimester placentas only) and smaller “pieces” (0.1 cm3, both first and third trimester placentas). Samples were then preserved using RNAlater, DNAgard or snap-freezing in liquid nitrogen, the method that has traditionally been considered the gold standard [19]. To assess which collection method for nucleic acid preservation was ideal, we performed the following tests on the extracted RNA and DNA: (i) determination of RNA integrity number (RIN) [20], (ii) microarray gene expression profiling, and (iii) microarray DNA methylation profiling.
Table 1.
Placenta samples and experimental conditions used to assess placental tissue banking. Each placenta sample name (e.g. Sample 3.1, refers to trimester (3rd trimester), followed by biological replicate number (replicate number 1). See Supplementary Table 1 for detailed information.
| Time (minutes) | 0 | 30 | 60 | 120 | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||
| Treatment | RIN | Gene expression | DNA methylation | RIN | RIN | Gene expression | RIN | Gene expression | DNA methylation |
| Snap-frozen | 1.1–1.4 | 1.1 | 1.1 | 1.1–1.4 | 1.1–1.4 | 1.1 | 1.1–1.4 | 1.1 | 1.1 |
| 3.1–3.4 | 3.1 | 3.1, 3.2 | 3.1–3.4 | 3.1–3.4 | 3.1 | 3.1–3.4 | 3.1 | 3.1, 3.2 | |
| RNAlater™ | 1.1–1.4 | 1.1–1.3 | 1.1 | 1.1–1.4 | 1.1–1.4 | 1.1, 1.2 | 1.1–1.4 | 1.1–1.3 | 1.1 |
| 3.1–3.4 | 3.1–3.3 | 3.1 | 3.1–3.4 | 3.1–3.4 | 3.1, 3.2 | 3.1–3.4 | 3.1–3.3 | 3.1 | |
| DNAgard® | N/A | N/A | 1.1, 1.2 3.1, 3.2 |
N/A | N/A | N/A | N/A | N/A | 1.1, 1.2 3.1, 3.2 |
RNA Integrity Number.
Fig. 1.

Overview of human placental tissue banking and collection methods. Human placenta from first and third trimester pregnancies were processed immediately after delivery. Samples were washed in PBS and cut into small pieces (0.1 cm3). The small samples were then placed into sterile 1.5 mL microfuge tubes and either snap-frozen in liquid nitrogen, placed into tubes containing 1 ml of RNAlater™, or DNAgard®. Samples that were snap-frozen were stored in a −80 °C freezer. Samples in DNAgard were stored at room temperature until the time of DNA extraction (within 1 month). Samples in RNAlater were immediately placed at 4 °C Larger samples, “chunks” (1 cm3) were processed with RNAlater only.
3.1. What is the optimal timing and collection method for RNA?
To determine which collection method was better for RNA analysis, we evaluated RNA quality and variability in gene expression profiles for two collection methods: RNAlater and snap-freezing. We also assessed the potential influence of the size of the tissue samples by comparing results from two different sample sizes collected in RNAlater.
3.1.1. RNA integrity and quality
Tissue from third trimester placentas was collected in RNAlater as small pieces and large chunks (Fig. 1), to determine whether larger samples could be initially collected and later divided into smaller pieces without compromising RNA quality. The RNA integrity number (RIN) [20] was used to assess RNA quality.
RINs obtained from the larger chunks collected in RNAlater ranged from 4.8 to 9.6 at time 0 and 2.5–9.1 at 60 min, while the RINs from the smaller pieces ranged from 6.8 to 8.8 at time 0 and 7.2–9.2 at 60 min. When comparing the RINs of the large samples to the small samples collected at the same time points in RNAlater, using the nonparametric Wilcoxon signed rank test there was no significant difference at either time 0 (P-value = 0.57) or 60 min (P-value = 0.46) (Supplementary Fig. 1 and Supplementary Table 1).
