Abstract
Fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), is a target species of transgenic corn (Zea mays L.) that expresses single and pyramided Bacillus thuringiensis (Bt) toxin. In 2014, S. frugiperda were collected from a light trap in North Carolina, and a total of 212 F1/F2 isofemale lines of S. frugiperda were screened for resistance to Bt and non-Bt corn. All of the 212 isolines were susceptible to corn tissue expressing Cry1A.105 + Cry2Ab, Cry1F + Cry1A.105 + Cry2Ab, and Cry1F + Cry1Ab + Vip3Aa20. Growth rate bioassays were performed to isolate non-recessive Bt resistance alleles. Seven individuals out of the 212 isofemale lines carried major non-recessive alleles conferring resistance to Cry1F. A pooled colony was created from the seven individuals. This colony was 151.21 times more resistant to Cry1F than a known-susceptible population and was also resistant to Cry1A.105, but was not resistant to Cry2Ab and Vip3Aa20. The results demonstrate that field populations of S. frugiperda collected from North Carolina are generally susceptible to Cry1F, but that some individuals carry resistant alleles. The data generated in this study can be used as baseline data for resistance monitoring.
Introduction
Corn, Zea mays (L.), expressing the Cry1F protein (Event TC1507, Herculex® I insect protection technology by Dow AgroSciences and DuPont Pioneer) was first registered in the United States during 2001. Many primary and secondary lepidopteran pest species, including fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) are targets of this event [1,2]. Field resistance of S. frugiperda to Cry1F corn was observed in Puerto Rico during 2006 and relatively high levels of Cry1F resistance were subsequently reported on this island following these initial observations [3,4]. Cry1F resistant S. frugiperda populations were documented on the U.S. mainland during 2011–2013 [5].
S. frugiperda is a widely distributed and well-known migrant pest of many crops throughout North and South America [6,7], and is a key pest of corn in the lower southeastern US [8]. It is also a sporadic pest of cotton, soybean and other crops, including many vegetables [9]. In North America, this insect has two distinct overwintering populations in Texas/Mexico and southern Florida that migrate north during spring [10,11]. These populations are separated by the Appalachian mountain range and overlap in Alabama/Georgia and in the mid-Atlantic [12,13]. Cry1F resistant S. frugiperda likely developed in 2006 in Puerto Rico [3]; although Cry1F resistant populations of S. frugiperda were not present on the U.S. mainland during 2012 [4], a 2013 study documented the first known Cry1F resistance in this geography. The highest Cry1F resistance ratios were located in Florida and North Carolina, but not in Georgia, Louisiana or Texas (excluding Louisiana and Florida lines isolated from populations using an F2 screen) [5]. In the U.S. mainland north of Florida, S. frugiperda has been effectively managed using corn hybrids expressing Cry1F [14,15]. The number of resistant and susceptible S. frugiperda individuals (frequency of resistant alleles) in the U.S. is unknown. With a migratory insect such as S. frugiperda, frequency of resistance in any location is likely influenced by fitness costs incurred by resistance [16], local selection, and influx of genes from immigrants.
To maintain the effectiveness of Bt corn, the frequency of resistance genes to Bt toxins should be monitored with a method that is appropriate for the pest and specific Bt crop [17,18]. Resistance evolution in field pest populations is complex and is influenced by a number of factors, including the extent of selection pressure exerted by Bt crops, the initial frequency of resistance alleles in the field population, and the migratory behavior of the adults [4,19]. Several methods have been developed to estimate recessive resistance frequencies to Bt corn in target insect pests [5,20–24]. Most methods focus on detecting the frequency of homozygous recessive Bt resistant individuals using specialized bioassays [25,26] and do not estimate the frequency of major non-recessive resistance alleles. Burd et al. [27] developed a bioassay to estimate the frequency of these major non-recessive resistance alleles using isofemale lines of F1/F2 generation. This method is appropriate when resistant alleles are not rare in the population.
One tactic that is important for maintaining insecticide resistance management is pyramiding Bt toxins, especially with pyramids that express proteins with dissimilar modes action, but that are effective against the same target pests [28]. Pyramided Bt corn hybrids (second generation) are being planted more frequently and are more effective for S. frugiperda management than single Bt protein hybrids (first generation) [14,15,28,29]. One reason pyramided Bt maize products are becoming more prevalent in North and South America is to manage Cry1F- resistant populations of S. frugiperda. Since resistance in North Carolina was detected during 2013 [5], we carried out experiments using the F1/F2 screening method that was proposed by Burd et al. [27] to monitor the frequency of non-recessive resistance alleles and larval susceptibility of S. frugiperda to Bt corn containing single or pyramided genes from eastern North Carolina, USA.
Materials and Methods
Bioassay of F1 generation on Bt and non-Bt corn tissue
During August through October 2014, a total of 400 S. frugiperda adult females were collected from a light trap at the Vernon G. James Research and Extension Center (N35.8750, W76.6606) in Plymouth, North Carolina, USA. Moths were individually placed into 350-ml clear plastic cups, covered with gauze, as oviposition substrate. Cups were kept at 27 ± 1°C, 70–80% RH, and L:D 14:10, and eggs were collected on a daily basis. If the moth produced more than 70 eggs daily, then the line was used in bioassays. We obtained sufficient eggs from 212 moths to use in bioassays.
The susceptibility of S. frugiperda was first evaluated on leaf tissue from two non-Bt and four Bt corn hybrids expressing single and pyramided traits (Table 1). Fully expanded leaf tissue of Bt and non-Bt corn hybrids was excised from greenhouse-grown V4–V10 stage plants. Pilot experiments indicated that larval response (growth and mortality) to Bt in leaf tissue of this age range was equivalent. Presence of Bt in the plants was confirmed using QuickStix kits (EnviroLogix, Portland, ME, data not presented), although each single transgenic leaf used in the study was not individually tested. In the bioassay, 2–3 pieces of leaf tissue from a single hybrid were placed in each well of a 32 well rearing tray (Frontier Agricultural Sciences, Newark, NJ). A single neonate (0–24 h old) was then placed into the well, with every S. frugiperda line tested on leaves from all corn hybrids. The total number of replicates for each S. frugiperda and hybrid combination was 32. Trays containing leaf tissues and neonates were placed in growth chambers maintained at 27°C, 70–80% RH, and L:D 14:10. After seven days, larval mortality was recorded, with larvae considered dead if they did not respond after being touched with a camel hair brush. The developmental stage of each surving larva was assessed using head capsule and body size as indicators; all growth stage values were converted to an ordinal ranking system where 0 = dead, 1 = first instar, 2 = early second, 3 = mid second, 4 = late second, 5 = early third, 6 = mid third, 7 = late third, 8 = early fourth, 9 = mid fourth. The weight of surviving larvae was also measured.
