Abstract
Phosphatidylinositol-3-kinase (PI3K) is a key regulator of diverse biological processes including cell proliferation, migration, survival, and differentiation. While a role of PI3K in chondrocyte differentiation has been suggested, its precise mechanisms of action are poorly understood. Here we show that PI3K signaling can down-regulate Nkx3.2 at both mRNA and protein levels in various chondrocyte cultures in vitro. In addition, we have intriguingly found that p85β, not p85α, is specifically employed as a regulatory subunit for PI3K-mediated Nkx3.2 suppression. Furthermore, we found that regulation of Nkx3.2 by PI3K requires Rac1–PAK1, but not Akt, signaling downstream of PI3K. Finally, using embryonic limb bud cultures, ex vivo long bone cultures, and p85β knockout mice, we demonstrated that PI3K-mediated suppression of Nkx3.2 in chondrocytes plays a role in the control of cartilage hypertrophy during skeletal development in vertebrates.
Keywords: Nkx3.2 (Bapx1), PI-3-Kinase, Chondrocyte differentiation, Cartilage development
1. Introduction
During vertebrate skeletal development, condensed mesenchymes first give rise to cartilage primordia, and later, hypertrophied cartilage tissues are replaced with mature bone cells. This multi-step process, collectively termed endochondral ossification, is responsible for forming the majority of the skeleton, including vertebral columns and limbs [1–4].
Nkx3.2, also known as Bapx1, belongs to the NK2 class of the homeo-box superfamily, and the NK family of homeobox genes has been shown to play an important role in cell fate determination during diverse organ development [5–9]. In particular, Nkx3.2 is initially expressed in chondrogenic progenitor cells and promotes chondrogenic cell fate [10–18]. Nkx3.2 has also been shown to play a role in protecting chondrocytes from programmed cell death [19,20]. Additionally, Nkx3.2 has been shown to up-regulate type II collagen [21] and down-regulate type X collagen [18]. Consistent with these observations, Nkx3.2 has been suggested to play a role in BMP7-induced inhibition of hypertrophy [22] and in hypoxia-mediated suppression of hypertrophy [23]. Taken together, these results strongly suggest that Nkx3.2 functions in early phase chondrocytes, while antagonizing terminal phase chondrocytes during chondrogenesis.
Homozygotic disruption of Nkx3.2 in mice severely reduces ventro-medial parts of vertebral bodies, leading to neonatal lethality [8,24,25]. Consistent with these knockout (KO) phenotypes, mutations of Nkx3.2 in humans have been identified for Spondylo-Megaepiphyseal-Metaphyseal Dysplasia (SMMD), a skeletal disorder resulting in a disproportionately short stature with a short and stiff neck and trunk [26].
Phosphatidylinositol-3-kinase (PI3K) is a ubiquitous lipid kinase that is well documented as a key regulator of a number of cellular processes including cell growth, differentiation, and survival [27–29]. While three catalytic subunits including p110α, p110β, and p110δ have demonstrated functions in various cell types [30], most previous studies identify p85α as the key regulatory subunit [31,32]. In addition, Akt, a protein kinase downstream of PI3K, has also been extensively investigated in conjunction with PI3K and shown to have diverse roles in multiple biological processes [33–35]. Rac1 GTPase is another well characterized downstream target of PI3K signaling that is involved in cyto-skeletal arrangement, endocytosis, cell cycle regulation, cancer migration, and ROS production by fibroblasts [36–39].
In chondrocyte regulation, PI3K and Rac1 signaling pathways have been implicated in chondrogenic differentiation and hypertrophic maturation [40], as well as in proteoglycan biosynthesis [40,41]. Most relevant to this study, LY294002, a pharmacological inhibitor of PI3K, has been shown to suppress cartilage hypertrophy in ex vivo long bone cultures [42]. Furthermore, in vivo gene ablation studies suggest that these pathways are necessary for appropriate skeletal development [43]. While these findings indicate that PI3K signaling plays a significant role in chondrocyte differentiation, the precise molecular mechanism of PI3K-associated pathway function during cartilage development remains unclear.
Here, we show that PI3K signaling can effectively suppress Nkx3.2, which in turn permits chondrocyte hypertrophy that is necessary for normal progression of cartilage maturation during endochondral bone development.
2. Materials and methods
2.1. Chemical reagents and antibodies
LY294002 (MERCK Millipore; Damstadt, Germany), NSC23766, BKM120, BYL719, and MK2206 (Selleckchem; Houston, TX, USA) were used as PI3 kinase signaling inhibitors. MG132 (A.G. Scientific; San Diego, CA, USA) was used as a proteasome inhibitor. Recombinant human BMP2 was obtained from Cellumed (Seoul, Korea) and Insulin–transferrin–selenium (ITS) supplements for chondrocyte maturation were purchased from Thermo Fisher (Waltham, MA, USA). Anti-HA polyclonal antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-V5 monoclonal- and anti-Myc polyclonal antibodies were obtained from Merck Millipore (Damstadt, Germany). Anti-Flag monoclonal antibody was purchased from Sigma (St. Louis, MO, USA). Anti-Type X collagen (COL 10) antibody (Cosmobio; Tokyo, Japan) was used for immunohistochemistry (IHC). Anti-GAPDH and anti-β-actin antibodies (AbFrontier; Seoul, Korea) were used as loading controls for Western blotting. Anti-Nkx3.2 antibody purchased from Abcam (Cambridge, UK) and custom-made anti-Nkx3.2 antibody was obtained from Cosmogenetech (Seoul, Korea). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG and anti-rabbit IgG were purchased from Cell Signaling (Danvers, MA, USA) and Cy3-conjugated anti-rabbit IgG was purchased from Jackson ImmunoResearch Inc. (West Grove, PA, USA).
2.2. Cell culture
ATDC5 (Riken Cell Bank; Ibaraki, Japan) cells were maintained in DMEM-Ham’s F-12 (1:1) (Thermo Fisher; Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS) (GE Healthcare; Wauwatosa, WI, USA) and 1% penicillin–streptomycin (PS) (Thermo Fisher; Waltham, MA, USA). HEK293T, C2C12, and C3H10T1/2 (ATCC; Manassas, VA, USA) were grown in DMEM supplemented with 10% FBS and 1% PS. NIH3T3 cells (ATCC; Manassas, VA, USA) were maintained in DMEM supplemented with 10% NCS and 1% PS. Human articular chondrocytes (HACs) (ScienCell; Carlsbad, CA, USA) were maintained in a chondrocyte medium containing 5% FBS, 1% chondrocyte growth supplement, and 1% PS according to the manufacturer’s instruction.
2.3. Micromass culture system
When ATDC5 cells reached at confluence, cells were detached using 0.05% trypsin-EDTA (Thermo Fisher; Waltham, MA, USA), centrifuged, and resuspended in chondrogenic media containing 10 ng/ml BMP2 and 1% ITS supplement. Cells (2 × 105) in 10 μl media were spotted onto 24 well plates. After 40 min, 500 μl chondrogenic media were gently added. Media were exchanged every three days.
2.4. Knockout mice and in vivo experiments
p85β knockout mice (p85β−/−) with a disruption of the first exon of the Pik3r2 gene [32] were housed in individually-ventilated microisolation cages in the specific pathogen-free facility of the Yonsei Laboratory Animal Research Center (YLARC). Mouse genotypes were determined by PCR using wild-type and common primers to PCR amplify the wild-type gene, and null and common primers to PCR amplify the knockout gene.
p85β null primer: 5′-TGT TAA GAA GGG TGA GAA CAG AGT ACC-3′.
p85β common primer: 5′-GTC GCC TGT GAC TTC TGG AAG T-3′.
p85β WT primer: 5′-GCA TCC AGC CCA CAT TGT GT-3′.
All animals were maintained on a 12 h:12 h light/dark cycle with access to food and water ad libitum. All behavioral procedures were conducted during the light phase of the cycle. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of YLARC and performed in accordance with the YLARC IACUC guidelines for the ethical use of animals.
2.5. Mouse embryo limb bud culture and chondrogenic differentiation
Wild-type and p85β knockout embryos were obtained at E11.5, and limb buds were dissected and subjected to micromass cultures. Limb bud mesenchymal cells were isolated by digestion with 0.1% dispase (Thermo Fisher; Waltham, MA, USA) at 37 °C for 1 h, dissociated by vigorous pipetting, and passed through a 40 μm nylon cell strainer (Thermo FIsher; Waltham, MA, USA). Next, cells were centrifuged and the supernatant was discarded. Cell pellets were resuspended in 2:3 DMEM/F12 media supplemented with 10% FBS and 1% PS. The resuspension volume was 5 μl for each limb bud, and a 10 μl cell droplet was spotted per well (24 well plates). Cell spots were maintained in 2:3 DMEM/F12 media supplemented with 10% FBS, and 1% PS, containing ITS supplement for chondrogenic differentiation.