We then compared the performance of RNAlater and snap-freezing for preservation of RNA quality over time, collecting smaller size (∼0.1 cm3) samples in the two conditions. First trimester RINs ranged from 6.3 to 9.4 for RNAlater and 2.2–8.9 for snap-frozen (Fig. 2A, Supplementary Table 1, Supplementary Fig. 2). Third trimester RINs ranged from 4.9 to 9.3 for RNAlater and 2.5–7.5 for snap-frozen (Fig. 2B, Supplementary Table 1, Supplementary Fig. 3). First trimester placental RINs were significantly different between snap-frozen and RNAlater at each time points (0, 30, 60, and 120 min) (Wilcoxon signed-rank test, P-value < 0.005). For term placentas, a significant difference (Wilcoxon signed-rank test, P-value ≤ 0.05) between RNAlater and snap-frozen samples was seen at time points 0, 30, 60 and 120 min. These results indicated that RNAlater was superior to snap-freezing for preservation of RNA quality as measured by RIN scores at all timepoints for first trimester placentas, and for the first 60 min for third trimester placentas.
Fig. 2.
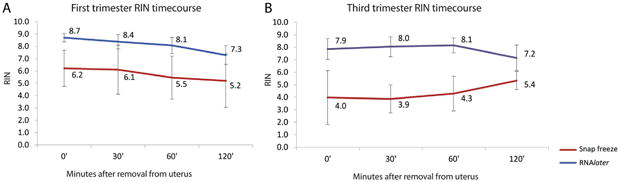
Comparison of RNA quality between RNAlater and snap-freezing treatment methods for tissue preservation: RNA integrity number (RIN) for small samples (0.1 cm3) from first (A) and third trimester (B) treated with RNAlater (blue) and snap-frozen in liquid nitrogen (red) at 0, 30, 60 and 120 min after removal from uterus. Technical triplicates samples were collected for each time point and RIN scores were averaged and standard deviation was calculated.
3.2. What is the optimal time to collect placental samples for RNA expression analysis?
To assess the impact of time and mode of collection on gene expression, microarray-based gene expression analysis was performed on three first and three third trimester placentas. For each placenta, samples from three independent RNA extractions per collection condition per time point (0, 60,120 min) were analyzed. After detection p-value filtering (see Methods), which yielded 27,719 analyzable probes, we performed 3D principal component analysis (PCA) for unsupervised clustering of the samples (Fig. 3). As expected, we observed that the principal component that accounted for the largest degree of variation in the data could be attributed to differences between first vs. third trimester placental samples (Fig. 3a). The next largest difference was associated with preservation method (Fig. 3b), although there was some overlap in samples collected using different methods. The time of collection showed a considerably weaker effect (Fig. 3d), and there was no detectable effect of size of sample (larger chunk vs. smaller pieces) (Fig. 3c).
Fig. 3.

3D principal component analysis (PCA) of placental samples for gene expression analysis. Unsupervised clustering of 27,719 probes based on (A) trimester (first and third); (B) treatment; (C) size (small pieces vs. large chunks); and (D) time. PCA was performed using Qlucore.
3.2.1. Gene expression
To identify probes that were impacted by time and/or mode of collection, we performed t-tests to identify genes/probes with expression levels that differed between early timepoints (0 vs. 60 min) and those that differed between later timepoints (60 vs. 120 min). If a gene was found to be significantly different (P-value < 0.01) across either of the two time intervals, it was grouped into one of nine possible patterns of change across time (Table 2, e.g. increased in the early time period and increased in the later time period, or increased in the early time period and no change in the later time period, etc.). No probes showed significant change over time if multiple testing correction was applied. We recognize that in future studies aimed at discovery of gene expression changes between placentas from different patient populations, multiple testing correction will be applied. However, in the present study, in order to identify subtle differences among the conditions tested, we did not use multiple testing correction.
Table 2.
