Abstract
In this study, mesoporous silica nanoparticles (MSNs) were functionalized with Cholera toxin subunit B (CTxB) protein to influence their intracellular trafficking pathways. The CTxB-MSN carrier was synthesized, and its chemical and structural properties were characterized. Endocytic pathway inhibition assays showed that the uptake of CTxB-MSNs in human cervical cancer (HeLa) cells was partially facilitated by both chlatrin- and caveolae-mediated endocytosis mechanisms. Laser scanning confocal microscopy (LSCM) experiments demonstrated that CTxB-MSNs were taken up by the cells and partially trafficked through the trans-Golgi network into to the endoplasmic reticulum in a retrograde fashion. The delivery abilities of CTxB-MSNs were evaluated using propidium iodide, an impermeable cell membrane dye. LSCM images depicted the release of propidium iodide in the endoplasmic reticulum and cell nucleus of HeLa cells.
Introduction
In recent years, many studies have been focusing on the synthesis of novel nanomaterials possessing distinct structural and functional features. Among them, mesoporous silica nanoparticles (MSNs) are highly attractive and intensively applied in biomedicine, biotechnology, separation, and catalysis.1–6 MSNs is an attractive platform due to its outstanding properties such as high surface area, pore volume, tunable pore diameter, easy modification, chemical stability and good biocompatibility.7 In particular, the use of MSNs as drug delivery system has been extensively explored in the past 15 years.1, 8 A wide variety of MSN-based platforms have been reported to deliver not only anticancer drugs but other type of therapeutic agents such as proteins, siRNA, DNA, photosensitizers, etc. The ability of MSNs to successfully deliver therapeutic agents has been demonstrated both in vitro and in vivo settings.8, 9 Several groups have investigated the possibility of increasing the release and concentration of therapeutic agents delivered by MSN platforms in vitro through changing the surface properties of MSNs. For that purpose, a wide variety of functional moieties have been attached to the surface of MSNs such as small molecules, antibodies, aptamers, peptides, sacharides and proteins.1, 7 Some of these strategies focus on improving the targeting properties of the MSN platform and others look for approaches to escape from endosomes and/or lysosomes.10 These tactics have shown partial success in increasing the delivery of therapeutic agents from MSNs.1, 7, 8, 11, 12 Nevertheless, another way to enhance the efficacy of MSNs as drug delivery system is by modifying their trafficking pathway inside the cells. By delivering the active compounds in specific compartments such as cytosol, nucleus or mitochondria the effect of the therapeutic agents can be enhanced.13 It has been shown that non-modified MSNs are mainly trafficked through the endolysosomal pathway. In principle, the MSN material is first transported to primary endosomes, followed by secondary endosomes, which fuse with lysosomes.7 Ideally, MSNs should escape the endolysosomes and enter the cytosolic compartment; however, non-modified MSNs are usually trapped in late endosomes and/or lysosomes. We hypothesize that alternative strategies to modify the trafficking pathway of MSN in cells can be explored. For example; biological toxins, such as shiga, ricin and cholera, have demonstrated the ability to escape degradation by exploiting alternative modes of intracellular trafficking upon internalization by the cell.14–16 Cholera toxin (CT), which is secreted by the aquatic bacterial pathogen Vibrio cholera, is the best characterized among these toxins. As a classic AB toxin, CT has been demonstrated to translocate from the plasma membrane through the trans-Golgi network into the endoplasmic reticulum (ER) in a retrograde fashion by binding the ganglioside GM1 via the B subunit of the native holotoxin.17, 18 Once in the ER lumen, the A-subunit of CT gains access to the cytosol through retro-translocation and subsequently induces toxicity by elevating cAMP levels. The CT-GM1 complex can enter cells via different clathrin-dependent and clathrin-independent mechanisms and this can vary by cell type.19, 20 However, not all endocytic pathways appear to lead to a toxic response. It has been estimated that only around 12% of the fraction of CT that enters the cells engages in the retrograde trafficking pathway.21
We postulate that by conjugating CTxB protein to MSNs, the endocytosis and trafficking pathways of regular MSN materials can be modified (Scheme 1). Interestingly, there are only few reports in the literature that use bacterial toxins to direct the intracellular localization of nanoparticles. Chakraborty et al. demonstrated the utility of cholera toxin B conjugated quantum dots (QDs) for live cell labeling.22 Nevertheless, the authors did not report a comprehensive study of the endocytosis and trafficking pathways of these materials. Iversen and co-workers conjugated the B subunits of shiga and ricin toxins to QDs. These authors speculated that the size of the subunit B-QDs platforms used in their study prevented them from following the same endocytic pathways as their respective B subunits, whether they were shiga or ricin.23
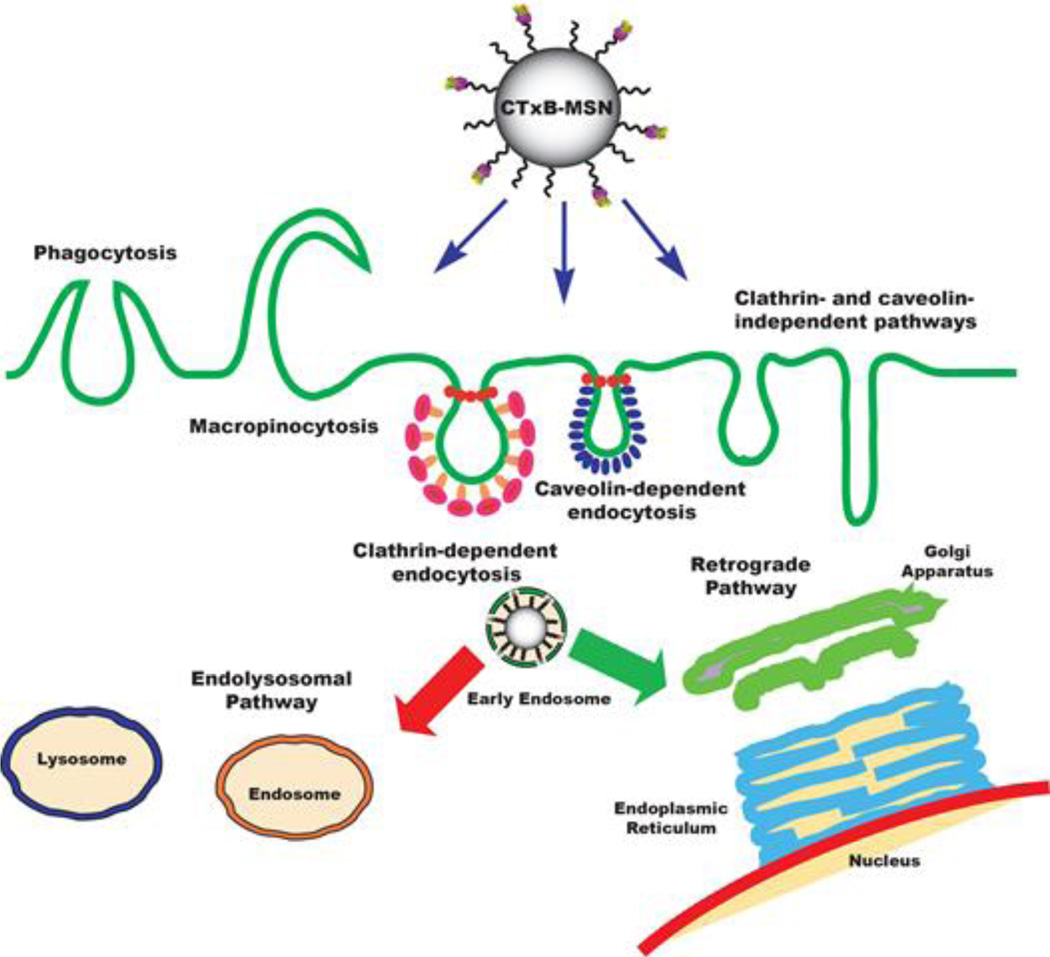
Scheme 1.
Schematic representation of the internalization and trafficking pathways of Cholera toxin subunit B-modified mesoporous silica nanoparticles (CTxB-MSNs).