2.6. Expression plasmids and transfection
An empty vector, pCS2, was used to adjust total DNA amounts. Expression plasmids of Nkx3.2, Cbfβ, RAC1, p85α, or p85β were sub-cloned into pCS4 or pCS5 vectors in our laboratory. The pCS4 and pCS5 epitope-tagging vectors were gifts from C. Y. Yeo (Ewha Women’s University, Seoul, Korea). The 2 kb proximal promoter of Nkx3.2 was cloned into a pGL3–Basic vector (Nkx3.2p-Luc) (Promega; Madison, WI, USA) for use in reporter assays. All newly generated constructs were verified by DNA sequencing. Short hairpin RNAs (shRNAs) of p85α and p85β were purchased from Thermo Scientific (Waltham, MA, USA). pCMV6 M–Pak1 WT and kinase-dead mutant were a gift from Jonathan Chernoff (Addgene plasmid # 12209) (Addgene; Cambridge, MA, USA). Cells were transiently transfected using Vivamagic (Vivagen; Seoul, Korea) according to the manufacturer’s instructions.
2.7. Lentiviral infection system
To effectively knock down endogenous p85α and p85β in cells, we generated lentiviral particles using the following protocol. pLKO-based shRNA plasmids were packaged into lentiviral particles by co-transfecting with packaging plasmids pMD2.G and pPAX2 into HEK293T cells in growing media without antibiotics. One day later, supernatants containing lentiviral particles were harvested and resupplied daily with complete growing media for three days. Lentiviral particles were used to infect ATDC5 cells with 8 μg/ul polybrene (Sigma; St. Louis, MO, USA).
2.8. RT-PCR and real-time qPCR
RNA was isolated using an Easy-spin™ Total RNA Extraction kit (Intron Biotechnology; Seongnam, Korea). cDNA was synthesized from isolated RNA using TOPscript™cDNA Synthesis kit (Enzynomics; Daejeon, Korea) and the level of Nkx3.2 expression was analyzed by conventional PCR followed agarose gel electrophoresis stained with ethidium bromide (EtBr) or real-time qPCR using SYBR® Premix Ex Taq (Tli RNaseH Plus) (TaKaRa; Shiga, Japan).
The following primers were used in conventional PCR:
p85α forward: 5′-ATTTCACCCCCTACTCCCAA-3′.
p85α reverse: 5′-GGCTGTCTCTCATTCCATTC-3′.
p85β forward: 5′-CGCAACACGGACAGACTGGT-3′.
p85β reverse: 5′-TAGCAGACGCACAGGGAAGT-3′.
Nkx3.2 forward: 5′-AGCCCCTAAACCGCGAAAGAA-3′.
Nkx3.2 reverse: 5′-GGTCATGCAGAGGCGAGCAGGTC-3′.
GAPDH forward: 5′-TTTGTGATGGGTGTGAACCACG-3′.
GAPDH reverse: 5′-TTGTGAGGGAGATGCTCAGTGTTG-3′.
The following quantitative real-time PCR primers were used:
p85α forward: 5′-CAGTTTGCCCCTCCTGATGT-3′.
p85α reverse: 5′-AATTCTGCAGGGTTGCTGGA-3′.
p85β forward: 5′-AGGACGAGTGGACGTACTCA-3′.
p85β reverse: 5′-GGTAGAGAAGGCGTGTGTCC-3′.
Nkx3.2 forward: 5′-AAAGTGGCCGTCAAGGTGCT-3′.
Nkx3.2 reverse: 5′-AGCCCGGGAGACAGTAGTAA-3′.
Type X collagen (COL10) forward: 5′-AGGGAGTGCAATCATGGAGC-3′.
Type X collagen (COL10) reverse: 5′-AGGACGAGTGGACGTACTCA-3′.
β-Actin forward: 5′-GATGTGGATCAGCAAGCAGGA -3′.
β-Actin reverse: 5′-AGGGTGTAAAACGCAGCTCAG-3′.
2.9. Immunoblotting
Cells were washed in phosphate-buffered saline (PBS) and lysed in a buffer containing 50 mM Tris (pH 6.8), 2% sodium dodecyl sulfate (SDS), 1 mM dithiothreitol (DTT), and 8.75% glycerol. Cells lysates were boiled and protein concentrations were measured using the Bio-Rad DC™protein assay kit (Bio-Rad; Hercules, CA, USA), and equal amounts of protein were analyzed by SDS-PAGE and Western blotting followed by ECL detection according to the manufacturer’s protocol (GE Healthcare; Wauwatosa, WI, USA and DoGen; Seoul, Korea).
2.10. Reporter assay
Nkx3.2p-Luc and pRL-TK normalization plasmids were transiently co-transfected into ATDC5 cells. After 16 h, DMSO and 20 μM LY294002 were added to the cultures for 24 h. Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega; Madison, WI, USA) with a 20/20n single-tube luminometer (Promega; Madison, WI, USA). The results were normalized to the expression of the renilla luciferase transfection control (pRL-Tk).
2.11. Immunohistochemistry
For immunohistological analysis, femurs of wild-type and p85β knockouts were fixed in 4% paraformaldehyde (PFA) in PBS for 24 h at room temperate (RT). Femurs were next embedded in paraffin, sectioned at 4 μm thickness, and mounted on silane-coated slides (Muto-Glass; Tokyo, Japan). Deparaffinization was performed in xylene, a xylene/ethanol mixture, and serial ethanol dilutions. To retrieve antigen, sections were incubated with 10 mM citrate (pH 6.0) containing 0.05% Tween-20 for 20 min at 80 °C, followed by a PBS wash containing 0.05% Tween-20. Sections were then pre-incubated with a blocking buffer containing 5% goat serum, 0.05% sodium azide, and 0.1% Triton-X-100 in PBS for 45 min at RT, and incubated overnight at 4 °C with anti-COL10 and anti-Nkx3.2 antibodies in PBS containing 1% goat serum, 0.05% sodium azide, and 0.02% Tween-20. Sections were then incubated with Cy3-conjugated anti-rabbit IgG in the dilution buffer described above for 30 min at RT.
2.12. Ex vivo experiment
Hind limbs of E15.5 mice were dissected and digested with 0.1% dispase (Thermo Fisher; Waltham, MA, USA) in Puck’s solution A containing 10% chicken serum at 37 °C for 1.5 h. Femurs were then isolated and incubated overnight in differentiation media containing 0.2% BSA Cohn fraction V (Sigma; St. Louis, MO, USA), 50 μg/ml gentamycine (Sigma; St. Louis, MO, USA), 300 μg/ml L-glutamate (Thermo Fisher; Waltham, MA, USA), 50 μg/ml ascorbic acid (Sigma; St. Louis, MO, USA), and 1 mM β-glycerophosphate (Sigma; St. Louis, MO, USA). Femurs were then cultured in this medium with 20 μM LY294002 or DMSO. After 6 days of treatment, femurs were fixed with 4% PFA. Sections (4 μm) of paraffin embedded femurs were analyzed by IHC. The differentiation media and inhibitor were exchanged every two days.
2.13. X-ray radiography analysis
X-ray radiographic analysis was performed using a DXS 4000 pro system (KODAK; Rochester, NY, USA). The distance from the 1st cervical vertebra to 1st coccyx vertebra (LCC) and femur length of p85β (+/+), p85β (−/+) and p85β (−/−) mice were measured at 4, 6 and 8 weeks of age.
2.14. Statistical analysis
Graphs were drawn with GraphPad Prism (GraphPad Software Inc.; San Diego, CA, USA) and Microsoft Excel (Microsoft; Redmond, WA, USA). Statistical data comparisons were analyzed using Student’s t-test with GraphPad Prism.
