Abstract
The thiopeptides are a family of ribosomally synthesized and posttranslationally modified peptide metabolites, and the vast majority of thiopeptides characterized to date possess one highly modified macrocycle. A few members, including thiostrepton A, harbor a second macrocycle that incorporates a quinaldic acid moiety and the four N-terminal residues of the peptide. The antibacterial properties of thiostrepton A are well established, and its recently discovered ability to inhibit the proteasome has additional implications for the development of antimalarial and anticancer therapeutics. We have conducted the saturation mutagenesis of Ala2 in the precursor peptide, TsrA, to examine which variants can be transformed into a mature thiostrepton analogue. Although the thiostrepton biosynthetic system is somewhat restrictive towards substitutions at the second residue, eight thiostrepton Ala2 analogues were isolated. The TsrA Ala2Ile and Ala2Val variants were largely channeled through an alternate processing pathway wherein the first residue of the core peptide, Ile1, is removed and the resulting thiostrepton analogues bear quinaldic acid macrocycles abridged by one residue. This is the first report revealing that quinaldic acid loop size is amenable to alteration during the course of thiostrepton biosynthesis. Both the antibacterial and proteasome inhibitory properties of the thiostrepton Ala2 analogues were examined. While the identity of the residue at the second position of the core peptide influences thiostrepton biosynthesis, our report suggests it may not be crucial for antibacterial and proteasome inhibitory properties of the full-length variants. In contrast, the contracted quinaldic acid loop can, to differing degrees, affect both types of biological activity.
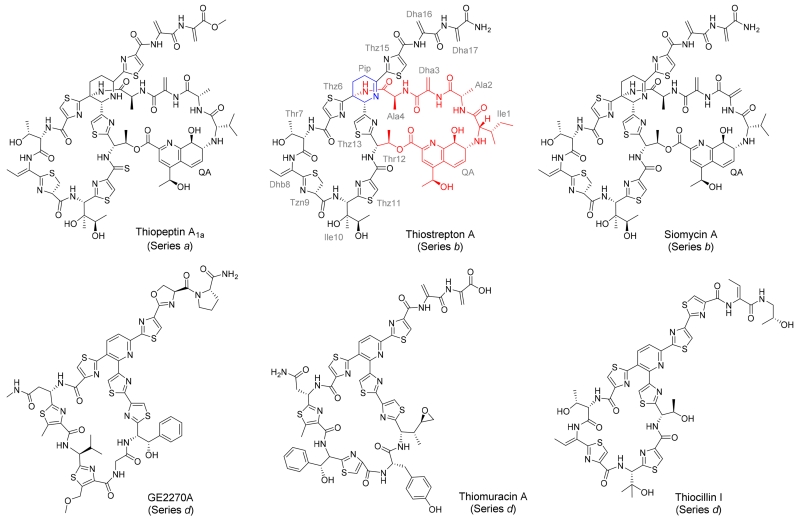
The introduction of antibiotics to the clinic during the last century revolutionized the treatment of infectious diseases. However, emerging microorganism resistance to the current pharmacopeia often compromises previously effective agents. There is therefore a pressing need for the development of new chemotherapeutics, especially those displaying new modes of action, that could be incorporated into human medicine to fight against ever-adapting pathogens.1 Great interest has been placed on the thiopeptides (Figure 1), a class of ribosomally synthesized and posttranslationally modified peptide (RiPP) metabolites.1-3 Thiopeptides are recognized for their remarkable combination of structural features; a heavily modified peptide backbone and a central piperidine, dehydropiperidine, or pyridine ring frame a core macrocycle that is observed among all members of this family.4 Multiple azol(in)e rings are observed in all thiopeptides, and several members of this family also present with dehydrated amino acid residues.4 The oxidation state of the core macrocycle’s six-membered nitrogeneous ring provides the primary criterion for classifying a thiopeptide into one of five structural series, a—e.4, 5 The antibacterial activity of a thiopeptide is typically accomplished through the disruption of protein synthesis by mechanisms not currently employed in any FDA-approved antibiotic. One common mode of action, exemplified by thiostrepton A and thiocillin I (Figure 1), is to bind the large ribosomal subunit near the GTPase-associated center and inhibit translocation along the mRNA transcript.6-8 GE2270A and thiomuracin A (Figure 1), on the other hand, form a complex with elongation factor Tu (EF-Tu) and derail the delivery of amino acyl-tRNAs to the ribosome, exemplifying a second major mechanism of antibacterial activity for the thiopeptides.9, 10
Figure 1.
Examples of thiopeptides. The thiostrepton A residues are abbreviated using a three-letter code and labeled in grey. The thiostrepton A quinaldic acid (QA) loop is highlighted in blue and the dehydropiperidine ring is in red. Dha and Dhb refer to dehydroalanine and dehydrobutyrine, respectively. Pip, Thz, and Tzn refer to dehydropiperidine, thiazole, and thiazoline, respectively.
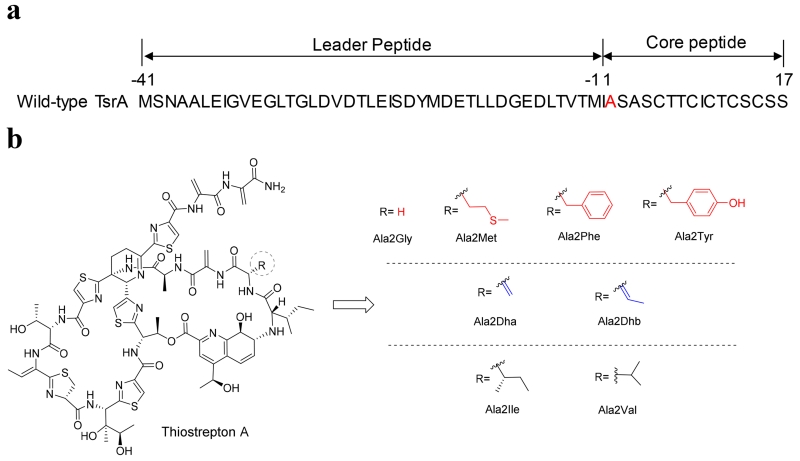
Thiostrepton A (Figure 1) has been isolated from multiple Streptomyces species, including Streptomyces laurentii ATCC 31255 (S. laurentii).4, 11 The S. laurentii thiostrepton A biosynthetic gene (tsr) cluster was reported in 2009 and the gene encoding the precursor peptide, tsrA, was identified.12, 13 The N-terminal leader peptide of TsrA is cleaved during maturation of the thiopeptide, and the 17 amino acid C-terminal core peptide provides the structural backbone for the final metabolite (Figure 2A).12 Thiostrepton A is a series b thiopeptide and contains a dehydropiperidine in its core macrocycle (Figure 1).4 Series b thiopeptides are also distinguished by the presence of a second macrocycle in which a separately biosynthesized quinaldic acid (QA) derivative forms a bridge between the α-amino group of the core peptide N-terminus and the β-hydroxyl group of a core macrocycle Thr residue (Figure 1). This second macrocycle is a feature shared with the piperidine-containing series a thiopeptides (Figure 1).4 In addition to the QA moiety and the components contributed by the thiopeptide core macrocycle, the four N-terminal amino acid residues of the peptide (Ile1, Ala2, Dha3, and Ala4) are contained within this second loop. In the inhibitory complex of thiostrepton A bound to the ribosome, those four residues are solvent-exposed, leaving the core macrocycle and the QA appendage to contribute the major binding interactions with the 50S subunit.7
Figure 2.