Differentially expressed genes based on placenta tissue preservation treatments across time. Nine possible patterns resulting from gene expression changes that could occur between early (0–60 min) and late collection (60–120 min) (t-test; P-value < 0.01).
| Pattern | 0-60 minutes (early response) | 60-120 minutes (late response) | Annotation | 1st Trimester | 3rd Trimester | ||||
|---|---|---|---|---|---|---|---|---|---|
| RNAlater 1.1 | RNAlater 1.2 | Snap Freeze 1.1 | RNAlater 3.1 | RNAlater 3.2 | Snap Freeze 3.1 | ||||
| 1 | Up, Up | 0 | 13 | 1 | 1 | 2 | 4 | ||
| 2 | Up, No Change | 110 | 211 | 99 | 115 | 266 | 402 | ||
| 3 | Up, Down | 9 | 2 | 3 | 10 | 15 | 28 | ||
| 4 | No Change, Up | 460 | 326 | 187 | 48 | 297 | 111 | ||
| 5 | No Change | 26503 (95.6%) | 26323 (94.9%) | 27145 (97.9%) | 27352 (98.6%) | 26258 (94.7%) | 26799 (96.6%) | ||
| 6 | No Change, Down | 561 | 517 | 177 | 85 | 453 | 117 | ||
| 7 | Down, Up | 6 | 13 | 2 | 3 | 37 | 16 | ||
| 8 | Down, No Change | 62 | 232 | 104 | 105 | 389 | 242 | ||
| 9 | Down, Down | 8 | 82 | 1 | 0 | 2 | 0 | ||
Total probes: 27719
Even without multiple testing correction, the large majority (>94.7% in all cases) of probes did not change over time (Table 2, pattern 5). For first trimester placentas, the bulk of the changes occurred in the second hour after removal from the uterus (patterns 4 and 6), suggesting that collection within the first hour will greatly minimize the risk of artifact. For third trimester placentas, most of the probes that changed did so in the first hour and then stabilized over the second hour (patterns 2 and 8). Since the overall percentage of probes that changed over the first 2 h was small for the third trimester placentas, and it is not possible to guarantee collection of placental samples within minutes of delivery, we suggest that it is acceptable to collect third trimester placenta samples up to 2 h after delivery.
We then evaluated the methodology for collection of placental samples for gene expression analysis. For the first trimester placenta that was preserved both using RNAlater and snap-freezing (Placenta 1.1) the number of probes that changed over the first hour was 195 for RNAlater and 210 for snap-freezing, not an important difference. For the third trimester placenta that was preserved using both methods (Placenta 3.1), the number of probes that changed over the first 2 h was 367 for RNAlater and 919 for snap-freezing, which suggests that RNAlater is the better collection method. Overall, these results support the conclusion that RNA from samples collected in RNAlater are as good or better than those collected by snap-freezing.
We then determined whether the same probes were unstable over time in different placentas. For each placenta, a Student's t-test was performed to compare the gene expression results at 0 and 120 min (P-value ≤0.01); for these experiments, RNAlater was used for all samples. As above, no probes changed over time if multiple testing correction was applied. For our analyses, we did not apply multiple testing correction. The number of differentially expressed probes varied across the three first trimester placentas samples and very few probes overlapped among them (Fig. 4A). The overlap between third trimester placentas samples was slightly higher than first trimester placentas, but generally followed the same trend (Fig. 4B). We concluded that the overlap in probes that changed expression level between 0 and 120 min among different placentas was not extensive enough, either in the first or third trimester, to allow identification of probes that should be systematically excluded from future analyses.
Fig. 4.