3. Results
3.1. PI3K signaling negatively regulates Nkx3.2 at protein level
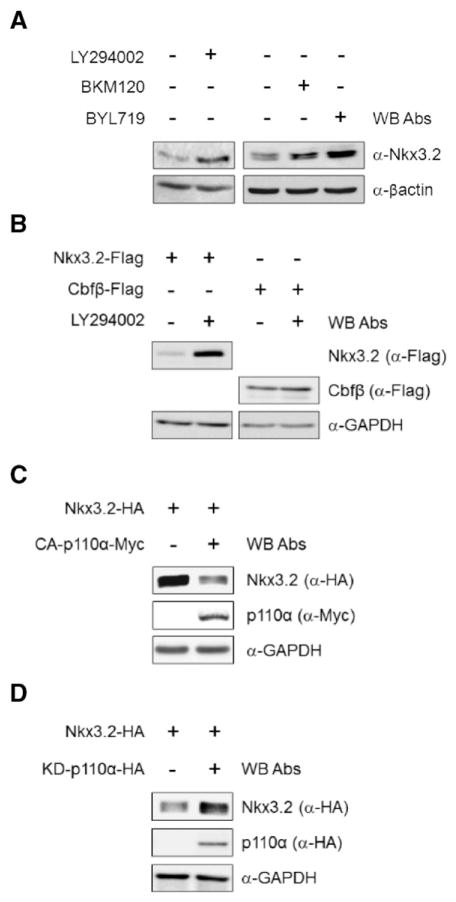
To understand the molecular mechanisms of PI3K-mediated control of chondrocyte differentiation, we first investigated whether PI3K signaling can modulate Nkx3.2 expression. To this end, using various PI3K inhibitors such as LY294002, BKM120, and BYL719 [44–46], we found that PI3K inhibition significantly elevated Nkx3.2 protein levels in human primary articular chondrocytes (HACs) (Fig. 1A). We next examined the effect of PI3K inhibition on the protein levels of ectopically expressed Nkx3.2. Similar to results obtained from endogenous Nkx3.2 proteins (Fig. 1A), pharmacological inhibition of PI3K also resulted in a remarkable increase in transiently overexpressed Nkx3.2 protein, whose expression is driven by a CMV promoter. Cbfβ expressed from the same vector was tested in parallel as a control (Fig. 1B).
Fig. 1.
Nkx3.2 protein levels can be suppressed by PI3K signaling. (A) Human articular chondrocytes (HACs) were cultured in the presence of 20 μM LY294002, 1 μM BKM120, and 10 μM BYL719, respectively, for 24 h. Cell lysates were subjected to Western blotting with the indicated antibodies. (B) Nkx3.2 and Cbfβ expression plasmids were overexpressed in 293T cells by transient transfection. After transfection, cells were treated with LY294002 for 24 h, harvested, and cell lysates were examined by Western blotting. Nkx3.2 and Cbfβ protein levels were analyzed with the indicated antibodies. (C, D) Nkx3.2 and the constitutively active form of p110α (CA-p110α-Myc), or kinase-dead form of p110α (KD-p110α-HA) expression plasmids were transiently transfected alone or in combination into 293T cells as indicated. After transfection, cell lysates were examined by Western blotting with the indicated antibodies.
We next asked whether a catalytic PI3K subunit is associated with this regulation. Since p110α is the most abundant catalytic subunit of PI3K in multiple chondrogenic cell lines that we have tested (unpublished observation), we first asked whether a constitutively active or dominant negative p110α can affect Nkx3.2 protein levels. Consistent with the results in Fig. 1A and B, in transient transfection assays Nkx3.2 protein levels were significantly reduced by co-expressing constitutively active p110α (CA-p110α; membrane-targeted CAAX-p110α) [47], and substantially increased by a kinase-dead form of p110α (KD-p110 α) [48] (Fig. 1C and D). Therefore, these results clearly indicate that PI3K signaling negatively regulates Nkx3.2 protein levels.
3.2. PI3K-mediated regulation of Nkx3.2 specifically employs p85β regulatory subunit
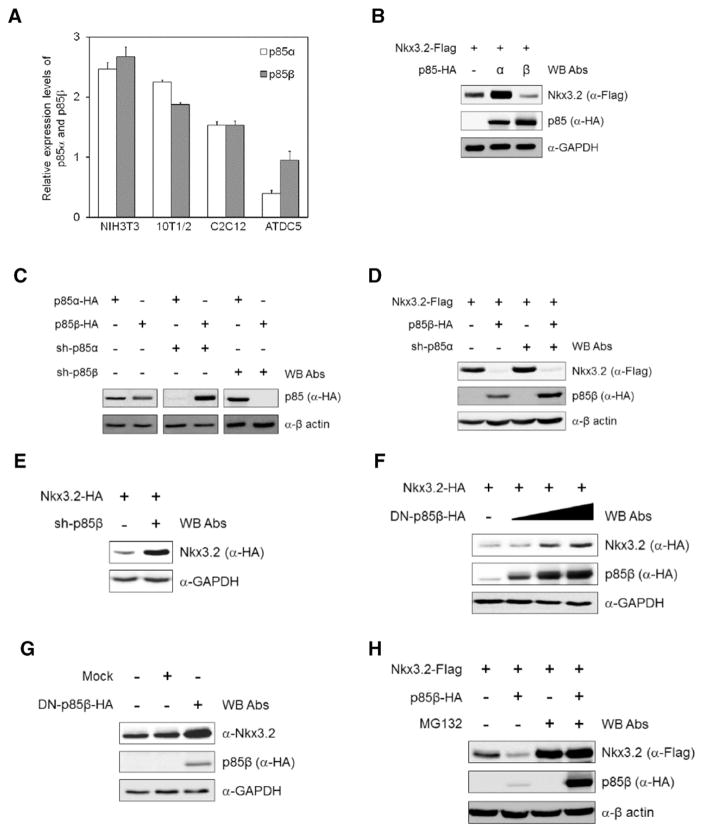
Having verified that p110α can be used as a catalytic subunit in PI3K-mediated Nkx3.2 regulation, we next wanted to determine which regulatory subunit of PI3K (i.e., p85α or p85β) was associated with this process. To do this, we first used RT-qPCR (reverse transcription-quantitative real-time PCR) to analyze the expression levels of p85α and p85β mRNA in mouse cell lines with chondrogenic (i.e., ATDC5) or non-chondrogenic (i.e., NIH3T3, C3H10T1/2 and C2C12) backgrounds, and found that absolute expression levels of p85 regulatory subunits (i.e., sum of p85α and p85β expression) are notably lower in chondrogenic ATDC5 cells than in non-chondrogenic cells (Fig. 2A, compare bars 1–6 and bars 7–8). We also noticed that that expression of p85β is 2.5-fold higher than p85α in ATDC5 cells (Fig. 2A, compare bars 7 and 8), while p85α and p85β had comparable expression levels in non-chondrogenic NIH3T3, C3H10T1/2 and C2C12 cells (Fig. 2A, see bars 1–6).
Fig. 2.
PI3K-mediated Nkx3.2 regulation employs p85β. (A) Expression of p85α and p85β in non-chondrogenic mouse cell lines, NIH3T3, C3H10T1/2 and C2C12, and in the chondrogenic mouse cell line ATDC5 were assessed by real-time quantitative PCR. (B) 293T cells were transfected with plasmids expressing Nkx3.2-Flag and isoforms of p85 (p85α-HA, p85β-HA). Cells were harvested and cell lysates were analyzed by Western blotting with the indicated antibodies. (C) p85α and p85β expression plasmids and their shRNA constructs were co-transfected into 293T cells. shRNA knockdown efficiency was validated by Western blotting with the indicated antibodies. (D) Nkx3.2 and p85β expression plasmids were transiently transfected with or without a p85α shRNA construct into ATDC5 cells. Cells were harvested and expression of Nkx3.2 and p85β was examined by Western blotting. (E) An Nkx3.2 expression plasmid (Nkx3.2-HA) and a p85β shRNA (sh-p85β) construct were transiently co-transfected into ATDC5 cells. Cell lysates were examined by Western blotting with the indicated antibodies. (F) An Nkx3.2 (Nkx3.2-HA) expression plasmid was transiently transfected into 293T cells alone, or in combination with increasing amounts of a plasmid expressing dominant negative (DN) p85β (DN-p85β-HA). Total cell lysates were analyzed by Western blotting. (G) ATDC5 cells were infected with lentiviruses expressing a control (Mock) or HA-tagged DN-p85β. Cell lysates were analyzed by Western blotting with the indicated antibodies. (H) Nkx3.2 (Nkx3.2-Flag) and p85β (p85β-HA) expression plasmids were transiently transfected into ATDC5 cells. After 34 h, indicated cells were incubated with 20 μM MG132 for 8 h. Cell lysates were analyzed by Western blotting with the indicated antibodies.