Thiostrepton A and the analogues generated by site-directed mutagenesis of TsrA Ala2. a) Wild-type TsrA sequence with Ala2 of the core peptide highlighted in red. b) Structures of the thiostrepton Ala2 analogues. The observed analogues possessing no posttranslational modification(s) of the variant residue are presented in the top panel and in red, while the ones bearing a posttranslational modification of the variant residue are presented in the middle panel and in blue. The variants that led to a contracted quinaldic acid loop are shown in the bottom panel and in black.
In addition to antibacterial activity, thiostrepton A exhibits antimalarial and anticancer properties. The antimalarial activity of thiostrepton A relies upon the dual inhibition of the cytosolic 20S proteasome and of organellar protein translation within the apicoplast of the Plasmodium parasite.14-17 The anticancer properties of thiostrepton A have been suggested to involve modulation of both the proteasome and the forkhead box M1 (FOXM1) transcription factor.18, 19 Current understanding of the structure-activity relationship (SAR) for thiostrepton A’s antibacterial activity has been enriched by the structural studies of this metabolite bound to the 50S ribosome and subsequent molecular dynamics simulations and docking studies of the complex.7, 8, 20, 21 In contrast, far less information is available to illustrate the details on the molecular interactions between thiostrepton A and its recently identified eukaryotic targets, the 20S proteasome and FOXM1. Thiostrepton action on eukaryotic cells appear to be complex and may involve multiple modes of action.22-24 The rational design of a thiopeptide-inspired derivative capable of selectively disrupting a single cellular process will therefore require the molecular features responsible for thiostrepton A’s association with the proteasome and FOXM1 to be deciphered.
The generation of thiopeptide analogues is therefore needed to develop the SARs for each biological target. Semisynthetic modifications of naturally occurring thiopeptides have contributed to structure-function studies for protein synthesis inhibition, yielding derivatives that retain potent antibacterial activity and are improved in aqueous solubility compared to the parent metabolite.3, 25-27 It is expected that similar strategies can be adopted for the other therapeutic targets of thiostrepton A. Although semisynthetic derivatization of a thiopeptide metabolite remains as a powerful tool for lead optimization, this methodology is confined by the chemical handles available in the naturally occurring scaffold. For example, semisynthetic approaches may not enable facile access to analogues of altered macrocycle size. As a result, it is currently unknown how side chain identity, core macrocycle size, or quinaldic acid loop size influence thiostrepton’s biological activities. Biosynthetic engineering offers an additional avenue to deliver thiopeptide derivatives to the development pipeline, and strides have already been made to this end by precursor peptide mutations.28-33
We previously established a platform for precursor peptide mutagenesis in S. laurentii to engineer thiostrepton A variants.33 Our initial efforts targeted the three residues of the TsrA core peptide (Ala2, Ala4, and Thr7) that are not subjected to posttranslational modification during thiostrepton A biosynthesis, and the amino acid residue substitutions explored were relatively conservative.31, 33 Substitution of Ala2 with Gly led to the successful production of thiostrepton Ala2Gly, an analogue that retained potent antibacterial activity.33 Herein, we report the saturation mutagenesis of the second residue in the TsrA core peptide to interrogate the permissiveness of the thiostrepton biosynthetic system toward structural alterations at this position. In the course of this effort, we identify two new thiostrepton analogues that contain a quinaldic acid macrocycle contracted by one amino acid residue. The resulting thiostrepton derivatives were evaluated for their antibacterial properties and abilities to inhibit the 20S proteasome in order to probe the effects of the incorporated structural modifications.
RESULTS AND DISCUSSION
Generation, production, and characterization of thiostrepton Ala2 analogues
A previously developed fosmid-based engineering platform in S. laurentii was used for the saturation mutagenesis of TsrA Ala2.33, 34 Central to this system is the fosmid int-3A100, an E. coli-Streptomyces shuttle fosmid containing the entire tsr gene cluster, in which the core peptide-encoding region of tsrA is substituted with a chloramphenicol resistance gene (chlR) and a levansucrase-encoding gene (sacB).33 An amplicon of each variant tsrA was used to replace the aforementioned dual-marker selection cassette in fosmid int-3A100 by λ-RED-mediated recombination.35, 36 The fosmids bearing the desired tsrA Ala2 mutations were individually introduced into a tsrA deletion strain of S. laurentii (S. laurentii NDS1) by intergeneric conjugation. Integration of the vector into the chromosome provided a total of 19 S. laurentii NDS1/int-A2X variants (“X” represents the introduced mutation using the one-letter amino acid code), including the previously constructed S. laurentii NDS1/int-A2G.33
Cultures of the S. laurentii tsrA mutant strains were performed as previously described and crude chloroform extracts were prepared prior to analysis by HPLC and HPLC-MS.33 Figure 2B includes the structures of the anticipated thiostrepton Ala2 analogues that could be generated from the 19 precursor peptide variants. When analyzed by HPLC-MS, the crude culture extracts of S. laurentii NDS1/int-A2D, A2E, A2H, A2K, A2L, A2N, A2Q, A2R, and A2W either did not contain any detectable levels or contained only a trace amount of a mature thiostrepton analogue. It therefore appears that amide side chains, charged amino acid residues, and the larger side chains of Leu and Trp at the second position of the core peptide are not well-tolerated by the collective thiostrepton biosynthetic machinery. Additionally, no species bearing a mass consistent with thiostrepton Ala2Pro was detected during HPLC-MS analysis of the S. laurentii NDS1/int-A2P crude extract, suggesting that the conformational restrictions imposed upon the polypeptide backbone by this cyclic residue may not be accepted by one or more of the thiostrepton biosynthetic enzymes.
Several low-abundance metabolites with UV-visible absorption spectra and masses consistent with thiostrepton Ala2Cys analogues were observed from the HPLC and HPLC-MS analyses of the S. laurentii NDS1/int-A2C culture extract, including multiple species bearing the expected mass for a mature thiostrepton (data not shown). The Ala4Cys substitution in TsrA similarly resulted in a mixture of minor metabolites following the substitution.34 In that case, two of the more abundant analogues, thiostreptons Ala4Cys F1 and F2, were isolated, structurally characterized, and revealed to be lanthionine (Lan)-containing diastereomers resulting from the nonenzymatic Michael addition of the Cys4 thiol on the β-carbon of Dha17.34 It is therefore possible that spontaneous Lan residue formation from the reaction of Cys2 with at least one dehydro residue yielded multiple products. Multiple stereoisomers that could ensue following this nonenzymatic event could be contributing to the complex metabolite mixture in the S. laurentii NDS1/int-A2C culture extract that may also include thiostreptons bearing a free, unmodified Cys2 thiol. Whether the Cys2- or Cys4-containing thioether bridge forms before or after assembly of either of the thiostrepton macrocycles remains to be determined. These observations suggest that, in contrast to the fate of all of the Cys residues in wild-type TsrA, Cys residues at the second and fourth positions of the TsrA core peptide are not preferred substrates for thiazol(in)e ring installation by the thiostrepton cyclodehydratase and dehydrogenase.12, 34 Alternatively, a heterocycle could be installed among the four N-terminal residues of a thiostrepton precursor but may not be efficiently processed due to the conformational restraints imparted by this modification. Despite our best efforts, the low titers of the various TsrA Ala2Cys metabolites, coupled with chemical instability and difficulties in separation of structurally similar metabolites, hindered further characterization of the thiostrepton-like Ala2Cys analogues.
