Abstract
Our recent study showed the important role of special AT-rich sequence binding protein 1 (SATB1) in the progression of human rectal cancer. However, the value of SATB1 in response to radiotherapy (RT) for rectal cancer hasn't been reported so far. Here, SATB1 was determined using immunohistochemistry in normal mucosa, biopsy, primary cancer, and lymph node metastasis from 132 rectal cancer patients: 66 with and 66 without preoperative RT before surgery. The effect of SATB1 knockdown on radiosensitivity was assessed by proliferation-based assay and clonogenic assay. The results showed that SATB1 increased from normal mucosa to primary cancer, whereas it decreased from primary cancer to metastasis in non-RT patients. SATB1 decreased in primary cancers after RT. In RT patients, positive SATB1 was independently associated with decreased response to preoperative RT, early time to metastasis, and worse survival. SATB1 negatively correlated with ataxia telangiectasia mutated (ATM) and pRb2/p130, and positively with Ki-67 and Survivin in RT patients, and their potential interaction through different canonical pathways was identified in network ideogram. Taken together, our findings disclose for the first time that radiation decreases SATB1 expression and sensitizes cancer cells to confer clinical benefit of patients, suggesting that SATB1 is predictive of response to preoperative RT and clinical outcome in rectal cancer.
Keywords: colorectal cancer, Ingenuity Pathway Analysis, preoperative radiotherapy, prognosis, SATB1
Abbreviations
- ATM
ataxia-telangiectasia mutated
- CRC
colorectal cancer
- DER
dose enhancement ratio
- DFS
disease-free survival
- ID50
50% growth inhibitory dose of radiation
- IPA
Ingenuity Pathway Analysis
- IPKB
Ingenuity Pathway Knowledge Base
- MAC30:
meningioma-associated protein
- OS
overall survival
- PBS
phosphate-buffered saline
- RT
radiotherapy
- RNAi
RNA interference
- SATB1
special AT-rich sequence binding protein 1
- SF:
surviving fraction
- siRNA
small interfering RNA
- XTT
2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl).
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the second leading cause of cancer deaths in many parts of the western world.1 Although short-term preoperative radiotherapy (RT) improves local control in patients with resectable rectal cancer, the overall survival rate of patients hasn't improved markedly. Since the response to RT and prognosis may differ between patients who have the same TNM stage, new markers need to be identified to more accurately predict the response of individual patients to RT and their prognosis. In this context, special AT-rich sequence binding protein 1 (SATB1) could have an important role.
A particular radiosensitivity can be exploited to improve oncologic outcome in appropriate cases. Recent research has revealed that resistance to DNA damage and repair, the hallmarks of cellular responses to ionizing radiation, is one of the major mechanisms of tumor resistance to RT.2 In vivo DNA damage and repair occurs on chromatin, where DNA is complexed with histones and compacted into higher-order structures.3 As a result, both the sensitivity of DNA to damage and the kinetics of repair are regulated by the underlying level of chromatin compaction.4 Recent study showed that condensed chromatin had more resistance to γ-rays than decondensed chromatin,5 indicating the existence of a DNA damage protection mechanism that is mediated by higher-order chromatin. As the chromatin organizer and transcription factor, SATB1 has emerged as a key factor in regulating global gene expression by controlling higher-order chromatin architecture. In thymocyte nuclei, SATB1 forms a characteristic “cage-like” network that presumably remodels the chromatin.6 SATB1 regulates genes by binding to the upstream regulatory elements and recruiting chromatin modifiers, 7,8 which regulate gene expression through histone modifications and nucleosome remodeling at the SATB1-bound association regions. 6,7 In T cells, the assembly of DNA damage sites appears to be mediated by SATB1.9 Therefore, SATB1 may recruit DNA repair proteins in response to DNA damage.
SATB1 in cancer was first described by Han et al.,10 where they discovered that SATB1 promotes the growth and metastasis of breast cancer by reprogramming chromatin organization and transcriptional profiles during tumorigenesis. Subsequent studies also found the tumor-promoting role of SATB1 in additional malignancies, such as laryngeal squamous cell carcinoma and gastric cancer.11–13 However, other studies yielded inconsistent or even opposite results.14–17 Recently, our study showed that SATB1 may play an important role in rectal carcinogenesis,18 which was consistent with the findings of Nodin et al.19 in CRC patients. Furthermore, most studies showed that positive SATB1 was related to poor prognosis in various malignancies.10,12,13,20 Nevertheless, some conflicting results were also found.14,21,22 In the CRC, especially, the prognostic value of SATB1 remains controversial. Nodin et al.19 found that positive SATB1 was a factor of poor prognosis in CRC with negative SATB2 expression, while Al-Sohaily et al.23 revealed that loss of SATB1 correlated with poor survival in CRC patients. Hence, further study is needed to address this issue.