In addition to this atypical enrichment of p85β, we have interestingly observed that Nkx3.2 protein levels can be significantly attenuated by co-expressing p85β, but not p85α (Fig. 2B). Because these results were unexpected, we next attempted to confirm whether p85β, but not p85α, is specifically involved in PI3K-mediated Nkx3.2 regulation. To this end, we first examined the specificity of pLKO-based shRNA reagents [49] to be used for p85α or p85β knockdown (KD) experiments and Western blotting analyses verified isoform-specific KD of p85 regulatory subunits using these reagents (Fig. 2C). We found that p85α KD did not alter Nkx3.2 protein levels (Fig. 2D, compare lanes 1 and 3), and also did not affect p85β-induced Nkx3.2 down-regulation (Fig. 2D, compare lanes 2 and 4). Furthermore, protein levels of Nkx3.2 in ATDC5 cells were effectively increased by p85β KD (Fig. 2E). Taken together, these results suggest that p85β, rather than p85α, plays a role as a regulatory subunit in PI3K-mediated Nkx3.2 suppression.
To further investigate the involvement of p85β in this process, we generated a dominant negative form of p85β (DN-p85β) that is defective in p110 binding [50,51], and found that protein levels of ectopically expressed Nkx3.2 significantly increased in a dose-dependent manner following DN-p85β co-transfection (Fig. 2F). In addition, endogenous Nkx3.2 protein levels in ATDC5 cells were elevated by lentiviral infection with DN-p85β (Fig. 2G, compare lanes 1 and 3). Finally, we found that p85β-mediated Nkx3.2 suppression can be abolished by treating with proteasome inhibitor, MG132, suggesting that Nkx3.2 down-regulation triggered by PI3K involves, at least in part, proteasome-dependent protein degradation (Fig. 2H).
3.3. Functional Rac1 and PAK1 are necessary for Nkx3.2 degradation mediated by PI3K
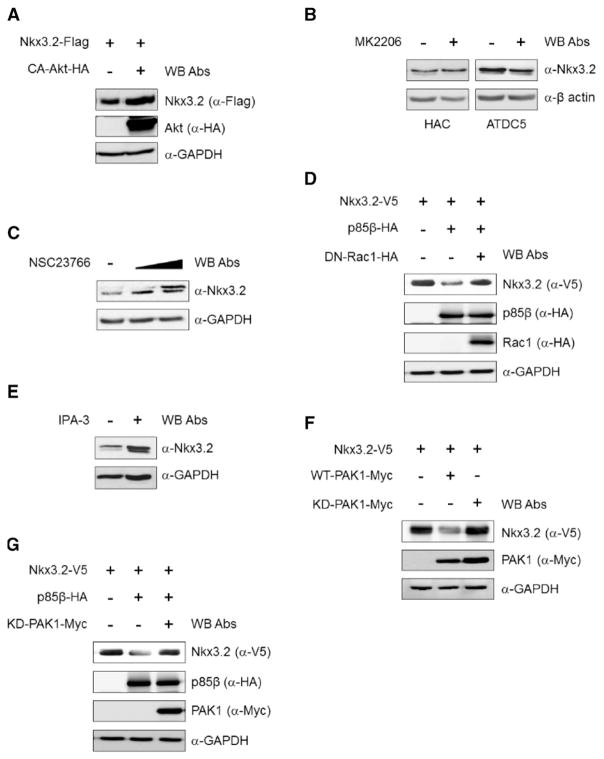
As we established that PI3K signaling is capable of inducing proteasomal degradation of Nkx3.2, we next wanted to identify downstream components of this pathway. Since Akt has been well documented as a key component in PI3K-associated signaling, we first asked whether Akt is involved in PI3K-mediated Nkx3.2 degradation. In this case, we found that constitutively active Akt [52] did not alter protein levels of Nkx3.2 in transient transfection assays (Fig. 3A). Consistent with these results, pharmacological inhibition of Akt using MK2206 [53] also failed to alter protein levels of endogenous Nkx3.2 in either HACs or ATDC5 cells (Fig. 3B). Therefore, these results indicate that Akt does not play a critical role in PI3K-triggered Nkx3.2 degradation.
Fig. 3.
Rac1 and PAK1 are required for PI3K-mediated Nkx3.2 regulation. (A) Nkx3.2 (Nkx3.2-Flag) and constitutively active Akt (CA–Akt–HA) expression plasmids were transiently transfected into 293T cells as shown. Cell lysates were analyzed by Western blotting with the indicated antibodies. (B) HACs and ATDC5 cells were cultured in the presence of 2.5 μM of the Akt inhibitor MK2206. After 24 h, cell lysates were analyzed by Western blotting with the indicated antibodies. (C) HACs were cultured in the presence of 5 or 10 μM Rac1 inhibitor (NSC23766) for 24 h. Total lysates were analyzed by Western blotting with the indicated antibodies. (D) Nkx3.2 (Nkx3.2-V5), p85β (p85β-HA) and the dominant negative form of Rac1 (DN–Rac1–HA) expression plasmids were transiently co-transfected into 293T cells. Cell lysates were analyzed by Western blotting with the indicated antibodies. (E) HACs were cultured in the presence or absence of 10 μM the PAK inhibitor, IPA-3. After 24 h, cells were harvested with lysis buffer and total lysates were analyzed by Western blotting with the indicated antibodies. (F) Nkx3.2 (Nkx3.2-V5) and wild-type (WT-PAK1-Myc) or kinase-dead (KD-PAK1-Myc) PAK1 expression plasmids were transiently transfected into 293T cells. Cell lysates were analyzed by Western blotting with the indicated antibodies. (G) Nkx3.2 (Nkx3.2-V5), p85β (p85β-HA), and kinase-dead PAK1 (KD-PAK1-Myc) expression plasmids were transiently co-transfected into 293T cells. Cell lysates were analyzed by Western blotting analysis with the indicated antibodies.
We next investigated the involvement of Ras-related C3 botulinum toxin substrate 1 (Rac1), which has been implicated in Akt-independent signaling pathways downstream of PI3K [54]. To this end, we used NSC23766, a Rac1 inhibitor [55], and interestingly found that Nkx3.2 protein levels were significantly elevated by pharmacological inhibition of Rac1 (Fig. 3C). Consistent with these results, overexpression of dominant negative Rac1 [56] abrogated p85β-triggered Nkx3.2 degradation (Fig. 3D).
Since Rac1 could mediate downstream signaling from PI3K leading to Nkx3.2 degradation, we next asked whether p21-activated kinase 1 (PAK1), which is downstream of Rac1 [57], is associated with this pathway. As shown in Fig. 3E, protein levels of Nkx3.2 in human primary chondrocytes were significantly augmented by administering IPA-3, a PAK1 inhibitor [58]. In addition, as seen in Fig. 3F, Nkx3.2 protein levels were remarkably attenuated by co-transfecting wild-type PAK1, but not kinase-dead PAK1 [59]. Overexpression of kinase-dead PAK1 gave rise to complete abrogation of p85β-induced Nkx3.2 degradation (Fig. 3G). Therefore, these results together clearly suggest that a Rac1–PAK1 axis is required to be intact for PI3K-triggered Nkx3.2 degradation.
3.4. PI3K signaling suppresses Nkx3.2 expression at mRNA level
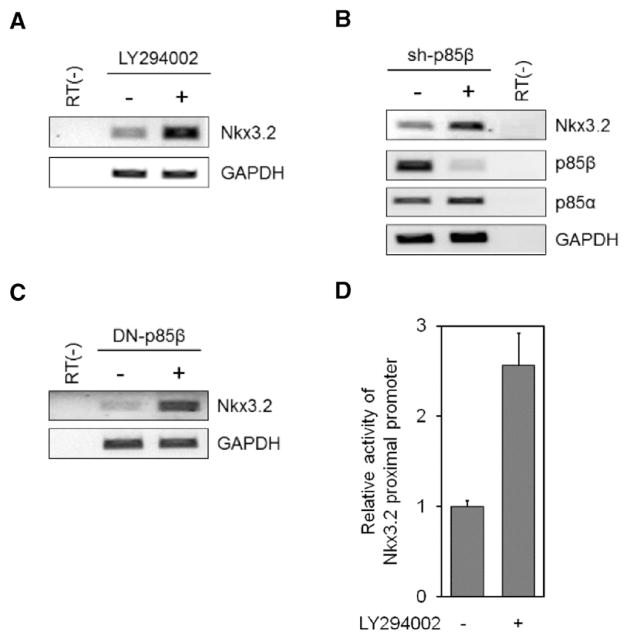
Since we established that PI3K signaling regulates Nkx3.2 protein expression, we next investigated whether PI3K signaling may also control Nkx3.2 transcription by testing the effect of inhibiting PI3K on Nkx3.2 mRNA levels in ATDC5 cells. Interestingly, pharmacological inhibition of PI3K by LY294002 treatment significantly augmented Nkx3.2 mRNA levels as judged by RT-PCR assays (Fig. 4A). Consistent with these results, RNA knockdown of p85β also notably elevated Nkx3.2 transcript levels (Fig. 4B). Conversely, DN-p85β overexpression via lentiviral infection caused a remarkable increase in Nkx3.2 mRNA levels (Fig. 4C). We also found that a luciferase reporter driven by the 2 kb proximal promoter of Nkx3.2 was activated upon pharmacological inhibition of PI3K using LY294002 (Fig. 4D). Together, these results revealed that PI3K signaling can down-regulate Nkx3.2 at the transcription level.