Table 1. Hybrids used to screen S. frugiperda in this experiment and the associated lepidopteran-specific Bt proteins.
| Trade name | Hybrid | Lepidopteran-specific insertion event | Lepidopteran-specific Bt proteins |
|---|---|---|---|
| Non-Bt1 | P1319Ra | - | - |
| Herculex®I | P1319HRa | TC1507 | Cry1F |
| Optimum® Leptra® | P1319VYHRa | TC1507 + MON810 + MIR162 | Cry1F + Cry1Ab + Vip3Aa20 |
| Non-Bt2 | DKC6482Rb | - | - |
| Genuity®VT Double Pro™ | DKC6489VT2Pb | MON89034 | Cry1A.105 + Cry2Ab |
| Genuity® Smart-Stax™ | DKC6487SSb | TC1507 + MON89304 | Cry1F + Cry1A.105 + Cry2Ab2 |
aDuPont Pioneer, Johnston, IA
bMonsanto Company, St. Louis, MO
Bioassay of F2 generation on Bt and non-Bt corn tissue
In order to determine the relationship between the F1 and F2 generation when feeding on Bt corn, F1 lines with a survivorship ≥50% were saved for testing during the F2 generation. F1 lines that developed ≥80% as well on Bt as they did on non-Bt corn were also saved for testing during the F2 generation. We hypothesized that F1 lines that developed relatively well on Bt corn carried at least one major non-recessive resistance allele in heterozygous form [30]. From each line, larvae that developed on non-Bt corn were reared to adult emergence and sib-mated. Resulting F2 neonates were subject to identical leaf tissue bioassays as described above. In this study, F2 larvae of 26 lines were tested using corn tissue expressing Cry1F, Cry1A.105 + Cry2Ab2 + Cry1F, and Cry1Ab + Cry1F + Vip3Aa20.
Confirmation of resistance to Cry1F protein and other Bt protein resistance
The number of individuals with major resistance genes in a given population is expected to be low, and heterozygotes are the most probable carriers of resistance alleles in field populations [25]. Even if a female carries a major dominant resistance gene, the mean growth rate of her F1 offspring on corn expressing Bt should still be reduced compared to what it would be on non-Bt corn. Therefore, we assumed that if the relative average development ratings of any F1 line and the resulting F2 lines producted from those populations were ≥0.8 (rather than 1.0), and the corrected survival rate was ≥50% on Bt corn, then the line carried a major resistance gene [30].
Seven lines were sourced from the F2 screening that had a relative average developmental rating of ≥0.8 on leaves expressing Cry1F; these lines were then pooled in the F3 generation. The protein Cry1F was provided by Dow AgroSciences (Indianapolis, IN) as a gift and stored in a dessicator at -20°C. The proteins Cry2Ab2 and Cry1A.105 were provided by Monsanto Company as a gift and stored at -80°C. Finally, the protein Vip3Aa20 was provided by Syngenta Crop Protection (Greensboro, NC) and stored in a freezer at -20°C. Larval susceptiblitity of the pooled F3 line was assayed using a meridic diet overlay procedure in 128-cell plastic trays with a 2 cm2 surface area (Frontier Agricultural Sciences). The meridic diet (WARD’S Stonefly Heliothis diet, Rochester, NY) rearing procedure followed Niu et al. [29]. One ml of Heliothis diet was dispensed into each 2 cm2 well and allowed to solidify. In the bioassay, ten concentrations, plus a control, were serially diluted using CAPS buffer. Forty μl of the formulated solution or control was overlaid on the diet using a pipette; control treatments were composed of both distilled water and buffer. When all of the wells within a tray were treated, each tray was tilted from left to right and front to back to ensure that the liquid sample completely coated the surface of the diet. After no liquid was visible on the diet surface, one S. frugiperda neonate larvae (0–24 h old) was added to each well using a fine brush. The trays were sealed with self-adhesive plastic sheets (BIO-CV-16, CD International Inc.) and placed in a climatic chamber (27±1°C, 60±10% relative humidity, and L:D 14:10h). The bioassays were repeated three times for each population, with each concentration repeated three times per bioassay (total of three replications of 16 neonates/concentration). Mortality (with larvae that remained as first instars throughout the experiment also considered dead) and the weight of surviving larvae were measured at seven days after treatment as described before. A Bt-susceptible colony, of S. frugiperda, originally obtained from non-Bt maize in Hidalgo Co., TX (SS-TX) [5], was used as a control.
Data analysis
To control for larval vigor effects, the growth rates of each line were compared on a Bt and a non-Bt hybrid from the same genetic background. Growth stage values were converted to an ordinal ranking system as described before. There were two hybrid groups: 1) P1319HR (Herculex®I), P1319VYHR (Optimum® Leptra®), P1319R (related non-Bt, non-Bt1), and 2) DKC64-89 (Genuity®VT Double Pro™), DKC64-87 (Genuity® Smart-Stax™), and DKC64-82 (related non-Bt, non-Bt2). The average developmental rating was defined as the the average ordinal ranking for a single female on each of the hybrids. The relative average developmental rating for a given line was calculated as the average developmental rating of a specific iso-female S. frugiperda line on given hybrid divided by the average developmental rating of the same iso-female S. frugiperda line on a given hybrid [31]. The corrected percentage survival was obtained using the Abbott’s formula [32]. Pearson’s correlation analysis was used to compare the average developmental rating, the mass weight and the survival rate of a given iso-female line to Bt and non-Bt corn hybrids, or to compare the relative average developmental rating and corrected survival rate of larvae of a given F1 generation with the F2 generation [33]. LC50 (GIC50) or LC90 values were obtained and compared using POLO-Plus [34], with significance defined as non-overlapping confidence limits.
Results
Bioassays on F1 generation test
A total of 212 female lines (isofemales) were screened on leaves expressing Cry1F. The relative average developmental ratings for most lines on Cry1F ranged from 0.00 (dead) to 1.00 with a mean relative average developmental rating of 0.15 (Fig 1a, Table 2). Twenty-six out of 212 lines had a corrected survival rate ≥50% and were saved for testing the F2 generation (Table 2). Among the 26 lines, there were eight lines with a relative average developmental rating ≥0.80 (Table 2).