Results
SATB1 increased from normal mucosa to primary cancer, whereas it decreased from primary cancer to metastasis in non-RT patients
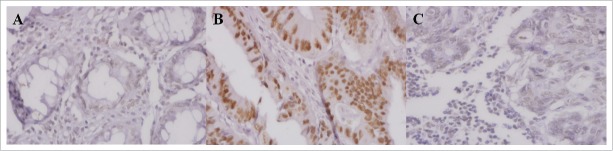
SATB1 protein was intensively localized in the nuclei of normal rectal epithelial cells, as well as primary and metastatic cancer cells (Fig. 1). The data of SATB1 expression in distant and adjacent normal mucosa was integrated because of no significant differences between them both in non-RT and RT patients (P>0.05). In non-RT patients, the frequency of positive SATB1 increased from normal mucosa (21% of 68) to primary cancer (38% of 66) (P=0.028), whereas it decreased from primary cancer to metastasis (9% of 22; P = 0.011; Fig. 2A). In RT patients, the difference of positive SATB1 in normal mucosa (19% of 80), primary cancer (20% of 66) and metastasis (18% of 17) wasn't significant (P>0.05; Fig. 2B).
Figure 1.
SATB1 expression in normal mucosa (A), primary cancer (B) and lymph node metastasis (C) determined by immunohistochemical staining (400× magnification).
Figure 2.
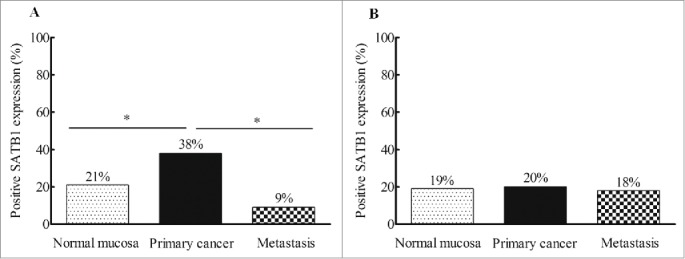
The frequency of positive SATB1 expression in patients without and with RT. (A) The frequency of positive SATB1 expression increased from normal mucosa to primary cancers and decreased from primary cancers to lymph node metastases in non-RT patients (*P<0.05). (B) The expression of positive SATB1 in RT patients was obviously higher in biopsies than in primary cancers (*P < 0.05).
Positive SATB1 in primary cancer correlated with worse survival and earlier distant recurrence in patients with preoperative RT
Because RT hasn't been shown to confer survival benefit for rectal cancer patients with stage IV, these patients were excluded from prognostic analysis. In RT patients, univariate analysis showed that positive SATB1 correlated with worse OS (P = 0.040) and DFS (P = 0.004, Figure 3A, B), which was independent of gender, age, stage and differentiation in multivariate analysis (OS: HR = 6.246, P = 0.006; 95% CI, 1.676–23.280; DFS: HR = 7.796, P < 0.001; 95% CI, 2.543–23.902; Table S2). Moreover, RT patients with positive SATB1 had earlier distant recurrence (P = 0.002; Fig. 3C) rather than local recurrence (P = 0.261; Fig. 3D), independent of gender, age, stage and differentiation (HR = 8.570, P < 0.001; 95% CI, 2.720–27.004; Table S3). These results suggested that positive SATB1 in primary cancer correlated with worse survival and earlier distant recurrence in patients with preoperative RT.
Figure 3.
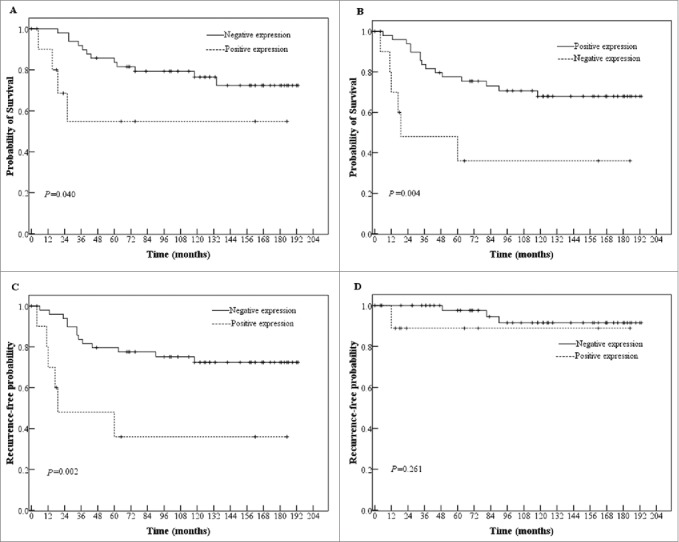
The relationship of SATB1 expression in primary rectal cancers with OS (A) and DFS (B), distant (C) and local recurrence (D) of RT patients.
In non-RT patients, however, no survival or recurrence were found in either univariate (Fig. S1) or multivariate analysis (P>0.05, data not shown). There was no significant association of SATB1 with other clinicopathological variables, including gender, age, stage or differentiation in non-RT and RT patients (P > 0.05; Table S4).