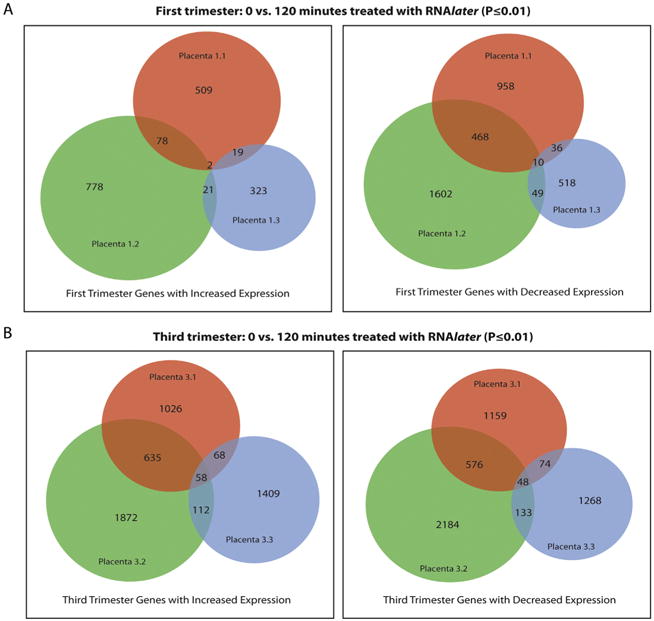
Venn Diagram of differential gene expression analysis probe overlap (A) Area-proportional Venn diagram of differentially expressed probes at 0 min and 120 min (P-value ≤0.01) that increase (right) and decrease (left) in expression across three first-trimester placental samples that were treated with RNAlater. (B) Area-proportional Venn diagram of differentially expressed probes at 0 min and 120 min (P-value ≤0.01) that increase (right) and decrease (left) in expression across three third-trimester placental samples that were treated with RNAlater.
3.3. What is the effect of collection method and time of collection on DNA methylation?
We evaluated the DNA methylation at 479,860 CpG sites using the HumanMethylation450 DNA Methylation BeadChip. All samples were normalized together using the SWAN method [18]. We compared the collection methods, snap-freeze and DNAgard for the first trimester samples; (snap-freeze, RNAlater, and DNAgard for the third trimester samples) at two timepoints, 0 and 120 min. We evaluated the first and third trimester placenta samples separately. Using PCA unsupervised clustering, the greatest difference in DNA methylation profiles was observed between first and third trimester samples (Fig. 5). As with the gene expression analysis above, preservation method (Fig. 5b) and time (Fig. 5c) appeared to have weaker effects compared to trimester (Fig. 5a).
Fig. 5.
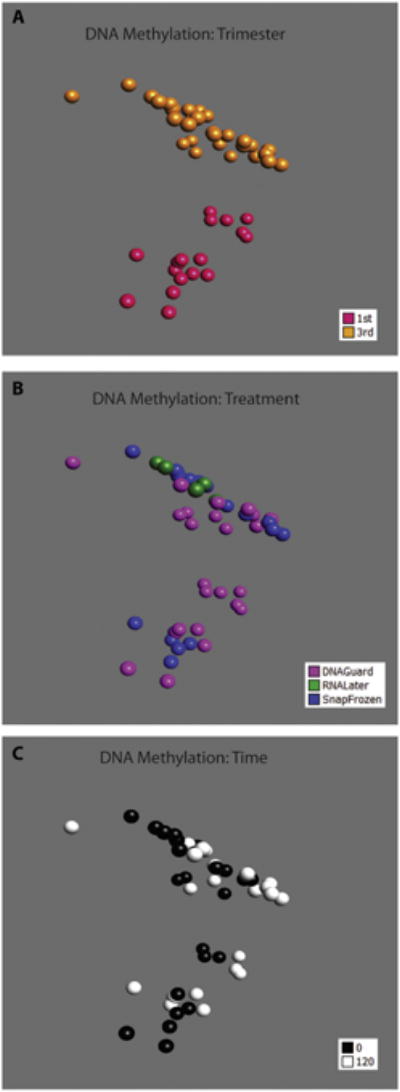
3D principal component analysis (PCA) of placental samples for DNA methylation profiling. Unsupervised clustering of 479,860 probes based on (A) trimester (first and third); (B) treatment; and (C) time.