Fig 1. Distribution of the relative average development rating for seven-day old F1 larvae of S. frugiperda female lines on corn leaf tissue expressing Cry1F (A) or Cry1A.105 + Cry2Ab (B).
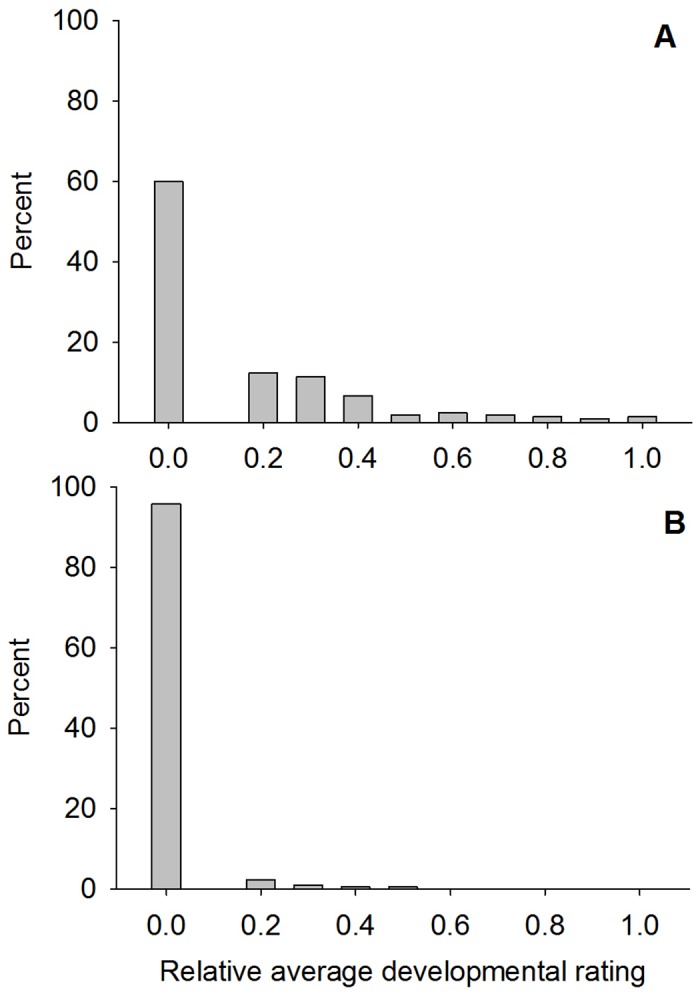
Table 2. Estimated resistance allele frequency for major non-recessive resistance alleles with Bayesian 95% credibility intervals for field-collected S. frugiperda individual female lines (n = 212) to Bt corn hybrids.
| Protein expressed in leaf tissue | F1 generation bioassay | F2 generation bioassay | Estimated r allele frequency (95% CI)b | |||||
|---|---|---|---|---|---|---|---|---|
| Number tested | Relative average development rating per line Mean (±SE) | % Corrected survival ≥50% | Number with relative average developmental rating ≥0.8 | Number tested | Relative average development rating per line Mean (±SE) | Number with relative average developmental rating ≥0.8a | ||
| Cry1F | 212 | 0.15 ± 0.02 | 26 | 8 | 26 | 0.67 ± 0.03 | 7 | 0.009346 (0.004048–0.016795) |
| Cry1A.105 + Cry2Ab | 212 | 0.01 ± 0.00 | 0 | 0 | - | - | - | <0.001168 (0.00003–0.004305) |
| Cry1F + Cry1A.105 + Cry2Ab2 | 212 | 0.00 ± 0.00 | 0 | 0 | 26 | 0.00 ± 0.00 | 0 | <0.001168 (0.00003–0.004305) |
| Cry1F + Cry1Ab + Vip3Aa20 | 212 | 0.00 ± 0.00 | 0 | 0 | 26 | 0.00 ± 0.00 | 0 | <0.001168 (0.00003–0.004305) |
aMoth was regarded as an individual with a resistance allele if, during the F1 and F2 generation, the relative average development ratings were ≥0.8.
bEach mated female carries two of her own alleles and two from her male counterpart (if she mated only once); by screening females on Bt corn to characterize (4 × total number) the genome, the estimated frequency for the resistance allele to each Bt hybrid would be: (the number of individual with resistance allele + 1) / 4 × (total number + 2)[26].
A total of 212 female lines (isofemales) were screened on leaves expressing Cry1A.105 + Cry2Ab2. The relative average developmental ratings for most lines on Cry1A.105 + Cry2Ab2 ranged from 0.00 (dead) to 0.50 with a mean relative average developmental rating of 0.01 (Fig 1b, Table 2). None of the 212 female lines had a relative average developmental rating ≥0.80 or a corrected surviorship ≥50% (Table 2). No larvae survived on leaves expresing Cry1F + Cry1A.105 + Cry2Ab2 or Cry1F + Cry1Ab + Vip3A; hence, relative average development ratings were 0.00 for these Bt pyramids.
Average development ratings were not correlated between leaves expressing Cry1F and leaves from a related non-Bt hybrid, or between leaves expressing Cry1A.105 + Cry2Ab2 and leaves from a related non-Bt hybrid (Table 3). Similarly, relative average development ratings were not correlated between leaves expressing Cry1F and leaves expressing Cry1A.105 + Cry2Ab2. Furthermore, there was no correlation for survival between leaves expressing Cry1F, or Cry1A.105 + Cry2Ab2, and leaves from their respective related non-Bt hybrids; finally, there was no correlation between corrected survival between leaves expressing Cry1F and leaves expressing Cry1A.105 + Cry2Ab2 (Table 3).
Table 3. Pearson correlation values of average developmental ratings, relative average developmental ratings, percent survival (corrected using Abbot’s formula [32]), and larval weight (after seven days) of F1 S. frugiperda lines selected on leaves expressing Bt and non-Bt leaves of closely related hybrids.