Herein, we report a comprehensive investigation of the endocytosis and cellular trafficking pathways of CTxB ligand conjugated to MSNs in human cervical cancer (HeLa) cells. The CTxB-FMSN platform consists of a fluorescein-labeled MSN (FMSN) material chemically functionalized with CTxB through a polyethylene glycol (MW = 2K) (PEG-2K) polymer linker (Scheme 2). The structural and chemical properties of this material were fully characterized by N2 sorption isotherms, dynamic light scattering (DLS), thermogravimetric analysis (TGA), and transmission electron microscopy (TEM). The endocytic pathways for the cellular internalization of CTxB-FMSNs were studied with fluorescence activated cell sorting (FACS) using different pharmacological inhibitors. The cellular trafficking of CTxB-FMSNs was investigated by laser scanning confocal microscopy (LSCM) in the presence of different organelle markers. The ability of CTxB-FMSN platform as delivery system was evaluated by the intracellular release of propidium iodide, an impermeable cell membrane dye, which is not internalized by healthy cells. We envision that this strategy of functionalizing MSNs with CTxB to circumvent the degradative endolysosomal pathway will enhance the delivery of therapeutic cargoes.
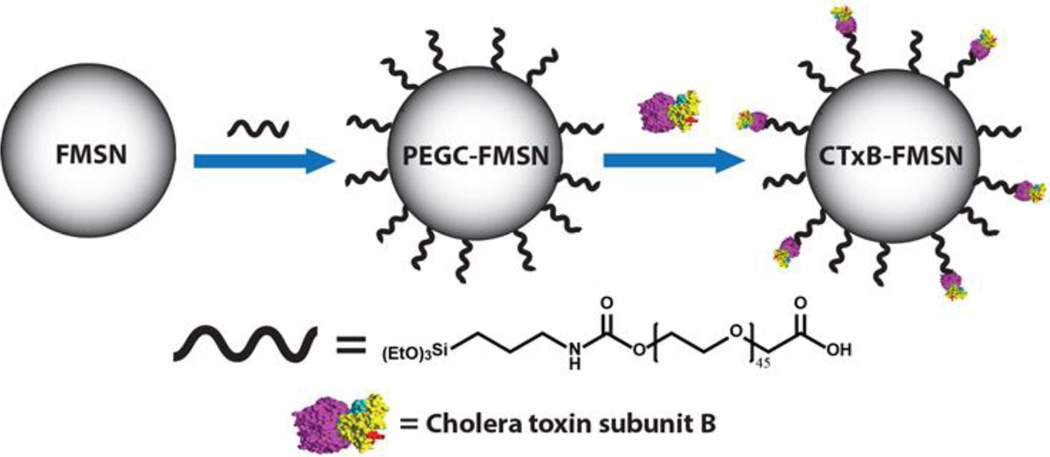
Scheme 2.
Synthesis of CTxB-FMSNs. The synthesis of fluorescein-labeled MSNs (FMSNs) was carried out using a surfactant-templated co-condensation approach. A carboxylic acid PEG silane derivative was grafted to the surface of FMSNs (PEGC-MSNs). Finally, Cholera toxin subunit B was conjugated to PEGC-FMSNs (CTxB-FMSNs).
2. Experimental
2.1. Synthesis of FITC-labeled mesoporous silica nanoparticles (FMSNs)
FMSNs were synthesized using a method from the literature with slight modifications.24 Briefly, 3.42 mL of ethanol (EtOH); 0.78 g of cetyltrimethylammonium bromide (2.14 mmol, CTAB), and 0.045 g diethanolamine (0.428 mmol, DEA) were added to nanopure water (21.6 mL). The resulting aqueous solution was stirred vigorously at 60 °C for 30 min. In addition; FITC silane derivative was prepared by adding 5 mg of fluorescein isothiocynate (FITC, 12.8 µmol) and 3 µL aminopropyl triethoxysilane (12.8 µmol, APTES) to 200 µL of acetonitrile. This organic solution was stirred at room temperature under dark conditions for 15–20 min. Tetramethylorthosilicate (2.19 mL, 14.7 mmol, TMOS) was added dropwise to the first aqueous solution over a period of 5 min. The FITC silane derivative solution (90 µL) was added midway through the addition of TMOS. The resulting suspension was stirred for an additional 18 h at 60 °C before being collected by centrifugation and resuspended in EtOH. The surfactant template CTAB was removed from the as-synthesized nanoparticles by washing the material in 1.0 M HCl methanolic solution at 60 °C with constant stirring for 48 h.
2.2. Grafting of carboxylic acid-PEG (PEGC) to FMSN (PEGC-FMSNs) materials
FMSNs were grafted with PEGC polymer silane by refluxing the materials in ethanol. The synthesis and characterization of the PEG silane heterobifunctional polymer is described in the electronic supporting information (ESI). PEGC silane material (25 mg) was mixed with FMSNs (50 mg) in a solution of 40 µL of NH4OH (35% v/v) in ethanol (50 mL). The dispersion was refluxed at 90 °C for 18 h with constant stirring. To obtain the final product, the PEGC-grafted MSNs were collected by centrifugation, washed three times with EtOH and finally re-suspended in EtOH. The amount of PEGC polymer grafted to FMSN particles was determined by TGA.
2.3. Functionalization of PEGC-FMSNs with Cholera Toxin subunit B (CTxB-FMSNs)
The chemical attachment of CTxB protein (Molecular Weight ~ 12KDa) was conducted through a coupling reaction between the carboxylic acid groups on the surface of PEGC-FMSNs and the amine groups of CTxB protein. PEGC-FMSN material (10 mg) was dispersed in 10 mL of DI water; later, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, 0.74 µg, 4.77 nmol) and CTxB protein (250 µg) were added to this solution. The resulting dispersion was stirred for 24 h at 4 °C under dark conditions. The CTxB-MSNs were obtained by centrifugation, washed three times with DI water and resuspended in DI water. The amount of CTxB protein chemically attached to PEGC-FMSNs was determined by calculating the difference between the initial amount of protein and the final amount present in the supernatant after the reaction and few washing solutions. The amount of protein was determined by the Pierce™ BCA protein quantification assay using the enhanced protocol.
2.4. Cell Culture
Human cervical carcinoma (HeLa) cells were cultured in the presence of RPMI 1640 medium supplemented with 10% (v/v) FBS and 1% (v/v) penicillin-streptomycin solution and incubated at 37 °C/5% CO2 environment. For endocytosis inhibition and receptor specificity assays, cells were seeded at 2×105 cells/well using 2 mL of cell media in 6-well tissue culture plates and incubated for 36 h. For fixed-cell confocal imaging and propidium iodide (PI) release experiments, cells were seeded at 1×105 cells/mL with 300 µL of cell media/well in 8-well chambered coverglass and incubated for 24 h.
2.5. Endocytosis inhibition assays
After growing HeLa cells as described in section 2.4, the cell medium was removed and the cultured cells were treated independently with 2 mL of each inhibiting reagent as follow: Wortmannin (240 nM, 30 min) to inhibit macropinocytosis; Chloropromazine (10.66 µg/mL, 30 min) to inhibit clathrin-mediated endocytosis; and Genstein (50 µg/mL, 30 min) to inhibit caveolae-mediated endocytosis. The cell medium was discarded and 2 mL of RPMI 1640 medium lacking FBS containing FMSNs, PEGC-FMSNs or CTxB-FMSNs in a concentration of 25 µg/mL for each material were added to the wells. The cells were incubated with each material for 1 h at 37 °C/5% CO2. After that, the medium was aspirated and replaced with RPMI 1640 medium supplemented with FBS. All cells were incubated for additional 2 h, trypsinized (1 mL of trypsin) and fixed in 2 mL of PBS solution of paraformaldehyde 4% (v/v) for 1 h. The cells were resuspended in 300 µL of PBS and transferred to 15 mL polystyrene round bottom tubes. Trypan blue solution (100 µL) was added to the centrifuge tubes to quench any FMSN material physically absorbed on the surface of the cell membrane.10, 25 Finally, 20 µL of PI staining solution (10 µg/mL) was added to each cell suspension to identify dead cells that will be excluded from the analysis. The cellular fluorescence resulting from internalization of each material was quantified using FACS.