Fig. 4.
Nkx3.2 mRNA levels can be diminished by PI3K signaling. (A) ATDC5 cells were incubated with or without 20 μM LY294002 for 24 h. RNA was isolated and then cDNA was generated from it. Levels of Nkx3.2 and GAPDH, as a house keeping gene, mRNA were assessed by RT-PCR. (B) p85β was knocked down by infecting ATDC5 cells with a sh-p85β shRNA lentivirus. After 72 h, RNA was isolated and expression of Nkx3.2, p85α, p85β and GAPDH were examined by RT-PCR. (C) Lentivirus expressing a DN-p85β or mock were infected into ATDC5 cells. RNA was isolated and Nkx3.2 and GAPDH mRNA levels were determined by RT-PCR. (D) An Nkx3.2p-Luc reporter plasmid containing 2 kb of the Nkx3.2 upstream proximal promoter region in the pGL3B vector was transiently transfected into ATDC5 cells. After 24 h, ATDC5 cells were cultured with or without 20 μM LY294002 for 24 h. Cells were harvested with 1× passive lysis buffer, and luciferase activity of the cell lysates was determined according to manufacturer’s protocol.
3.5. Inhibition of PI3K during chondrogenesis delays hypertrophic maturation via Nkx3.2 up-regulation
After demonstrating that PI3K signaling can effectively suppress Nkx3.2 in chondrocyte monolayer cultures in vitro, we next evaluated the biological significance of this regulation using ex vivo and in vivo assays.
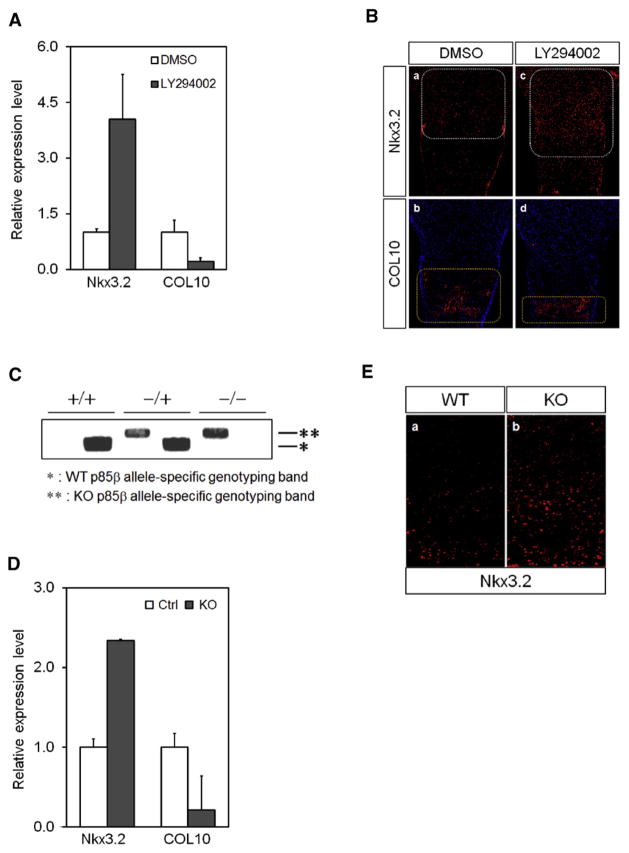
In chondrocyte maturation assays using 3D micromass culture [60], we found that LY294002 treatment notably elevated the levels of Nkx3.2 mRNA expression, and that this increase was accompanied by a significant reduction of type X collagen (COL10) expression (Fig. 5A). Since type X collagen is the most reliable hypertrophy marker indicating chondrocyte maturation, these results strongly imply that inhibition of PI3K up-regulates Nkx3.2 expression, which, in turn, suppresses hypertrophic chondrocyte maturation.
Fig. 5.
PI3K inhibition up-regulates Nkx3.2 and suppresses hypertrophic maturation. (A) ATDC5 cells were micromass cultured in chondrogenic media containing 1% ITS and 10 ng/ml BMP2 for 6 days. During chondrogenic differentiation, cells were cultured with or without 20 μM LY294002 or DMSO. Total RNA was isolated and expression of type X collagen and Nkx3.2 mRNA was determined by real-time qPCR. (B) Femurs of E15.5 mice were cultured in differentiation media in the presence of LY294002 or DMSO. After 6 days, femurs were fixed, embedded in paraffin blocks and sectioned at 4 μm. Regions of Nkx3.2 and type X collagen expression were identified by immunohistological analysis. (C) Mouse genotypes were validated by PCR. An asterisk indicates the wild-type and a double-asterisk indicates the knockout (KO) genotype. (D) Mesenchymal cells from E11.5 embryos carrying heterozygotic or homozygotic KO of p85β were micromass cultured in differentiation media containing 1% ITS for 7 days. Total RNA was isolated and type X collagen and Nkx3.2 expression levels were examined by real-time qPCR. (E) Wild-type and homozygotic p85β knockout mice were sacrificed and dissected at P0. Fixed femurs were embedded in paraffin and sectioned at 4 μm. Regions of Nkx3.2 and type X collagen expression in growth plates were identified by immunohistological analysis.
Consistent with these results, ex vivo organ cultures using femora isolated from E15.5 mouse embryos [42] also demonstrated that pharmacological inhibition of PI3K can remarkably increase Nkx3.2 expression in proliferating tissues of the growth plate (Fig. 5B, compare white-dotted rectangles in panels a and c). This augmentation is associated with a significantly reduced type X collagen (COL10) expression in hypertrophic tissues (Fig. 5B, compare yellow dotted rectangles in panels b and d).
To confirm the relationship between PI3K signaling and Nkx3.2 with respect to cartilage hypertrophy, we next compared Nkx3.2 expression levels in p85β knockout and wild-type littermates. We assessed Nkx3.2 and type X collagen expression from micromass cultures using E11.5 limb buds [61] dissected from either wild-type or p85β knockout embryos. Typical results of PCR genotyping of p85β (+/+), p85β (−/+), and p85β (−/−) are shown in Fig. 5C. Similar to the results from ATDC5 micromass cultures with pharmacological inhibition of PI3K, real-time-qPCR assays revealed that Nkx3.2 mRNA levels are significantly increased, and type X collagen levels are significantly decreased, in p85β KO embryos (Fig. 5D). We next compared Nkx3.2 protein levels in femur growth plates of wild-type and KO mice by immunohisto-chemistry (IHC) and found substantially more Nkx3.2 expression and expansion of Nkx3.2 expressing cells in the femoral growth plates of new-born (P0) p85β KO mice than in wild-type mice (Fig. 5E). Taken together, these results indicate that PI3K-mediated Nkx3.2 suppression plays a significant role in proper progression of chondrocyte hypertrophy during cartilage development.
