Abstract
The T-box transcription factor TBX18 is essential to mesenchymal cell differentiation in several tissues and Tbx18 loss-of-function results in dramatic organ malformations and perinatal lethality. Here we demonstrate for the first time that Tbx18 is required for the normal development of periductal smooth muscle stromal cells in prostate, particularly in the anterior lobe, with a clear impact on prostate health in adult mice. Prostate abnormalities are only subtly apparent in Tbx18 mutants at birth; to examine postnatal prostate development we utilized a relatively long-lived hypomorphic mutant and a novel conditional Tbx18 allele. Similar to the ureter, cells that fail to express Tbx18 do not condense normally into smooth muscle cells of the periductal prostatic stroma. However, in contrast to ureter, the periductal stromal cells in mutant prostate assume a hypertrophic, myofibroblastic state and the adjacent epithelium becomes grossly disorganized. To identify molecular events preceding the onset of this pathology, we compared gene expression in the urogenital sinus (UGS), from which the prostate develops, in Tbx18-null and wild type littermates at two embryonic stages. Genes that regulate cell proliferation, smooth muscle differentiation, prostate epithelium development, and inflammatory response were significantly dysregulated in the mutant urogenital sinus around the time that Tbx18 is first expressed in the wild type UGS, suggesting a direct role in regulating those genes. Together, these results argue that Tbx18 is essential to the differentiation and maintenance of the prostate periurethral mesenchyme and that it indirectly regulates epithelial differentiation through control of stromal-epithelial signaling.
Introduction
During middle and late gestation of the mouse, the T-box transcription factor (TF) TBX18 is expressed in a population of mesenchymal cells in the lower embryonic abdomen. These cells contribute to the stromal layer of nearly every organ in the urogenital system but with differing affects in each of them [1]. In the ureter, Tbx18 is essential to the formation of a coordinated smooth muscle layer that can conduct urine from the kidney to the bladder. Beginning at embryonic day 11.5 (E11.5) Tbx18-expressing mesenchymal cells begin to coalesce around the nascent ureter epithelial duct [2]. Secreted SHH and WNT signals from the ureter epithelium maintain the proliferation and eventual differentiation of these Tbx18-positive condensing mesenchymal cells [3–5]. However, in the absence of Tbx18, the mesenchymal cells fail to respond to the epithelial signals and subsequently retire to a fibrocytic fate [2]. Consequently, a still unknown signal acting downstream of Tbx18 in the mesenchyme, which normally reciprocates the proliferation signal to the ureter epithelium, fails to be activated. Due to the loss of these interdependent signaling mechanisms, neither the ureter epithelium nor the stroma proliferate sufficiently resulting in a ureter of reduced length, thickness, and elasticity. The consequent fluid build-up leads to a grotesque enlargement of both the ureters and kidneys in Tbx18 mutants [2,6].
Toward the posterior end of the urogenital system, the prostate is an organ essential to male fertility that arises developmentally from the urogenital sinus (UGS) [7]. Beginning at E16.5, the urogenital sinus mesenchyme (UGS-M) begins differentiating under the influence of testicular androgens and then induces the adjacent urogenital sinus epithelium (UGS-E) towards a path of prostate committal [8,9]. The naïve UGS-E, responding to signals emanating from the UGS-M, begins to invade the adjacent undifferentiated mesenchyme beginning around E17.5 [10–13]. As the epithelial buds extend into and beyond the UGS-M, mesenchymal cells condense around the buds producing the early rudiments of the prostate stromal layer, comprised mostly of smooth muscle cells and fibroblasts [14]. Paracrine signals including Wnt [15,16], Notch [17], Bmp [12,18], Tgfb [19,20] and Shh [21,22] play critical roles in this process of prostate bud induction and differentiation, each with regional expression patterns and differential affects on the formation of the various lobes. Furthermore, abnormal expression of these factors and their downstream receptor pathways are indicative, and sometimes directly causative, of lobe specific prostate pathologies such as fibrosis and neoplasias [19,23–25].
In the adult mouse prostate, the prostatic lobe stroma adjacent to the urethra consists of a core of smooth muscle cells (SMCs) with a few VIM+ fibroblasts positioned at the lobe perimeter [26,27]. The well-organized smooth muscle bundles produce large amounts of Tgfb1, which inhibits proliferation of the adjacent epithelial stem cell population; the loss of Tgfb1 signaling induces epithelial neoplasias [28,29]. Paradoxically, in prostatic models of “reactive stroma”, SMCs and fibroblasts in the stroma experience up-regulation of Tgfb1 production resulting in progressive induction of the stroma towards a diseased myofibroblast phenotype [20,30]. These myofibroblasts, distinguished by co-expression of SMA and Vimentin (VIM), exhibit elevated production of ECM components (collagens), Transforming growth factors (Tgfb1), and matrix remodeling enzymes (Serpines and Cathepsins) to create a growth-promoting microenvironment [31]. As a consequence of these changes in the stromal microenvironment, the adjacent epithelium experiences hyper-proliferation and can become invasive [17,20,29,30]. A common outcome from this situation is the formation of Prostatic Intraepithelial Neoplasia (PIN) [32].
A number of studies have indicated that Tbx18 contributes to urogenital structures aside from the ureter and bladder [1,33,34]. However, since Tbx18-null animals die perinatally [35], phenotypic effects in the postnatally developing prostate have been impossible to discern. We recently reported a novel hypomorphic cis-regulatory mutation of Tbx18, called 12Gso [6], and the relative longevity of these animals offered the opportunity to examine Tbx18-related postnatal phenotypes. Using this mutant in combination with a novel Tbx18 conditional allele, here we demonstrate that Tbx18 regulates differentiation of a SMC subpopulation that contribute to the periductal prostate stroma, particularly within the mouse anterior lobe. In contrast to the ureter, which displays reduced stromal and epithelial thickness in mutant animals, periductal stroma proximal to the urethra is hypertrophied in Tbx18 mutant adult prostates, composed primarily of enlarged and disoriented SMCs and myofibroblasts. Furthermore, prostatic epithelial cells surrounded by this abnormal stroma are significantly disorganized by early adulthood, and the abnormal ductal regions also contain unusually large numbers of disorganized, Vimentin-positive cells. Together, our data indicate an important role for Tbx18 in regulating the reciprocal epithelial-stromal signaling from the earliest stages of prostate development, with implications for human prostate disease.
Methods
Histopathology and Immunohistochemistry
Tissues were dissected at the appropriate gestational or postnatal stage. Tissues were fixed in 4% PFA at 4°C, dehydrated, and embedded in paraffin wax for sectioning. 4–6μm sections were used in all experiments. TBX18-2 antibody is used at 1:800 in normal goat serum blocking solution and incubated overnight at 4°C. Following washes, a Goat anti-Rabbit:HRP (Jackson ImmunoResearch 111-035-003 1:500) secondary antibody was applied for 1hr at room temperature. Sections were covered in TSA-Rhodamine for 6 minutes then washed and counter stained with Hoecsht 33342. Commercial antibodies are VIM (Abcam ab92547 1:500), CK8/CK18 (ab53280 1:100), CK5 (ab24647 1:750), and SMA (ab7817 1:75, ab5694 1:1000) with secondary antibodies (from Thermo-Fisher Scientific), Goat anti-Rabbit IgG, Alexa Fluor 488, A-11008 1:200 and Goat anti-Mouse IgG, Alexa Fluor 568, A-11004 1:200 and Alexa Fluor 647, A-31573, 1:200.
RT-qPCR
Total RNA was collected by homogenizing tissues in Invitrogen Trizol reagent. RNA was DNase I treated with NEB DNase I, and cleaned on Zymo Research RNA Clean & Concentrate 25 columns. NEB M-MULV reverse transcriptase was used to generate cDNA. Primer sequences were obtained from PrimerBank [36] and validated prior to use. Primer sequences are listed in S3 Table. An Applied Biosciences QuantStudio Flex 6 thermocycler was used for amplification. The delta-delta-Ct method was employed to establish quantity differences.
RNA-Seq
Whole urogenital sinuses were dissected from male mouse embryos at embryonic day E16.5 and E18.5. Tissues were homogenized in Trizol and RNA was isolated. RNA was treated with NEB DNase I, then analyzed on a Bioanalyzer for quality. RNA-Seq libraries were prepared with KAPA Biosystems KAPA Stranded mRNA-Seq Kit for Illumina Platform (KR0960—V2.14). Single-end reads were performed on Illumina Hi-Seq 2000 sequencers. Single stranded sequencing reads were processed with Tophat 2.1.0 (using Bowtie 2.1.0 and Samtools 0.1.19) with all Tophat options at default. Cufflinks 2.2.1 was used to determine differential expression. Results were filtered to consider only those genes with at least 1 FPKM in either the mutant or control samples for the final selection of differentially expressed genes. Datasets reported in this study have been deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE80083.