Radiation decreased SATB1 expression both in cancer patients and cell lines
There was no effect of RT on SATB1 in the normal mucosa and metastasis except primary cancer (Fig. 4A). Positive SATB1 in primary cancer of RT patients significantly decreased when compared with the corresponding biopsy which didn't undergo radiation (43% vs. 20%; P = 0.025). Likewise, SATB1 is significantly lower in primary cancer of RT patients, compared with that of non-RT patients (20% vs. 38%; P = 0.034). Similarly, SATB1 in cells after 2Gy of radiation significantly decreased at 24h, 48h and even 72h when compared with non-irradiated cells (P < 0.05; Fig. 4B). These results suggested that radiation could decrease SATB1 expression both in cancer patients and cell lines.
Figure 4.
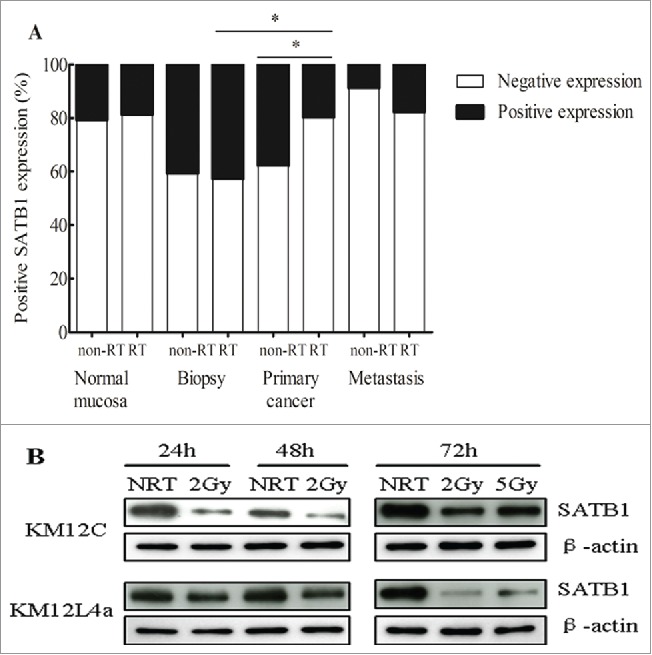
The effect of RT on SATB1 expression. (A) After RT, the frequency of positively expressed SATB1 was significantly decreased from 38% to 20% in primary cancers (*P<0.05), and from 43% in biopsies to 20% in primary cancers (*P<0.05). (B) The expression of SATB1 protein in cells after 2Gy of radiation at 24h, 48h and 72h or 5Gy of radiation at 72h was significantly decreased when compared with the cells without radiation (NRT) (*P < 0.05).
Knockdown of SATB1 increased radiosensitivity of cancer cells
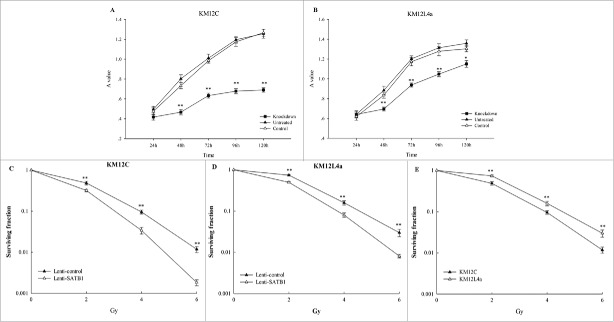
After SATB1 knockdown by siRNA, both cell lines exposed to 2Gy radiation exhibited reduced viability in proliferation-based assay from 48h to 120h compared with non-irradiated cells (P < 0.001). There were no difference of cell viability between control and untreated cells at each time point after 2Gy of radiation (P > 0.05; Figure 5A, B).
Figure 5.
The effect of SATB1 knockdown on the radioresponse of cancer cells. (A, B) Short-term radiosensitivity of cells after SATB1 knockdown was measured by XTT assay with mean and SE shown in KM12C and KM12L4a cells (*P<0.05, **P<0.001). (C, D, E) Long-term radiosensitivity was determined by clonogenic assay in KM12C cells, KM12L4a cells and the both cell lines (*P < 0.05, **P < 0.001).
Furthermore, we assessed the radiosensitivity of cells with SATB1 knockdown using lentiviral vectors in clonogenic assay and found a significant increase in radiosensitivity of cells compared with the control (P < 0.001). The DER was 0.75 for KM12C cells and 0.97 for KM12L4a cells, suggesting different radiosensitizing effect between the cells (Fig. 5C, D), which was consistent with the difference between non-irradiated KM12C and KM12L4a cells with different basal SATB1 expression (Fig. 5E). These results showed that knockdown of SATB1 increased radiosensitivity of cancer cells.
SATB1 correlated with ATM, pRb2/p130, Ki-67 and Survivin expression in primary cancer after RT
To further understand the role of SATB1 in response to radiation, at first, the relationship of SATB1 with radiation-associated factors was evaluated. The results showed that SATB1 negatively correlated with ATM and pRb2/p130, and positively correlated with Ki-67 and Survivin (P < 0.05; Table 1). The association of SATB1 with other biological variables in RT patients was also shown in Table 1.