2.5.1. FACS data analysis
The FACS data was analyzed with BD FACSDiva software. To determine the extent of changes in the uptake of the different materials, the mean fluorescence of the population due to cells negative for nanoparticle uptake was subtracted from the positive population and normalized in relation to an untreated control experiment. This control follows the same protocol as described above; however, the cells were not incubated in the presence of inhibiting agents. Prior to harvesting, all cells were washed 2× with PBS between each treatment. The experiments were run in triplicate for each endocytosis inhibition assay.
2.6. Fixed-cell LSCM experiments
2.6.1. Lysosomal Colocalization
After growing HeLa cells as described in section 2.4, the cell medium was removed and replaced with 300 µL of serum-free medium containing FMSN, PEGC-FMSN, or CTxB-FMSN materials at a concentration of 25 µg/mL. The, cells were incubated for 30 min at 37 °C/5% CO2. Afterwards, the medium was removed, cells were stained with 300 µL of Lysotracker Red DND-99 (500 nM) to selectively label lysosomes, and incubated for another 30 min. Cells were washed with PBS and fixed with paraformaldehyde solution (4% (v/v)) for 1 h. After that, the fixing solution was removed and wells were washed with PBS. A nuclei-staining dye, Hoescht 33342 (300 µL, 5µg/ml) was added and the cells were incubated for 30 min 37 °C/5% CO2. Finally, the staining solution was aspirated and the cells were washed with PBS. After the addition of 300 µL PBS to each well, the intracellular localization of all materials was determined by LSCM.
2.6.2. Endoplasmic Reticulum Colocalization
After growing HeLa cells as described in section 2.4, the cell medium was removed and replaced with 300 µL of serum-free medium containing FMSN, PEGC-FMSN, or CTxB-FMSN materials at a concentration of 25 µg/mL. The, cells were incubated for 30 min at 37 °C/5% CO2. Afterwards, the medium was removed; cells were stained with 300 µL of ERtracker™Blue-White DPX (5 µM) to selectively label the endoplasmic reticulum and incubated for another 30 min. Cells were washed with PBS and fixed with paraformaldehyde solution (4% (v/v)) for 1 h. After that, the fixing solution was removed and cells were washed with PBS. After the addition of 300 µL PBS to each well, the intracellular localization of all materials was determined by LSCM.
2.6.3. Golgi Apparatus Colocalization
After growing HeLa cells as described in section 2.4, the cell medium was removed and replaced with 300 µL of serum-free medium containing FMSN, PEGC-FMSN, or CTxB-FMSN materials at a concentration of 25 µg/mL. The, cells were incubated for 30 min at 37 °C/5% CO2. Afterwards, the medium was removed; cells were stained with 300 µL of BODIPY-TR™ Ceramide (25 µM) to selectively label the Golgi apparatus and incubated for another 30 min. Cells were washed with PBS and fixed with paraformaldehyde solution (4% (v/v)) for 1 h. After that, the fixing solution was removed and wells were washed with PBS. A nuclei-staining dye, Hoescht 33342 (300 µL, 5µg/ml) was added and the cells were incubated for 30 min 37 °C/5% CO2. Finally, the staining solution was aspirated and the cells were washed with PBS. After the addition of 300 µL PBS to each well, the intracellular localization of all materials was determined by LSCM.
2.6.4. Colocalization analysis
Quantitative colocalization analysis was performed using a method similar to that used by Pollock et al.26 Briefly, a region of interest (ROI) was defined for each possible colocalization event with the cell (10 cells per condition) with events scoring coefficients greater than 0.5 being counted as positive for colocalization. Colocalization is reported as the frequency of colocalization events for all possible colocalization events averaged over 10 cells. The defined ROI was filtered using a median filter set to 3×3 pixels, and Manders correlation coefficients calculated using thresholds set at the mean + 1 SD intensity for each image channel. All analysis was done through ImageJ using the JaCoP plug-in.27
2.7. Propidium Iodide Release
PI was loaded in each FMSN, PEGC-FMSN and CTxB-FMSN materials using the following procedure: 25 µg of FMSN, PEGC-FMSN, or CTxB-FMSN material was resuspended in 1 mL of DI aqueous solution (250 µg/mL) and stirred at RT under dark conditions for 18 h. The materials were collected by centrifugation and resuspended in 1 ml of serum-free media. The cell medium was removed and replaced with 300 µL of serum-free medium containing FMSN, PEGC-FMSN, or CTxB-FMSN materials (25 µg/mL). After inoculating for 1 h at 37 °C/5% CO2, the medium was removed and the wells were washed with PBS. Cell medium (300 µL) was added to each well and the cells were incubated for another 24 h at 37 °C/5% CO2. The medium was then aspirated and the cells washed with PBS. HeLa cells were stained with 300 µL of ERtracker Blue-White DPX (5 µM) for 30 min at 37 °C/5% CO2. The staining solution was removed and the cells were fixed for 1 h with a paraformaldehyde solution (4% (v/v)). After that, the fixing solution was removed and wells were washed with PBS. After the addition of 300 µL PBS to each well, the intracellular localization of all materials was determined by LSCM.
3. Results and discussion
3.1. Preparation and Characterization of CTxB-FMSNs
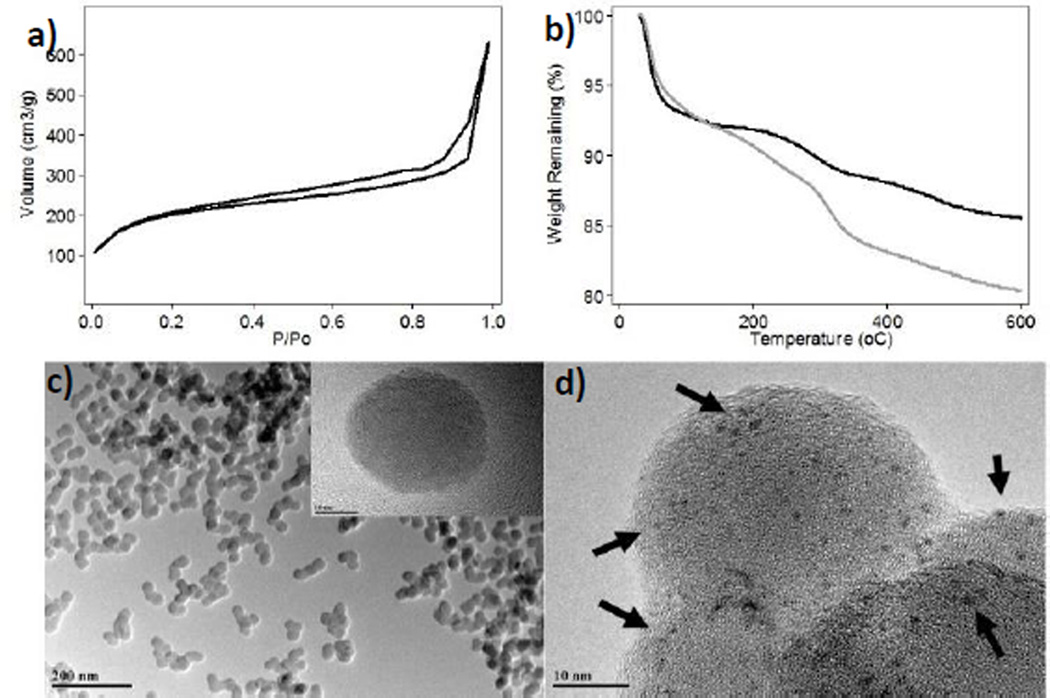
The synthesis of FMSNs was carried out by using a surfactant-templated co-condensation approach, which was modified from the literature.24 The surfactant was removed by an acid wash in methanolic solution under reflux. The structural properties of the FMSN material were analyzed by N2 sorption isotherms (BET method), dynamic light scattering (DLS), ζ-potential, transmission electron microscopy (TEM), and thermogravimetric analysis (TGA). The BET analysis shows that the FMSN material has a surface area of 725.8 m2/g (Fig. 1a). The hydrodynamic diameter of this material is 450.2 nm in simulated physiological environment (phosphate buffer solution (PBS), pH 7.4, 1.0 mM) and 152.5 nm in water. The surface of the FMSNs is negatively charged due to the presence of deprotonated silanols on the surface of the nanoparticles as it is corroborated by the ζ–potential (−16.2 ± 0.45 mV) (Table 1). TEM micrographs show that the FMSNs have diameters of 35.8 ± 5.8 nm (Fig. 1c). The amount of fluorescein dye chemically attached to the interior channels of the FMSNs was approximately 7.0 wt.%, according to TGA (Fig. 1b).