Mouse breeding
All mouse lines used were bred onto B6C3-F1 hybrid background because genetic background appears to influence Tbx18 phenotype severity [6]. All animal work described in this study, including euthanasia, was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Illinois (Animal Assurance Number: A3118-01; approved IACUC protocol number 15245)
Generation of the Tbx18Flox allele
The conditional Tbx18 allele was generated by inserting LoxP sites flanking the second exon of the endogenous Tbx18 gene. In crosses with mice carrying the epibliastic Meox2-Cre knockin allele [B6.129S4-Meox2tm1(cre)Sor/J [37]; obtained from the Jackson Laboratory, Bar Harbor, ME] homozygous Tbx18 flox/flox mice displayed phenotypes identical to those previously published for global Tbx18-null mice, confirming the efficient ablation of Tbx18. A more detailed description of phenotypes associated with this allele will be forthcoming (Guimarães-Camboa et al., manuscript in preparation).
Results
Tbx18 is expressed in the early urogenital sinus mesenchyme
Previous reports of the Tbx18 expression pattern in the urogenital system have not included analysis of the urogenital sinus or prostate [1,2]. However, the prostate develops at the base of the bladder in near proximity to the ureter, and the obvious parallels between the development of these two structures prompted a careful re-evaluation.
We set out to determine the quantity and location of native Tbx18 expression in prostate through the late stages of gestation and the postnatal days through puberty (Fig 1). First we used quantitative reverse transcript PCR (RT-qPCR) to detect Tbx18 mRNA in the UGS beginning at E14.5, before prostate development initiates, through gestation and the first few days after birth. Tbx18 mRNA was barely detectable at E14.5, rose dramatically by E16.5, and dropped slowly throughout the first week postnatal. Shortly after P3 and continuing over the first two weeks of postnatal development, Tbx18 mRNA levels dropped towards the limits of qPCR detection (Fig 1A).
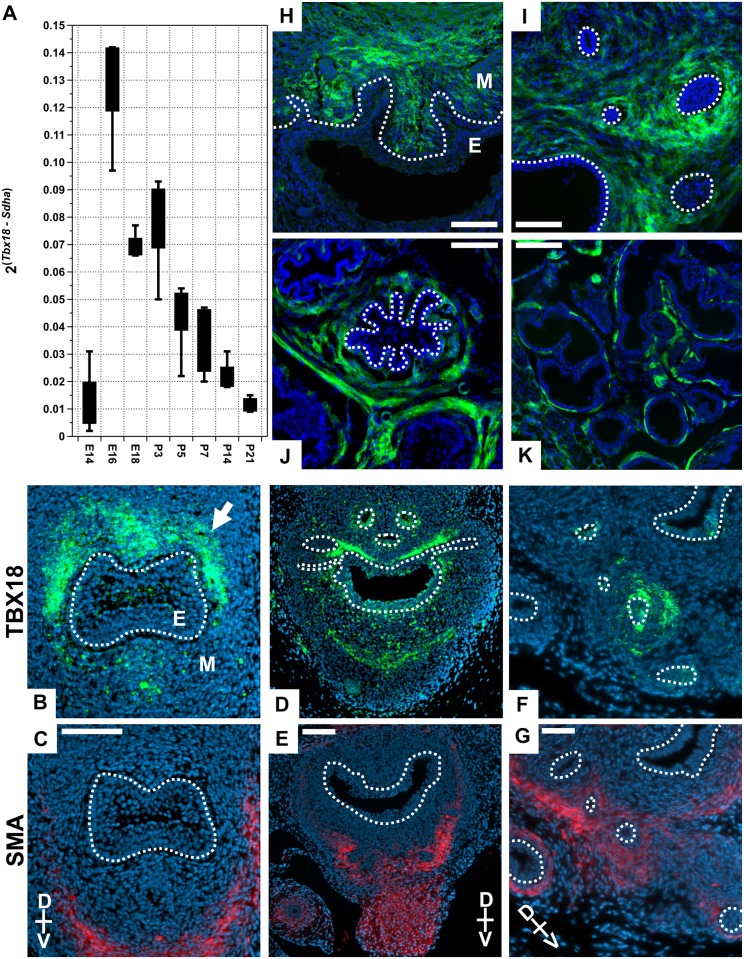
Fig 1. Tbx18 expression in the urogenital sinus mesenchyme.
(A) Quantitative reverse transcription PCR detects Tbx18 mRNA as early as E14.5, with expression peaking around embryonic day 16.5 (for each stage n ≥ 4). (B, D, F) TBX18 immunohistochemistry shows TBX18 is expressed in the dorsal aspect of the UGS in the region of the forming anterior prostate buds at E16.5 (B), P0 (D), and P3 (F). (C, E, G) Smooth Muscle Actin (SMA) IHC on sections adjacent to TBX18 stains showing the proximity of these two expression domains; TBX18 is detected in only a small subset of SMA-positive cells. (H-K) Lineage tracing analysis of Tbx18 expressing cells using the Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J reporter. Green cells express or are descended from Tbx18 expressing cells (in the images presented here, the Tomato signal was excluded for clarity). (H-I) P0. (J-K) P35 sections show Tbx18 descendants primarily occupy the anterior prostate stroma (J), and dorsal prostate stroma (K). M is Mesenchyme, and E is Epithelium. Scale bars are 100μm B and C, 50μm in D-K.
To provide spatial resolution of TBX18 protein localization, we used immunohistochemistry (IHC) with a tested TBX18 antibody [6] in sectioned E16.5 to P3 animals. At E16.5, TBX18 staining was observed primarily on the dorsal aspect of the UGS-M, proximal to the epithelium in regions from which the dorsal and anterior prostate buds will eventually emerge (Fig 1B). At P0, TBX18 was also detected adjacent to the dorsal UGS-E with staining around the proximal regions of the epithelial prostate buds (Fig 1D) and a few isolated cells ventral to the UGS-E. By P3, TBX18 was detected in clusters of cells in the UGS-M proximal to the prostate epithelial buds (Fig 1F). Due to the known role of Tbx18 in smooth muscle formation, we also examined the relationship between TBX18 expression and smooth muscle formation by evaluating alpha-Smooth Muscle Actin (SMA) antibody staining on sections adjacent to those stained for TBX18. Consistent with previous reports of TBX18 and SMA expression, the expression of these two proteins were mostly spatially and temporally separated [2]. SMA staining was frequently observed in cells in the vicinity of TBX18-positive cells, but the two proteins were largely detected in non-overlapping domains (Fig 1C, 1E and 1G). Together these analyses indicated that Tbx18 UGS expression is primarily limited to late gestation and early postnatal stages. The position of the TBX18+ cells adjacent to nascent prostate buds suggested they may contribute to the adult prostate stroma.
We used a lineage-tracing system to identify the adult prostate regions and cell types to which TBX18+ embryonic cells contribute, crossing mice carrying a Cre recombinase knock-in to the Tbx18 locus [38] with the Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG) reporter line. In these animals, only Cre-expressing cells and their descendants will express eGFP [39]. We observed GFP expression, indicating the presence of Tbx18-driven Cre activity, first at E16.5 in the UGS-M (not shown) and at P0 with the same patterns as that detected in IHC (Fig 1H and 1I). In prostates of 5 week-old animals, we observed GFP expression in the periductal stromal smooth muscle layer of the anterior and the dorsolateral lobes confirming that Tbx18-expressing cells contribute significantly to prostate stromal SMCs (Fig 1J and 1K).
Subtle abnormalities in the nascent prostate of Tbx18 mutant mice
Tbx18 expression in the UGS during embryonic and postnatal stages suggested that Tbx18 might play a role in prostate development. However, most prostate development occurs postnatally precluding the use of Tbx18-null mutations to identify later prostate phenotypes. Since ureter expression and development are disrupted in 12Gso mice [6], we reasoned that this hypomorphic allele might be associated with Tbx18 gene expression differences and phenotypes in the prostate as well.
We first evaluated the affects of Tbx18 abrogation by comparing UGS morphology in Tbx18GFP/GFP null mutants [38], hypomorphic 12Gso/Tbx18LacZ [6,35] compound heterozygotes, and wild type littermates at two prenatal stages: E16.5, before the time that prostate budding begins, and E18.5, after the process has initiated. At both of these prenatal stages we observed no morphological differences between animals of the three genotypes (data not shown).