Table 1.
Correlation of biological factors and SATB1 expression in rectal cancer patients with RT
| SATB1 expression |
|||
|---|---|---|---|
| Biological factors | Negative (%) | Positive (%) | P value |
| Ki-67 (mean) | |||
| <45% | 32(94) | 2(6) | 0.002 |
| ≥45% | 9(53) | 8(47) | |
| Survivin | |||
| Negative | 16(94) | 1(6) | 0.034 |
| Positive | 11(58) | 8(42) | |
| ATM | |||
| Negative | 9(53) | 8(47) | 0.017 |
| Positive | 15(94) | 1(6) | |
| pRb2/p130 | |||
| Negative | 8(50) | 8(50) | 0.003 |
| Positive | 31(91) | 3(9) | |
| Necrosis | |||
| Negative | 34(81) | 8(19) | 1 |
| Positive | 16(80) | 4(20) | |
| p73 | |||
| Negative | 33(85) | 6(15) | 0.167 |
| Positive | 5(63) | 3(37) | |
| p53 | |||
| Negative | 44(83) | 9(17) | 0.386 |
| Positive | 7(70) | 3(30) | |
| FXYD-3 | |||
| Weak | 15(71) | 6(29) | 0.503 |
| Strong | 28(82) | 6(18) | |
| MAC30 | |||
| Weak | 16(84) | 3(16) | 0.505 |
| Strong | 24(75) | 8(25) | |
Next, the associating networks between SATB1 and these proteins were predicted using IPA (Fig. 6). The pathways involved in Ki-67, pRb2/p130, ATM, and Survivin were mapped. The network ideogram showed the potential interaction of SATB1 with these radiation-associated factors through different canonical pathways.
Figure 6.
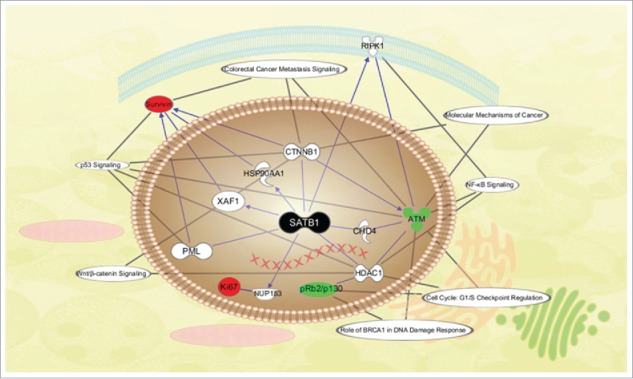
Schematic diagram of the molecular network of SATB1. Important pathways and cellular localization of proteins were shown. Nodes with different shapes represented the molecular class of proteins and lines represented the relationships between the proteins.
Discussion
The features of SATB1 expression differ considerably in various malignancies. Han et al.10 showed that SATB1 reprogrammed gene expression to promote growth and metastasis of breast cancer. Subsequent studies showed that elevated SATB1 participated in the pathogenesis of other malignancies.11-13 However, some studies yielded inconsistent or even opposite results.14-17 These results suggest that SATB1 seems to be tissue-specific. In the present study, SATB1 increased from normal mucosa to primary cancer, which is consistent with our recent study in rectal cancer18 and also corroborated with the study by Nodin et al.19 and Al-Sohaily et al.23 in CRC, indicating the role of SATB1 in the promotion of malignant transformation of colorectal mucosa. Consistent with our findings, a recent study also provided evidence to support the tumor-promoting role of SATB1 in colorectal carcinogenesis.34
There are few and contradictory studies on SATB1 as an independent prognostic factor for CRC. Nodin et al.18 found that positive SATB1 was a factor of poor prognosis in CRC patients with negative SATB2, a close homolog of SATB1. Our current findings in rectal cancer showed that positive SATB1 independently correlated with worse survival and earlier distant recurrence in RT patients. In fact, oncologic outcome in appropriate cases could be improved by a particular radiosensitivity. To further explore if the less benefit in patients with positive SATB1 than negative SATB1 is attribute to the response of SATB1 to RT, we determined the radiosensitivity in cells with or without SATB1 by proliferation-based assay and clonogenic assay, and found that SATB1 knockdown significantly increased radiosensitivity of cancer cells. Thus, decreased SATB1 would lead to reduced growth rates and resistance to radiation, which could in turn lead to improved clinical outcomes. Although the underlying mechanism isn't yet known, it is tempting to hypothesize that different genes involved in different functions and pathways are modulated by SATB1 after radiation.
In the present study, we discovered a negative correlation between SATB1 with ATM and pRb2/p130, and a positive correlation with Ki-67 and Survivin in patients with preoperative RT. ATM, Survivin and pRb2/p130 were regulated by SATB1 in breast cancer,10 and have actually been implicated in the response of human cancer cells to radiation.35,36 Therefore, we speculate that these proteins may be involved in the process of SATB1 in response to radiation. In order to test this hypothesis, we used IPA to identify documented molecular interactions between SATB1 and these proteins. The data determined by IPA depicts functional networks in which SATB1 was of paramount importance in the modulation of these key proteins.