Thiostreptons Ala2Met, Ala2Phe, and Ala2Tyr, on the other hand, were produced at appreciable levels by the corresponding S. laurentii TsrA Ala2 variants. These three analogues were purified by semi-preparative HPLC and their structures were verified by HR-MS and MALDI-MS/MS (Figures S26–S28 and Table S5). MALDI MS/MS of thiostrepton analogues typically leads to loss of the QA moiety followed by the successive fragmentation of the peptide’s N-terminal residues generating ions that are diagnostic for the identity of the second residue (Figure S1).31, 34 Although replacement of TsrA Ala2 with the linear or aromatic side chains of Met, Phe, and Tyr did permit the successful production of mature thiopeptides, these variants were not strongly favored by the thiostrepton biosynthetic system. These three analogues yielded only 1–3 mg L−1 whereas wild-type thiostrepton A and thiostrepton Ala2Gly are produced at 115 ± 35 mg L−1 and 19 ± 3 mg L−1, respectively, in this production platform (Table S6).33
When culture extracts of S. laurentii NDS1 expressing TsrA Ala2Ser and Ala2Thr were inspected by HPLC-MS, the anticipated Ala2Ser and Ala2Thr derivatives were not observed. Instead, each extract harbored a major metabolite reduced in mass by 18 Da from the expected analogues, suggesting the presence of the corresponding dehydration products, thiostrepton Ala2Dha (Dha: dehydroalanine), and Ala2Dhb (Dhb: dehydrobutyrine; Figure 2B). A TsrA Ala2Ser variant generated by Liu and coworkers also led to the production of thiostrepton Ala2Dha.37 The two analogues generated here were both purified and analyzed by HR-MS, MALDI-MS/MS, and NMR (Figures S2–S17; Tables S1–S2 and S5). MALDI MS/MS revealed daughter ions diagnostic for the presence of Dha and Dhb at the second position of the two peptide metabolites (Figures S1, S2, and S10). The parent compound, thiostrepton A, contains three Dha residues and one Dhb moiety within its framework: Dha3, Dha16, Dha17, and Dhb8 (Figure 1). Comparison of the NMR spectra of thiostrepton Ala2Dha to that of thiostrepton A supported the presence of a fourth Dha residue, revealing an additional quaternary carbon (δC: 134.2) and an unsaturated CH2 (δC: 100.1 and δH: 6.27, 5.01; Table S1). For thiostrepton Ala2Dhb, additional resonances consistent with an unsaturated quaternary carbon (δC: 128.4) and CH (δC: 119.4 and δH: a multiplet at 5.62 ppm) groups (Table S2) confirmed the presence of a second Dhb residue.
In our previous report, we demonstrated that when TsrA Ala4Ser and Ala4Thr were utilized by the S. laurentii production system, the thiostreptons generated were also predominantly dehydrated at the fourth position.34 The TsrA Ala4Thr variant, as observed here for Ala2Thr, led to the exclusive production of a Dhb residue at the substituted site.34 The Ala2Ser substitution in TsrA reported here and by Liu and coworkers, led solely to the maturation of thiostrepton Ala2Dha.37 TsrA Ala4Ser, on the other hand, yielded a mixture of unmodified and dehydrated products, with the Ala4Dha derivative accounting for 93% of the thiostreptons generated.34 These observations suggest that the thiostrepton dehydratases are relatively efficient at dehydrating both a Ser2 and a Ser4 in the precursor peptide.34 Siomycin A, another series b thiopeptide (Figure 1), naturally harbors a Dha2 residue, and the dehydratases from at least the QA macrocycle-containing thiopeptide biosynthetic systems appear to have a propensity to recognize a dehydration substrate at this position of the core peptide.38 Indeed, comparison of the thiostrepton production levels from S. laurentii NDS1 complemented with wild-type and Ala2Ser tsrA reveals that TsrA Ala2Ser is robustly processed to a mature thiopeptide: the titer for this analogue in the crude extract is increased over 2.5-fold (at 301 ± 7 mg L−1) relative to wild-type thiostrepton A (Table S6). The Thr2 residue of TsrA is also well-tolerated by the biosynthetic system, though not as well as Ser2, given the Ala2Dhb derivative is produced at 39 ± 9 mg L−1 (Table S6). LanB-type dehydratases are responsible for the dehydration of Ser and Thr residues during the biosynthesis of class I lanthipeptides, a separate family of RiPP metabolites.12, 39 TsrC and TsrD, homologues to which are present in all reported thiopeptide biosynthetic systems, are similar to the LanB-type dehydratases and are expected to install Dha and Dhb residues during the thiopeptide maturation process.32 The lanthipeptide dehydratases can demonstrate broad substrate specificity, catalyzing the dehydration of non-native Ser and Thr residues.40-42 Likewise, a growing body of work based on the outcomes from precursor peptide engineering suggests that thiopeptide dehydratases are flexible with respect to their substrate peptides.30, 34, 37
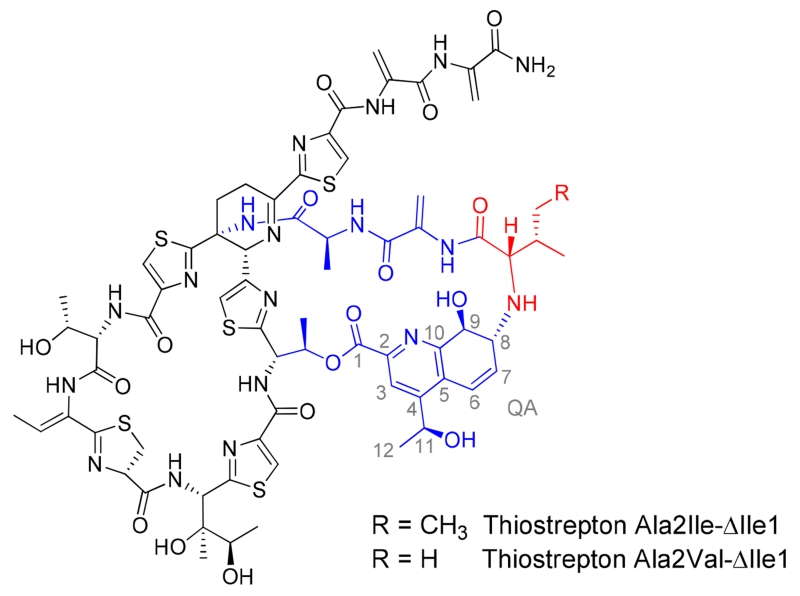
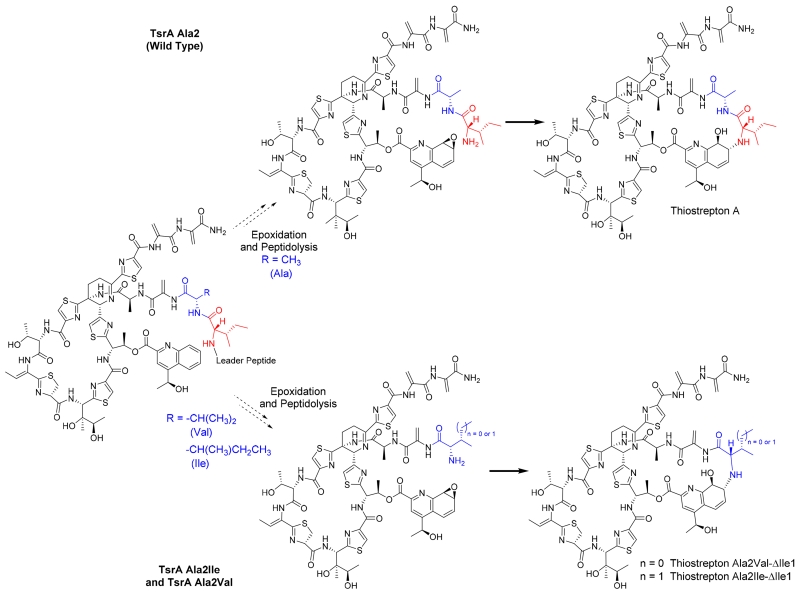
A major metabolite displaying a UV-visible absorption spectrum comparable to that of thiostrepton A was identified from HPLC analysis in each of the culture extracts of the S. laurentii NDS1/int-A2I and int-A2V mutants. Examination of the crude extracts by HPLC-MS, however, revealed that the expected mature metabolites, thiostreptons Ala2Ile and Ala2Val, were either absent entirely or were present only in trace amounts (data not shown). Instead, the predominant thiostrepton-like products of both strains appeared to be reduced by 113 Da, consistent with the absence of an isoleucine residue (Figures S18 and S29). The HR-MALDI-MS and MALDI-MS/MS analyses of the purified Ala2Ile and Ala2Val metabolites revealed two key fragments corresponding to the loss of QA and QA plus Ile (in the Ala2Ile variant) or QA plus Val (in the Ala2Val variant), suggesting that the quinaldic acid macrocycle was still present in each analogue, albeit missing Ile1 and therefore contracted by one amino acid residue (Figures S18 and S29 and Table S5). One- and two-dimensional NMR analyses were performed to verify the structures proposed for thiostreptons Ala2Ile-ΔIle1 and Ala2Val-ΔIle1 (Figures 3, S19–S25, S30–S36, and Tables S3–S4). As expected following the MS analysis of the two analogues, the resonances corresponding to Ile1 of wild-type thiostrepton A were absent (Tables S3 and S4). The chemical shifts corresponding to QA-8 and QA-9 in both ΔIle1 metabolites (δC-8: ~63; δH-8: ~3.4; and δC-9: ~74; δH-9: ~4.5) resembled the resonances observed in other thiostrepton analogues bearing an intact QA macrocycle (Tables S3 and S4).31, 33, 34, 43 The resonance for QA-9H in thiostreptons bearing a wild-type QA loop size typically appear as a doublet (J = 1.5 Hz), a broad singlet, or a singlet, and the magnitude of the splitting is likely due to subtle alterations in the dihedral angle between QA-9H and QA-8H in each derivative.31, 33, 34, 43 In the ΔIle1 variants, the doublets bear J = 12.4 Hz (for Ala2Ile-ΔIle1) and 12.3 Hz (for Ala2Val-ΔIle1), suggesting the truncated macrocycle imposes a QA-9H/QA-8H dihedral angle that approaches 180°. For thiostrepton Ala2Val-ΔIle1, a heteronuclear multiple bond correlation was observed between the α-proton of Val2 and the C-8 of the QA moiety, verifying the covalent link between the engineered residue and the quinaldic acid (Table S4).