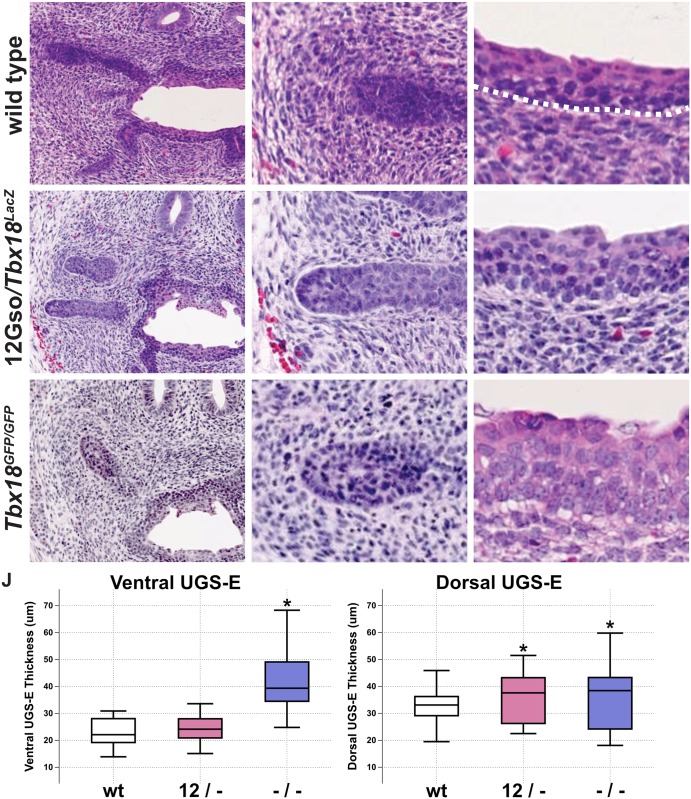
However, at P0, we observed two subtle but consistent changes (Fig 2). We sectioned through the entire abdominal region including the developing prostate in multiple animals (n = 7 of each genotype) and stained slides at regular intervals to compare morphology in a large sampling of sectioning planes. The differences between mutant and wild type prostates at this stage were subtle but consistently observed throughout the organs. First, in animals of both mutant genotypes, the apical surface of the urethral epithelium displayed increased convolutions and irregularities compared to wild type (Tbx18+/+) littermate controls (Fig 2A, 2D and 2G). In the Tbx18-null animals, we also noticed that epithelial cells displayed increased cell size and nuclear volume (arrows in Fig 2C, 2F and 2I). In addition, the epithelium in both mutant genotypes was relatively thicker, with additional layers protruding into the lumen from the dorsal and ventral sides, than was observed in wild type littermate controls (Fig 2C, 2F and 2I). We quantified the differences by measuring the thickness of the urethral epithelium (luminal surface to basement membrane) at ten positions on both the dorsal and ventral aspect in wild type and Tbx18 mutants, and confirmed that the ventral urethral epithelium was increased in Tbx18GFP/GFP mutants, while the dorsal epithelium thickness was increased in both mutants (Fig 2J). We also observed that the characteristic pattern of higher UGS-M cell density around the forming prostate buds of wild type mice was absent in Tbx18GFP/GFP mutant animals and reduced in 12Gso/Tbx18LacZ compared to wild type litter mates (Fig 2B, 2E and 2H).
Fig 2. Tbx18 LOF phenotype in P0 UGS.
(A-I) H&E stains of P0 urogenital sinuses. (A-C) Wild type histology shows high cell density in the mesenchyme surrounding the epithelial prostate buds (A, B), and urethral epithelium is composed of 4–6 cell layers with a smooth apical surface (C). (D-F) 12Gso/Tbx18LacZ compound heterozygotes present an intermediate phenotype in the UGS mesenchyme and the urethral epithelium. (G-I) Tbx18GFP/GFP mutants have very low mesenchymal cell density surrounding epithelial prostate buds. The urethral epithelium in these mutants is increased in thickness with larger cell volumes (compare arrows in C, F, and I). (J) Measurements of the epithelial thickness in the urethral epithelium. The epithelium on the dorsal side is significantly increased in thickness compared to wild type littermates.
Loss of Tbx18 affects the structure and integrity of the anterior prostate lobe
Most of the prostate develops postnatally, with a significant amount of growth and differentiation during adolescent and pubertal stages up to five weeks after birth [7]. We therefore reasoned that more striking morphological differences might be detected in mutant animals at 5 weeks. Although 12Gso/Tbx18LacZ compound heterozygotes only occasionally survive to adulthood, we did observe a striking phenotype in two 5 week-old males we examined (Fig 3). Subtle abnormalities were detected in the dorsolateral lobes, but the most striking phenotypes were limited to the anterior prostate.
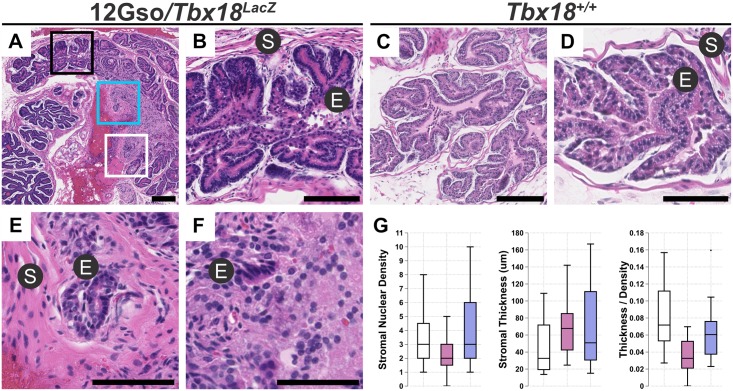
Fig 3. Tbx18 LOF in 12Gso/Tbx18LacZ mutants leads to prostate abnormalities in adults.
The anterior prostates of five week-old 12Gso/Tbx18LacZ mutants include stromal hypertrophy and epithelial disorganization, as revealed by Hematoxylin and Eosin (H&E) stains. Sectioned anterior prostates of 12Gso/Tbx18LacZ mutants (panel A, with areas highlighted in black, blue, and white boxes shown at high resolution in B, E, F respectively) include distal regions with relatively normal morphology (B) indistinguishable from that of wild type littermates (C, D). However, the same sections of mutant prostates show significant stromal hypertrophy (E) and epithelial disorganization and lack a clear boundary between stromal and epithelial compartments (F) in regions proximal to the urethra. (G) Measurement of stromal thickness and numbers of nuclei within the stromal regions confirm that both measurements are significantly different in mutant proximal anterior prostates than in wild type littermate controls. S is stroma; E is epithelium. Scale bars correspond to 100μm in all panels.
When the anterior prostates of these mutants were sectioned along the axis proximal-distal to the urethra, we observed a very similar phenotype in both animals. Specifically, the regions of the anterior ducts located distal of the urethra displayed overtly normal structure, including organized epithelial folds encased by thin layer of condensed smooth muscle stromal cells (Fig 3A and 3B). We could discern no obvious difference between these distal regions of mutant anterior prostates and those of wild type littermate controls (Fig 3C and 3D). However, in the more proximal regions of the same ducts, the mutant anterior prostates were significantly different from those in wild type animals in two respects. First, we noted a marked increase in the thickness of the stromal smooth muscle layer, containing cells that were significantly increased overall in size and misshapen, compared to age-matched wild type prostates (Fig 3A, 3E, 3C and 3D). Second, we observed that epithelium directly adjacent to the disorganized stroma in the mutants lacked the typical well-organized morphology and cell layering seen in comparable wild type sections. Rather, cells in the epithelial compartments of these proximal regions were disorganized and appeared to be loosely associated in the mutant mice (arrowhead in Fig 3E and 3F).
To quantify cell number and cell size we measured the stromal thickness (the distance from the epithelial basement membrane to the distal stromal periphery) at twenty positions around the anterior prostate lobes and counted the number of nuclei along the same linear path of measurement (Fig 3G). We found that the median stromal width was significantly larger in 12Gso/Tbx18LacZ mutants than in wild type littermates on average: 32μm in wild type, versus 67μm and 50.8μm for 12Gso/Tbx18LacZ mutant prostates respectively. By measuring the ratio of the number of nuclei per stromal width, we observed a decrease in the number of nuclei per width in the prostate stroma of 12Gso/Tbx18LacZ mutants (Fig 3G), supporting a hypertrophy model. Thus it appears the abnormal stromal thickness is due to cells occupying the periductal space normally reserved for stromal SMC were enlarged and lacked the condensed, parallel organization observed in wild type mice. In these morphological characteristics, the abnormal stromal cells in Tbx18 mutant prostates resemble myofibroblasts, which are frequently associated with prostate pathologies [30,40,41].