Using the Student's t-test (P-value ≤0.01), we compared the number of differentially methylated probes between timepoints for each preservation method (Table 3a). There were no significant changes in any condition over the first 2 h when multiple testing was applied. Without multiple testing correction, the number of DNA methylation changes over the first 2 h was highest using snap-freezing as the preservation method and lowest using DNAgard for both first and third trimester.
Table 3a.
Differentially methylated probes. Differentially methylated probes over time (0 vs. 120 min) across different preservation treatment (P-value < 0.01). Numbers reflect total probes found during t-test of triplicates per placenta sample. Snap:Snap-frozen.
| Placenta sample | 0 vs. 120 minutes | ||
|---|---|---|---|
|
| |||
| Snap-frozen | DNAgard | RNAlater | |
| 1.1 | 14,352 | 9632 | – |
| 1.2 | – | 6344 | – |
| 3.1 | 23,157 | 5288 | 15,274 |
| 3.2 | 4233 | 2679 | – |
RNAlater was studied for the third trimester collection, and the number of differences between the RNAlater and DNAgard samples at time zero is extremely small (Table 3b). Therefore, if a sample can be collected immediately after removal from the uterus, RNAlater is an acceptable alternative to DNAgard, allowing for simplification of the collection process to a single condition for both RNA expression and DNA methylation analyses.
Table 3b.
Differentially methylated probes. Differentially methylated probes between different treatments at time 0 and 120 min (P < 0.01). Numbers reflect total probes found during t-test of triplicates per placenta sample. Snap:Snap-frozen.
| Placenta sample | Snap vs. DNAgard | Snap vs. RNAlater | DNAgard vs. RNAlater | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| 0 | 120 | 0 | 120 | 0 | 120 | |
| 1.1 | 6933 | 14,277 | – | – | – | – |
| 3.1 | 3328 | 6107 | 12,921 | 25,426 | 18,026 | 12,768 |
| 3.2 | 34,287 | 27,153 | – | – | – | – |
We compared the DNA methylation profiles for samples collected at 0 and 120 min from two different first trimester placentas in DNAgard (Fig. 6a), two third trimester placentas in DNAgard (Fig. 6b), and two snap-frozen third trimester placentas (Fig. 6c). There was little overlap in the sets of probes that changed between 0 and 120 min across different placentas. Therefore, we did not identify probes that should be systematically excluded from placental DNA methylation analyses. From these results, we conclude that the DNA methylation profiles of both first and third trimester placentas preserved in DNAgard were quite stable over the first 2 h after removal from the uterus.
Fig. 6.
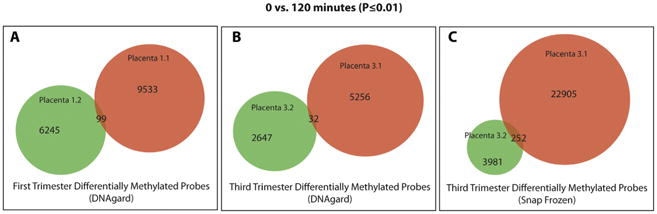
Area-proportional Venn diagram of differentially methylated probes at 0 min and 120 min (P-value ≤0.01) for (A) first-trimester placentas, (B) third-trimester placentas treated with DNAgard, and (C) third trimester placentas that were snap-frozen.
4. Discussion
Previous studies have sought to determine appropriate methods for placental banking. Fajardy et al. reported adequate RNA stability as measured using the RIN and transcript levels of a small number of stress-responsive genes using RNAlater with human placenta [21]. They compared two methods of collection in RNAlater, either direct tissue transfer or dissection of placental villi at 24 h intervals over a 4 day time course. They concluded placental collection method was important, with direct transfer being superior to dissection. They also concluded that placental samples could be stored at 4 °C for up to 48 h prior to collection without significant decline in RNA quality. Subsequently, Avila et al. looked at a 24 h processing interval and determined a more rapid degradation post-delivery of in the five specific genes they evaluated [22]. These conflicting results led us to postulate that genome-wide assessments of nucleic acid stability were necessary for such studies. More recently, in an opinion piece, Burton et al. highlighted several factors that may affect molecular analyses of placental tissue such as mode of delivery, placenta weight and ideal methods for storage and banking of tissues [23]. Other studies incorporating other tissues [24,25] used snap-freezing and RNAlater to preserve samples for microarray profiling, but have not systematically compared the quality of the nucleic acid or the data derived from samples collected using these methods. In agreement with our study, the authors recommended using RNAlater over snap-freezing and initially obtaining larger samples (1–2 cm diameter) and cutting these into smaller pieces. Our findings here provide evidence for these recommendations, demonstrating the molecular impact on the tissue using these methods.