Fig. 1.
Structural characterization of FMSN materials synthesized in this project. a) Nitrogen sorption isotherm of FMSNs. b) Thermogravimetric analysis of FMSNs (black) and PEGC-MSNs (gray). TEM images of c) FMSNs and d) CTxB-FMSNs. The dark spots confirmed the presence of CTxB protein onto MSNs.
Table 1.
Structural properties of FMSN, PEGC-FMSN and CTxB-FMSN materials.
| Material | HD (H2O) (nm)/PdI |
HD (PBS) (nm)/PdI |
ZP (mV) | OC (%) |
SA (m2/g) |
|---|---|---|---|---|---|
| FMSNs | 152.5/0.3 | 450.2/0.4 | −16.2 ± 0.5 | 7.0 | 725.8 |
| PEGC-FMSNs | 139.7/0.1 | 1442.0/0.9 | −18.0 ± 0.3 | 11.4 | 486.3 |
| CTxB-FMSNs* | 164.3/0.2 | 759.8/0.4 | −19.3 ± 1.1 | --- | --- |
HD=Hydrodynamic diameter; PdI=Polydispersity index; ZP=ζ-potential; OC=Organic content; SA=Surface area;
The organic content and surface area of CTxB-FMSNs were not characterized due to the small amount of material that was obtained after the conjugation with CTxB protein.
Carboxylic acid-PEG(PEGC)-silane polymer was grafted to the FMSNs to afford the corresponding PEGC-FMSN material. The BET analyses of the PEGylated FMSN particles shows a reduction in the surface area indicating the presence of the PEG molecules that block the entrance of the pores from MSN particles (Table 1). The PEGC-FMSN material show poor colloidal stability in PBS; however, the nanoparticles exhibit improved stability in water with hydrodynamic diameters of 139.7 and 294.4 nm, respectively. The grafting of PEG chains into MSN materials usually results in the reductions of the absolute values of the ζ–potentials.28 However, in the case of PEGC-FMSNs the ζ–potential remains negative (−18.0 ± 0.31 mV) because of the presence of carboxylic acid groups, which under simulated physiological conditions (PBS 1.0 mM, pH = 7.4), will be deprotonated to form negatively charged carboxylate groups. TGA data further confirmed the presence of PEG moieties on the surface of FMSNs; as anticipated, the organic content of PEGC-FMSNs increased 4.4 wt.% in comparison with FMSNs. TEM images confirm that the grafting conditions did not change the morphology or size of the nanoparticles (Fig. S1, ESI).
PEGC-FMSN particles were further functionalized with CTxB protein to afford CTxB-FMSN materials. CTxB was chemically attached to PEGC-MSNs by a coupling reaction mediated by EDC coupling agent. The amount of CTxB protein was quantified using the BCA protein quantification assay, which determines the difference between the starting amount of protein added to the conjugation reaction and the unreacted protein in the supernatant and washing solutions. Based on this method, we determined that the amount of CTxB protein chemically attached to PEGC-FMSNs is 8.2 µg CTxB/mg of material.
This amount was further corroborated by quantifying the fluorescence of a FITC-labeled CTxB protein attached to a Rhodamine B-MSNs following the protocol described in the ESI. The FITC-CTxB Rhodamine B-labeled MSNs (FCTxB-RMSNs) were also used to demonstrate that the combo CTxB-MSNs can be internalized by HeLa cells, see section 3.3. The presence of CTxB on the surface of FMSNs was also confirmed directly by negative-staining TEM. TEM grids containing CTxB-FMSN material were prepared and stained with Nano-W™.29 Fig. 1d shows the TEM image of CTxB-FMSNs negatively-stained with Nano-W. The dark spots (black arrows) depict the presence of CTxB proteins, which are fairly homogeneously distributed throughout the nanoparticles (Figs. 1d and S1c). A control CTxB-MSN sample that was not stained with Nano-W did not show the presence of the dark spots in the TEM image (Fig. S1b, ESI).
3.2. Study of the Internalization Pathways of CTxB-FMSN Material in HeLa Cells
The endocytosis of CTxB protein in several cell types occurs by different endocytic mechanisms, both clathrin-/caveolae-dependent and clathrin-independent mechanisms.17, 19, 20, 30 Some pathways appear more or less prominently depending on the cell type studied. For example; in hippocampal neurons, BHK and HeLa cells, CTxB can bind GM1 and enter the cell by clathrin-mediated mechanisms, but this not occur in motor neurons or appear to be a major entry pathway for CTxB protein in certain other cell types.20 To distinguish between the various mechanisms of endocytosis potentially utilized by the FMSN materials synthesized in this project, a series of endocytic pathways assays were performed in the presence of inhibitors and resultant changes in internalization quantified using flow cytometry (Fig. 2a–b).
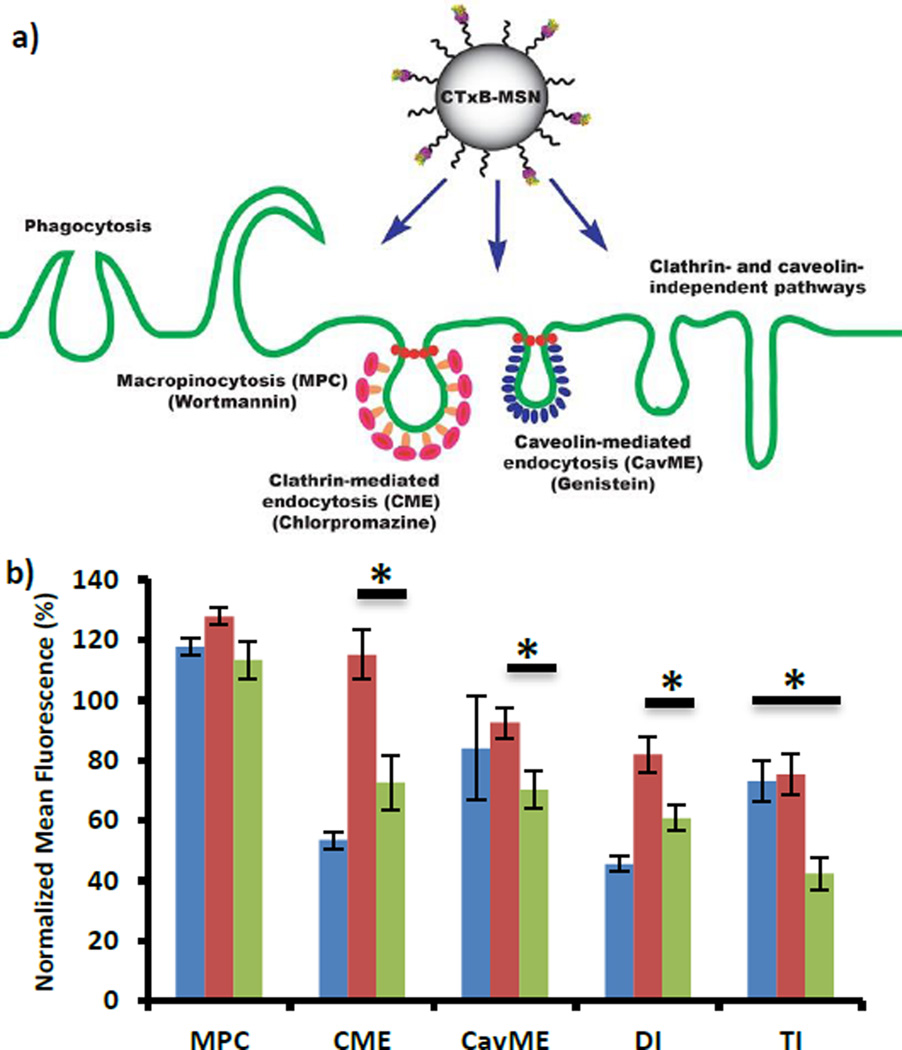
Fig. 2.