Figure 3.
Structures of thiostrepton Ala2Ile-ΔIle1 and thiostrepton Ala2Val-ΔIle1. The quinaldic acid (QA) loop is highlighted in blue, except for the mutated residue shown in red. The numbering system used for the QA moiety is labeled in grey.
During thiostrepton A maturation, the Thr12 β-hydroxyl is acylated with the QA derivative, 4-(1-hydroxyethyl)quinoline-2-carboxylic acid (HEQ) (Scheme 1).44 Once attached, the HEQ ligand is proposed to undergo epoxygenation at the C-8 to C-9 double bond, preparing the QA derivative for further modification (Scheme 1).44 Peptidolysis of the TsrA leader peptide, either before or after the QA moiety elaborations, deprotects the N-terminal amine of the core peptide and permits nucleophilic attack upon the nascent QA epoxy group and formation of the second macrocycle that distinguishes series a and b thiopeptides (Scheme 1).31, 44, 45 Similar to Ile1 in wild-type thiostrepton A, Ile2 and Val2 of the two variants also confer β-branched, hydrophobic side chains. In fact, replacement of Ala2 with Ile and Val still permitted robust production of the truncated QA-loop analogues, providing 34 ± 4 mg L−1 and 51 ± 16 mg L−1 of thiostreptons Ala2Ile-ΔIle1 and Ala2Val-ΔIle1, respectively (Table S6). A species bearing a mass consistent with thiostrepton Ala2Leu-ΔIle1 was present in the crude culture extract of S. laurentii NDS1/int-A2L, however, this was only observed in trace amounts by HPLC-MS (Figure S37). The crude culture extracts of S. laurentii NDS1/int-A2M and NDS1/int-A2F also revealed the presence of metabolites consistent with the contracted ΔIle1 analogs (Figures S38 and S39). The corresponding ΔIle1 thiostrepton derivatives in extracts of the strains expressing wild-type TsrA or the remaining Ala2 variants either were not observed or were not produced at appreciable levels under the conditions examined in this study (data not shown). While thiostrepton A (series b thiopeptide) possesses an Ile1, both siomycin A (series b thiopeptide) and thiopeptin A1a (series a thiopeptide) possess a Val1 in their QA loops (Figure 1).5, 38 Furthermore, a TsrA Ile1Val substitution is well-tolerated by the thiostrepton biosynthetic enzymes, and is processed to thiostrepton Ile1Val.37 The presence of a Val or Ile at the first position of the core peptide, immediately C-terminal to the site of leader peptide cleavage, may therefore provide a key recognition element for the peptidase involved in maturation of series a and b thiopeptides. The identity of the thiostrepton peptidase, however, is not yet known. A gene encoding a dedicated thiopeptide leader peptidase is not readily identifiable in either the tsr or sio biosynthetic loci, and it is possible a housekeeping or general protease expressed in these thiopeptide producers instead executes leader peptide cleavage.12, 13
Scheme 1.
Proposed biosynthesis of thiostreptons A, Ala2-Ile-ΔIle1 and Ala2Val-ΔIle1. The top route reflects the maturation of thiostrepton A, when the second residue is Ala. The bottom route depicts the maturation of thiostreptons Ala2Ile-ΔIle1 and Ala2Val-ΔIle1, when the second residue is either Ile or Val.
Mutation of TsrA Thr7 to Ala and Val revealed the accumulation of shunt metabolites SL105-1 and SL106-1, respectively.31 In these species, the QA moiety is attached, but nonspecific proteolysis removed the leader peptide and the first two residues of the core peptide, leaving an N-terminal pyruvoyl residue.31 HPLC-MS analysis of the crude extracts from wild-type and the Ala2 mutants in this study also suggest the presence of an analogous shunt metabolite (SL-1, Figure S40), at least in trace levels, in nearly all of the variants. Interestingly, a species bearing a mass consistent with the shunt metabolite containing a C-8/C-9 epoxide on the quinaldic acid moiety (SL-2, Figure S40) was also observed in nearly all strains, lending support to the existence of an epoxygenated intermediate in the biosynthesis of thiostrepton A and the ΔIle1 analogs (Scheme 1). Related shunt metabolites, however, were not detected in the crude culture extracts from S. laurentii NDS1 expressing the Arg, Asp, Glu, Lys, and Pro variants at the second position, suggesting these peptides are poorly tolerated by the thiostrepton biosynthetic system (data not shown). Once the appropriate peptidase is identified, further biochemical and structural studies with this enzyme would permit detailed understanding of substrate recognition and the stage of biosynthesis at which the leader peptide is removed. Likewise, the catalyst responsible for QA loop closure has not yet been identified, and the parameters involved in directing the installation of this second macrocycle remain cryptic at this time. Ultimately, more detailed insights into the enzymes and factors governing formation of thiostrepton’s second macrocycle would aid in the rational design of a thiostrepton analogue decorated with either a “normal” or a “contracted” QA loop.
Overall, the thiostrepton biosynthetic machinery appears to be somewhat restrictive toward substitutions at the second position of the TsrA core peptide, but eight thiostrepton variants were obtained from this single-site saturation mutagenesis study. This observation contrasts against the wider range of amino acid substitutions tolerated in place of Ala4 in TsrA.32 Ala2 of TsrA is one residue removed from the site of peptidolysis during leader peptide cleavage. Structural variations at this position could influence peptidase affinity for the precursor peptide, alter the preferred conformation of the precursor peptide, or simply render the substrate peptide less favored by one or more of the maturation enzymes, including those involved in QA macrocycle formation. Any combination of these factors could account for the reduced tolerance of the biosynthetic system toward modifications at the second residue of the core peptide, in contrast to the more mutable fourth residue of TsrA.