3.6. Homozygotic deletion of p85β results in post-natal dwarfism
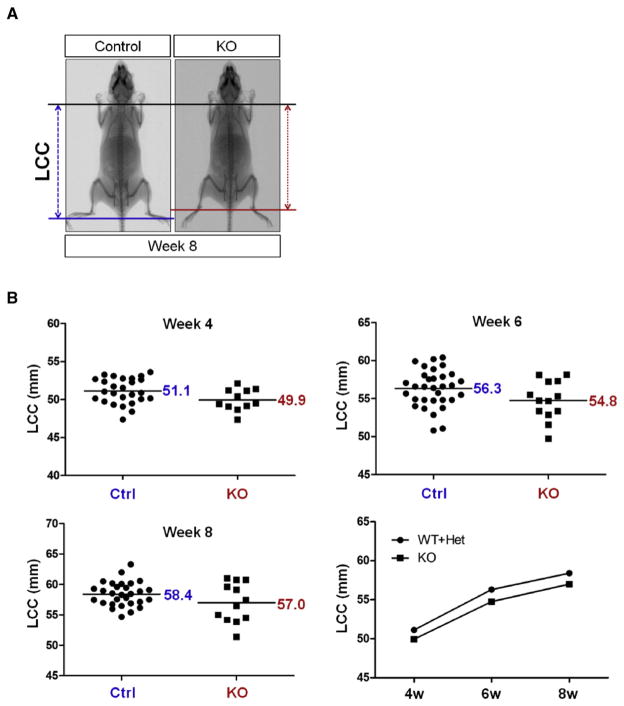
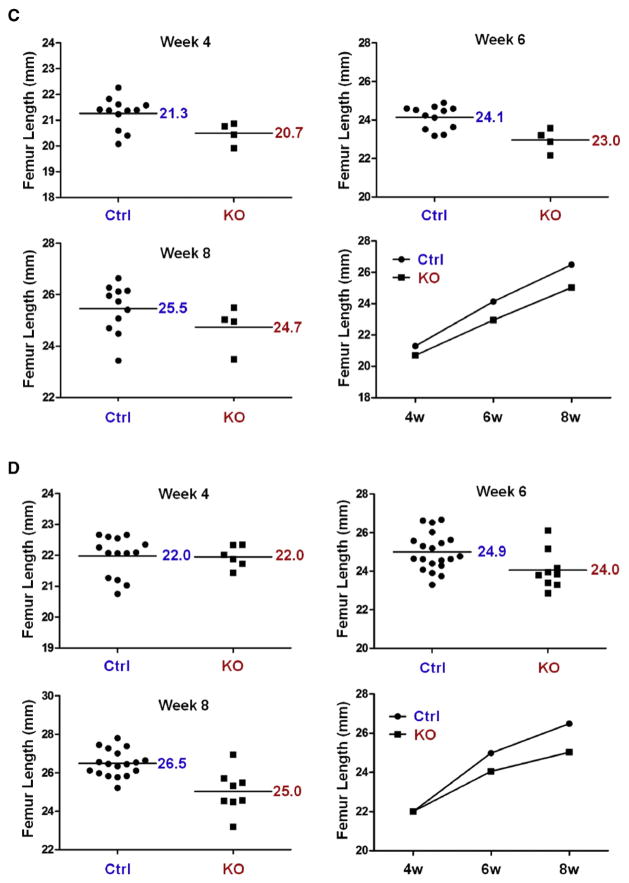
As we have validated PI3K-mediated suppression of Nkx3.2 using various in vitro, ex vivo, and in vivo experiments, we next sought to assess the physiological significance of this regulation by analyzing skeletal phenotypes of p85β knockout mice. p85β KO mice showed no apparent defects in overall skeletal patterning; however, skeletal size in p85β KO mice was significantly reduced in the post-natal period. Representative radiographic images of this skeletal dwarfism are shown in Fig. 6A. Quantitative LCC analysis (Length from 1st Cervical vertebrae to 1st Coccyx vertebrae) values of control (i.e., p85β (+/+) and p85β (−/+)) and KO (i.e., p85β (−/−)) mice at 4, 6 and 8 weeks of age revealed consistently less skeletal growth in KO mice than in control mice (Fig. 6B). Analysis of femur length in control (i.e., p85β (+/+) and (−/+)) and KO (i.e., p85β (−/−)) mice showed that femurs from both female (Fig. 6C) and male (Fig. 6D) KO mice were considerably shorter than those from control littermates. These results suggest that deletion of p85β regulatory subunit of PI3K causes post-natal attenuation of skeletal growth in both genders.
Fig. 6.
Ablation of p85β results in skeletal growth retardation. (A) Skeletal growth of control and homozygotic p85β knockout (KO) mice were analyzed by X-ray radiography. (B) The length from 1st cervical vertebrae to 1st coccyx vertebra (LCC) of control and homozygotic p85β knockout (KO) mice were examined at 4, 6, and 8 weeks by X-ray radiographic analysis. (C, D) Femur length of control and KO female (C) and male (D) mice were measured at 4, 6 and 8 weeks by X-ray radiographic analysis. All quantitative data were analyzed with GraphPad PRISM. P values of each analysis were less than 0.05.
4. Discussion
While PI3K-mediated signaling has been shown to regulate chondrocyte functions [34,42], its precise mode of action is poorly understood. In this work, we have elucidated molecular mechanisms of chondrocyte hypertrophy control triggered by PI3K signaling. Our findings revealed that PI3K triggers a signaling pathway employing Rac1 and PAK1 to modulate chondrocyte hypertrophy. We also found that Akt is not a critical component of this pathway. In addition, our results showed that Nkx3.2 suppression, a typical event that advances the initiation of hypertrophic chondrocyte maturation [14,18,62], is mediated by signaling through the PI3K–Rac1–PAK1 axis.
Indian hedgehog (Ihh) signaling has been shown to control chondrocyte hypertrophy during skeletal development [18]. Along with Ihh, a variety of Wnt family members function as key regulators of chondrocyte differentiation [63–65]. In particular, Wnt5a has been shown to play a critical role in regulating chondrocyte hypertrophy [18,66]. Interestingly, our current findings suggest that PI3K signaling also plays a role in hypertrophic chondrocyte maturation. Interestingly, apart from PI3K-mediated Nkx3.2 suppression shown in this work, we previously showed that a pathway triggered by Ihh and Wnt5a can also effectively down-regulate Nkx3.2 [18], and that this inhibition of Nkx3.2 by the Ihh–Wnt5a signaling axis plays a role in controlling cartilage hypertrophy [18]. These multiple lines of evidence suggest that elimination of Nkx3.2 might be a common goal of signaling pathways that control chondrocyte hypertrophy. Thus, Nkx3.2 can be considered as a key factor that must be eliminated in a timely fashion for appropriate chondrocyte hypertrophy to be achieved during endochondral ossification.
The catalytic subunit of PI3K comprises p110α, p110β, and p110δ. p110α and p110β are expressed ubiquitously, while the expression of p110δ is restricted to immune cells [67]. We have repeatedly observed that p110α is most abundantly expressed in multiple chondrogenic lineage cells, while other p110 subunits (i.e., p110β and p110δ) are barely detectable (data not shown). In addition, the ability of BYL719, a p110α-specific PI3K inhibitor, to suppress Nkx3.2 is similar to the broader specificity PI3K inhibitors, LY294002 and BKM120 (Fig. 1A). Taken together, these results demonstrate that p110α is the catalytic subunit responsible for PI3K-mediated inhibition of Nkx3.2.
In most cellular contexts investigated to date, p85α has been shown to be the dominant regulatory subunit of PI3K [31]. However, we have interestingly found that p85β, and not p85α, is specifically involved in PI3K-mediated Nkx3.2 suppression (Fig. 2). We also found that, in contrast to many non-chondrogenic lineage cells, p85β expression is at least 2-fold greater than p85α in ATDC5, a murine chondrogenic cell line, (Fig. 2A) as well as in various human primary chondrocytes (data not shown). Since p85α and p85β share a highly conserved p110α binding domain (i.e., iSH2 domain), the fact that p85β is specifically associated with p110α-dependent inhibition of Nkx3.2 is indeed unusual. Nonetheless, p85α and p85β have been previously shown to execute discrete functions under certain circumstances. For example, a single amino acid difference (i.e., alanine vs. serine) in the p110α binding region of different p85 isoforms can cause distinct oncogenic fusions with HUMORF8, a deubiquitinating enzyme [68]. Furthermore, due to different phosphorylated residues of p85 isoforms, p85α and p85β function differently during T cell activation [69]. Along with these rare examples, our current findings provide additional evidence supporting isoform-specific functions of PI3K signaling pathways.
In conclusion, this study uncovered a novel PI3K-dependent pathway specifically using p85β and p110α, which effectively suppresses Nkx3.2 to promote chondrocyte hypertrophy. Our results also support the idea that a Rac1–PAK1 axis may have a significant role in cartilage hypertrophy control by mediating signals from PI3K to Nkx3.2.
Acknowledgments
This work was supported by the grants (NRF-2012M3A9A9055078; NRF-2008-0062422; 2015 K000180) funded by the Korean Ministry of Science, ICT and Future Planning, and the grant R01-GM041890 funded by NIH, USA.