Study of the mechanism of internalization for CTxB-FMSN material in HeLa cells. a) Schematic representation of the possible endocytosis pathways for CTxB-FMSNs. b) Flow cytometry results for the inhibition of macropinocytosis (MPC), clathrin-mediated endocytosis (CME), caveolin-mediated endocytosis (CavME), double (DI) (MPC/CME) and triple (TI) (MPC/CME/CavME) inhibition for FMSNs (blue), PEGC-MSNs (red) and CTxB-FMSNs (green). Data represent ± SEM of triplicates from 3 independent experiments. Data are represented in relation to untreated control (100%). Asteriks indicate p < 0.02.
Macropinocytosis (MPC) is a common clathrin-independent route of cellular entry. To investigate whether MPC plays a role in the internalizations of MSNs, we tested the uptake of these materials in the presence of the macropinocytosis-inhibiting agent, Wortmannin. This agent irreversibly binds to the catalytic subunit of phosphatidylinositol-3 (PI-3) kinase, which regulates the arrangements of actin filaments.31, 32 The presence of Wortmannin resulted in a decrease in the uptake of FMSNs (117.7 ± 3.0%), and in an increase in the internalization of PEGC-FMSNs (127.8 ± 2.7%) and CTxB-FMSN materials (113.3 ± 6.1%). The values obtained for these experiments higher than 100% indicate an increase in fluorescence higher that the control experiment, which is directly proportional to the internalization of MSN materials. This data corroborates that MPC is not a major internalization pathway for the MSNs materials. Previous reports have shown that MPC can be an important internalization pathway for silica nanoparticles in other types of cells such as pulmonary epithelial cells (NCI-H292)33 or human dermal fibroblasts;34 however, in the case of HeLa cells only MSNs with different aspect ratio has been reported to be endocytosed by MPC.35
The effect of clathrin inhibition on MSN particle uptake was tested by using the cationic amphiphilic drug chlorpromazine, which causes clathrin to accumulate in late endosomes, thereby inhibiting the formation of clathrin-coated pits.32 The uptake of FMSNs (53.5 ± 2.8%) and CTxB-FMSNs (72.6 ± 9.1%) in chlorpromazine-treated cells was significantly reduced.
Previous reports have shown that FMSN particles are mainly internalized by clathrin-mediated endocytosis (CME).7, 10, 36 Moreover, this endocytic pathway has been proposed as one of the mechanisms for the internalization of CTxB protein in HeLa cells.19, 20 The uptake of PEGC-FMSN (115.0 ± 8.2%) is increased in the presence of chlorpromazine, which indicates that this material is not internalized through this mechanism. Caveolae-mediated endocytosis (CavME) is a common non-clathrin route of cell entry.37 To determine whether this mechanism is involved in the endocytosis of FMSN materials, we tested genistein, a caveolae-inhibiting agent. Genistein, a tyrosine kinase inhibitor, has been shown to inhibit the caveolae-mediated endocytosis of SV40 virus and cholera toxin.32 The presence of genistein inhibitor slightly decreases the uptake of FMSNs (83.9 ± 17.2%). In a similar way, the uptake of PEGC-FMSN particles exhibited a minor decrease in cells treated with genistein (92.5 ± 4.9%). These results suggest that CavME mechanisms may play a minor role in the endocytosis of FMSNs or PEGC-FMSNs.38 Interestingly, genistein (70.2 ± 6.0%) did reduce the internalization of CTxB-FMSNs, indicating that CavME mechanisms are partially responsible for the internalization of this material.19 To determine whether the inhibition of more than one endocytic mechanism makes a difference in the internalization of MSN materials, we performed the simultaneous inhibition of two (MPC/CME) and three (MPC/CME/CavME) internalization pathways. Double inhibition in the presence of Wortmannin and chlorpromazine reduced the internalization of FMSNs (45.5 ± 2.6%) and CTxB-FMSNs (60.8 ± 4.3%) mainly due to the inhibition of CME. Triple inhibition in the presence of Wortmannin, chlorpromazine and genistein reduced the internalization of FMSNs (73.1 ± 7.0%) and CTxB-FMSNs (42.3 ± 5.5%) as an indication that indeed CME and CavME are endocytic routes for the internalization of CTxB-FMSNs. There was also a slight inhibition in the uptake of PEGC-FMSNs for double (81.8 ± 6.1%) and triple (75.3 ± 6.9%) inhibition. The study of the endocytic pathways for the internalization of CTxB-FMSNs shows that, similar to the CTxB protein, this material is endocytosed by different endocytic mechanisms, both clathrin- and caveolin-dependent mechanisms. Moreover, the endocytosis of this material is different (Student’s t-Test, p <0.02) from PEGC-FMSNs as an indication that the presence of CTxB protein on the surface of FMSNs influence this process.
The internalization of the native cholera holotoxin is initiated by the binding of the B subunit to the cell surface ganglioside GM1.17 We tested the internalization of FMSNs, PEGC-FMSNs and CTxB-FMSNs by receptor competition in the presence of unlabeled CTxB protein (Fig. S2 in the ESI). The control experiment, which tests the internalization of FITC-labeled CTxB protein, demonstrates that the GM1 receptors in HeLa cells can be blocked in the presence of CTxB protein (18.5 ± 0.2%). The flow cytometry data show a slight decrease on the uptake of CTxB-FMSNs (91.8 ± 4.0%) in the presence of CTxB protein. However, this data is not statistically different from the uptake PEGC-FMSNs (95.9 ± 2.9%). The internalization of FMSNs is not affected by the presence of CTxB protein (102.6 ± 1.8%).
3.3. Investigation of the Internalization and Intracellular Trafficking of CTxB-MSN Materials by Confocal Microscopy
To demonstrate that the combo CTxB-MSN is indeed internalized as a whole material by HeLa cells, FCTxB-RMSNs were fabricated, see ESI for details of the synthesis. FCTxB-RMSN material allowed us to localize the presence of both CTxB (FITC-channel) and MSNs (TRITC-channel) once the material has been endocytosed by HeLa cells using LSCM. Figs. S3a–d shows that FCTxB-RMSNs have been internalized by HeLa cells. In addition, the fluorescence emission of FCTxB protein (green) co-localized with the emission of RMSNs (red) as an indication that the protein is chemically attached to the material and the combo CTxB-MSN is endocytosed together. The control experiment (RMSNs) is also internalized by HeLa cells, but only showed fluorescence emission in the TRITC-channel (Figs. S3e–h).