4.1. RNA
Reproducible genomic analysis depends on the quality of the relevant nucleic acid in the banked tissue. In regard to gene expression, a RIN greater than 7 is suitable for microarray analysis, giving the fewest false positive results [26]. When comparing methods, collecting placental samples in RNAlater yielded better RNA integrity numbers and fewer changes in gene expression, particularly for third trimester placenta samples, (as assessed with a commonly used microarray platform compared to standard snap-freezing). We also showed that larger (1 cm3) and smaller (0.1 cm3) samples collected in RNAlater yielded equivalent RNA quality. The ability to collect larger samples in RNAlater allows for a procedure that involves collection of a single sample per placenta at room temperature without sacrificing quality or integrity. This simplified procedure can be performed quickly and easily by clinical personnel, enabling consistent sample collection at clinical sites without the need to have on-site research personnel at all times.
First and third trimester placental samples differed in their sensitivity to time and method of collection. First trimester placental samples showed more changes in gene expression over time, while third trimester placental samples showed larger differences between collection methods. Therefore, it is important to collect samples from first trimester placentas within 1 h, while it is acceptable to collect third trimester placentas within 2 h. The longer window for third trimester placentas allows research personnel to be “on-call” off-site and travel to the Labor and Delivery site after delivery, or clinical personnel to finish their clinical duties before collecting samples for research.
4.2. DNA
DNA is more stable than RNA. While there are no studies that have observed global DNA methylation changes in human placental samples over time, Avila and colleagues showed that DNA methylation was less susceptible to alterations due to processing time in the few sites that were analyzed [22].
In our study, DNA methylation profiles were more stable than gene expression profiles over time of collection. For samples collected immediately after removal from the uterus, the differences among collection methods were quite small, particularly for third trimester placenta comparing RNAlater and DNAgard. However, the number of DNA methylation changes between the 0 and 120 min timepoints were much higher for samples collected by snap-freezing or in RNAlater compared to DNAgard; we therefore concluded that DNAgard is the optimal method for collection of placental samples for DNA methylation analysis.
4.3. RNA and DNA
The superior performance of RNAlater and DNAgard over snap-freezing for preservation of RNA and DNA, respectively, both simplifies the collection of high-quality placental samples for genomic analysis and removes the need for liquid nitrogen and freezer space at clinical sites.
Simplistically, one might suppose that each collection method imposes a specific set of changes, while a delay in collection imposes a different set of changes. If this were the case, the number of changes between samples collected at different timepoints should be the same regardless of the method of collection. However, our results and others [27] have shown that the number of differences between 0 and 120 min was quite different for the three collection methods and that RNA quality decreases with delay in processing. The mechanisms by which RNAlater and DNAgard prevent changes in gene expression and DNA methylation, respectively, are not known (in samples collected at different timepoints after placental removal from the uterus). It is possible that over time, cells produce nucleases sequestered in vesicles which are disrupted upon freezing, and thus activated at the time samples are thawed for nucleic acid extraction. If the RNAlater and DNAgard are able to suppress nuclease function, this may explain why the samples collected using these products are more stable. However, a full description of these changes would require analysis of additional placentas.