Footnotes
Disclosure statement
The authors declare no conflict of interest.
References
- 1.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- 4.Onyekwelu I, Goldring MB, Hidaka C. Chondrogenesis, joint formation, and articular cartilage regeneration. J Cell Biochem. 2009;107:383–392. doi: 10.1002/jcb.22149. [DOI] [PubMed] [Google Scholar]
- 5.Faure S, Georges M, McKey J, Sagnol S, de Santa Barbara P. Expression pattern of the homeotic gene Bapx1 during early chick gastrointestinal tract development. Gene Expr Patterns. 2013;13:287–292. doi: 10.1016/j.gep.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Hecksher-Sorensen J, Watson RP, Lettice LA, Serup P, Eley L, De Angelis C, Ahlgren U, Hill RE. The splanchnic mesodermal plate directs spleen and pancreatic laterality, and is regulated by Bapx1/Nkx3.2. Development. 2004;131:4665–4675. doi: 10.1242/dev.01364. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen C, Murtaugh LC, Chyung JC, Lassar A, Roberts DJ. Gizzard formation and the role of Bapx1. Dev Biol. 2001;231:164–174. doi: 10.1006/dbio.2000.0151. [DOI] [PubMed] [Google Scholar]
- 8.Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 9.Tucker AS, Watson RP, Lettice LA, Yamada G, Hill RE. Bapx1 regulates patterning in the middle ear: altered regulatory role in the transition from the proximal jaw during vertebrate evolution. Development. 2004;131:1235–1245. doi: 10.1242/dev.01017. [DOI] [PubMed] [Google Scholar]
- 10.Church V, Yamaguchi K, Tsang P, Akita K, Logan C, Francis-West P. Expression and function of Bapx1 during chick limb development. Anat Embryol (Berlin) 2005;209:461–469. doi: 10.1007/s00429-005-0464-z. [DOI] [PubMed] [Google Scholar]
- 11.Lettice L, Hecksher-Sorensen J, Hill R. The role of Bapx1 (Nkx3.2) in the development and evolution of the axial skeleton. J Anat. 2001;199:181–187. doi: 10.1046/j.1469-7580.2001.19910181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murtaugh LC, Zeng L, Chyung JH, Lassar AB. The chick transcriptional repressor Nkx3.2 acts downstream of Shh to promote BMP-dependent axial chondrogenesis. Dev Cell. 2001;1:411–422. doi: 10.1016/s1534-5807(01)00039-9. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigo I, Hill RE, Balling R, Munsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130:473–482. doi: 10.1242/dev.00240. [DOI] [PubMed] [Google Scholar]
- 14.Provot S, Kempf H, Murtaugh LC, Chung UI, Kim DW, Chyung J, Kronenberg HM, Lassar AB. Nkx3.2/Bapx1 acts as a negative regulator of chondrocyte maturation. Development. 2006;133:651–662. doi: 10.1242/dev.02258. [DOI] [PubMed] [Google Scholar]
- 15.Zeng L, Kempf H, Murtaugh LC, Sato ME, Lassar AB. Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev. 2002;16:1990–2005. doi: 10.1101/gad.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol Cell Biol. 2003;23:8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DW, Kempf H, Chen RE, Lassar AB. Characterization of Nkx3.2 DNA binding specificity and its requirement for somitic chondrogenesis. J Biolumin Chemilumin. 2003;278:27532–27539. doi: 10.1074/jbc.M301461200. [DOI] [PubMed] [Google Scholar]
- 18.Choi SW, Jeong DU, Kim JA, Lee B, Joeng KS, Long F, Kim DW. Indian hedgehog signalling triggers Nkx3.2 protein degradation during chondrocyte maturation. Biochem J. 2012;443:789–798. doi: 10.1042/BJ20112062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park M, Yong Y, Choi SW, Kim JH, Lee JE, Kim DW. Constitutive RelA activation mediated by Nkx3.2 controls chondrocyte viability. Nat Cell Biol. 2007;9:287–298. doi: 10.1038/ncb1538. [DOI] [PubMed] [Google Scholar]
- 20.Yong Y, Choi SW, Choi HJ, Nam HW, Kim JA, Jeong DU, Kim DY, Kim YS, Kim DW. Exogenous signal-independent nuclear IkappaB kinase activation triggered by Nkx3.2 enables constitutive nuclear degradation of IkappaB-alpha in chondrocytes. Mol Cell Biol. 2011;31:2802–2816. doi: 10.1128/MCB.00253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawato Y, Hirao M, Ebina K, Shi K, Hashimoto J, Honjo Y, Yoshikawa H, Myoui A. Nkx3.2 promotes primary chondrogenic differentiation by upregulating Col2a1 transcription. PLoS One. 2012;7:e34703. doi: 10.1371/journal.pone.0034703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caron MM, Emans PJ, Cremers A, Surtel DA, Coolsen MM, van Rhijn LW, Welting TJ. Hypertrophic differentiation during chondrogenic differentiation of progenitor cells is stimulated by BMP-2 but suppressed by BMP-7. Osteoarthr Cartil. 2013;21:604–613. doi: 10.1016/j.joca.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Kawato Y, Hirao M, Ebina K, Tamai N, Shi K, Hashimoto J, Yoshikawa H, Myoui A. Nkx3.2-induced suppression of Runx2 is a crucial mediator of hypoxia-dependent maintenance of chondrocyte phenotypes. Biochem Biophys Res Commun. 2011;416:205–210. doi: 10.1016/j.bbrc.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Akazawa H, Komuro I, Sugitani Y, Yazaki Y, Nagai R, Noda T. Targeted disruption of the homeobox transcription factor bapx1 results in lethal skeletal dysplasia with asplenia and gastroduodenal malformation. Genes Cells. 2000;5:499–513. doi: 10.1046/j.1365-2443.2000.00339.x. [DOI] [PubMed] [Google Scholar]
- 25.Herbrand H, Pabst O, Hill R, Arnold HH. Transcription factors Nkx3.1 and Nkx3.2 (Bapx1) play an overlapping role in sclerotome development of the mouse. Mech Dev. 2002;117:217–224. doi: 10.1016/s0925-4773(02)00207-1. [DOI] [PubMed] [Google Scholar]
- 26.Hellemans J, Simon M, Dheedene A, Alanay Y, Mihci E, Rifai L, Sefiani A, van Bever Y, Meradji M, Superti-Furga A, Mortier G. Homozygous inactivating mutations in the NKX3-2 gene result in spondylo-megaepiphyseal-metaphyseal dysplasia. Am J Hum Genet. 2009;85:916–922. doi: 10.1016/j.ajhg.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 28.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 29.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 30.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, Inukai K, Asano T, Kaburagi Y, Ueki K, Nakajima H, Hanafusa T, Matsuzawa Y, Sekihara H, Yin Y, Barrett JC, Oda H, Ishikawa T, Akanuma Y, Komuro I, Suzuki M, Yamamura K, Kodama T, Suzuki H, Koyasu S, Aizawa S, Tobe K, Fukui Y, Yazaki Y, Kadowaki T. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 32.Ueki K, Yballe CM, Brachmann SM, Vicent D, Watt JM, Kahn CR, Cantley LC. Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2002;99:419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL, Franklin RA, McCubrey JA. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 34.Hidaka K, Kanematsu T, Takeuchi H, Nakata M, Kikkawa U, Hirata M. Involvement of the phosphoinositide 3-kinase/protein kinase B signaling pathway in insulin/IGF-I-induced chondrogenesis of the mouse embryonal carcinoma-derived cell line ATDC5. Int J Biochem Cell Biol. 2001;33:1094–1103. doi: 10.1016/s1357-2725(01)00067-x. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama K, Kimoto K, Itoh Y, Nakatsuka K, Matsuo N, Yoshioka H, Kubota T. The PI3K/Akt pathway mediates the expression of type I collagen induced by TGF-beta2 in human retinal pigment epithelial cells. Graefes Arch Clin Exp Ophthalmol (Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie) 2012;250:15–23. doi: 10.1007/s00417-011-1766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton PA, Dudek SM, Chiang ET, Garcia JG. Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P1 receptor, PI3 kinase, Tiam1/Rac1, and alpha-actinin. FASEB J. 2005;19:1646–1656. doi: 10.1096/fj.05-3928com. [DOI] [PubMed] [Google Scholar]