* = P <0.05, ** = P <0.001.
| Correlation | r | P |
|---|---|---|
| Average developmental rating on non-Bt1 and Cry1F | 0.0142 | 0.8374 |
| Average developmental rating on non-Bt2 and Cry1A.105 + Cry2Ab2 | -0.0526 | 0.4465 |
| Relative average developmental rating on Cry1F and Cry1A.105 + Cry2Ab2 | 0.0962 | 0.1629 |
| Percent survival on non-Bt1 and Cry1F | 0.0614 | 0.3735 |
| Percent survival on non-Bt2 and Cry1A.105 + Cry2Ab2 | 0.0641 | 0.3529 |
| Percent corrected survival on Cry1F and Cry1A.105 + Cry2Ab2 | -0.0201 | 0.7715 |
| Percent survival and average developmental rating on non-Bt1 | 0.0781 | 0.2577 |
| Percent survival and average developmental rating on Cry1F | 0.7522** | <0.0001 |
| Percent survival and average developmental rating on non-Bt2 | 0.1533* | 0.0256 |
| Percent survival and average developmental rating on Cry1A.105 + Cry2Ab2 | 0.7518** | <0.0001 |
| Larval weight on Cry1F and Cry1A.105 + Cry2Ab2 | -0.0379 | 0.5828 |
| Percent survival and larval weight on non-Bt1 | 0.2516** | 0.0002 |
| Percent survival and larval weight on Cry1F | 0.5402** | <0.0001 |
| Percent survival and larval weight on non-Bt2 | 0.5310** | <0.0001 |
| Percent survival and larval weight on Cry1A.105 + Cry2Ab2 | 0.8047** | <0.0001 |
| Average developmental rating and larval weight on non-Bt1 | 0.4552** | <0.0001 |
| Average developmental rating and larval weight on Cry1F | 0.4986** | <0.0001 |
| Average developmental rating and larval weight on non-Bt2 | 0.3224** | <0.0001 |
| Average developmental rating and larval weight on Cry1A.105+ Cry2Ab2 | 0.8808** | <0.0001 |
There was a correlation between average developmental rating and survival on leaves expressing Cry1F, larval weight and survival on leaves expressing Cry1F, and average development rating and larval weight on leaves expressing Cry1F (Table 3). In contrast, there was no correlation between average development rating and survival on non-Bt leaves from the same hybrid family as the Cry 1F leaves (non-Bt1). Larval weight was correlated with survival, as was larval weight and average developmental rating, on non-Bt leaves from the same hybrid family as the Cry1F leaves (non-Bt1).
Average developmental rating was correlated with survival, as well as larval weight on leaves expressing Cry1A.105 + Cry2Ab2 (Table 3). Similarly, these factors were correlated for larvae that developed on non-Bt corn leaves from the same hybrid family as the Cry1A.105 + Cry2Ab2 leaves (non-Bt2).
Bioassay on F2 generation test
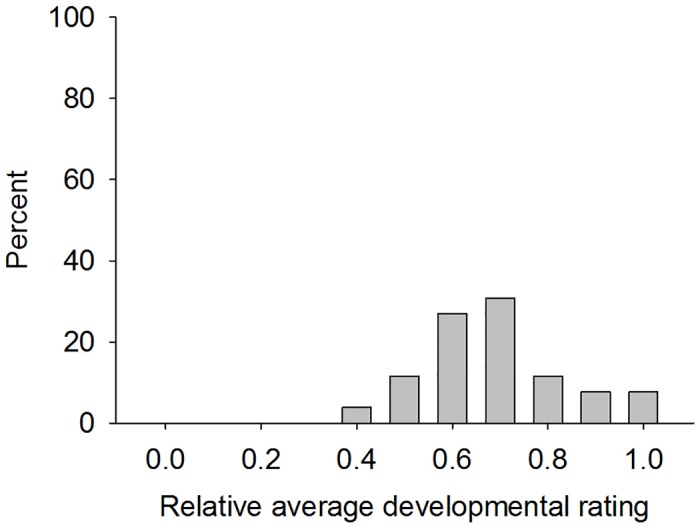
A total of 26 isofemale lines that survived to the F2 generation were screened on corn leaves expressing Cry1F, Cry1F + Cry1A.105 + Cry2Ab2, and Cry1F + Cry1Ab + Vip3A20. The distribution of the relative average developmental ratings on Cry1F leaves of seven-day old larvae from all these lines on Cry1F is presented in Fig 2. These relative average development ratings for most lines on Cry1F ranged from 0.40 to 1.0 with a mean relative average development rating of 0.67 (Table 2). No larvae survived on Cry1F + Cry1A.105 + Cry2Ab2 and Cry1F + Cry1Ab + Vip3A; consequently relative average development ratings were 0.00.
Fig 2. Distribution of the relative average development rating for seven-day old F2 larvae of S. frugiperda female lines on corn leaf tissue expressing Cry1F.
There was a positive correlation between relative average development ratings of these 26 lines between the F1 and F2 generation on Cry1F leaves (r = 0.4323, P = 0.0274, Fig 3a). There was also a positive correlation between corrected seventh day survival of these 26 lines between the F1 and F2 generation on Cry1F leaves (r = 0.5022, P = 0.0089, Fig 3b). Since these results indicate a genetically-based variation in response to Cry1F, the frequency of major resistance alleles to this protein was calculated as 0.009346 (Table 2).
Fig 3. Correlations between the relative average developmental ratings (A) and corrected percent survival (B) for S. frugiperda lines during the F1 generation and their offspring during the F2 generation.