LSCM was also used to investigate the intracellular trafficking pathway of the FMSN materials synthesized in this project. Like many other nanomaterials, unmodified and PEGylated MSN materials usually follow the endolysosomal trafficking pathway. As reported in the literature and confirmed by our own experiments, these particles are mainly internalized through the CME mechanism, encapsulated in early endosomes and follow the maturation process until they become late endosomes, and finally ending as lysosomes.7 It has been shown previously that the surface functionalization of MSNs influences whether some of the nanoparticles will escape late endosomes or lysosomes.10 On the contrary, CTxB proteins follow a retrograde pathway, which takes the protein from an early endosome through the trans-Golgi network into to the endoplasmic reticulum.17, 20
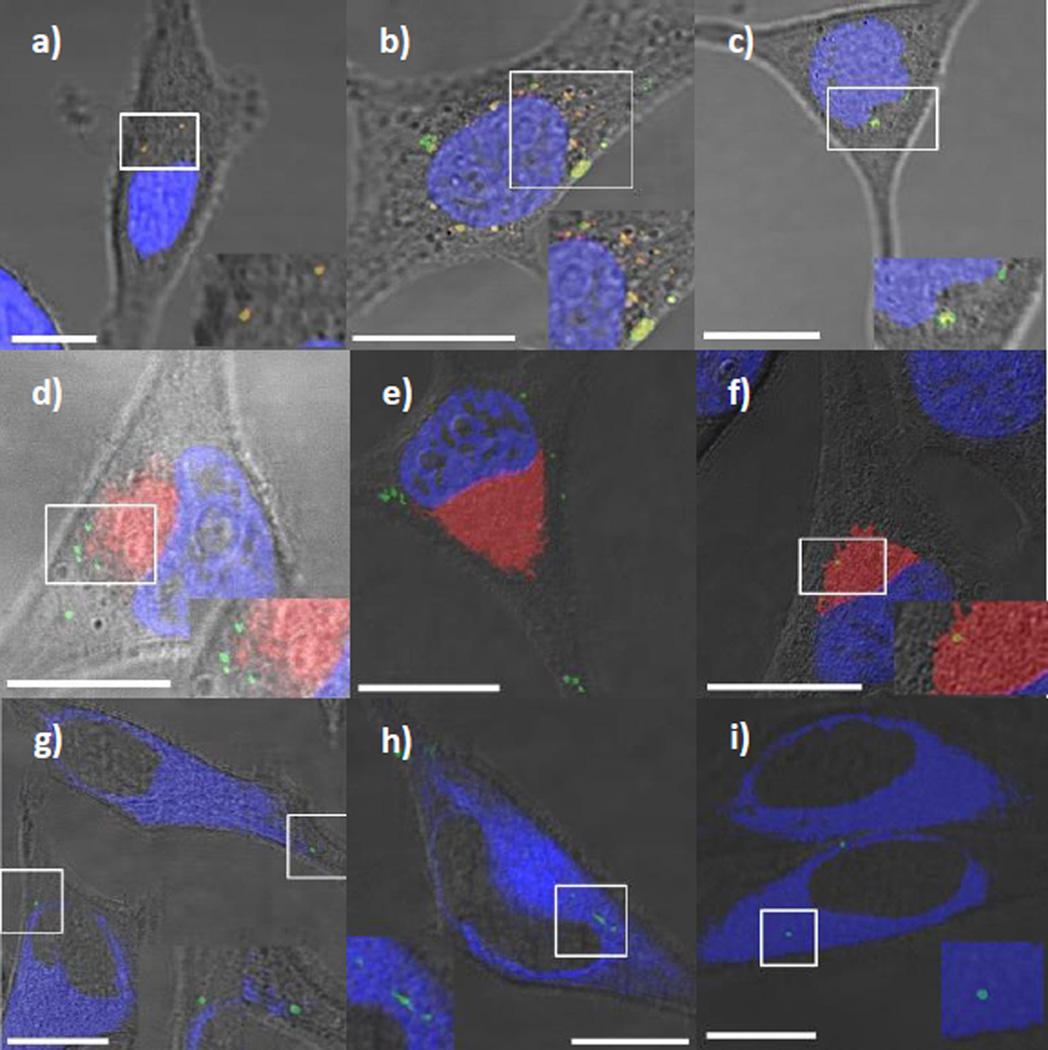
To study this trafficking mechanism, we selectively stained different organelles of HeLa cells, such as the lysosomes, endoplasmic reticulum (ER), Golgi apparatus (GA) and nucleus. Lysosomes were stained with Lysotracker™ Red (DND-99), a fluorescent probe that selectively accumulates in acidic organelles. LSCM images show that FMSNs colocalized exclusively with the lysosome, in agreement with previous reports (Fig. 3a). PEGC-FMSNs were also shown to colocalize with cellular lysosomes following internalization (Fig. 3b). Interestingly, a significantly lower amount of CTxB-FMSNs were found to colocalize with lysosomes, suggesting that this material either escapes endosomes/lysosomes or follows a different route of internalization (Fig. 3c). To determine if CTxB-FMSNs colocalize with the GA organelle, a GA-staining dye was used (BODIPY-Ceramide TR conjugated to BSA™). Ceramides are the biological building blocks of more complex sphingolipids. Metabolism of ceramides typically occurs in GA and ER membranes, and fluorescent ceramide analogs are important probes for measuring the intracellular distribution and transport of the labeled molecules in live cells.39 Ceramide LSCM images show that FMSN and PEGC-FMSN materials do not colocalize with the GA (Fig. 3d–e). Meanwhile, the colocalization of CTxB-FMSNs with the GA was seen to be higher than that observed for the other materials (Fig. 3f). These data confirm that a certain number of CTxB-FMSN particles exist transiently within the GA following uptake.
Fig. 3.
LSCM images of HeLa cells inoculated with FMSNs (a,d,g), PEGC-FMSNs (b,e,h) and CTxB-FMSNs (c,f,i) in the presence of Lysotracker Red (DND-99) (red) (a–c), BODIPY-Ceramide TR conjugated to BSA™ (red) (d–f) and ER-Tracker™ Blue-White DPX (blue) (g–i). All FMSN materials are shown as green spots. The yellow spots result from the colocalization of FMSN materials with either Lysotracker Red (DND-99) or BODIPY-Ceramide TR conjugated to BSA™. The white spots resulted from the colocalization of FMSN materials with ER-Tracker™ Blue-White DPX. Insets on the bottom right corner (left corner for Fig. 3h) of the micrographs show a close-up of the highlighted region in the white square. All the scale bars are 10 µm in size.
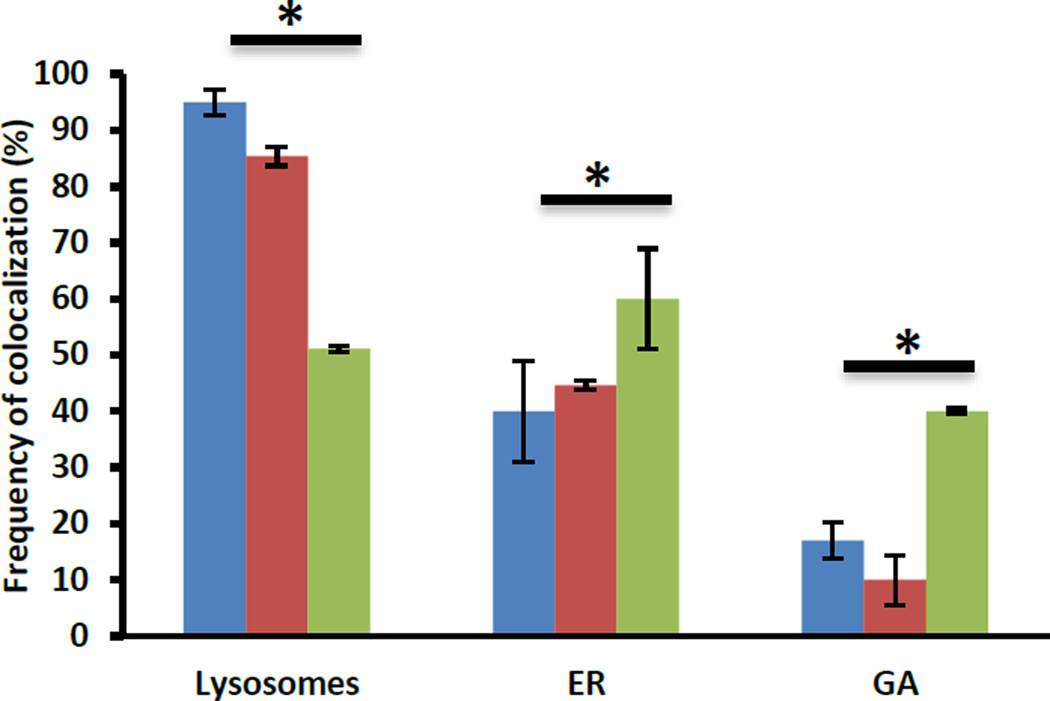
The colocalization of these materials with the ER was studied by labeling this organelle with ER-Tracker™ Blue-White DPX. ER-Tracker dyes are cell-permeant, live-cell stains that are highly selective for the ER.40 LSCM images show that FMSNs, PEGC-FMSNs and CTxB-FMSNs do not exhibit consistent levels of colocalization with the ER (Fig. 3g–h). To confirm our observations from the previous LSCM images, the amount of FMSN material colocalized in specific organelles was determined by following a quantitative analysis on LSCM micrographs.26 Colocalization analyses of single organelle-stained experiments confirmed that both FMSNs and PEGC-FMSNs colocalized exclusively with the lysosome (95.0 ± 2.2% and 85.3 ± 1.7%, respectively) (Fig. 4). Interestingly, a significantly lower amount of CTxB-FMSNs were found to colocalize with lysosomes (51.2 ± 0.5%; p < 0.05). In the case of the GA, the colocalization analyses show that FMSN and PEGC-FMSN materials slightly colocalize with this organelle (17.0 ± 3.1% and 10.0 ± 4.5%, respectively). On the contrary, the colocalization of CTxB-FMSNs with the GA was seen to be significantly higher than that observed for the other materials (40.0 ± 0.5%; p < 0.05). Finally, these analyses show that there is also a statistically significant difference in the colocalization percentage between FMSN, PEGC-FMSN and CTxB-FMSN materials (40.0 ± 8.9%; 44.7 ± 0.9%; and 60.0 ± 8.9%, respectively) and the ER (Fig. 4).