- 37.Basquin C, Sauvonnet N. Phosphoinositide 3-kinase at the crossroad between endocytosis and signaling of cytokine receptors. Commun Integr Biol. 2013;6:e24243. doi: 10.4161/cib.24243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page K, Li J, Wang Y, Kartha S, Pestell RG, Hershenson MB. Regulation of cyclin D(1) expression and DNA synthesis by phosphatidylinositol 3-kinase in airway smooth muscle cells. Am J Respir Cell Mol Biol. 2000;23:436–443. doi: 10.1165/ajrcmb.23.4.3953. [DOI] [PubMed] [Google Scholar]
- 39.Baumer AT, Ten Freyhaus H, Sauer H, Wartenberg M, Kappert K, Schnabel P, Konkol C, Hescheler J, Vantler M, Rosenkranz S. Phosphatidylinositol 3-kinase-dependent membrane recruitment of Rac-1 and p47phox is critical for alpha-platelet-derived growth factor receptor-induced production of reactive oxygen species. J Biolumin Chemilumin. 2008;283:7864–7876. doi: 10.1074/jbc.M704997200. [DOI] [PubMed] [Google Scholar]
- 40.Harrington EK, Coon DJ, Kern MF, Svoboda KK. PTH stimulated growth and decreased col-X deposition are phosphotidylinositol-3,4,5 triphosphate kinase and mitogen activating protein kinase dependent in avian sterna. Anat Rec (Hoboken) 2010;293:225–234. doi: 10.1002/ar.21072. [DOI] [PubMed] [Google Scholar]
- 41.Hubchak SC, Sparks EE, Hayashida T, Schnaper HW. Rac1 promotes TGF-beta-stimulated mesangial cell type I collagen expression through a PI3K/Akt-dependent mechanism. Am J Physiol Ren Physiol. 2009;297:F1316–F1323. doi: 10.1152/ajprenal.00345.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulici V, Hoenselaar KD, Gillespie JR, Beier F. The PI3K pathway regulates endochondral bone growth through control of hypertrophic chondrocyte differentiation. BMC Dev Biol. 2008;8:40. doi: 10.1186/1471-213X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G, Woods A, Agoston H, Ulici V, Glogauer M, Beier F. Genetic ablation of Rac1 in cartilage results in chondrodysplasia. Dev Biol. 2007;306:612–623. doi: 10.1016/j.ydbio.2007.03.520. [DOI] [PubMed] [Google Scholar]
- 44.Tsurutani J, West KA, Sayyah J, Gills JJ, Dennis PA. Inhibition of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway but not the MEK/ERK pathway attenuates laminin-mediated small cell lung cancer cellular survival and resistance to imatinib mesylate or chemotherapy. Cancer Res. 2005;65:8423–8432. doi: 10.1158/0008-5472.CAN-05-0058. [DOI] [PubMed] [Google Scholar]
- 45.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, Demanse D, De Buck SS, Ru QC, Peters M, Goldbrunner M, Baselga J. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol Off J Am Soc Clin Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 46.Azab F, Vali S, Abraham J, Potter N, Muz B, de la Puente P, Fiala M, Paasch J, Sultana Z, Tyagi A, Abbasi T, Vij R, Azab AK. PI3KCA plays a major role in multiple myeloma and its inhibition with BYL719 decreases proliferation, synergizes with other therapies and overcomes stroma-induced resistance. Br J Haematol. 2014;165:89–101. doi: 10.1111/bjh.12734. [DOI] [PubMed] [Google Scholar]
- 47.Egawa K, Sharma PM, Nakashima N, Huang Y, Huver E, Boss GR, Olefsky JM. Membrane-targeted phosphatidylinositol 3-kinase mimics insulin actions and induces a state of cellular insulin resistance. J Biolumin Chemilumin. 1999;274:14306–14314. doi: 10.1074/jbc.274.20.14306. [DOI] [PubMed] [Google Scholar]
- 48.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 49.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 50.Klippel A, Escobedo JA, Hu Q, Williams LT. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol Cell Biol. 1993;13:5560–5566. doi: 10.1128/mcb.13.9.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hara K, Yonezawa K, Sakaue H, Ando A, Kotani K, Kitamura T, Kitamura Y, Ueda H, Stephens L, Jackson TR, et al. 1-Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc Natl Acad Sci U S A. 1994;91:7415–7419. doi: 10.1073/pnas.91.16.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohn AD, Takeuchi F, Roth RA. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biolumin Chemilumin. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 53.Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, Sanchez-Martin M, Perez-Garcia A, Rigo I, Castillo M, Indraccolo S, Cross JR, de Stanchina E, Paietta E, Racevskis J, Rowe JM, Tallman MS, Basso G, Meijerink JP, Cordon-Cardo C, Califano A, Ferrando AA. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell. 2013;24:766–776. doi: 10.1016/j.ccr.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang LJ, Tao BB, Wang MJ, Jin HM, Zhu YC. PI3K p110alpha isoform-dependent Rho GTPase Rac1 activation mediates H2S-promoted endothelial cell migration via actin cytoskeleton reorganization. PLoS One. 2012;7:e44590. doi: 10.1371/journal.pone.0044590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwivedi S, Pandey D, Khandoga AL, Brandl R, Siess W. Rac1-mediated signaling plays a central role in secretion-dependent platelet aggregation in human blood stimulated by atherosclerotic plaque. J Transl Med. 2010;8:128. doi: 10.1186/1479-5876-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffiing. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 57.Lozano E, Frasa MA, Smolarczyk K, Knaus UG, Braga VM. PAK is required for the disruption of E-cadherin adhesion by the small GTPase Rac. J Cell Sci. 2008;121:933–938. doi: 10.1242/jcs.016121. [DOI] [PubMed] [Google Scholar]
- 58.Neveu G, Ziv-Av A, Barouch-Bentov R, Berkerman E, Mulholland J, Einav S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J Virol. 2015;89:4387–4404. doi: 10.1128/JVI.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y, Chen Z, Ambrose D, Liu J, Gibbs JB, Chernoff J, Field J. Kinase-deficient Pak1 mutants inhibit ras transformation of rat-1 fibroblasts. Mol Cell Biol. 1997;17:4454–4464. doi: 10.1128/mcb.17.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoogendam J, Parlevliet E, Miclea R, Lowik CW, Wit JM, Karperien M. Novel early target genes of parathyroid hormone-related peptide in chondrocytes. Endocrinology. 2006;147:3141–3152. doi: 10.1210/en.2006-0075. [DOI] [PubMed] [Google Scholar]
- 61.Zuscik MJ, Ma L, Buckley T, Puzas JE, Drissi H, Schwarz EM, O’Keefe RJ. Lead induces chondrogenesis and alters transforming growth factor-beta and bone morphogenetic protein signaling in mesenchymal cell populations. Environ Health Perspect. 2007;115:1276–1282. doi: 10.1289/ehp.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kempf H, Ionescu A, Udager AM, Lassar AB. Prochondrogenic signals induce a competence for Runx2 to activate hypertrophic chondrocyte gene expression. Dev Dyn. 2007;236:1954–1962. doi: 10.1002/dvdy.21205. [DOI] [PubMed] [Google Scholar]
- 63.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 64.Church V, Nohno T, Linker C, Marcelle C, Francis-West P. Wnt regulation of chondrocyte differentiation. J Cell Sci. 2002;115:4809–4818. doi: 10.1242/jcs.00152. [DOI] [PubMed] [Google Scholar]
- 65.Dong Y, Drissi H, Chen M, Chen D, Zuscik MJ, Schwarz EM, O’Keefe RJ. Wnt-mediated regulation of chondrocyte maturation: modulation by TGF-beta. J Cell Biochem. 2005;95:1057–1068. doi: 10.1002/jcb.20466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradley EW, Drissi MH. WNT5A regulates chondrocyte differentiation through differential use of the CaN/NFAT and IKK/NF-kappaB pathways. Mol Endocrinol (Baltimore, MD) 2010;24:1581–1593. doi: 10.1210/me.2010-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janssen JW, Schleithoff L, Bartram CR, Schulz AS. An oncogenic fusion product of the phosphatidylinositol 3-kinase p85beta subunit and HUMORF8, a putative deubiquitinating enzyme. Oncogene. 1998;16:1767–1772. doi: 10.1038/sj.onc.1201695. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Anderson DH, Liu Q, Zhou Y. Mechanism of influenza A virus NS1 protein interaction with the p85beta, but not the p85alpha, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J Biolumin Chemilumin. 2008;283:23397–23409. doi: 10.1074/jbc.M802737200. [DOI] [PubMed] [Google Scholar]