5. Conclusion
A well-functioning placenta is critical to a healthy pregnancy. Detection of molecular differences between normal placentas and those complicated by an adverse pregnancy outcome can be used to identify biomarkers of placental dysfunction, gleaning insights into the mechanisms underlying adverse pregnancy outcomes. Such studies require the collection of high-quality placental samples, in which the molecular profiles are well-preserved. Our study demonstrates that time-dependent changes in the placental gene expression and DNA methylation profiles start to occur immediately after delivery, with the method of tissue collection impacting the magnitude of these changes. Based on our results, we recommend tissue collection within the first hour in RNAlater for gene expression studies. For DNA methylation studies, collection in DNAgard within the first 2 h is optimal. If samples can be collected immediately after delivery, collecting a larger sample in RNAlater is valid for both RNA and DNA analyses. The latter process is highly efficient, allowing clinical personnel to collect placental samples immediately after placental removal from the uterus for research without adversely impacting clinical care. This means that effective tissue collection of high-quality placental tissue samples for genomic analysis can be accomplished at all times, even when a member of the research team is not on-site. This study will serve as a foundation that will enable future studies that include a sufficient number of placental samples from normal and complicated pregnancies, enabling detection of changes in placental gene expression or DNA methylation associated with pregnancy complications.
Supplementary Material
Acknowledgments
LMW, RDT, FB, WKK, LCL, and the work described in this manuscript were supported by funds from the Department of Reproductive Medicine at UCSD. LCL was also supported by a WRHR Career Development Award from NIH/NICHD (K12 HD001259). LMW and VT were supported by a CIRM Clinical Fellow Training Award. RC and JFL were supported by CIRM (RM1-01717 and TR1-01250). MP was supported by funds from a New Faculty Award California Institute for Regenerative Medicine (RN2-00931-1) and NIH/NICHD (R01HD071100). The work described here was supported by funds from the Department of Reproductive Medicine at UCSD and CIRM (TR1-01250).
Footnotes
Conflict of Interest: We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome. 5/2/2014.
The authors have no financial conflict of interest. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of Planned Parenthood Federation of America, Inc.
Appendix A. Supplementary data: Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.placenta.2014.05.005.
References
- 1.Zhou Y, McMaster M, Woo K, Janatpour M, Perry J, Karpanen T, et al. Vascular endothelial growth factor ligands and receptors that regulate human cyto-trophoblast survival are dysregulated in severe preeclampsia and hemolysis, elevated liver enzymes, and low platelets syndrome. Am J Pathol. 2002 Apr;160(4):1405–23. doi: 10.1016/S0002-9440(10)62567-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basso O, Rasmussen S, Weinberg CR, Wilcox AJ, Irgens LM, Skjaerven R. Trends in fetal and infant survival following preeclampsia. JAMA. 2006 Sep 20;296(11):1357–62. doi: 10.1001/jama.296.11.1357. [DOI] [PubMed] [Google Scholar]
- 3.Habli M, Levine RJ, Qian C, Sibai B. Neonatal outcomes in pregnancies with preeclampsia or gestational hypertension and in normotensive pregnancies that delivered at 35, 36, or 37 weeks of gestation. Am J Obstet Gynecol. 2007 Oct;197(4):406. doi: 10.1016/j.ajog.2007.06.059. [DOI] [PubMed] [Google Scholar]
- 4.Baschat AA, Gembruch U, Reiss I, Gortner L, Weiner CP, Harman CR. Relationship between arterial and venous Doppler and perinatal outcome in fetal growth restriction. Ultrasound Obstet Gynecol. 2000 Oct;16(5):407–13. doi: 10.1046/j.1469-0705.2000.00284.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein IM, Horbar JD, Badger GJ, Ohlsson A, Golan A. Morbidity and mortality among very-low-birth-weight neonates with intrauterine growth restriction. The Vermont Oxford Network. Am J Obstet Gynecol. 2000 Jan;182(1 Pt 1):198–206. doi: 10.1016/s0002-9378(00)70513-8. [DOI] [PubMed] [Google Scholar]