Our model also revealed that these proteins specifically correlated with the NF-κB signaling pathways, BRCA1-mediated DNA damage responsive pathway and G1/S checkpoint pathways in cell cycle regulation, and these pathways play an important role in the regulatory mechanisms of radiation response. There is, furthermore, growing evidence that cellular response of these proteins to DNA damage induced by radiation was associated with large-scale remodeling in chromatin structures. For instance, radiation induced DNA damage signaling is initiated by the DNA double strand breaks sensor―ATM, which rapidly causes changes in higher-order chromatin structures.37 Activated ATM mediates phosphorylation of various nuclear proteins, such as p53, CHK2/Cds1, BRCA1.38 The bi-directional relationship between ATM and SATB1 shown in our model suggested that negative feedback may exist, further studies are needed to determine it. Besides, Rb2/p130-E2F4 complexes accumulate on promoters in response to DNA damage and function as local and global regulators of gene expression and chromatin dynamics through recruitment to CpG-rich regions in the genome to shape the chromatin landscape.39 Therefore, we believe that SATB1 architecture may provide a platform for assembling these radiation-associated proteins following DNA damage. Further studies focusing on SATB1-centered pathway are likely to uncover other crucial signal transducers in the complex signaling network responsible for radiation.
In summary, this study demonstrates that the effect of SATB1 on the radiation response, and clinical outcome of CRC using in vivo, in vitro and in silico studies. The results suggest that positive SATB1 was independently associated with decreased response to preoperative RT, early time to metastasis, and worse survival of rectal cancer patients undergoing RT. Therefore, our findings highlight a specific value of SATB1 as a biomarker to predict the response to preoperative RT and clinical outcome in rectal cancer. It is the first study, to our knowledge, on the value of SATB1 in response to RT for rectal cancer.
Materials and Methods
Patients
The study included 132 patients with rectal cancer that participated in a randomized clinical trial (Stockholm Trial I) from the Southeast Swedish Health Care region.24 Locally curative resection was performed in all patients. Sixty-six patients received tumor resection alone, and 66 received RT before surgery. RT was given at a total of 25Gy in 5 fractions over a median of 6 d (5–12 days). Surgery was carried out in a median of 3 d (range, 1–13 days) after RT. Specimens were collected from distant (n = 97), adjacent normal mucosa (n = 65), biopsy (n = 84), primary cancer (n = 132) and lymph node metastasis (n = 39). “Biopsy” represents the primary cancer before surgery and RT, while “primary cancer” refers to the tissue of primary tumor collected at the time of surgery, regardless of RT. Tissue collection and microarray preparation were described in our previous study.25 No patients received adjuvant chemotherapy before or after surgery. There were no statistical differences between non-RT and RT patients regarding the characteristics of patients and tumors (P>0.05; Table S1). The median follow-up period was 75 months (0–193 months). Informed consent was obtained from all patients. All experiments were performed in accordance with relevant guidelines and regulations.
Immunohistochemical assay
Immunohistochemistry was performed on 4μm tissue microarray sections from paraffin-embedded surgical specimens. A detailed protocol was described in Supplementary material. For the immunohistochemical score, both the staining intensity and positive cell proportion were estimated. The former was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong), respectively. The latter was defined as 0 (no positive cell); 1 ( < 25% positive cells); 2 (25–50% positive cells), and 3 (>50% positive cells), respectively. Multiplication by the 2 indexes yields an overall score of 0–9 for each specimen: 0, negative; 1–2, weak; 3–6, moderate; and 9, strong, respectively. Sections were reviewed and scored by 2 investigators independently. The negative cases were considered as negative expression, and the weak, moderate or strong cases were considered as positive expression.
Cell culture
Human colon cancer KM12C and KM12L4A cells were chosen in the present study because of prominently higher SATB1 expression than other cells, such as SW480 and HCT116, shown in our recent study.18 Cells were maintained in Eagle's MEM medium supplemented with 10% fetal bovine serum at 37°C in 5% CO2.
SATB1 knockdown
Small interfering RNA (siRNA) against SATB1 (knockdown) and siRNA without RNA interference (RNAi) effect (control) were transfected into cells by DharmaFECT Transfection Reagent-2 (Thermo Scientific, MA), according to the manufacture's protocol. Cells without siRNA treatment (untreated) were included in each assay. The efficacy of RNAi was assessed by Western blot at 72h after transfection. A detailed Western blot protocol was described in the Supplementary Material.
Besides, the lentivirus carrying short hairpin RNA (shRNA), targeting SATB1 or without RNAi effect, was constructed by cotransfecting 293T cells with lentiviral plasmids designated as Lenti-shSATB1 and Lenti-control, respectively. The transduction of KM12C and KM12L4a cells with lentivirus was described in Supplementary material. Pooled populations of knockdown cells, obtained after 10 d of drug selection without subcloning, were used for clonogenic assay.