Antibacterial activities of thiostrepton analogues
A microbroth dilution method was used to determine the antibacterial activities of the isolated thiostrepton Ala2 analogues against three indicator strains of Staphylococcus, Bacillus, and Enterococcus.31, 33 Most of the thiostrepton Ala2 analogues demonstrated either comparable or modestly reduced abilities to inhibit the growth of these test strains relative to thiostrepton A, which provided MIC values between 0.012 and 0.025 μg mL−1 (Table 1). The antibacterial activities of thiostreptons Ala2Dhb and Ala2Tyr did decrease by up to 20-fold, resulting in MIC values ranging from 0.1 to 0.3 μg mL−1 (Table 1). Thiostreptons Ala2Ile-ΔIle1 and Ala2Val-ΔIle1, however, were no longer able to inhibit bacterial growth in the concentration range tested (MIC >5.0 μg mL−1, Table 1). Consistent with the inability of thiopeptides to inhibit growth of Gram-negative bacteria, none of the Ala2 analogues were able to inhibit the growth of E. coli ATCC 27856 (Data not shown).4, 33 To determine whether the loss of antibacterial activity observed of the ΔIle1 thiostrepton variants reflected weakened binding interactions with the ribosome or an inability to reach the intracellular target, protein synthesis inhibition was interrogated using an in vitro coupled transcription-translation assay. Thiostrepton A provided a half-maximal inhibitory concentration (IC50) of 0.63 ± 0.01 μM (Table 2), consistent with previously reported values.34, 46 With the exception of the two ΔIle1 variants, the IC50 parameters obtained for the thiostrepton Ala2 analogues were comparable or slightly improved relative to that for wild-type thiostrepton A (Table 2). The potency of thiostrepton Ala2Ile-ΔIle1 was decreased over 10-fold relative to thiostrepton A (Table 2). Although the IC50 for the Ala2Val-ΔIle1 variant could not be determined due to solubility limitations, the activity of this compound was diminished by at least 5-fold (Table 2). Thiostreptons Ala2Dhb and Ala2Tyr were equivalent to thiostrepton A in the inhibition of in vitro protein synthesis (Table 2), suggesting off-target factors, such as cell permeability, may account for the reduced activities of these two analogues against bacterial growth in liquid culture (Table 1). Thus far, only Ala or Dha residues have been observed at the second residue of naturally-occurring series a and b thiopeptides, all of which, due to close structural resemblances, are presumed to inhibit protein synthesis by a similar binding mode to the ribosome.4 We have expanded the chemical entities presented at the second position to include additional hydrophobic residues and demonstrated that the thiostrepton analogues harboring the full-length QA macrocycle retained their in vitro potency against protein synthesis. The X-ray crystal structure and docking studies of thiostrepton A bound to the ribosome reveal that the four N-terminal residues of the QA loop, including the Ala2 residue, are solvent-exposed and do not contribute to any interactions in the complex.7, 21 Our results suggest that the second residue of a thiostrepton analogue could accommodate additional structural modifications without negatively impacting 50S ribosomal subunit binding. In contrast, the size of the QA-containing macrocycle does influence thiostrepton’s antibacterial potency, as indicated by the reduced in vivo and in vitro inhibitory properties of the two ΔIle1 variants. The QA moiety is involved in key hydrophobic interactions that stabilize the thiostrepton-ribosome complex.7, 20, 21 Subtle modifications to the QA moiety itself, such as a hydroxyethyl side chain or fluorinated derivatives, lead to comparable or improved antibacterial properties, suggesting these alterations can be well-tolerated by the bacterial ribosome.24, 47 The contracted QA loop of the ΔIle1 variants likely impart additional conformational restraints on the two macrocycles of thiostrepton, and, as a result, prevent optimal contacts needed for high-affinity binding of the thiopeptide.
Table 1.
MIC values of thiostrepton Ala2 analogues
| Compound | MICa (μg mL−1) |
||
|---|---|---|---|
| MRSAb | VREc | Bacillus d | |
| Thiostrepton A | 0.012 | 0.012 | 0.025 |
| Ala2Dha | 0.010 | 0.008 | 0.042 |
| Ala2Dhb | 0.11 | 0.11 | 0.11 |
| Ala2Gly | 0.023 | 0.019 | 0.19 |
| Ala2Ile-ΔIle1 | >5.0 | >5.0 | >5.0 |
| Ala2Met | 0.014 | 0.007 | 0.086 |
| Ala2Phe | 0.022 | 0.011 | 0.087 |
| Ala2Tyr | 0.22 | 0.11 | 0.27 |
| Ala2Val-ΔIle1 | >5.0 | >5.0 | >5.0 |
| Vancomycin | 0.39 | NDe | ND |
| Chloramphenicol | ND | 3.9 | 0.98 |
Minimum inhibitory concentration.
Staphylococcus aureus ATCC 10537.
Enterococcus faecium ATCC 12952.
Bacillus sp. ATCC 27859.
Not determined.
Table 2.
In vitro translation inhibition by thiostrepton Ala2 analogues
| Compound | IC50 (μM) |
|---|---|
| Thiostrepton A | 0.63 ± 0.01 |
| Ala2Dha | 0.58 ± 0.03 |
| Ala2Dhb | 0.35 ± 0.04 |
| Ala2Gly | 0.46 ± 0.05 |
| Ala2Ile-ΔIle1 | 8.1 ± 1.0 |
| Ala2Met | 0.33 ± 0.03 |
| Ala2Phe | 0.34 ± 0.02 |
| Ala2Tyr | 0.58 ± 0.08 |
| Ala2Val-ΔIle1a | >3.28 |
IC50 not determined due to solubility limitations.
20S proteasome inhibitory activities of thiostrepton Ala2 analogues
Thiostrepton A and structurally related thiopeptide derivatives inhibit the 20S proteasome, a property that, at least partially, accounts for the thiopeptide’s antimalarial activity.14, 17 Chymotrypsin-, trypsin-, and caspase-like proteolytic activities are imparted by three different types of β-subunits to the multicomponent 20S proteasome complex.48 The thiostrepton analogues generated here were examined for their abilities to inhibit the three proteolytic functions of the 20S proteasome using fluorogenic substrates specific to each type of activity. Many of the thiostrepton Ala2 analogues isolated and assessed here revealed IC50 values comparable to those of thiostrepton A (Table 3), which in turn provided IC50 values in line with those previously reported.17, 34 The presence of an aromatic side chain in thiostreptons Ala2Phe and Ala2Tyr contributed to an improvement (3- to 5-fold) in inhibiting the trypsin-like activity of the proteasome (Table 3). The longer, linear side chain introduced in thiostrepton Ala2Met led to enhanced potency for all three of the proteasome’s proteolytic properties, an effect most pronounced for the trypsin-like function (Table 3). The efficacy of thiostrepton Ala2Dhb as a proteasome inhibitor was mostly unaffected, except for a 3-fold IC50 reduction against the chymotrypsin-like activity relative to that of thiostrepton A. Thiostreptons Ala2Ile-ΔIle1 and Ala2Val-ΔIle1 did retain inhibitory activities against the proteasome, however, the potencies for the two ring-contracted analogues were predominantly reduced (Table 3). Inhibition of the chymotrypsin-like function was most perturbed, by up to a 6-fold increase in IC50. Although QA loop size may not be absolutely critical to inhibit all proteasome activities, the ΔIle1 analogues appear to differ in inhibition profiles against the proteasome relative to the parent analogue. The thiopeptides that have been reported thus far to inhibit the 20S proteasome harbor a quinaldic acid loop, such as thiostrepton and siomycin (Figure 1).19, 49 Most, but not all, of the thiostrepton derivatives reported to date that lack the enclosed QA loop are less effective 20S proteasome inhibitors, suggesting that the orientation of the QA moiety relative to the core macrocycle may be key for optimal contacts with the proteasome.17, 34
Table 3.