Conditional abrogation of Tbx18 in the prostate
The survival of 12Gso/Tbx18LacZ animals to adulthood was sufficiently rare to impede efficient further investigation. To overcome this problem, we obtained a conditional allele of Tbx18 (Tbx18flox). To remove Tbx18 function only in the developing prostate, we required a tissue specific Cre that would not interfere with Tbx18 function in other tissues that have previously been associated with loss-of-function lethality. We therefore chose to abrogate Tbx18 in the prostate utilizing a well-tested transgenic mouse line expressing Cre-recombinase from a transgenic rat Probasin gene promoter (PB-Cre4) [42] (Fig 4).
Fig 4. Conditional ablation of Tbx18 in the prostate.
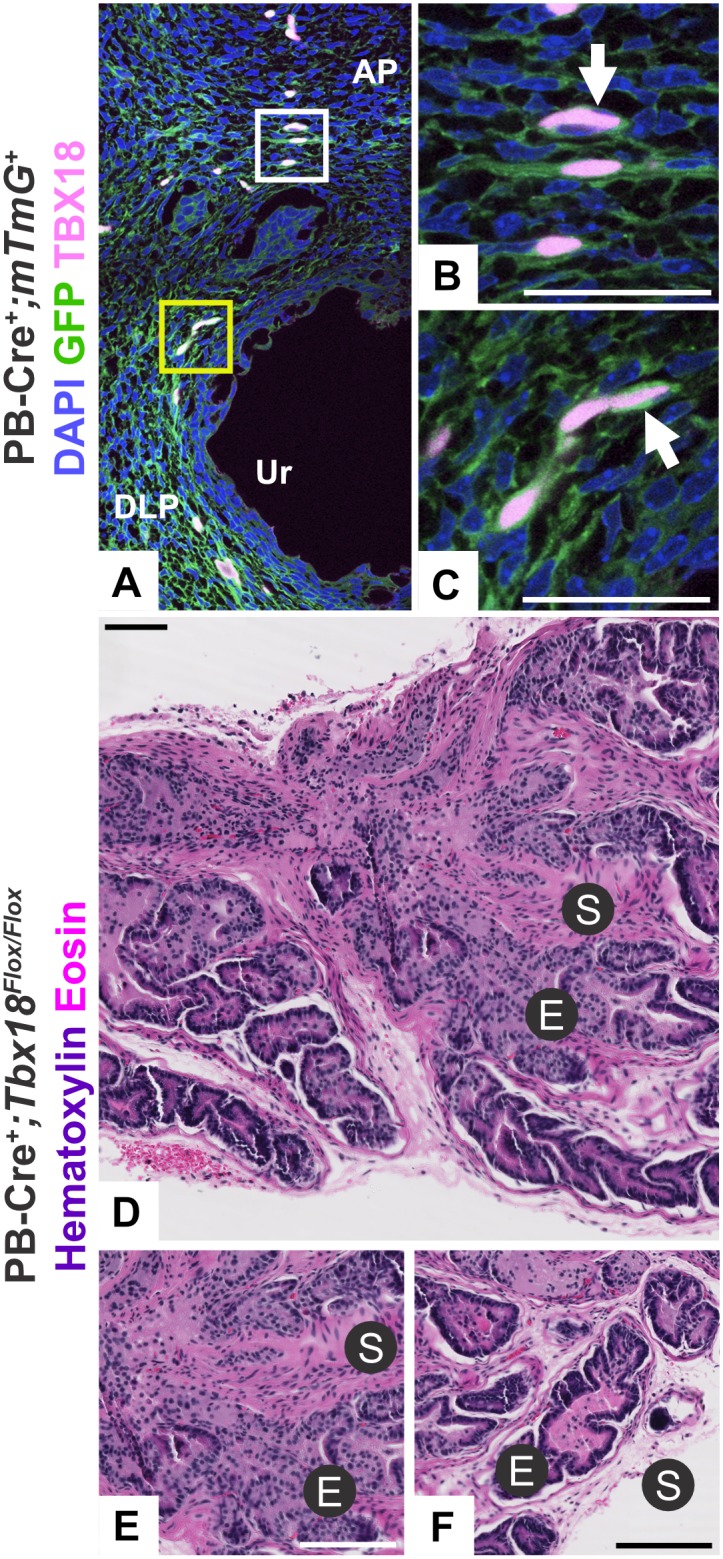
(A) The PB-Cre4 transgene is active at P0 in the prostatic mesenchyme as shown by the mT/mG reporter line, and (B, C) cells with Cre expression co-stain with TBX18 around the anterior buds. (D-F) PB-Cre+; Tbx18Flox/Flox animals display similar pathology to the 12Gso/Tbx18Lacz animal at P35, with (E) urethra-proximal regions of stromal hypertrophy and epithelial disorganization and (F) distal regions of the same ducts displaying normal pathology. Scale bars correspond to 100μm in figures (A, D-F) and 50μm in figure (B,C). AP is anterior prostate mesenchyme. DLP is dorsolateral prostate mesenchyme. Ur is urethra.
PB-Cre4 is expressed at highest levels in the postnatal mouse prostate epithelium and is most commonly used to study epithelial development. However, this transgene is also expressed within the mesenchyme adjacent to the dorsal and anterior lobes, with activity in the mesenchyme reported as early as P1 [17,43]. We confirmed the mesenchymal expression of PB-Cre4 and the overlap between Cre and TBX18 expression in prostate mesenchyme using mT/mG reporter strain co-stained with TBX18 antibody. In PB-Cre4+; mT/mG+ mice, we observed Cre activity in UGS-M cells surrounding the emerging prostate buds, as early as P0 (Fig 4A). In sections co-stained with the TBX18 antibody we confirmed that Tbx18 and PB-Cre4 are co-expressed in a subset of the same stromal cells around the emerging prostate buds at this stage, with TBX18 protein (stained with far red and shown in pink) localized clearly in the nucleus of cells that were also expressing membrane-targeted GFP (green in Fig 4A–4C).
To evaluate the conditional loss of Tbx18 in these cells, we produced PB-Cre4+, Tbx18flox/flox animals and collected prostates at 5.5–6.5 weeks of age (n = 6 animals). The phenotype we observed in these animals was highly consistent with that of the 12Gso/Tbx18LacZ phenotype, characterized by patches of tissue with a thickened prostate stroma surrounding a highly disorganized epithelium. As in 12Gso/Tbx18LacZ prostates, this phenotype was focused primarily in the anterior prostate lobes and in regions proximal to the urethra, with obviously abnormal ducts adjoined to normally structured acini in the same tissue sections (Fig 4D–4F). Therefore, the inheritance of the 12Gso/Tbx18LacZ genotype and deletion of Tbx18 in PB-Cre4-expressing cells yield very similar effects on prostate development.
Immunohistochemical markers confirm the presence of myofibroblasts and loss of epithelial cell identity in mutant prostates
To further characterize these abnormal prostate phenotypes, we stained mutant and control prostate sections for expression of diagnostic molecular markers. We used slides directly adjacent to those highlighted in Figs 3 and 4 for immunohistochemical staining, comparing staining patterns from both types of Tbx18 mutants to wild type age-matched controls, with very similar results (Fig 5). First we asked whether epithelial cell-type balance was being maintained by examining mutant and wild type sections with cytokeratin markers of basal or luminal epithelial cell identity (CK5 and CK8/CK18, respectively) [44]; these slides were co-stained with the SMA antibody to delineate stromal and epithelial layers clearly. Next, we examined prostate sections adjacent to those stained with CK markers, after co-staining with Vimentin (VIM) and SMA. These proteins separately mark fibroblasts and smooth muscle cells, respectively, but when expressed together are characteristic of the myofibroblastic phenotype [30].
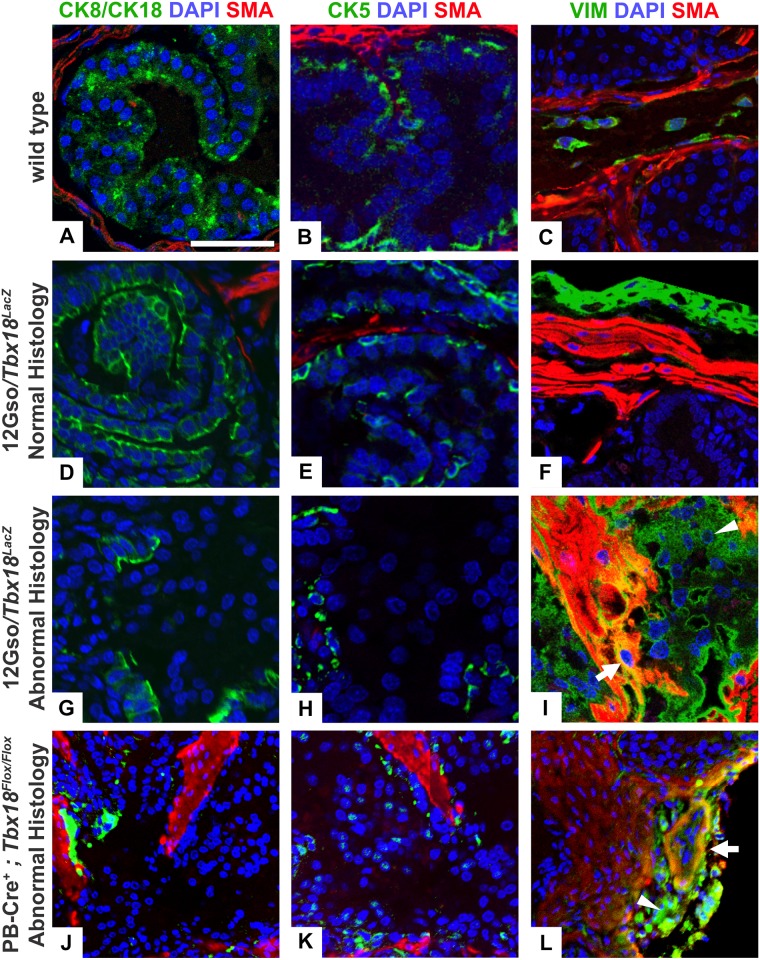
Fig 5. Immunohistochemical analysis with stromal and epithelial markers.