Fig. 4.
Statistical analyses of the frequency of colocalization of FMSN (blue), PEGC-FMSN (red) and CTxB-FMSN (green) materials in lysosomes, GA and ER. Data represent ± SEM of triplicates from 10 independent experiments. Asteriks indicate p < 0.05.
These results suggest that CTxB-FMSNs are taken up by the HeLa cells and partially trafficked through the GA network into to the ER. As mentioned before, it has been estimated that only around 12% of the fraction of CT that enters the cells engages in the retrograde trafficking pathway.21 Therefore, it is expected that only a partial amount of the CTxB-FMSN material will be trafficked through this pathway. Nevertheless, it was recently reported that even an increase of 1–2% in the effective intracellular release of siRNA can make a significant impact in the final therapeutic outcome.41 The fact that the intracellular trafficking of some of the CTxB-MSN particles has been shifted from the endolysosomal to the retrograde pathway, allow us to believe that there is potential for this system to improve the delivery of sensitive cargoes such as therapeutic proteins, siRNA or DNA.
3.5. Intracellular Release of Propidium Iodide
To test the delivery abilities of the CTxB-FMSN material, propidium iodide (PI) was physically loaded into CTxB-FMSNs (PI-loaded CTxB-FMSNs). PI is a DNA intercalating agent and a fluorescent cell membrane-impermeable molecule that is generally excluded from viable cells. PI is commonly used for identifying dead cells in a population and as a counterstain in multicolor fluorescent techniques.42 HeLa cells were inoculated with PI-loaded MSN materials (FMSNs, PEGC-FMSNs and CTxB-FMSNs) for 1 hour and incubated for another 24 hours. To investigate whether the FMSN particles were colocalized with the ER, the cells were stained with ER-Tracker™ Blue-White DPX. For this set of experiments, the fluorescence of PI molecules is shown in the red channel.
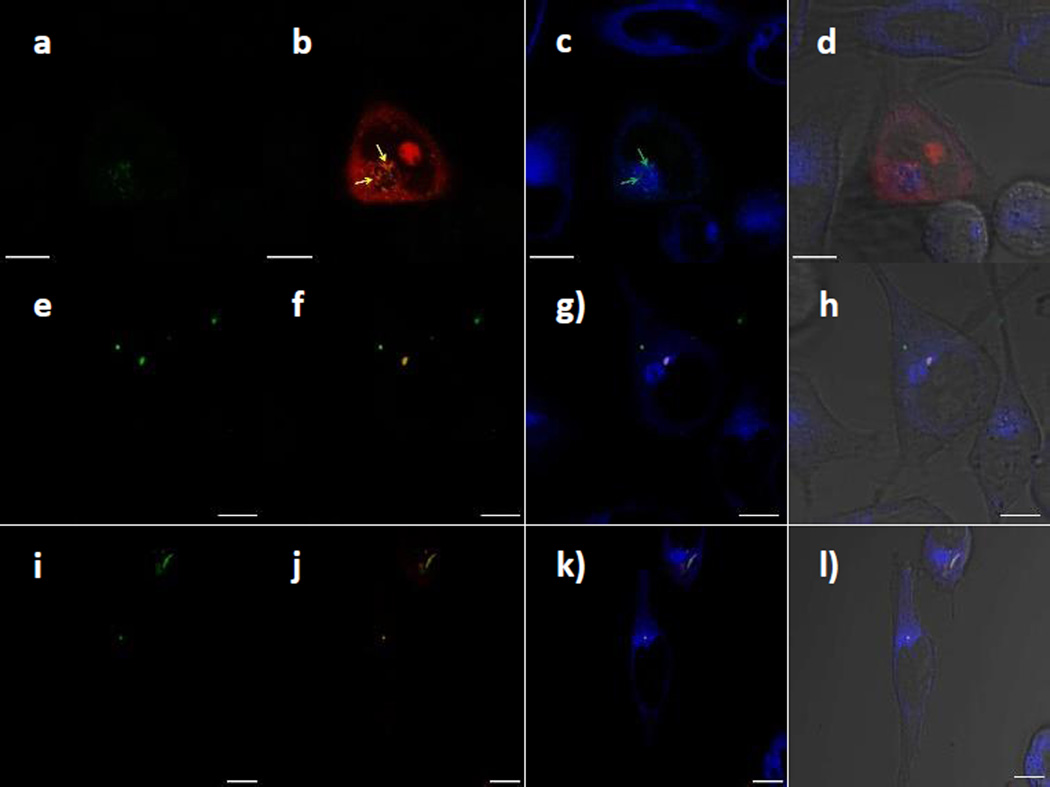
LSCM images show that PI-loaded CTxB-MSNs are internalized by HeLa cells (Fig. 5a) and that the PI dye has been efficiently released inside some of this cells (Fig. 5b). ER staining shows the diffusion of PI molecules colocalized with the ER organelle (Fig. 5c), indicating that the membrane impermeable dye has been trapped in this organelle. Interestingly, some of the PI molecules have diffused to the cell nucleus and localized in what appears to be the nucleolus (Fig. 5d).
Fig. 5.
LSCM images of HeLa cells inoculated with PI-loaded CTxB-FMSNs, FMSNs and PEGC-FMSNs. (a,e,i) CTxB-FMSNs, FMSNs and PEGC-FMSNs (green). (b,f,j) CTxB-FMSNs, FMSNs or PEGC-FMSNs super-imposed with PI molecules (red); yellow arrows in Figure 5b indicate the colocalization of CTxB-FMSNs and PI molecules. (c,g,k) ER-stained images super-imposed with CTxB-FMSNs, FMSNs or PEGC-FMSNs; white arrows in Figure 5c indicate the colocalization of CTxB-FMSNs and the ER organelle. (d,h,l) Super-imposed micrographs of the previous images with the DIC channel. All the scale bars are 10 µm in size.
These results show that CTxB-MSNs have carried PI molecules inside HeLa cells through the retrograde pathway and released them in the ER and cell nucleus. This observation is supported by other LSCM images shown in the supporting information (Fig. S3, ESI). In the case of PI-loaded FMSNs and PEGC-FMSNs, LSCM images show that both are internalized by HeLa cells. However, the intracellular release of the PI molecules from MSN materials was not observed (Fig. 5e–l). The data indicate that these materials are still trapped in the endolysosomal pathway, which prevents the diffusion of PI molecules away from the MSN particles. Overall, these results demonstrate that by modifying the surface of MSNs with CTxB protein, this platform can transport a cell membrane-impermeable molecule through the retrograde trafficking pathway.
4. Conclusions
We have successfully designed and characterized MSNs functionalized with the CTxB protein. The results obtained from the endocytic pathway inhibition assays demonstrated that the endocytosis of CTxB-FMSNs in HeLa cells is facilitated by both clathrin- and caveolin-dependent mechanisms. In addition, LSCM experiments showed that CTxB-FMSNs are taken up by the cells and partially trafficked through the trans-Golgi network into the endoplasmic reticulum in a retrograde fashion. Finally, we successfully demonstrated the subcellular targeted specific release of propidium iodide in the ER and the cell nucleus. All of these results are consistent with the hypothesis that by functionalizing MSNs with cholera toxin subunit B, the internalization and trafficking pathways can be modified. We envision that this novel MSN platform can be used to target and enhance the efficient release of a variety of sensitive cargoes, including therapeutic proteins, DNA and siRNA at the subcellular level.