20S proteasome inhibitory activities of thiostrepton Ala2 analogues
| IC50 (μM) |
|||
|---|---|---|---|
| Compound | Trypsin- like |
Chymotrypsin- like |
Caspase- like |
| Thiostrepton A | 0.95 ± 0.15 | 1.0 ± 0.1 | 0.59 ± 0.06 |
| Ala2Dha | 0.71 ± 0.07 | 0.99 ± 0.01 | 0.51 ± 0.03 |
| Ala2Dhb | 1.3 ± 0.2 | 3.1 ± 0.3 | 0.92 ± 0.06 |
| Ala2Gly | 1.2 ± 0.1 | 0.76 ± 0.12 | 0.37 ± 0.04 |
| Ala2Ile-ΔIle1 | 2.1 ± 0.4 | 6.3 ± 0.4 | 1.6 ± 0.3 |
| Ala2Met | 0.09 ± 0.01 | 0.62 ± 0.01 | 0.14 ± 0.01 |
| Ala2Phe | 0.18 ± 0.02 | 0.96 ± 0.03 | 0.46 ± 0.04 |
| Ala2Tyr | 0.34 ± 0.03 | >0.92a | 0.58 ± 0.09 |
| Ala2Val-ΔIle1 | 0.84 ± 0.04 | 3.9 ± 0.2 | 1.2 ± 0.04 |
| Bortezomib | 1.6 ± 0.2 | 0.005 ± 0.001 | 0.049 ± 0.003 |
IC50 not determined due to solubility limitation.
For the Ala2 variants adopting the native thiostrepton scaffold, it appears that the second residue may not necessarily be a decisive factor in 20S proteasome inhibition. We note, however, that the current study only examines six thiostrepton Ala2 analogues, all harboring a hydrophobic residue at the variable site. The generation of additional derivatives would be necessary to develop deeper insight into the types of modifications, including polar moieties, that still support proteasome inhibition. Semisynthetic thiostrepton derivatives revealed that alterations to the C-terminus of the peptide and increasing the oxidation state of the core macrocycle thazoline ring still support potent proteasome inhibition.17 Substitution of the Ala4 position of the QA loop and Thr7 of the core macrocycle also yielded engineered thiopeptides that are potent proteasome inhibitors.34 The efficacy of the Ala2 and Ala4 variants suggest that the peptidic portion of the full-length QA loop does not engage in essential contacts with the proteasome, and may therefore be amenable to further alterations. The varied effects thiostrepton analogues exert on the 20S proteasome’s proteolytic activities suggest that either multiple thiostrepton binding sites exist or that inhibition could be due to an allosteric interaction. Structural information is required to fully articulate the nature by which thiostrepton A engages the 20S proteasome, and to offer a firm explanation for the activities of the various thiostrepton analogue activities reported to date.
All naturally-occurring series a and b thiopeptides reported thus far contain a four-residue peptidic arm, comprised of the first four residues of the core peptide, in the QA loop in which the second residue is either Ala or Dha. We expanded the chemical entities encountered at this position to include other hydrophobic residues and added eight new thiostrepton variants, including two that harbor a contracted QA macrocycle, to a growing library of thiostrepton derivatives. Our results show that the thiostrepton A biosynthetic machinery is moderately permissive toward amino acid substitutions at the second position. Furthermore, Ile2 and Val2 derivatives are channeled through an alternate processing route that requires modified activities in at least two biosynthetic steps. First, the peptidase responsible for leader peptide removal must recognize a new cleavage site. Second, the catalyst involved in closure of the QA loop must be able to efficiently generate a macrocycle reduced in size by one amino acid residue, or three atoms. For thiostrepton derivatives bearing the native-sized QA macrocycle, we demonstrate that the identity of the second residue may not be essential for either ribosome or proteasome inhibition. On the other hand, the size of the QA loop, and likely the relative placement of the quinaldic acid moiety, does influence the biological activities of a series b-like thiopeptide. Additional analogues that vary to a greater extent in side chain identity, including polar functional groups, are needed to fully explore the structure-function relationships in this region of the thiopeptide. Potentially, the reactive side chains of Dha and Dhb could be parlayed as substrates for semisynthetic modification to accomplish greater structural diversity at the second residue by adapting previously reported strategies.14, 18, 50 Prior to this work, a majority of the efforts taken to obtain thiostrepton analogues relied upon the derivatization of a naturally-occurring thiopeptide scaffold, none of which could efficiently alter the size or composition of the QA-containing loop. Our current study has expanded the variations accessible for a thiostrepton-like scaffold and identified a new site that could support additional thiopeptide engineering efforts.
METHODS
Engineering TsrA Ala2 Variants in S. laurentii
A synthetic ultramer was designed that contained a degenerate codon (NNS) at the second codon in the tsrA core peptide-encoding region and contained regions homologous to fosmid int-3A100 (Table S9). This ultramer provided the template for amplification of mutant tsrA genes by PCR with the primers Amp TsrA-SP-F/R (Table S9). The amplicons were cloned into pSC-B-amp/kan, and the resulting plasmids were analyzed by DNA sequencing. Plasmids with the tsrA inserts containing the highest percentage of synonymous codon usage for Streptomyces were chosen as the template for amplification of each mutant tsrA gene with the primers Amp-TsrA-SP-F/R (Table S9).51 The PCR products were used to substitute the dual-marker disruption cassette in fosmid int-3A100 by PCR-targeted gene replacement in E. coli BW25113/pKD46.35, 36 Fosmids were isolated from the chloramphenicol-sensitive and sucrose-tolerant E. coli colonies and transformed into chemically competent E. coli EPI300 to induce high copy numbers of the fosmids. The isolated fosmids were digested by EcoRI and samples displaying the same restriction digestion pattern as the control (fosmid int-3A10) were analyzed by DNA sequencing to confirm the allelic replacement of wild-type tsrA by an Ala2-encoding mutant of tsrA. Mutant int-A2X fosmids (Table S8) were first transformed into E. coli ET12567/pUZ8002, and then introduced into S. laurentii NDS1 by intergeneric conjugation.35, 36 Apramycin-resistant colonies were confirmed by PCR using the primers SD3-F/R and DNA sequencing analysis (Table S9). The engineered S. laurentii strains were designated as S. laurentii NDS1/int-A2X, according to the introduced mutation (Table S8).
Evaluation of Thiostrepton Ala2 Analogue Production in S. laurentii
The fermentations of S. laurentii mutant strains were carried out in a three-step process as described previously.12 First, 50 mL tryptic soy broth (TSB) with apramycin was inoculated with 50 μL of glycerolic mycelium stock of a S. laurentii mutant strain and grown at 28 °C and 220 rpm for 24 h in a 250-mL Erlenmeyer flask. Next, 500 μL of this S. laurentii preculture was used to inoculate 50 mL seed medium (15 g L−1 soybean flour, 50 g L−1 glucose, 15 g L−1 soluble starch, pH 7.2) in a 250-mL Erlenmeyer flask. After a 48 h incubation at 28 °C and 220 rpm, 10 mL of the seed culture was used to inoculate 100 mL fermentation medium (11 g L−1 yeast extract, 50 g L−1 glucose, 15 g L−1 TSB, 1 mL L−1 trace elements solution (5 g L−1 CoCl2·6H2O, 0.5 g L−1 Na2MoO4, 0.5 g L−1 H3BO3, 1.0 g L−1 CuSO4·2H2O, 1.0 g L−1 ZnSO4·7H2O)) in a 500-mL Erlenmeyer flask. The fermentation culture was incubated at 28 °C and 220 rpm for 4 days and extracted twice with an equal volume of chloroform. The chloroform layers were pooled together and solvent was removed in vacuo. The solid residue was dissolved in 4 mL chloroform for HPLC and HPLC-MS analyses. HPLC analyses of the crude culture extracts were performed using a Phenomenex Luna C18(2) column (250 × 4.6 mm, 5 μm) developed with a gradient of 0 to 100% (v/v) acetonitrile in water over 30 min at 1 mL min−1. Absorbance was monitored at 254 nm.