Radiation procedure
Radiation was performed at room temperature with photons X generated by a 6MV linear accelerator Varian Clinac 600C/D (Varian Medical Systems, CA). The field size was 30cm×30cm, and the distance between sources and cells was 100cm. Acrylic glass plates were placed above (3cm thick) and underneath (10cm thick) the cells. Cells were exposed to different doses of radiation (0, 1, 2, 4, 6 and 10Gy) after 48h of seeding at 1.0×103 cells/well of 96-well plate to determinate the 50% growth inhibitory dose of radiation (ID50). Furthermore, cell survival was determined using XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl) assay at 24h and 48h after radiation by Cell Proliferation Kit II (Roche, Germany) according to the manufacture's protocol, and then the dose-survival curve was plotted. The ID50 obtained was about 2Gy for both cell lines. All experiments were repeated 3 times.
Effect of radiation on SATB1 expression in cancer cells
To measure the effect of radiation on SATB1 in cancer cells, cells at 1.0×105 cells/well in 6-well plate were exposed to 2Gy or 5Gy of radiation and harvested at 24h, 48h and 72h, respectively, for Western blot analysis. All experiments were repeated 3 times.
Short-term radiosensitivity assay
Cells were seed at 1.0×103 cells/well in 96-well plate and exposed to 2Gy of radiation at 72h after siRNA-mediated RNAi. Cell viability (A value = A492-A690) was measured from 24h to 120h after radiation by XTT assay. All experiments were performed in triplicate.
Long-term radiosensitivity assay
To determine the long-term radiosensitivity, an in vitro clonogenic assay was performed. Cells treated with control or SATB1 lentiviral vectors were plated at numbers appropriate for different doses of radiation. After 20h, cells were exposed to 0, 2, 4, or 6Gy of radiation and further incubated at 37 °C for 7 d. Colonies were fixed for 15min with methanol and stained for 20min with 10% Giemsa, then counted by microscope with a cutoff of 50 viable cells. Surviving fraction (SF) for transfected cells was normalized to the respective plating efficiencies for transfection alone. Using Sigmaplot 12.0, survival curve parameters D0 (mean lethal dose) and N (extrapolation number) were fitted to the multi-target single-hit model: SF = 1-(1-e-D/D0)N. Dose enhancement ratio (DER) was calculated as the dose (Gy) for control divided by the dose for SATB1-knockdown cells, at a SF of 0.37. Error bars were calculated as ± SE by pooling the results of 3 independent experiments.
Evaluation of the radiation-associated factors
Furthermore, on the same series of the patient materials as used in the present study, we have previously found that necrosis, Ki-67, p53, p73, ataxia telangiectasia mutated (ATM), FXYD-3, pRb2/p130, meningioma-associated protein (MAC30), and Survivin had, more or less, certain relationships with RT.25-33 Hence, in the present study, we also studied their relationships with SATB1 in RT patients.
Network analysis
The functional significance of SATB1 was evaluated by Ingenuity Pathway Analysis (IPA, www.ingenuity.com). GenBank IDs including SATB1 were uploaded into the IPA program, and mapped into predicted functional networks available in the Ingenuity Pathway Knowledge Base (IPKB). Networks are comprised of biological functions assigned to focus gene functions compared with the whole IPKB.
Ethics statement
The study was approved by the Institutional Review Board of Linköping University, Sweden.
Statistical analysis
Fisher's exact test or Pearson χ2 method was used to test the differences of SATB1 among the distant/adjacent normal mucosa, biopsy, primary cancer and metastasis, as well as the association of SATB1 with clinicopathological and biological variables. The relationship of SATB1 with overall survival (OS), disease-free survival (DFS), or recurrence was tested by Cox regression analysis. The Student's t-test was used for the quantitative analysis of cell lines. P-values of <0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
Funding
This work was supported by the grants from the Swedish Cancer Foundation, Swedish Research Council and the Health Research Council in the South-East of Sweden.