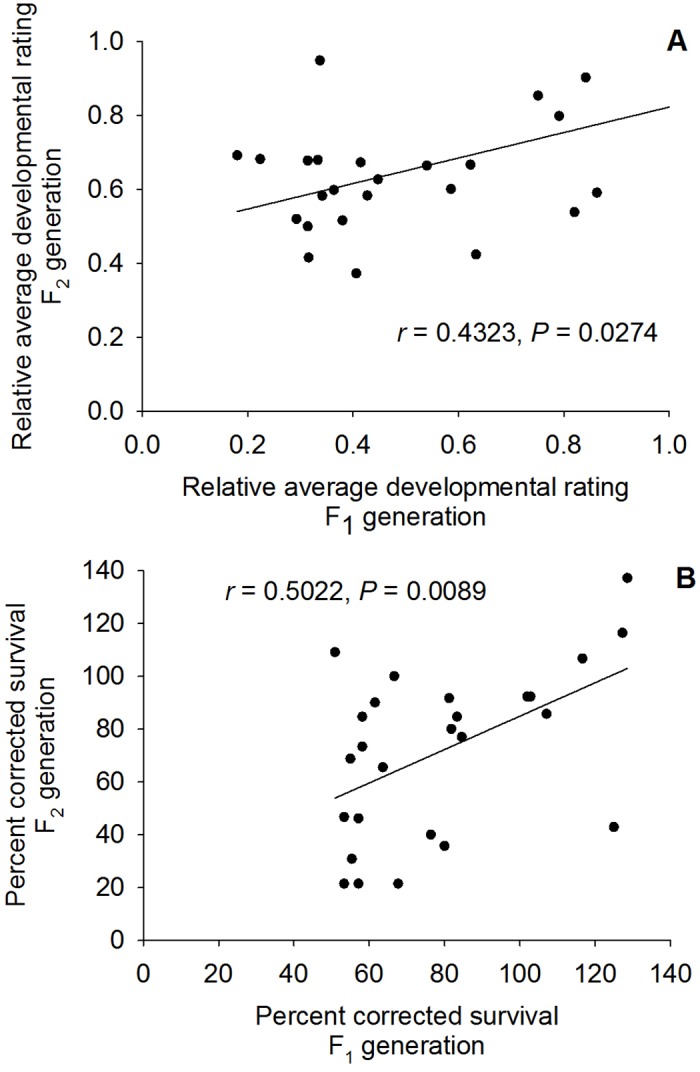
Larvae were reared on corn tissue expressing Cry1F. Pearson correlation results reported as r and P values.
Confirmation of resistance to Cry1F protein and other Bt protein resistance
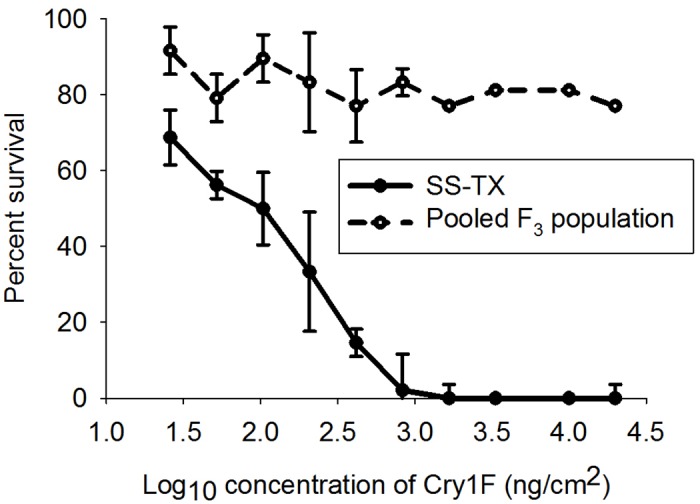
There were seven F2 generation lines with relative average developmental ratings ≥0.8 on Cry1F leaf tissue; this relative average developmental rating was used as a threshold, above which the development of these larvae on Cry1F leaf tissue was considered to be substantially higher compared to other lines in the F2 generation (Table 2). The seven lines with a relative average developmental rating ≥0.8 on Cry1F leaves in the F2 generation were pooled, mated, and tested for their resistance level to Cry1F, Cry2Ab2, Cry1A.105, and Vip3Aa20 protein during the F3 generation (pooled population). The corrected percent surviorship after seven days of a Cry1F-susceptible control line (SS-TX) was zero at 1667 ng/cm2, while the corrected surviorship after seven days of the pooled-population was 78.72% at the same concentration of protein (Fig 4). The LC50 of the SS-TX population to Cry1F was 131.601 ng/cm2. No significant larval mortality (≤21.28%, corrected mortality) of the pooled population was observed across any of the Cry1F concentrations assayed; thus, the LC50 value of this population was estimated to be greater than the highest Cry1F concentrations tested (>19900 ng/cm2), which corresponded to a resistance ratio of >151.21.
Fig 4. Concentration-mortality response (ng/cm2) of S. frugiperda neonates of a known susceptible population (SS-TX) and a population selected on Cry1F corn leaf tissue (pooled F3) to a diet overlay assay using Cry1F protein.
The LC50 of the SS-TX population to Cry1A.105 was 0.241 ng/cm2, 4.4 times lower than that of the pooled population (1.070 ng/cm2). Since the 95% confidence limits overlapped, this difference was not significant. In contrast, the confidence limits of the LC90 values did not overlap. The LC90 of the SS-TX population was 30.5 times lower (1.292 ng/cm2) than that of the pooled population (39.382 ng/cm2, Table 4). The growth inhibition curve after seven days (GIC50), was 0.027 ng/cm2 with 95% CL of 0.004–0.072 for the SS-TX population and 0.880 ng/cm2 with 95% CL of 0.306–3.426 for the pooled population. This indicates that the pooled population was 32.59 times more resistant to Cry1A.105 compared to the known Cry1F susceptible control population.
Table 4. Concentration-mortality response (ng/cm2) of S. frugiperda neonates of a known susceptible population (SS-TX) and a population selected on Cry1F corn leaf tissue (pooled F3) to diet overlay assays using Cry1F, Cry2Ab2, Cry1A.105 and Vip3Aa20 proteins.
SE = standard error; CL = confidence limits; LC50 = concentration of protein (ng/cm2) required to kill 50% of larvae in the observation period of seven days; LC90 = concentration of protein (ng/cm2) required to kill 90% of larvae in the observation period of seven days.
| Protein | Population | n | Slope ± SE | LC50 (95% CL)a | LC90 (95% CL)a |
|---|---|---|---|---|---|
| Cry1F | SS-TX | 336 | 1.986 ± 0.370 | 131.601 (66.591–193.520) | 581.459 (417.808–950.041) |
| Pooled F3 | 512 | 0.146 ± 0.088 | >19900 | - | |
| Cry2Ab2 | SS-TX | 336 | 1.851 ± 0.441 | 160.351 (35.346–263.473)a | 790.135 (491.267–3178.453) a |
| Pooled F3 | 336 | 1.135 ± 0.401 | 408.462 (93.176–836.623)a | 5487.257 (1924.345–906567.387)a | |
| Cry1A.105 | SS-TX | 336 | 1.759 ± 0.410 | 0.241 (0.079–0.420)a | 1.292 (0.781–2.951)a |
| Pooled F3 | 336 | 0.818 ± 0.181 | 1.070 (0.205–3.144)a | 39.382 (9.896–2456.091)b | |
| Vip3Aa20 | SS-TX | 336 | 2.328 ± 0.469 | 156.496 (87.152–217.960)a | 555.817 (404.762–945.182)a |
| Pooled F3 | 336 | 1.376 ± 0.235 | 33.913 (6.172–71.768)b | 289.402 (145.516–1015.551)b |
aValues designated by different letters within a column are significantly different from each other. Values were significant when 95% fiducial limits did not overlap.