Antibacterial Activity Assay
Minimum inhibitory concentrations (MICs) of thiostrepton analogues against different biological indicator strains (MRSA, VRE, Bacillus, and E. coli ATCC 27854) were determined using the liquid microdilution method.33 Thiostrepton A and its analogues were prepared in DMSO and quantified by their absorbances at 280 nm using an extinction coefficient of 0.027 cm−1μM−1, assuming similar spectral properties for the compounds.52 Chloramphenicol was included as the positive control for Bacillus and VRE, and vancomycin was used as the positive control for MRSA. The negative control in all assays was DMSO. Cell growth was determined by comparing the optical density at 600 nm (OD600) at the time of inoculation and after 18 h incubation at 37 °C. A difference in OD600 defined growth, and the lowest concentration able to completely inhibit bacterial growth is the MIC.
In vitro Transcription-Translation Coupled Assay
In vitro transcription-translation coupled assays were completed using the E. coli S30 Extract System for Circular DNA and the Luciferase Assay System from Promega. The experiments were performed according to the manufacturer’s protocol, except that the reaction was reduced to a 10 μL volume containing 1 μL amino acid mixture, 4 μL S30 premix, 3 μL S30 extract, 1.25 μL nuclease-free water, 0.25 μL thiostrepton analogue, and 0.5 μL pBESTLuc™ template. Thiostrepton A and its analogues were prepared in a range of concentrations in DMSO and quantified as described above. The reactions were incubated at 37 °C for 1 h, at which time 5 μL of the mixture was transferred to a well in a 96-well plate. Luciferase substrate was prepared as recommended by the manufacturer. Immediately before luminescence was measured, 50 μL of the luciferase substrate was added to each well. Relative activity was obtained by normalizing luminescence to that of a DMSO control and plotted against compound concentration. Each assay was performed in triplicate and the half maximal inhibitory concentration (IC50) was calculated for each compound by fitting the data to the Hill equation using GraphPad Prism 5.
20S Proteasome Inhibition Assay
The 20S proteasome inhibition assay components were obtained from Enzo Life Sciences, including the purified human 20S proteasome and the 7-amino-4-methylcoumarin (AMC) fluorogenic substrates. The proteolytic activities of the 20S proteasome were measured using the substrates Boc-Leu-Arg-Arg-AMC for trypsin-like activity, Suc-Leu-Leu-Val-Tyr-AMC for chymotrypsin-like activity, and Ac-Nle-Pro-Nle-Asp-AMC for caspase-like activity. Thiostrepton A and its analogues were prepared in DMSO and quantified as described above. The 20S proteasome was incubated with thiostrepton A or its analogue for 15 min at 37 °C before the addition of the fluorogenic substrate. The assay was performed in a volume of 50 μL in a 384-well plate consisting of: 5 μL thiostrepton analogue, 10 μL 1 μg mL−1 20S proteasome, 10 μL 50 μM fluorogenic substrate and 25 μL buffer (20 mM Tris-HCl, 1 mM EDTA, pH 7.5). Each analysis was performed at least in triplicate and DMSO was used as a control. Fluorescence was measured using an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Emissions were recorded every 50 s for 1 h and the arbitrary fluorescence units were plotted against time for each compound to acquire the slope of the linear fit. Relative activity, obtained by normalizing the compound slope to the DMSO control slope, was plotted against compound concentration and fit to the Hill equation using GraphPad Prism 5 to calculate IC50.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the Defense Advanced Research Projects Agency (N6601-09-2086) and the National Institutes of Health (R01GM090327). We thank C. Hsiao (currently at National Taiwan University) for his guidance with the modeling studies. We thank D. Bostwick for performing mass spectrometry, and E. Yasi and G. Castro are acknowledged for their assistance with bacterial cultures. We are grateful to L. Gelbaum and J. Glushka (Complex Carbohydrate Research Center, University of Georgia) for helpful suggestions with NMR experiments.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available
Tables of strains, plasmids, primers, NMR spectra, mass spectra, and supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline in 2013. J. Antibiot. 2013;66:571–591. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]
- (2).Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Just-Baringo X, Albericio F, Alvarez M. Thiopeptide engineering: A multidisciplinary effort towards future drugs. Angew. Chem. Int. Ed. Engl. 2014;53:6602–6616. doi: 10.1002/anie.201307288. [DOI] [PubMed] [Google Scholar]
- (4).Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- (5).Hensens OD, Albers-Schonberg G. 13C NMR study of thiostrepton and thiopeptin components. J. Antibiot. 1983;36:832–845. doi: 10.7164/antibiotics.36.832. [DOI] [PubMed] [Google Scholar]
- (6).Porse BT, Leviev I, Mankin AS, Garrett RA. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- (7).Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT, Fucini P. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- (8).Lentzen G, Klinck R, Matassova N, Aboul-ela F, Murchie AIH. Structural basis for contrasting activities of ribosome binding thiazole antibiotics. Chem. Biol. 2003;10:769–778. doi: 10.1016/s1074-5521(03)00173-x. [DOI] [PubMed] [Google Scholar]
- (9).Heffron SE, Jurnak F. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 A resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry. 2000;39:37–45. doi: 10.1021/bi9913597. [DOI] [PubMed] [Google Scholar]
- (10).Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- (11).Trejo WH, Dean LD, Pluscec J, Meyers E, Brown WE. Streptomyces laurentii, a new species producing thiostrepton. J. Antibiot. 1977;30:639–643. doi: 10.7164/antibiotics.30.639. [DOI] [PubMed] [Google Scholar]
- (12).Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- (13).Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, Pradel G. Thiostrepton and derivatives exhibit antimalarial and gametocytocidal activity by dually targeting parasite proteasome and apicoplast. Antimicrob. Agents Chemother. 2011;55:1338–1348. doi: 10.1128/AAC.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Clough B, Strath M, Preiser P, Denny P, Wilson IR. Thiostrepton binds to malarial plastid rRNA. FEBS Letters. 1997;406:123–125. doi: 10.1016/s0014-5793(97)00241-x. [DOI] [PubMed] [Google Scholar]
- (16).McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 1997;272:2046–2049. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- (17).Schoof S, Pradel G, Aminake MN, Ellinger B, Baumann S, Potowski M, Najajreh Y, Kirschner M, Arndt HD. Antiplasmodial thiostrepton derivatives: proteasome inhibitors with a dual mode of action. Angew. Chem. Int. Ed. Engl. 2010;49:3317–3321. doi: 10.1002/anie.200906988. [DOI] [PubMed] [Google Scholar]