Supplementary Material
Acknowledgments
We acknowledge financial support from UNC Charlotte (start-up funding and the Junior Faculty Development Award), the Nanoscale Science Program at UNC Charlotte, ORAU Ralph E. Powe Junior Faculty Enhancement Award, and the National Institutes of Health AREA grant 1R15CA192160-01. We thank the Department of Biological Sciences at UNC-Charlotte for allowing us to use their FACS and LSCM facilities, and Mr. Zhiguang Cui for help with some of the TEM images. We are grateful with Dr. Merlis Alvarez-Berrios and Dr. Richard Jew for critical reading of the manuscript and helpful suggestions.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental description for the synthesis of PEGC polymer, CTxB protein quantification, double and triple endocytosis inhibition assays, and Cholera Toxin Subunit B receptor specificity assays are provided in the electronic supporting information. See DOI: 10.1039/x0xx00000x
References
- 1.Giret S, Man MWC, Carcel C. Chem. - Eur. J. 2015;21:13850–13865. doi: 10.1002/chem.201500578. [DOI] [PubMed] [Google Scholar]
- 2.Vivero-Escoto JL, Huxford-Phillips RC, Lin W. Chemical Society Reviews. 2012;41:2673–2685. doi: 10.1039/c2cs15229k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douroumis D, Onyesom I, Maniruzzaman M, Mitchell J. Crit. Rev. Biotechnol. 2013;33:229–245. doi: 10.3109/07388551.2012.685860. [DOI] [PubMed] [Google Scholar]
- 4.Popat A, Hartono SB, Stahr F, Liu J, Qiao SZ, Lu GQ. Nanoscale. 2011;3:2801–2818. doi: 10.1039/c1nr10224a. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Xiao Z, Wu L. Curr. Org. Chem. 2013;17:1325–1333. [Google Scholar]
- 6.Malgras V, Ji Q, Kamachi Y, Mori T, Shieh F-K, Wu KCW, Ariga K, Yamauchi Y. Bull. Chem. Soc. Jpn. 2015;88:1171–1200. [Google Scholar]
- 7.Vivero-Escoto JL, Slowing II, Trewyn BG, Lin VSY. Small. 2010;6:1952–1967. doi: 10.1002/smll.200901789. [DOI] [PubMed] [Google Scholar]
- 8.Argyo C, Weiss V, Braeuchle C, Bein T. Chemistry of Materials. 2014;26:435–451. [Google Scholar]
- 9.Chen Y, Chen H, Shi J. Advanced Materials (Weinheim, Germany) 2013;25:3144–3176. doi: 10.1002/adma.201205292. [DOI] [PubMed] [Google Scholar]
- 10.Slowing I, Trewyn BG, Lin VSY. J. Am. Chem. Soc. 2006;128:14792–14793. doi: 10.1021/ja0645943. [DOI] [PubMed] [Google Scholar]
- 11.He Q, Shi J. Journal of Materials Chemistry. 2011;21:5845–5855. [Google Scholar]
- 12.Morelli C, Maris P, Sisci D, Perrotta E, Brunelli E, Perrotta I, Panno ML, Tagarelli A, Versace C, Casula MF, Testa F, Ando S, Nagy JB, Pasqua L. Nanoscale. 2011;3:3198–3207. doi: 10.1039/c1nr10253b. [DOI] [PubMed] [Google Scholar]
- 13.Shete HK, Prabhu RH, Patravale VB. Journal of Nanoscience and Nanotechnology. 2014;14:460–474. doi: 10.1166/jnn.2014.9082. [DOI] [PubMed] [Google Scholar]
- 14.Tarrago-Trani MT, Storrie B. Advanced Drug Delivery Reviews. 2007;59:782–797. doi: 10.1016/j.addr.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina-Kauwe LK. Advanced Drug Delivery Reviews. 2007;59:798–809. doi: 10.1016/j.addr.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruenberg J, van der Goot FG. Nature Reviews Molecular Cell Biology. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 17.Wernick NLB, Chinnapen DJF, Cho JA, Lencer WI. Toxins. 2010;2:310–325. doi: 10.3390/toxins2030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlandi PA, Fishman PH. Journal of Cell Biology. 1998;141:905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinnapen DJF, Chinnapen H, Saslowsky D, Lencer WI. FEMS Microbiol. Lett. 2007;266:129–137. doi: 10.1111/j.1574-6968.2006.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torgersen ML, Skretting G, Van Deurs B, Sandvig K. J. Cell Sci. 2001;114:3737–3747. doi: 10.1242/jcs.114.20.3737. [DOI] [PubMed] [Google Scholar]
- 21.Guimaraes CP, Carette JE, Varadarajan M, Antos J, Popp MW, Spooner E, Brummelkamp TR, Ploegh HL. Journal of Cell Biology. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty SK, Fitzpatrick JAJ, Phillippi JA, Andreko S, Waggoner AS, Bruchez MP, Ballou B. Nano Letters. 2007;7:2618–2626. doi: 10.1021/nl0709930. [DOI] [PubMed] [Google Scholar]
- 23.Tekle C, van Deurs B, Sandvig K, Iversen T-G. Nano Letters. 2008;8:1858–1865. doi: 10.1021/nl0803848. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Z-A, Zhang L, Guo M, Liu Y, Huo Q. Chemistry of Materials. 2009;21:3823–3829. [Google Scholar]
- 25.Hed J, Hallden G, Johansson SG, Larsson P. J. Immunol Methods. 1987;101:119–125. doi: 10.1016/0022-1759(87)90224-9. [DOI] [PubMed] [Google Scholar]
- 26.Pollock S, Antrobus R, Newton L, Kampa B, Rossa J, Latham S, Nichita NB, Dwek RA, Zitzmann N. FASEB Journal. 2010;24:1866–1878. doi: 10.1096/fj.09-145755. 1810 1096/fj 1809-145755. [DOI] [PubMed] [Google Scholar]
- 27.Bolte S, Cordelieres FP. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 28.He Q, Zhang J, Shi J, Zhu Z, Zhang L, Bu W, Guo L, Chen Y. Biomaterials. 2010;31:1085–1092. doi: 10.1016/j.biomaterials.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 29.Beales PA, Geerts N, Inampudi KK, Shigematsu H, Wilson CJ, Vanderlick TK. J. Am. Chem. Soc. 2013;135:3335–3338. doi: 10.1021/ja311561d. [DOI] [PubMed] [Google Scholar]
- 30.Massol RH, Larsen JE, Fujinaga Y, Lencer WI, Kirchhausen T. Molecular Biology of the Cell. 2004;15:3631–3641. doi: 10.1091/mbc.E04-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araki N, Johnson MT, Swanson JA. The Journal of cell biology. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iversen T-G, Skotland T, Sandvig K. Nano Today. 2011;6:176–185. [Google Scholar]
- 33.Vranic S, Boggetto N, Contremoulins V, Mornet S, Reinhardt N, Marano F, Baeza-Squiban A, Boland S. Part. Fibre Toxicol. 2013;10:2. doi: 10.1186/1743-8977-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Hu L, Yu D, Gao C. Biomaterials. 2010;31:8465–8474. doi: 10.1016/j.biomaterials.2010.07.060. [DOI] [PubMed] [Google Scholar]
- 35.Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, Zhang H, Wang X, Lin S, Huang C, Zhou ZH, Zink JI, Nel AE. ACS Nano. 2011;5:4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao N, Li L, Zhang Q, Huang X, Meng X, Zhang Y, Chen D, Tang F, Li L. Microporous and Mesoporous Materials. 2012;162:14–23. [Google Scholar]
- 37.Conner SD, Schmid SL. Nature (London, United Kingdom) 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Liao L, Zhu L, Zhang P, Guo K, Kong J, Ji C, Liu B. Talanta. 2013;107:408–415. doi: 10.1016/j.talanta.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 39.Cornell-Bell AH, Otake LR, Sadler K, Thomas PG, Lawrence S, Olsen K, Gumkowski F, Peterson JR, Jamieson JD. Methods in Cell Biology. 1993;38:221–240. doi: 10.1016/s0091-679x(08)61005-5. [DOI] [PubMed] [Google Scholar]
- 40.Cole L, Davies D, Hyde GJ, Ashford AE. Journal of Microscopy (Oxford) 2000;197:239–248. doi: 10.1046/j.1365-2818.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- 41.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoeter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Nature Biotechnology. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 42.Moore A, Donahue CJ, Bauer KD, Mather JP. Methods Cell Biol. 1998;57:265–278. doi: 10.1016/s0091-679x(08)61584-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.