We used immunohistochemistry to examine the distribution of stromal and epithelial markers in sections of 5 week-old (A-C) wild-type, (D-I) 12Gso/Tbx18LacZ, and (J-L) PB-Cre4+; Tbx18flox/flox anterior prostates. IHC was carried out on slides adjacent to those imaged in Figs 3 and 4 and include the same regions of the tissue. (A) Luminal epithelial cells stained with an antibody detecting Cytokeratins 8 and 18 (CK8/CK18, green) form a continuous layer on the apical surface of the epithelial compartment in wild type prostate and (D) in distal ductal regions with normal pathology in the 12Gso/Tbx18LacZ compound heterozygotes. (B, E) Basal epithelial cells stained with Cytokeratin 5 (CK5, green) also stain similarly in the wild type and distal regions of the mutant ducts, forming a discontinuous layer between the apical and basement membrane. (G, H, J, K) In contrast, CK8/CK18 and CK5 staining is significantly reduced in the epithelial compartments of the proximal anterior prostate regions in both 12Gso/Tbx18LacZ (G, H) and PB-Cre4+; Tbx18flox/flox mice (J, K). SMA (red) was co-stained with the Cytokeratin markers to define the smooth muscle layer of the stroma in each section. (C, F) Smooth muscle cells, identified with SMA (red), and fibroblasts, stained with the VIM antibody (green) form apposed but separate layers around the epithelial compartments in wild type animals and in distal regions of normal histology in the anterior prostate of 12Gso/Tbx18LacZ mice. (I, L) However, in both types of Tbx18 mutants we found cells positive for both the VIM and SMA markers (arrow in Fig I, L), indicative of myofibroblasts. In addition, compared to wild type littermates (C), the mutant anterior lobes contained larger numbers of VIM+ cells (green; arrowheads in I, L). Scale bars correspond to 100μm.
Tbx18-mutant anterior lobes contained stromal smooth muscle cells and epithelium at the distal portions of the lobes that matched the organization and staining patterns of wild type lobes at equivalent positions (Fig 5A–5C). In these normally structured regions, a layer of flattened, SMA-positive cells surrounded an organized epithelium. In the epithelium, a basal layer marked by CK5, and a luminal layer of CK8/CK18+ cells was apparent (shown in Fig 5D and 5E). In contrast, cells within the epithelial compartments of the abnormally structured proximal ductal regions in both 12Gso/Tbx18LacZ (Fig G,H) and PB-Cre4+;Tbx18flox/flox mutants (Fig 5J and 5K) displayed significantly reduced expression of both epithelial markers.
We also found that the distribution of SMA and VIM-positive cells was very similar in the normally structured distal regions of mutant anterior prostates and the prostates of wild type littermates. Specifically, VIM+ cells were distributed around the outside of the normally structured ducts in mutant mice, and VIM staining did not co-occur with SMA (Fig 5F). In contrast, in the disorganized proximal regions of the mutant anterior prostates we observed clusters of stromal cells co-staining with VIM and SMA (arrows in Fig 5I and 5L). This pattern was observed in multiple sections across proximal regions of the anterior prostate in both types of Tbx18 mutants (not shown). In addition, we observed unusually large numbers of cells marked by VIM but not with SMA, in the periductal space surrounding the myofibroblastic cells and the abnormal epithelium in the mutant anterior prostate lobes. This staining pattern suggests an abnormal increase in numbers and organization of stromal fibroblasts in the mutant prostates.
Significant gene expression differences presage Tbx18 mutant prostate pathology
The subtle UGS phenotype observed in newborn animals (Fig 3), in conjunction with the fact that Tbx18 transcription was first detected at significant levels in UGS at E16.5 (Fig 1), indicated that the earliest detectable molecular changes associated with Tbx18 deficiency would be discovered between E16.5 and P0. This time period is also critical for the formation of the first prostate buds and marks the beginning of mesenchymal condensation around the nascent prostate buds [7,45]. To determine potential gene expression changes in Tbx18-null UGS, we collected whole UGS from E16.5 and E18.5 Tbx18GFP/GFP mutant male embryos and littermate wild type controls. For each stage, we isolated RNA and performed RNA-Seq, comparing gene expression in mRNA from pooled tissues from each genotype at each stage. Gene expression analysis identified 324 differentially expressed genes (DEGs) at E16.5 and 6101 DEGs at E18.5 with an absolute fold change greater than or equal to 2.0; including genes with absolute fold change above 1.5 at the earlier stage identified a total of 694, many of which were further dysregulated at the later stage (S1 Table). To determine which major functional categories were differentially expressed at these two stages, we performed Gene Ontology (GO) analysis on the DEG sets using the DAVID functional analysis program (Table 1) [46].
Table 1. Functional categories enriched in RNA-seq comparison of E16.5 and E18.5 Tbx18 mutant compared to wild type UGS.
| Enriched Functional Categories | ||
|---|---|---|
| E16.5 | E18.5 | |
| Down-Regulated DEGs | Myofibril | Regulation of transcription |
| Muscle organ development | Zinc finger / zinc ion binding | |
| Muscle system process | Chromosome organization | |
| Muscle cell differentiation | mRNA processing | |
| Sarcoplasmic reticulum | Microtubule organizing center | |
| Cytoskeletal protein binding | Cell cycle | |
| Myosin complex | Ribonucleoprotein complex | |
| Cardiac muscle tissue development | Cellular response to stress | |
| Myogenesis | Programmed cell death | |
| Establishment of protein localization | ||
| Regulation of mesenchymal cell proliferation | ||
| Regulation of epithelial cell proliferation | ||
| Prostate gland growth | ||
| Skeletal system development | ||
| Serine threonine kinase activity | ||
| Regulation of apoptosis | ||
| Up-Regulated DEGs | Peptidase inhibitor activity | Ribosomal protein / ribosome |
| Extracellular region | Mitochondrion | |
| Defense response to bacterium | ncRNA metabolism | |
| Complement and coagulation cascades | Proteinaceous extracellular matrix | |
| Nucleosome assembly | Response to nutrient levels | |
| High density lipoprotein particle | Wnt superfamily | |
| NF-KB binding | ||
| Oxidative phosphorylation | ||
| Regulation of smooth muscle proliferation | ||
| Regulation of Ras GTPase activity | ||
The functions of genes that were significantly up-regulated or down-regulated in E16.5 and E18.5 Tbx18GFP/GFP compared to wild type urogenital sinus (S1 Table) were analyzed separately using the DAVID functional clustering algorithm [46]. The leading GO term in significantly enriched functional clusters are shown in order of their enrichment.
At E16.5 nearly every enriched Functional Category was related to myogenesis and muscle structure. The most highly enriched of these categories included genes that encode structural components of myofibrils including Smooth muscle actins (Acta2, Actg2), Calponin 1 (Cnn1), smooth muscle Myosin heavy chain 11 (Myh11), and Transgelin (Tagln) all of which were significantly down-regulated in mutants at E16.5. Genes encoding protease inhibitors (particularly the Stephin family of Cathepsin inhibitors; Stfa1, Stfa2, Stfa2l1, Stfa3), extracellular components (Col10a1, Gdf15, Fgg, and Fgb) and genes involved in defense response, the complement cascade, and inflammatory response (e.g. S100a8, S100a9, C3, Cd52, Masp2) were significantly up-regulated at this early stage (Table 1).