- (18).Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 2011;3:725–731. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- (19).Pandit B, Bhat U, Gartel AL. Proteasome inhibitory activity of thiazole antibiotics. Cancer Biol. Ther. 2011;11:43–47. doi: 10.4161/cbt.11.1.13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wolf A, Baumann S, Arndt HD, Kirschner KN. Influence of thiostrepton binding on the ribosomal GTPase associated region characterized by molecular dynamics simulation. Bioorg. Med. Chem. 2012;20:7194–7205. doi: 10.1016/j.bmc.2012.09.025. [DOI] [PubMed] [Google Scholar]
- (21).Wolf A, Schoof S, Baumann S, Arndt HD, Kirschner KN. Structure-activity relationships of thiostrepton derivatives: implications for rational drug design. J. Comput. Aided Mol. Des. 2014;28:1205–1215. doi: 10.1007/s10822-014-9797-0. [DOI] [PubMed] [Google Scholar]
- (22).Newick K, Cunniff B, Preston K, Held P, Arbiser J, Pass H, Mossman B, Shukla A, Heintz N. Peroxiredoxin 3 is a redox-dependent target of thiostrepton in malignant mesothelioma cells. PloS One. 2012;7:e39404. doi: 10.1371/journal.pone.0039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sandu C, Chandramouli N, Glickman JF, Molina H, Kuo CL, Kukushkin N, Goldberg AL, Steller H. Thiostrepton interacts covalently with Rpt subunits of the 19S proteasome and proteasome substrates. J. Cell. Mol. Med. 2015;19:2181–2192. doi: 10.1111/jcmm.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zheng Q, Wang Q, Wang S, Wu J, Gao Q, Liu W. Thiopeptide antibiotics exhibit a dual mode of action against intracellular pathogens by affecting both host and microbe. Chem. Biol. 2015;22:1002–1007. doi: 10.1016/j.chembiol.2015.06.019. [DOI] [PubMed] [Google Scholar]
- (25).LaMarche MJ, Leeds JA, Amaral A, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Ding J, Dzink-Fox J, Gamber G, Jain A, Lee K, Lee L, Lister T, McKenney D, Mullin S, Osborne C, Palestrant D, Patane MA, Rann EM, Sachdeva M, Shao J, Tiamfook S, Trzasko A, Whitehead L, Yifru A, Yu D, Yan W, Zhu Q. Discovery of LFF571: an investigational agent for Clostridium difficile infection. J. Med. Chem. 2012;55:2376–2387. doi: 10.1021/jm201685h. [DOI] [PubMed] [Google Scholar]
- (26).Myers CL, Hang PC, Ng G, Yuen J, Honek JF. Semi-synthetic analogues of thiostrepton delimit the critical nature of tail region modifications in the control of protein biosynthesis and antibacterial activity. Bioorg. Med. Chem. 2010;18:4231–4237. doi: 10.1016/j.bmc.2010.04.098. [DOI] [PubMed] [Google Scholar]
- (27).Schoof S, Baumann S, Ellinger B, Arndt HD. A fluorescent probe for the 70S-ribosomal GTPase-associated center. ChemBioChem. 2009;10:242–245. doi: 10.1002/cbic.200800642. [DOI] [PubMed] [Google Scholar]
- (28).Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J. Am. Chem. Soc. 2010;132:7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bowers AA, Acker MG, Young TS, Walsh CT. Generation of thiocillin ring size variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J. Am. Chem. Soc. 2012;134:10313–10316. doi: 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Li C, Zhang F, Kelly WL. Mutagenesis of the thiostrepton precursor peptide at Thr7 impacts both biosynthesis and function. Chem. Commun. 2012;48:558–560. doi: 10.1039/c1cc14281j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhang F, Kelly WL. In vivo production of thiopeptide variants. Methods Enzymol. 2012;516:3–24. doi: 10.1016/B978-0-12-394291-3.00022-8. [DOI] [PubMed] [Google Scholar]
- (33).Li C, Zhang F, Kelly WL. Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs. Mol. BioSyst. 2011;7:82–90. doi: 10.1039/c0mb00129e. [DOI] [PubMed] [Google Scholar]
- (34).Zhang F, Kelly WL. Saturation mutagenesis of TsrA Ala4 unveils a highly mutable residue of thiostrepton A. ACS Chem. Biol. 2015;10:998–1009. doi: 10.1021/cb5007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Guo H, Wang J, Yeming L, Yu Y, Zheng Q, Wu J, Liu W. Insight into bicyclic thiopeptide biosynthesis benefited from development of a uniform approach for molecular engineering and production improvement. Chem. Sci. 2014;5:240–246. [Google Scholar]
- (38).Tori K, Tokura K, Okabe K, Ebata M, Otsuka H. Carbon-13 NMR studies of peptide antibiotics, thiostrepton and siomycin A: the structure relationship. Tetrahedron Lett. 1976;17:185–188. [Google Scholar]
- (39).Garg N, Salazar-Ocampo LMA, van der Donk WA. In vitro activity of the nisin dehydratase NisB. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7258–7263. doi: 10.1073/pnas.1222488110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kuipers OP, Rollema HS, Yap WM, Boot HJ, Siezen RJ, de Vos WM. Engineering dehydrated amino acid residues in the antimicrobial peptide nisin. J. Biol. Chem. 1992;267:24340–24346. [PubMed] [Google Scholar]
- (41).Bierbaum G, Szekat C, Josten M, Heidrich C, Kempter C, Jung G, Sahl HG. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 1996;62:385–392. doi: 10.1128/aem.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJM, Moll GN, Kuipers OP. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry. 2005;44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- (43).Mocek U, Beale JM, Floss HG. Reexamination of the 1H and 13C NMR spectral assignments of thiostrepton. J. Antibiot. 1989;42:1649–1652. doi: 10.7164/antibiotics.42.1649. [DOI] [PubMed] [Google Scholar]
- (44).Priestley ND, Smith TM, Shipley PR, Floss HG. Studies on the biosynthesis of thiostrepton: 4-(1-hydroxyethyl)quinoline-2-carboxylate as a free intermediate on the pathway to the quinaldic acid moiety. Bioorg. Med. Chem. 1996;4:1135–1147. doi: 10.1016/0968-0896(96)00126-5. [DOI] [PubMed] [Google Scholar]
- (45).Mocek U, Zeng Z, O’Hagan D, Zhou P, Fan L-DG, Beale JM, Floss HG. Biosynthesis of the modified peptide antibiotic thiostrepton in Streptomyces azureus and Streptomyces laurentii. J. Am. Chem. Soc. 1993;115:7992–8001. [Google Scholar]
- (46).Jonker HRA, Baumann S, Wolf A, Schoof S, Hiller F, Schulte KW, Kirschner KN, Schwalbe H, Arndt HD. NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site. Angew. Chem. Int. Ed. Engl. 2011;50:3308–3312. doi: 10.1002/anie.201003582. [DOI] [PubMed] [Google Scholar]
- (47).Duan L, Wang S, Liao R, Liu W. Insights into quinaldic acid moiety formation in thiostrepton biosynthesis facilitating fluorinated thiopeptide generation. Chem. Biol. 2012;19:443–448. doi: 10.1016/j.chembiol.2012.02.008. [DOI] [PubMed] [Google Scholar]
- (48).Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- (49).Bhat UG, Halasi M, Gartel AL. Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PloS One. 2009;4:e5592. doi: 10.1371/journal.pone.0005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Naidu BN, Sorenson ME, Bronson JJ, Pucci MJ, Clark JM, Ueda Y. Synthesis, in vitro, and in vivo antibacterial activity of nocathiacin I thiol-Michael adducts. Bioorg. Med. Chem. Lett. 2005;15:2069–2072. doi: 10.1016/j.bmcl.2005.02.046. [DOI] [PubMed] [Google Scholar]
- (51).Wright F, Bibb MJ. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- (52).Ryan PC, Lu M, Draper DE. Recognition of the highly conserved GTPase center of 23 S ribosomal RNA by ribosomal protein L11 and the antibiotic thiostrepton. J. Mol. Biol. 1991;221:1257–1268. doi: 10.1016/0022-2836(91)90932-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.