The LC50 and LC90 response to Cry2Ab was not different between the SS-TX and pooled populations. A significant difference in mortality response was observed to Vip3Aa20 between the two populations in both the LC50 and the LC90 (Table 4); the pooled population was more sensitive to Vip3A20 than SS-TX population (sensitivity ratio = 4.61).
Discussion
In this study, we used the F1/F2 larvae of isofemale lines to test the frequency of non-recessive resistance alleles of S. frugiperda collected in North Carolina. The bioassays of F1/F2-generation individuals used in this study were specifically designed to test the fitness of individuals with resistance genes, to detect the presence of dominant resistance genes in the heterozygous form, or to detect the presence of recessive resistance genes in the homozyous form. A total of 212 isofemale lines were successfully screened on leaves expressing Cry1F; of these, seven individuals were identified as carrying major alleles conferring resistance to Cry1F. The major resistance allele frequency to Cry1F was 0.009346. According to the categories and patterns of field-evolved resistance to Bt crops described by Tabashink et al. [35], the resistance level of S. frugiperda to Cry1F in this location could be characterized as an early warning of resistance, as 3% of the individuals were resistant, with good development on Cry1F corn leaves compared to non-Bt. The relatively low frequency of resistance alleles in this single location also confirmed the observations of previous investigators, who characterized corn hybrids expressing Cry1F as effective to manage S. frugiperda in the mainland US north of Florida [14, 15]. It should be noted that this estimation is conservative because it did not estimate the frequency of recessive alleles in the heterozygous form.
In addition, we did not detect resistance to Bt corn pyramids expressing Cry1A.105 + Cry2Ab, Cry1F + Cry1A.105 + Cry2Ab, and Cry1F + Cry1Ab + Vip3Aa20. Pyramided Bt crops can delay the evolution of resistance by producing two or more distinct toxins that kill the same pest, although the effect of the pyramid is reduced when there is cross resistance [36] and there is resistance to one or more of the toxins in the pyramid [37]. Our findings of increased lethal concentration values to Cry1A.105 protein in the population selected on Cry1F leaves (F3 pooled population) compared to a known Cry1F susceptible population (SS-TX) indicate that, based on the current available pyramided Bt corn hybrids, pyramiding without consideration for cross-resistance may not be the best tactic to delay the development of resistance. However, we did not clearly show cross resistance, as there were no correlations between Cry1F and Cry1A.105 + Cry2Ab2 for relative average developmental ratings, percent survival, and larval weight (after seven days) of F1 S. frugiperda lines. Furthermore, diverse mechanisms of resistance to Bt toxins and diverse mutations of resistance alleles associated with Bt toxins have been reported for many insect species [38–43]. Hence, the genetic basis for resistance to pyramided Bt crops may involve multiple loci. As a result, the method proposed by Andow and Alstad [26] to estimate the frequency of resistance may overestimate this frequency, since it is based on the assumption of a single resistant allele. A appropriate method should be developed to estimate the frequency of resistant alleles to Bt crop pyramids in the future.
The most probable carrier for a resistance gene in field populations is a heterozygote, since homozygous resistant individuals are rarer. Also the most probable mating is between heterozygote and homozygote susceptible individuals [44]. Thus, the most likely offspring from this cross would be one-half heterozygote and one-half homozygote susceptible. Hence, the Cry1F resistance gene we observed is dominant or incompletely dominant, since 26 out of the 212 isofemale lines had a corrected survival ≥ 50% and relative average developmental rating ≥ 0.8 on Cry1F leaves in the F1 generation. Results in this paper are not consistent with those reported previously [3,18,23], which indicated that the Cry1F resistance gene of S. frugiperda in Puerto Rico was recessive or incompletely recessive. Likely the inheritance of resistance to Cry1F in populations of S. frugiperda is complex.
In our study, resistance was characterized by F1/F2 screening using corn leaves expressing Cry1F. After the F2 generation, seven resistant individuals were pooled and their offpsring were assayed in the F3 generation. These displayed a >151.21-fold resistance to Cry1F compared to a known susceptible population. Based on the growth inhibition bioassay, the selected pooled population also displayed a 32.59-fold resistance to Cry1A.105. These results are consistent with a previous study which demonstrated resistance to both Cry1F and Cry1A.105 in a Florida S. friguperda population [5]. Since both Cry1F and Cry1A.105 have a high affinity and compete for the same binding sites, cross-resistance is likely between these two proteins [45]. Furthermore, Cry1F-resistant S. frugiperda was susceptible to Cry2Ab and Vip3Aa20, consistent with populations tested from Florida [5] and Puerto Rico [23]. Since these proteins do not share midgut binding sites, cross-resistance among these toxins would be unexpected [23,45,46]. However, the population selected on Cry1F leaves (pooled population in this study) was more susceptible to Vip3Aa20 than a known Cry1F susceptible population (SS-TX). The explanation for this finding is unclear. It is possible that there is antagonism or negative cross-resistance between Cry1F and Vip3Aa20. Another possibility is that there is inherent variation in susceptibility to Vip3Aa20 between these two strains that are from two geographically distinct and genetically distinct populations [13].
Nonetheless, our results also suggest that deploying Cry2Ab and Vip3Aa20 alone or in a pyramid is an effective tactic to manage S. frugiperda in the southern US. Moreover, the high frequency of Cry1F resistant S. frugiperda populations in Puerto Rico, the tropical climate, the year-round cultivation of maize, extensive prior use of Bt as an insecticidal foliar spray, abundant pest populations, drought conditions, and minimal use of non-Bt refuge are likely the main factors that led to resistance evolution in S. frugiperda [3,4, 24]. Most of these conditions are not present in the North Carolina environment, with the exception of short-term drought and minimal use of non-Bt refuge. Huang et al. [5] speculated that because S. frugiperda is a polyphagous insect with a wide host range, selection pressure in North Carolina does not appear to be a major factor driving the development of field resistance. In our North Carolina study, we documented resistance allele frequency to Cry1F as 0.009346. It is unclear whether this resistance is a result of immigrants from other areas or from local selection in North Carolina. More work should be done to document the host range of this insect and the interplay of local movement and long-range dispersal to improve resistance management in Bt crops.