Despite the fact that major morphological differences were not observed in mutant UGS until P0, RNA-seq analysis of E18.5 mutant UGS detected a massive difference in gene expression between mutant and wild type mice with highly significant functional enrichments. Most notably, down-regulated DEGs were enriched for transcription factors including the key muscle differentiation factors including Mef2a, Mef2c, and Mef2d. At this stage, we also observed significant differential expression of TF genes including Sox9, Hoxd13, Pten and Trp63, all genes with important roles in prostate development and pathologies [17,26,47,48]. Genes regulating apoptosis and the proliferation of mesenchymal and epithelial cells were also down-regulated (Table 1); here we should note that because of its action in other tissues, Tbx18 itself is annotated in both the apoptosis and cell proliferation categories, as are down-regulated DEGs Shh, Trp63 and Fgfr2.
Tellingly, components of the extracellular matrix including eight Collagen, nine Matrix metalloprotease, as well as Wnt genes were also significantly up-regulated (Table 1), all of which have known roles in myofibroblast formation. DEGs associated with regulation of smooth muscle proliferation (including Bmp4, Notch4) were also enriched in the up-regulated genes. We also observed at this stage a massive up-regulation of genes directly involved in the formation of the ribosomes and mitochondrial components which are also up-regulated in myofibroblasts [49]. To confirm the results of the RNA-Seq study, we validated several genes at both developmental time points. Using RT-qPCR on RNA from individual UGS samples, we found that the results were highly consistent with the differential expression values from the pooled tissues used in sequencing (S1 Fig).
To assign particular genes and functions to mesenchymal or epithelial compartments, we cross-referenced our DEG sets with genes enriched in dissected UGS-M or UGS-E in a published microarray study (overlaps marked as M or E in S2 Table) [34]. Through this intersection, E16.5 functions related to regulation of myogenesis, muscle structural proteins, and inflammation were assigned to UGS-M; at E18.5 functions related to mitosis, mesenchymal cell development, and negative regulators of apoptosis were significantly down-regulated, while the C1Q complement system, regulation of cell shape, and Hedgehog signaling were up-regulated in the mesenchymal compartment. At the later stage, functions related to epithelial cell development, apoptosis, apical junctions, and prostate gland development were assigned to the UGS-E, while up-regulated functions related to cell adhesion and cell migration, NADP metabolism, secretion, and Wnt superfamily members were also associated with in the epithelial compartment (S2 Table).
Discussion
This study provides the first evidence that Tbx18 is required for normal differentiation of the prostatic stroma and that it indirectly influences the organization, proliferative control, and cellular identity of prostate epithelium. Using three different mutant genotypes, we show that Tbx18 loss-of-function in the UGS during late embryogenesis and early postnatal life results in grossly disorganized stromal and epithelial layers of the anterior prostate in young adults, with many of the periductal stromal cells displaying a myofibroblastic phenotype.
Analysis of gene expression in mutant embryos revealed a cohort of functionally related, differentially expressed genes that may presage this later pathology. Most notably, genes involved in muscle development were already significantly down-regulated, while genes associated with extracellular matrix remodeling and inflammatory response were up-regulated in mutants as early as E16.5. These up-regulated functions are the same as those known to be associated with the appearance of myofibroblasts and epithelial dysplasia in adults [30], indicating a predisposition for the formation of later pathologies at this very early age. Since E16.5 is also the time when Tbx18 is first robustly detected in wild type prostates, the data suggest an essential role for Tbx18 in the regulatory hierarchy controlling expression of those genes.
By P0, we observed the first signs of morphological differences in developing mutant prostates including decreased density of UGS-M cells surrounding the prostate buds, possibly linked to proliferative failure of Tbx18-deficient mesenchymal cells as observed for ureter development [3]. In addition to these stromal abnormalities, the epithelium displayed increased thickness and subtle loss of organization in Tbx18 mutants at this stage. However, these minor morphological changes belie the significant molecular and genetic changes that have already taken place within these cells.
Prominent among down-regulated DEGs in E18.5 embryos were transcription factors and signaling pathways with important roles in mediating cell-type specific differentiation in prostate. For example Hoxd13 loss-of-function mutations result in reduced anterior prostate size with increased mesenchymal thickness [50]. Foxa1 directly promotes cell growth in the prostate via an AR-dependent pathway and indirectly regulates other genes that are essential to prostate development, including Shh and Bmp4 [51] which were also dysregulated in Tbx18GFP/GFP UGS at E18.5. Sox9, which interacts with Tbx18 in ureter development [52], was also significantly dysregulated at E18.5; this gene is already well established for its critical role in prostate development. For example, inactivation of Sox9 before Androgen Receptor activation results in complete agenesis of the prostate [47,53]; at later developmental stages, Sox9 is required to promote basal epithelial proliferation and is regulated by Wnt signaling through β-catenin [54,55]. Interestingly, many of these same developmental regulators are dysregulated in prostate cancer [23,47,51,55], and their early mis-expression may be linked to the cellular changes we detected in young adult mutant mice.
Although the phenotype of newborn mutant animals does not reflect the massive shift in the prostate gene expression that is already evident at E18.5, the failure of developmental and signaling pathways and the signature of inflammation are manifested clearly in 5 week-old mice. In contrast to the ureter, where the epithelium is reduced in thickness and cell numbers as well as organization in Tbx18-/- mice [2,56], the cells that populate the adult prostate epithelium are not obviously reduced in number and instead, the numbers of cells in the epithelial compartment appear to have increased. These differences may stem from the distinct developmental origins of the epithelial layers and the paracrine factors they produce, as ureter epithelium is of mesodermal origin, and UGS-E is endodermal.
However, these differences in epithelial phenotype may also reflect different fates of Tbx18-deficient stroma in ureter and prostate. In particular, whereas Tbx18-deficient cells in ureter undergo apoptosis and are replaced by a thin layer of fibroblasts (Airik 2006), in prostate these cells appear to be replaced by a myofibroblasts (MFBs), which support the proliferation of epithelial cells [57]. The origins of these MFBs and the role of Tbx18 in their formation are still unclear and will require an in-depth developmental study. MFBs are most commonly thought to differentiate from fibroblasts, and it is possible that Tbx18-prospective cells are first replaced by fibroblasts that further differentiate to MFBs in the prostate environment. We did detect an increase of Vimentin-positive cells in the lumen of mutant glands, and these might serve as a reservoir for MFBs.
However, other possibilities should be considered. For example, Vimentin is expressed widely throughout the prostate stroma during prenatal development, and VIM+SMA+ MFBs are also present during these stages, presumably reflecting an intermediate differentiation state [58,59]. Therefore, one possibility is that in the absence of Tbx18, mesenchymal cells destined for the SMC fate may simply fail to differentiate beyond this transitional stage. Alternatively, MFBs can also arise directly from mesenchymal stem cells that reside in human prostate, a process that is inhibited by the TGFB1-regulated TF, RUNX1 [60]. Runx1 was significantly down-regulated in Tbx18-mutant UGS at E18.5, raising the possibility that Tbx18-deficient mesenchymal precursors are redirected in mutant prostate anterior lobes to differentiate into MFBs. Using the Tbx18flox allele in combination with PB-Cre4 and other mesenchymally-expressed Cre alleles, these possibilities can now be tested.
Whatever their origins, the appearance of MFBs in the Tbx18 mutant stroma provides a potentially novel link between the developmental activities of this T-box factor and predisposition to adult prostate disease. MFBs are associated with multiple types of human prostate pathologies, including prostate cancer as well as benign prostate hypoplasia, and prostatic fibrosis [30,57,61]. Most of the factors that have been linked to MFB differentiation are expressed in adult prostate, either normally or in response to injury, infection, or other inflammatory events. However, as far as we can discern from RT-qPCR, IHC, and lineage tracing studies, Tbx18 is expressed only transiently during early development and is not reactivated in the prostates of either healthy or mutant adults. These data suggest that a deficiency of Tbx18 during a limited period of perinatal development is sufficient to predispose animals to frank prostate disease later in life. We speculate that the loss of Tbx18 sets the stage for later prostate dysfunction through loss of a functional SMC stroma, but also due to the early creation of an inflammatory stromal environment that supports differentiation of MFBs.
Finally, from these data it remains unclear which stromal factors are key to the abnormalities in epithelial proliferation, differentiation and organization we observed since a large number of pathways predicted to influence these processes were dysregulated together in Tbx18 mutant mice. Nevertheless, it is clear from our observation of these phenotypes in several different Tbx18 genotypes that these factors are under the control, either directly or downstream and indirectly, of Tbx18 activity. Future studies will focus on pinpointing the mechanisms of Tbx18 activities in prostate development, and examine in depth the long-term effects of Tbx18 insufficiency during the earliest postnatal stages on prostate health in later adult life.