References
- 1.Gellad ZF, Provenzale D. Colorectal cancer: national and international perspective on the burden of disease and public health impact. Gastroenterology 2010; 138(6):2177-90; PMID:20420954; http://dx.doi.org/ 10.1053/j.gastro.2010.01.056 [DOI] [PubMed] [Google Scholar]
- 2.Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res 2009; 15(20):6314-20; PMID:19808869; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-0096 [DOI] [PubMed] [Google Scholar]
- 3.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 2009; 43:559-99; PMID:19886812; http://dx.doi.org/ 10.1146/annurev.genet.032608.103928 [DOI] [PubMed] [Google Scholar]
- 4.Cann KL, Dellaire G. Heterochromatin and the DNA damage response: the need to relax. Biochem Cell Biol 2011; 89(1):45-60; PMID:21326362; http://dx.doi.org/ 10.1139/O10-113 [DOI] [PubMed] [Google Scholar]
- 5.Takata H, Hanafusa T, Mori T, Shimura M, Iida Y, Ishikawa K, Yoshikawa K, Yoshikawa Y, Maeshima K. Chromatin compaction protects genomic DNA from radiation damage. PLoS One 2013; 8(10):e75622; PMID:24130727; http://dx.doi.org/ 10.1371/journal.pone.0075622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue specific nuclear architecture and gene expression regulated by SATB1. Nat Genet 2003; 34(1):42-51; PMID:12692553; http://dx.doi.org/ 10.1038/ng1146 [DOI] [PubMed] [Google Scholar]
- 7.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 2002; 419(6907):641-5; PMID:12374985; http://dx.doi.org/ 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- 8.Kumar PP, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell 2006; 22(2):231-43; PMID:16630892; http://dx.doi.org/ 10.1016/j.molcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 9.Kumar PP, Mehta S, Purbey PK, Notani D, Jayani RS, Purohit HJ, Raje DV, Ravi DS, Bhonde RR, Mitra D, Galande S. SATB1-binding sequences and Alu-like motifs define a unique chromatin context in the vicinity of human immunodeficiency virus type 1 integration sites. J Virol 2007; 81(11):5617-27; PMID:17376900; http://dx.doi.org/ 10.1128/JVI.01405-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008; 452(7184):187-93; PMID:18337816; http://dx.doi.org/ 10.1038/nature06781 [DOI] [PubMed] [Google Scholar]
- 11.Zhao XD, Ji WY, Zhang W, He LX, Yang J, Liang HJ, Wang LL. Overexpression of SATB1 in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec 2010; 72(1):1-5; PMID:20110741; http://dx.doi.org/ 10.1159/000264777 [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Cheng C, Zhu S, Yang Y, Zheng L, Wang G, Shu X, Wu K, Liu K, Tong Q. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep 2010; 24(4):981-7; PMID:20811679 [DOI] [PubMed] [Google Scholar]
- 13.Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu S, Liu K, Wu K, Tong Q. Expression of SATB1 and heparanase in gastric cancer and its relationship to clinicopathologic features. APMIS 2010; 118(11):855-63; PMID:20955458; http://dx.doi.org/ 10.1111/j.1600-0463.2010.02673.x [DOI] [PubMed] [Google Scholar]
- 14.Iorns E, Hnatyszyn HJ, Seo P, Clarke J, Ward T, Lippman M. The role of SATB1 in breast cancer pathogenesis. J Natl Cancer Inst 2010; 102(16):1284-96; PMID:20595686; http://dx.doi.org/ 10.1093/jnci/djq243 [DOI] [PubMed] [Google Scholar]
- 15.Li K, Cai R, Dai BB, Zhang XQ, Wang HJ, Ge SF, Xu WR, Lu J. SATB1 regulates SPARC expression in K562 cell line through binding to a specific sequence in the third intron. Biochem Biophys Res Commun 2007; 356(1):6-12; PMID:17343824; http://dx.doi.org/ 10.1016/j.bbrc.2007.01.201 [DOI] [PubMed] [Google Scholar]
- 16.Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell 2009; 16(4): 507-16; PMID:19386260; http://dx.doi.org/ 10.1016/j.devcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, et al.. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest 2007; 117(9):2611-20; PMID:17694175; http://dx.doi.org/ 10.1172/JCI30525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng WJ, Yan H, Zhou B, Zhang W, Kong XH, Wang R, Zhan L, Li Y, Zhou ZG, Sun XF. Correlation of SATB1 overexpression with the progression of human rectal cancer. Int J Colorectal Dis 2012; 27(2):143-50; PMID:21870058; http://dx.doi.org/ 10.1007/s00384-011-1302-9 [DOI] [PubMed] [Google Scholar]
- 19.Nodin B, Johannesson H, Wangefjord S, O'Connor DP, Lindquist KE, Uhlén M, Jirström K, Eberhard J. Molecular correlates and prognostic significance of SATB1 expression in colorectal cancer. Diagn Pathol 2012; 7:115; PMID:22935204; http://dx.doi.org/ 10.1186/1746-1596-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, He J, Han P, Tian D. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int 2012; 32(7): 1064-78; PMID:22583549; http://dx.doi.org/ 10.1111/j.1478-3231.2012.02815.x [DOI] [PubMed] [Google Scholar]