- 6.Garite TJ, Clark R, Thorp JA. Intrauterine growth restriction increases morbidity and mortality among premature neonates. Am J Obstet Gynecol. 2004 Aug;191(2):481–7. doi: 10.1016/j.ajog.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Lackman F, Capewell V, Richardson B, daSilva O, Gagnon R. The risks of spontaneous preterm delivery and perinatal mortality in relation to size at birth according to fetal versus neonatal growth standards. Am J Obstet Gynecol. 2001 Apr;184(5):946–53. doi: 10.1067/mob.2001.111719. [DOI] [PubMed] [Google Scholar]
- 8.McIntire DD, Bloom SL, Casey BM, Leveno KJ. Birth weight in relation to morbidity and mortality among newborn infants. N Engl J Med. 1999 Apr 22;340(16):1234–8. doi: 10.1056/NEJM199904223401603. [DOI] [PubMed] [Google Scholar]
- 9.Lapaire O, Grill S, Lalevee S, Kolla V, Hosli I, Hahn S. Microarray screening for novel preeclampsia biomarker candidates. Fetal Diagn Ther. 2012;31(3):147–53. doi: 10.1159/000337325. [DOI] [PubMed] [Google Scholar]
- 10.Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):5169–74. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003 Mar;111(5):649–58. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourque DK, Avila L, Penaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not pre-eclampsia. Placenta. 2010 Mar;31(3):197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Yuen RK, Penaherrera MS, von Dadelszen P, McFadden DE, Robinson WP. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 2010 Sep;18(9):1006–12. doi: 10.1038/ejhg.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Baumann M, Nikitina L, Wenger F, Surbek D, Korner M, et al. RNA degradation differentially affects quantitative mRNA measurements of endogenous reference genes in human placenta. Placenta. 2013 Jul;34(7):544–7. doi: 10.1016/j.placenta.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 15.R Development Core Team. R: a language and environment for statistical computing. 3.0.1. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 16.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007 Jan;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 17.Hulsen T, de Vlieg J, Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maksimovic J, Gordon L, Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perlmutter MA, Best CJ, Gillespie JW, Gathright Y, Gonzalez S, Velasco A, et al. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. J Mol Diagn. 2004 Nov;6(4):371–7. doi: 10.1016/S1525-1578(10)60534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fajardy I, Moitrot E, Vambergue A, Vandersippe-Millot M, Deruelle P, Rousseaux J. Time course analysis of RNA stability in human placenta. BMC Mol Biol. 2009;10:21. doi: 10.1186/1471-2199-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avila L, Yuen RK, Diego-Alvarez D, Penaherrera MS, Jiang R, Robinson WP. Evaluating DNA methylation and gene expression variability in the human term placenta. Placenta. 2010 Dec;31(12):1070–7. doi: 10.1016/j.placenta.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Burton GJ, Sebire NJ, Myatt L, Tannetta D, Wang YL, Sadovsky Y, et al. Optimising sample collection for placental research. Placenta. 2014 Jan;35(1):9–22. doi: 10.1016/j.placenta.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Hatzis C, Sun H, Yao H, Hubbard RE, Meric-Bernstam F, Babiera GV, et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J Natl Cancer Inst. 2011 Dec 21;103(24):1871–83. doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdary D, Lathrop J, Skelton J, Curtin K, Briggs T, Zhang Y, et al. Prognostic gene expression signatures can be measured in tissues collected in RNAlater preservative. J Mol Diagn. 2006 Feb;8(1):31–9. doi: 10.2353/jmoldx.2006.050056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson KL, Pine PS, Rosenzweig BA, Turpaz Y, Retief J. Characterization of the effect of sample quality on high density oligonucleotide microarray data using progressively degraded rat liver RNA. BMC Biotechnol. 2007;7:57. doi: 10.1186/1472-6750-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jobarteh ML, Moore SE, Kennedy C, Gambling L, McArdle HJ. The effect of delay in collection and processing on RNA integrity in human placenta: experiences from rural Africa. Placenta. 2014 Jan;35(1):72–4. doi: 10.1016/j.placenta.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