Supporting Information
In the RNA-Seq experiment we analyzed pools of tissues from three individuals for each genotype and each stage. To validate that the pooled averages reflect individual samples we performed RT-qPCR on individual samples. The Fold Changes observed in RNA-Seq (no error bars) are presented next to the FCs obtained from qPCR on individual animals, with error bars showing the degree of variation between the three samples.
(TIFF)
Differentially expressed genes from comparisons of mRNA from whole urogenital sinus (UGS) dissected from Tbx18Gfp/Gfp null embryos and wild-type littermates at E16.5 and E18.5, respectively (as designated in column A). Column B indicates genes assigned as being enriched in mesenchymal (M) or epithelial (E) UGS compartments by Blum and colleagues [34]; only genes that were assigned unambiguously in that study are marked. Log2 fold change (column L) and other data columns were taken directly from the output of CuffDiff analysis, as described in Methods.
(XLSX)
DEGs marked as M (for UGS-M) or E (UGS-E) in S1 Table were analyzed with the DAVID Functional Annotation Clustering algorithm. Up- and Down-regulated DEGS were analyzed separately. Clusters of categories are represented here by the most highly enriched category in the cluster. Only functional category clusters with DAVID Enrichment Score >1.3 [thus with a geometric mean of combined P values ≥ 0.05, (46)] are listed, in order of enrichment for each class of genes.
(DOCX)
The primer sets listed were used to validate expression in individual E16.5 and E18.5 animals comprising a sequenced pool, with results reported in S1 Fig. Primer sets are named after the target gene, with “-f “after each gene name denoting “forward”, and “-r” denoting “reverse” primer designs.
(XLSX)
Acknowledgments
We thank Dina Leiding and Seeon Lee for expert assistance with mouse husbandry, dissections and tissue collection. This work was supported by grant number DK095685 from the U.S. National Institute of Diabetes and Digestive and Kidney Disorders (awarded to L.S.). The production of the Tbx18flox, Tbx18Cre, and Tbx18GFP alleles was supported by U.S. NIH National Heart, Lung, and Blood Institute R01 grants HL123747, HL117649, and HL074066 (awarded to S.M.E).
Data Availability
Datasets reported in this study have been deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE80083.
Funding Statement
This work was supported by grant number DK095685 from the U.S. National Institute of Diabetes and Digestive and Kidney Disorders (awarded to L.S.). The production of the Tbx18flox, Tbx18Cre, and Tbx18GFP alleles was supported by U.S. NIH National Heart, Lung, and Blood Institute R01 grants HL123747, HL117649, and HL074066 (awarded to S.M.E). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bohnenpoll T, Bettenhausen E, Weiss A-C, Foik AB, Trowe M-O, Blank P, et al. Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Developmental Biology. Elsevier; 2013;380: 25–36. 10.1016/j.ydbio.2013.04.036 [DOI] [PubMed] [Google Scholar]
- 2.Airik R, Bussen M, Singh MK, Petry M, Kispert A. Tbx18 regulates the development of the ureteral mesenchyme. Journal of Clinical Investigation. 2006;116: 663–674. 10.1172/JCI26027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trowe MO, Trowe MO, Airik R, Airik R, Weiss AC, Weiss AC, et al. Canonical Wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development. 2012;139: 3099–3108. 10.1242/dev.077388 [DOI] [PubMed] [Google Scholar]
- 4.Haraguchi R, Matsumaru D, Nakagata N, Miyagawa S, Suzuki K, Kitazawa S, et al. The Hedgehog Signal Induced Modulation of Bone Morphogenetic Protein Signaling: An Essential Signaling Relay for Urinary Tract Morphogenesis. PLoS ONE. 2012;7: e42245–12. 10.1371/journal.pone.0042245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129: 5301–5312. [DOI] [PubMed] [Google Scholar]
- 6.Bolt CC, Elso CM, Lu X, Pan F, Kispert A, Stubbs L. A distant downstream enhancer directs essential expression of Tbx18 in urogenital tissues. Developmental Biology. Elsevier; 2014;392: 483–493. 10.1016/j.ydbio.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimura Y, Cunha GR, Donjacour AA. Morphogenesis of ductal networks in the mouse prostate. Biol Reprod. 1986;34: 961–971. [DOI] [PubMed] [Google Scholar]
- 8.Donjacour AA, Cunha GR. Induction of prostatic morphology and secretion in urothelium by seminal vesicle mesenchyme. Development. 1995;121: 2199–2207. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y, Kregel S, Vander Griend DJ. Formation of human prostate epithelium using tissue recombination of rodent urogenital sinus mesenchyme and human stem cells. J Vis Exp. 2013;: e50327–e50327. 10.3791/50327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu G, Zhau HE, He H, Zhang L, Shehata B, Wang X, et al. Sonic and desert hedgehog signaling in human fetal prostate development. Prostate. 2007;67: 674–684. 10.1002/pros.20563 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang J, Lin Y, Lan Y, Lin C, Xuan JW, et al. Role of epithelial cell fibroblast growth factor receptor substrate 2 in prostate development, regeneration and tumorigenesis. Development. 2008;135: 775–784. 10.1242/dev.009910 [DOI] [PubMed] [Google Scholar]
- 12.Grishina IB, Kim SY, Ferrara C, Makarenkova HP, Walden PD. BMP7 inhibits branching morphogenesis in the prostate gland and interferes with Notch signaling. Developmental Biology. 2005;288: 334–347. 10.1016/j.ydbio.2005.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai K-P, Yamashita S, Vitkus S, Shyr C-R, Yeh S, Chang C. Suppressed Prostate Epithelial Development with Impaired Branching Morphogenesis in Mice Lacking Stromal Fibromuscular Androgen Receptor. Molecular Endocrinology. 2012;26: 52–66. 10.1210/me.2011-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemeth JA, Lee C. Prostatic ductal system in rats: regional variation in stromal organization. Prostate. Wiley Subscription Services, Inc., A Wiley Company; 1996;28: 124–128. [DOI] [PubMed] [Google Scholar]
- 15.Mehta V, Abler LL, Keil KP, Schmitz CT, Joshi PS, Vezina CM. Atlas of Wnt and R-spondin gene expression in the developing male mouse lower urogenital tract. Dev Dyn. 2011;240: 2548–2560. 10.1002/dvdy.22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simons BW, Hurley PJ, Huang Z, Ross AE, Miller R, Marchionni L, et al. Wnt signaling though beta-catenin is required for prostate lineage specification. Developmental Biology. Elsevier; 2012;371: 246–255. 10.1016/j.ydbio.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Xu K, Zhang L, Deng Y, Lee P, Shapiro E, et al. Differentiation of the ductal epithelium and smooth muscle in the prostate gland are regulated by the Notch/PTEN-dependent mechanism. Developmental Biology. Elsevier Inc; 2011;356: 337–349. 10.1016/j.ydbio.2011.05.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin T-M, et al. Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Developmental Biology. Elsevier; 2013;376: 125–135. 10.1016/j.ydbio.2013.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, et al. Activation of canonical Wnt signalling is required for TGFβ-mediated fibrosis. Nature Communications. Nature Publishing Group; 2012;3: 735–12. 10.1038/ncomms1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Untergasser G, Gander R, Lilg C, Lepperdinger G, Plas E, Berger P. Profiling molecular targets of TGF-beta1 in prostate fibroblast-to-myofibroblast transdifferentiation. Mech Ageing Dev. 2005;126: 59–69. 10.1016/j.mad.2004.09.023 [DOI] [PubMed] [Google Scholar]
- 21.Podlasek CA, Barnett DH, Clemens JQ, Bak PM, Bushman W. Prostate development requires Sonic hedgehog expressed by the urogenital sinus epithelium. Developmental Biology. 1999;209: 28–39. 10.1006/dbio.1999.9229 [DOI] [PubMed] [Google Scholar]
- 22.Doles J, Cook C, Shi X, Valosky J, Lipinski R, Bushman W. Functional compensation in Hedgehog signaling during mouse prostate development. Developmental Biology. 2006;295: 13–25. 10.1016/j.ydbio.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 23.Schweizer L, Rizzo CA, Spires TE, Platero JS, Wu Q, Lin T-A, et al. The androgen receptor can signal through Wnt/β-Catenin in prostate cancer cells as an adaptation mechanism to castration levels of androgens. BMC Cell Biol. 2008;9: 4–15. 10.1186/1471-2121-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauman TM, Vezina CM, Huang W, Marker PC, Peterson RE, Ricke WA. Beta-catenin is elevated in human benign prostatic hyperplasia specimens compared to histologically normal prostate tissue. Am J Clin Exp Urol. 2014;2: 313–322. [PMC free article] [PubMed] [Google Scholar]
- 25.Freestone SH, Marker P, Grace OC, Tomlinson DC, Cunha GR, Harnden P, et al. Sonic hedgehog regulates prostatic growth and epithelial differentiation. Developmental Biology. 2003;264: 352–362. [DOI] [PubMed] [Google Scholar]