- 21.Hanker LC, Karn T, Mavrova-Risteska L, Ruckhäberle E, Gaetje R, Holtrich U, Kaufmann M, Rody A, Wiegratz I. SATB1 gene expression and breast cancer prognosis. Breast 2011; 20(4):309-13; PMID:20980149; http://dx.doi.org/ 10.1016/j.breast.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 22.Selinger CI, Cooper WA, Al-Sohaily S, Mladenova DN, Pangon L, Kennedy CW, McCaughan BC, Stirzaker C, Kohonen-Corish MR. Loss of special AT-rich binding protein 1 expression is a marker of poor survival in lung cancer. J Thorac Oncol 2010; 6(7):1179-89; http://dx.doi.org/ 10.1097/JTO.0b013e31821b4ce0 [DOI] [PubMed] [Google Scholar]
- 23.Al-Sohaily S, Henderson C, Selinger C, Pangon L, Segelov E, Kohonen-Corish MR, Warusavitarne J. Loss of special AT-rich sequence-binding protein 1 (SATB1) predicts poor survival in patients with colorectal cancer. Histopathology 2014; 65(2):155-63; PMID:24118100; http://dx.doi.org/ 10.1111/his.12295 [DOI] [PubMed] [Google Scholar]
- 24.No authors listed . Improved survival with preoperative radiotherapy in resectable rectal cancer. Swedish Rectal Cancer Trial. N Engl J Med 1997; 336(14):980-7; PMID:9091798; http://dx.doi.org/ 10.1056/NEJM199704033361402. [DOI] [PubMed] [Google Scholar]
- 25.Knutsen A, Adell G, Sun XF. Inflammatory infiltration, fibrosis, necrosis and mucinous content in relation to clinicopathological and molecular factors in rectal cancers with or without preoperative radiotherapy. Oncol Rep 2006; 16(2):321-7; PMID:16820910 [PubMed] [Google Scholar]
- 26.Jansson A, Sun XF. Ki-67 expression in relation to clinicopathological variables and prognosis in colorectal adenocarcinomas. APMIS 1997; 105(9):730-4; PMID:9350218; http://dx.doi.org/ 10.1111/j.1699-0463.1997.tb05078.x [DOI] [PubMed] [Google Scholar]
- 27.Adell G, Sun XF, Stål O, Klintenberg C, Sjödahl R, Nordenskjöld B. p53 status: an indicator for the effect of preoperative radiotherapy of rectal cancer. Radiother Oncol 1999; 51(2):169-74; PMID:10435809; http://dx.doi.org/ 10.1016/S0167-8140(99)00041-9 [DOI] [PubMed] [Google Scholar]
- 28.Sun XF. p73 overexpression is a prognostic factor in patients with colorectal adenocarcinoma. Clin Cancer Res 2012; 8(1):165-70. [PubMed] [Google Scholar]
- 29.Arlehag L, Adell G, Knutsen A, Thorstenson S, Sun XF. ATM expression in rectal cancers with or without preoperative radiotherapy. Oncol Rep 2005; 14(2):313-7; PMID:16012708 [PubMed] [Google Scholar]
- 30.Loftås P, Onnesjö S, Widegren E, Adell G, Kayed H, Kleeff J, Zentgraf H, Sun XF. Expression of FXYD-3 is an independent prognostic factor in rectal cancer patients with preoperative radiotherapy. Int J Radiat Oncol Biol Phys 2009; 75(1):137-42; http://dx.doi.org/ 10.1016/j.ijrobp.2008.10.076 [DOI] [PubMed] [Google Scholar]
- 31.Moparthi SB, Bergman V, Adell G, Thorstensson S, Sun XF. pRb2/p130 protein in relation to clinicopathological and biological variables in rectal cancers with a clinical trial of preoperative radiotherapy. Int J Colorectal Dis 2009; 24(11):1303-10; PMID:19597825; http://dx.doi.org/ 10.1007/s00384-009-0767-2 [DOI] [PubMed] [Google Scholar]
- 32.Moparthi SB Arbman G, Wallin A, Kayed H, Kleeff J, Zentgraf H, Sun XF. Expression of MAC30 protein is related to survival and biological variables in primary and metastatic colorectal cancers. Int J Oncol 2007; 30(1):91-5; PMID:17143516 [PubMed] [Google Scholar]
- 33.Knutsen A, Adell G, Sun XF. Survivin expression is an independent prognostic factor in rectal cancer patients with and without preoperative radiotherapy. Int J Radiat Oncol Biol Phys 2004; 60(1):149-55; PMID:15337550; http://dx.doi.org/ 10.1016/j.ijrobp.2004.02.007 [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Zhang B, Zhang X, Sun Y, Wei X, McNutt MA, Lu S, Liu Y, Zhang D, Wang M, Lin Z, Niu N. SATB1 expression is associated with biologic behavior in colorectal carcinoma in vitro and in vivo. PLoS One 2013; 8(1):e47902; PMID:23326301; http://dx.doi.org/ 10.1371/journal.pone.0047902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell 2010; 18(6):619-29; PMID:21156285; http://dx.doi.org/ 10.1016/j.ccr.2010.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carson-Walter EB, Winans BN, Whiteman MC, Liu Y, Jarvela S, Haapasalo H, Tyler BM, Huso DL, Johnson MD, Walter KA. Characterization of TEM1/endosialin in human and murine brain tumors. BMC Cancer 2009; 9:417; PMID:19948061; http://dx.doi.org/ 10.1186/1471-2407-9-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421(6922):499-506; PMID:12556884; http://dx.doi.org/ 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- 38.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003; 3(3):155-68; PMID:12612651; http://dx.doi.org/ 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- 39.Montoya-Durango DE, Ramos KS. Retinoblastoma family of proteins and chromatin epigenetics: a repetitive story in a few LINEs. BioMol Concepts 2011; 2(4):233-45; PMID:25962032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.