Acknowledgments
We thank Dow AgroSciences for providing Cry1F protein, Monsanto for providing Cry2Ab2 and Cry1A.105 protein, and Syngenta for providing Vip3Aa20 protein. We would like to acknowledge the help of Emily Goldsworthy and Donna Michel for assistance in the laboratory and the research staff at the Vernon James Center in Plymouth for assisting with greenhouse cultivation of the corn plants. This research was supported by ad-hoc funding of the Reisig laboratory, and the Key project for Breeding Genetically modified Organisms (No. 2014ZX08012-004, 2016ZX08012-004), China scholarship Council (No. 2011841039) and Science-Technology Foundation for Outstanding Young Scientists of Henan Academy of Agricultural Sciences (Grant no. 2016YQ15).
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by ad-hoc funding of the Reisig laboratory, and the Key project for Breeding Genetically modified Organisms (No. 2014ZX08012-004, 2016ZX08012004-007), China scholarship Council (No. 2011841039) and Science-Technology Foundation for Outstanding Young Scientists of Henan Academy of Agricultural Sciences (Grant no. 2016YQ15). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siebert MW, Babcock JM, Nolting S, Santos AC, Adamczyk JJ, Neese PA Jr., et al. Efficacy of Cry1F insecticidal protein in maize and cotton for control of fall armyworm (Lepidoptera: Noctuidae). Fla Entomol. 2008; 91: 555–565. [Google Scholar]
- 2.Siebert MW, Tindall KV, Leonard BR, Van Duyn JW, Babcock JM. Evaluation of corn hybrids expressing Cry1F (Herculex® I Insect Protection) against fall armyworm (Lepidoptera: Noctuidae) in the southern United States. J Entomol Sci. 2008; 43: 41–51. [Google Scholar]
- 3.Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW, et al. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J Econ Entomol. 2010; 103: 1031–1038. [DOI] [PubMed] [Google Scholar]
- 4.Storer NP, Kubiszak ME, King JE, Thompson GD, Santo AC. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J Invert Pathol. 2012; 110: 294–300. [DOI] [PubMed] [Google Scholar]
- 5.Huang F, Qureshi J CA, Meagher RL, Reisig DD, Head GP, Andow DA, et al. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. PLoS ONE. 2014; 10.1371/journal.pone.0112958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparks AN. A review of the biology of the fall armyworm. Fla Entomol. 1979. 62: 82–87. [Google Scholar]
- 7.Mitchell ER, McNeil JN, Westbrook JK, Silvain JF, Lalanne-Cassou B, Chalfant RB, et al. Seasonal periodicity of fall armyworm (Lepidoptera: Noctuidae) in the Caribbean basin and northward to Canada. J Entomol Sci. 1991; 26: 39–50. [Google Scholar]
- 8.Buntin GD. A review of the plant response to fall armyworm, Spodoptera frugiperda (J. E. Smith), injury in selected field and forage crops. Fla Entomol. 1986; 69: 549–559. [Google Scholar]
- 9.Ali A, Luttrell RG, Pitre HN, Davis FM. Distribution of fall armyworm (Lepidoptera: Noctuidae) egg masses on cotton. Environ Entomol. 1989; 18: 881–885. [Google Scholar]
- 10.Pair SD, Raulston JR, Rummel DR, Westbrook JK, Wolf WW, Sparks AN, et al. Development and production of corn earworm and fall armyworm in the Texas High Plains: Evidence for reverse fall migration. Southwest Entomol. 1987; 12: 89–99. [Google Scholar]
- 11.Mitchell ER, McNeil JN, Westbrook JK, Silvain JF, Lalanne-Cassou B, Chalfant RB, et al. Seasonal periodicity of fall armyworm, (Lepidoptera: Noctuidae) in the Caribbean basin and northward to Canada. J Entomol Sci. 1991; 26: 39–50. [Google Scholar]
- 12.Pashley DP. The current status of fall armyworm host strains. Fla Entomol. 1988; 71: 227–234. [Google Scholar]
- 13.Nagoshi RN, Meagher RL, Hay-Roe M. Inferring the annual migration patterns of fall armyworm (Lepidoptera: Noctuidae) in the United States from mitochondrial haplotypes. Ecol Evol. 2012; 10.1002/ece3.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siebert MW, Nolting SP, Hendrix W, Dhavala S, Craig C, Leonard BR, et al. Evaluation of corn hybrids expressing Cry1F, Cry1A.105, Cry2Ab2, Cry34Ab1/Cry35Ab1, and Cry3Bb1 against southern United States insect pests. J Econ Entomol. 2012; 105: 1825–1834. [DOI] [PubMed] [Google Scholar]
- 15.Reisig DD, Akin DS, Bessin RT, Brewer MJ, Buntin DG, Catchot AL, et al. Lepidoptera (Crambidae, Noctuidae, and Pyralidae) injury on corn containing pure single and pyramided Bt traits and non-Bt hybrids compared to a refuge blend with non-Bt and pyramided Bt hybrids, in the southern United States. J Econ Entomol. 2015; 108: 157–165. 10.1093/jee/tou009 [DOI] [PubMed] [Google Scholar]
- 16.Dangal V, Huang F. Fitness costs of Cry1F resistance in two populations of fall armyworm, Spodoptera frugiperda (J. E. Smith), collected from Puerto Rico and Florida. J Invert Pathol. 2015; 127: 81–86. [DOI] [PubMed] [Google Scholar]
- 17.Storer NP, Peck SL, Gould F, Van Duyn JW, Kennedy GG. Spatial processes in the evolution of resistance in Helicoverpa zea (Lepidoptera: Noctuidae) to Bt transgenic corn and cotton in a mixed agroecosystem: a biology-rich stochastic simulation model. J Econ Entomol. 2003; 96: 156–172. [DOI] [PubMed] [Google Scholar]
- 18.Storer NP, Peck SL, Gould F, Van Duyn JW, Kennedy GG. Sensitivity analysis of a spacially-explicit stochastic simulation model of the evolution of resistance in Helicoverpa zea (Lepidoptera: Noctuidae) to Bt transgenic corn and cotton. J Econ Entomol. 2003; 96: 173–187. [DOI] [PubMed] [Google Scholar]
- 19.Tabashnik BE, Dennehy TJ, Carriere Y. Delayed resistance to transgenic cotton in pink bollworm. Proc Natl Acad Sci. 2005; 10.1073/pnas.0507857102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegfried BD, Spencer T, Crespo AL, Storer NP, Head GP, Owens ED, et al. Ten years of Bt resistance monitoring in the European corn borer. American Entomol. 2007; 53: 208–214. [Google Scholar]