- 26.Peng Y-C, Joyner AL. Hedgehog signaling in prostate epithelial–mesenchymal growth regulation. Developmental Biology. Elsevier; 2015;400: 94–104. 10.1016/j.ydbio.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng YC, Levine CM, Zahid S. Sonic hedgehog signals to multiple prostate stromal stem cells that replenish distinct stromal subtypes during regeneration. Proceedings of the National Acaday of Science. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsujimura A. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J Cell Biol. 2002;157: 1257–1265. 10.1083/jcb.200202067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303: 848–851. 10.1126/science.1090922 [DOI] [PubMed] [Google Scholar]
- 30.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Can Res. 2002;8: 2912–2923. [PubMed] [Google Scholar]
- 31.Yang X, Chen B, Liu T, Chen X. Reversal of myofibroblast differentiation: A review. European Journal of Pharmacology. Elsevier; 2014;734: 83–90. 10.1016/j.ejphar.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 32.Grabowska MM, DeGraff DJ, Yu X, Jin RJ, Chen Z, Borowsky AD, et al. Mouse models of prostate cancer: picking the best model for the question. Cancer Metastasis Rev. 2014;33: 377–397. 10.1007/s10555-013-9487-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Tripathi P, Guo Q, Coussens M, Ma L, Chen F. Cre/lox recombination in the lower urinary tract. genesis. 2009;47: 409–413. 10.1002/dvg.20515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum R, Gupta R, Burger PE, Ontiveros CS, Salm SN, Xiong X, et al. Molecular Signatures of the Primitive Prostate Stem Cell Niche Reveal Novel Mesenchymal-Epithelial Signaling Pathways. PLoS ONE. 2010;5: e13024–14. 10.1371/journal.pone.0013024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bussen M, Petry M, Schuster-Gossler K, Leitges M, Gossler A, Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes & Development. 2004;18: 1209–1221. 10.1101/gad.300104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Research. 2011;40: D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. genesis. 2000;26: 113–115. [DOI] [PubMed] [Google Scholar]
- 38.Cai C-L, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454: 104–108. 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. genesis. 2007;45: 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 40.Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Can Res. 2003;9: 4792–4801. [PubMed] [Google Scholar]
- 41.Shaw A, Attia S, Bushman W. Prostate stromal and urogenital sinus mesenchymal cell lines for investigations of stromal–epithelial interactions. Differentiation. 2008;76: 599–605. 10.1111/j.1432-0436.2008.00275.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X, Wu J, Huang J, Powell WC, Zhang JF. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech of Dev. 2001;101: 61–69. 10.1016/S0925-4773(00)00551-7 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141: 4698–4710. 10.1210/endo.141.12.7837 [DOI] [PubMed] [Google Scholar]
- 44.Niu Y, Wang J, Shang Z, Huang S-P, Shyr C-R, Yeh S, et al. Increased CK5/CK8-Positive Intermediate Cells with Stromal Smooth Muscle Cell Atrophy in the Mice Lacking Prostate Epithelial Androgen Receptor. Agoulnik I, editor. PLoS ONE. 2011;6: e20202–9. 10.1371/journal.pone.0020202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C, Sensibar JA, Dudek SM, Hiipakka RA, Liao ST. Prostatic ductal system in rats: regional variation in morphological and functional activities. Biol Reprod. 1990;43: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 46.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 47.Huang Z, Hurley PJ, Simons BW, Marchionni L, Berman DM, Ross AE, et al. Sox9 is required for prostate development and prostate cancer initiation. Oncotarget. 2012;3: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurita T. Role of p63 and basal cells in the prostate. Development. 2004;131: 4955–4964. 10.1242/dev.01384 [DOI] [PubMed] [Google Scholar]
- 49.Negmadjanov U, Godic Z, Rizvi F, Emelyanova L, Ross G, Richards J, et al. TGF-β1-Mediated Differentiation of Fibroblasts Is Associated with Increased Mitochondrial Content and Cellular Respiration. Ling F, editor. PLoS ONE. 2015;10: e0123046–12. 10.1371/journal.pone.0123046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podlasek CA, Duboule D, Bushman W. Male accessory sex organ morphogenesis is altered by loss of function of Hoxd13. Dev Dyn. 1997;208: 454–465. [DOI] [PubMed] [Google Scholar]
- 51.Jin HJ, Zhao JC, Ogden I, Bergan RC, Yu J. Androgen Receptor-Independent Function of FoxA1 in Prostate Cancer Metastasis. Cancer Research. 2013;73: 3725–3736. 10.1158/0008-5472.CAN-12-3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Airik R, Trowe M-O, Foik A, Farin HF, Petry M, Schuster-Gossler K, et al. Hydroureternephrosis due to loss of Sox9-regulated smooth muscle cell differentiation of the ureteric mesenchyme. Human Molecular Genetics. 2010;19: 4918–4929. 10.1093/hmg/ddq426 [DOI] [PubMed] [Google Scholar]
- 53.Thomsen MK, Butler CM, Shen MM, Swain A. Sox9 is required for prostate development. Developmental Biology. 2008;316: 302–311. 10.1016/j.ydbio.2008.01.030 [DOI] [PubMed] [Google Scholar]
- 54.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 Is Expressed in Normal Prostate Basal Cells and Regulates Androgen Receptor Expression in Prostate Cancer Cells. Cancer Research. 2007;67: 528–536. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Leav I, Ibaragi S, Wegner M, Hu G-F, Lu ML, et al. SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Research. 2008;68: 1625–1630. 10.1158/0008-5472.CAN-07-5915 [DOI] [PubMed] [Google Scholar]
- 56.Nie X, Sun J, Gordon RE, Cai CL, Xu PX. SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development. 2010;137: 755–765. 10.1242/dev.045757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schauer IG, Rowley DR. The functional role of reactive stroma in benign prostatic hyperplasia. Differentiation. Elsevier; 2011;82: 200–210. 10.1016/j.diff.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayward SW, Baskin LS, Haughney PC, Foster BA, Cunha AR, Dahiya R, et al. Stromal development in the ventral prostate, anterior prostate and seminal vesicle of the rat. Acta Anat (Basel). 1996;155: 94–103. [DOI] [PubMed] [Google Scholar]
- 59.Bierhoff E, Walljasper U, Hofmann D, Vogel J, Wernert N, Pfeifer U. Morphological analogies of fetal prostate stroma and stromal nodules in BPH. Prostate. 1997;31: 234–240. [DOI] [PubMed] [Google Scholar]
- 60.Kim W, Barron DA, San Martin R, Chan KS, Tran LL, Yang F, et al. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proceedings of the National Academy of Sciences. 2014;111: 16389–16394. 10.1073/pnas.1407097111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gharaee-Kermani M, Kasina S, Moore BB, Thomas D, Mehra R, Macoska JA. CXC-Type Chemokines Promote Myofibroblast Phenoconversion and Prostatic Fibrosis. Singh SR, editor. PLoS ONE. 2012;7: e49278–14. 10.1371/journal.pone.0049278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the RNA-Seq experiment we analyzed pools of tissues from three individuals for each genotype and each stage. To validate that the pooled averages reflect individual samples we performed RT-qPCR on individual samples. The Fold Changes observed in RNA-Seq (no error bars) are presented next to the FCs obtained from qPCR on individual animals, with error bars showing the degree of variation between the three samples.
(TIFF)
Differentially expressed genes from comparisons of mRNA from whole urogenital sinus (UGS) dissected from Tbx18Gfp/Gfp null embryos and wild-type littermates at E16.5 and E18.5, respectively (as designated in column A). Column B indicates genes assigned as being enriched in mesenchymal (M) or epithelial (E) UGS compartments by Blum and colleagues [34]; only genes that were assigned unambiguously in that study are marked. Log2 fold change (column L) and other data columns were taken directly from the output of CuffDiff analysis, as described in Methods.
(XLSX)
DEGs marked as M (for UGS-M) or E (UGS-E) in S1 Table were analyzed with the DAVID Functional Annotation Clustering algorithm. Up- and Down-regulated DEGS were analyzed separately. Clusters of categories are represented here by the most highly enriched category in the cluster. Only functional category clusters with DAVID Enrichment Score >1.3 [thus with a geometric mean of combined P values ≥ 0.05, (46)] are listed, in order of enrichment for each class of genes.
(DOCX)
The primer sets listed were used to validate expression in individual E16.5 and E18.5 animals comprising a sequenced pool, with results reported in S1 Fig. Primer sets are named after the target gene, with “-f “after each gene name denoting “forward”, and “-r” denoting “reverse” primer designs.
(XLSX)
Data Availability Statement
Datasets reported in this study have been deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE80083.