- 21.Bourguet D, Chaufaux J, Seguin M, Buisson CJL, Hinton TJ, Stodola P, et al. Frequency of alleles conferring resistance to Bt maize in French and US corn belt populations of the European corn borer, Ostrinia nubilalis. Theor Appl Genet. 2003; 106: 1225–1233. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Qureshi JA, Leonard BR, Head GP, Niu Y, Huang F. Susceptibility of Louisiana and Florida Populations of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Pyramided Bt Corn Containing Genuity®VT Double Pro™ and Smartstax™ Traits. Fla Entomol. 2013; 96:714–723. [Google Scholar]
- 23.Vélez AM, Spencer TA, Alves AP, Moellenbeck D, Meagher RL, Chirakkal H, et al. Inheritance of Cry1F resistance, cross-resistance and frequency of resistant alleles in Spodoptera frugiperda (Lepidoptera: Noctuidae). Bull Entomol Res. 2013; 10.1017/S0007485313000448 [DOI] [PubMed] [Google Scholar]
- 24.Farias JR, Andow DA, Horikoshi RJ, Sorgatto RJ, Fresia P, Santos AC, et al. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 2014; 64: 150–158. [Google Scholar]
- 25.Gould F, Anderson A, Jones A, Sumerford D, Heckel DG, Lopez J, et al. Initial frequency of alleles for resistance to Bacillus thuringiensis toxin in field populations of Heliothis virescens. Proc Natl Acad Sci USA. 1997; 94: 3519–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andow DA, Alstad DN. F2 screen for rare resistance alleles. J Econ Entomol. 1998; 91: 572–578. [Google Scholar]
- 27.Burd AD, Gould F, Bradley JR, Vanduyn JW, Moar WJ. Estimated frequency of non-recessive Bt resistance genes in bollworm, Helicoverpa zea (Bolddie) (Lepidoptera: Noctuidae) in eastern North Carolina. J Econ Entomol. 2003; 96: 137–142. [DOI] [PubMed] [Google Scholar]
- 28.Bates SL, Zhao JZ, Roush RT, Shelton AM. Insect resistance management in GM crops: past, present and future. Nat Biotech. 2005; 10.1038/nbt1056 [DOI] [PubMed] [Google Scholar]
- 29.Niu Y, Meagher R Jr., Yang F, Huang F. Susceptibility of field populations of the fall armyworm (Lepidoptera: Noctuidae) from Florida and Puerto Rico to purified Cry1F protein and corn leaf tissue containing single and pyramided Bt genes. Fla Entomol. 2013; 96: 701–713. [Google Scholar]
- 30.Li GP, Wu KM, Gould F, Wang JK, Mao J, Gao XW, et al. Increasing tolerance to Cry1Ac cotton from cotton bollworm, Helicoverpa armigera, was confirmed in Bt cotton farming area of China. Ecol Entomol. 2007; 32: 366–375. [Google Scholar]
- 31.Li GP, Wu KM, Gould F, Feng HQ, He YZ, Guo YY. Frequency of Bt resistance genes in Helicoverpa armigera populations from the Yellow River cotton-farming region of China. Entomol Exp Appl. 2004; 112: 135–143. [Google Scholar]
- 32.Abbott WS. A method of computing the effectiveness of an insecticide. J Econ Entomol. 1925; 18: 265–267. [Google Scholar]
- 33.SAS Institute. SAS/STAT user guide, release 6.03. SAS Institute, Cary, NC; 1998. [Google Scholar]
- 34.LeOra Software. POLO-PC: a User’s Guide to Probit and Logit Analysis. Berkeley, CA; 2003. [Google Scholar]
- 35.Tabashnik BE, Motasanchez D, Whalon ME, Hollingworth RM. Defining terms for proactive management of resistance to Bt crops and pesticides. J Econ Entomol. 2014; 107: 496–507. [DOI] [PubMed] [Google Scholar]
- 36.Carrière Y, Crickmore N, Tabashnik BE. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015; 33: 161–168. 10.1038/nbt.3099 [DOI] [PubMed] [Google Scholar]
- 37.Brévault T, Heuberger N, Zhang M, Ellers-Kirk C, Ni X, Masson L, et al. Potential shortfall of pyramided Bacillus thuringiensis cotton for resistance management. PNAS. 2013; 10.1073/pnas.1216719110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu YD. Detection and mechanisms of resistance evolved in insects to Cry toxins from Bacillus thuringiensis. Adv Insect Physiol. 2014; 47: 297–342. [Google Scholar]
- 39.Adang MJ, Crickmor N, Jurat-Fuentes JL. Diversity of Bacillus thuringiensis crystal toxins and mechanisms of action. Adv Insect Physiol. 2014; 47: 39–87. [Google Scholar]
- 40.Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ, et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS ONE. 2014; 10.1371/journal.pone.0097900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coates BS, Siegfried BD. Linkage of an ABCC transporter to a single QTL that controls Ostrinia nubilalis larval resistance to the Bacillus thuringiensis Cry1Fa toxin. Insect Bio-chem Mol Biol. 2015; 63: 86–96. [DOI] [PubMed] [Google Scholar]
- 42.Tay WT, Mahon RJ, Heckel DG, Walsh TK, Downes S, Jame WJ, et al. Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily Aprotein. PLoS Genet. 2015; 10.1371/journal.pgen.1005534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabashnik BE. ABCs of insect resistance to Bt. PLoS Genet. 2015; 10.1371/journal.pgen.1005646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gould F. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Ann Rev Entomol. 1998; 43: 701–726. [DOI] [PubMed] [Google Scholar]
- 45.Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. Shared Midgut Binding Sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa Proteins from Bacillus thuringiensis in Two Important Corn Pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS ONE. 2013; 10.1371/journal.pone.0068164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sena JDA, Hernández-Rodríguez CS, Ferré J. Interaction of Bacillus thuringiensis Cry1 and Vip3A proteins with Spodoptera frugiperda midgut binding sites. App Environ Microbiol. 2009; 75: 2236–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


