Abstract
Speciation and allopolyploidization in cereals may be accompanied by dramatic changes in abundance of centromeric repeated transposable elements. Here we demonstrate that the reverse transcriptase part of Ty3/gypsy centromeric retrotransposon (RT-CR) is highly conservative in the segmental hexaploid Thinopyrum intermedium (JrJvsSt) and its possible diploid progenitors Th. bessarabicum (Jb), Pseudoroegneria spicata (St) and Dasypyrum villosum (V) but the abundance of the repeats varied to a large extent. Fluorescence in situ hybridization (FISH) showed hybridization signals in centromeric region of all chromosomes in the studied species, although the intensity of the signals drastically differed. In Th. intermedium, the strongest signal of RT-CR probe was detected on the chromosomes of Jv, intermediate on Jr and faint on Js and St subgenome suggesting different abundance of RT-CR on the individual chromosomes rather than the sequence specificity of RT-CRs of the subgenomes. RT-CR quantification using real-time PCR revealed that its content per genome in Th. bessarabicum is ~ 2 times and P. spicata is ~ 1,5 times higher than in genome of D. villosum. The possible burst of Ty3/gypsy centromeric retrotransposon in Th. intermedium during allopolyploidization and its role in proper mitotic and meiotic chromosome behavior in a nascent allopolyploid is discussed.
Introduction
Centromeres are responsible for kinetochore assembly that links chromosome to microtubes and thus play a key role in equal chromosome segregation and transmission during cell division. In plants, the centromere sequences are usually represented by DNA satellites interspersed with long terminal repeat (LTR) centromeric retrotransposons (CR), both are associated with CENH3, centromere specific variant of histone H3 [1–7]. CRs were found as highly conserved centromere-specific sequences in grasses [2, 3] and are represented by Cereba in barley, CRW in wheat, CRR in rice, CRM in maize, CRBd1 in Brachypodium distachyon [3–5, 8–11]. As revealed by analysis of the reverse transcriptase (RT) sequences, CRs belong to chromoviruses (Chromoviridae), a lineage of Ty3/gypsy retrotransposons forming a phylogenetically distinct clade designated CRM [12–15].
The copy number of centromeric repeats varies among different species and even ecotypes [16–18]. The abundance of centromeric repeat may be associated with the ploidy level and their amplification and transposition may occur in response to polyploidization event [19]. In bread wheat (Triticum aestivum), a recently formed polyploid (BBAADD), two centromere-specific elements have been found: CRW (also called Cereba) and Quinta that are the youngest elements at the centromeres of common wheat and its diploid ancestors. It was shown that the D subgenome chromosomes contained fewer copies of CRW than either the A or B subgenome chromosomes [18].
The close relative of wheat is intermediate wheatgrass (Thinopyrum intermedium (Host) Barkworth & D.R. Dewey), an important forage crop and a valuable source of genes used for wheat improvement through wide hybridization [20, 21]. Thinopyrum intermedium is a segmental allohexpaloid (2n = 42) with complex genomic constitution. The following symbols were proposed for the designation of Th. intermedium subgenomes: Jr (= J = E), Jvs (= Js = (V-J-R)s = (V-J-H)s), and St (= S = X) [22–31] with J related to Th. bessarabicum (Jb), St to Pseudoroegneria spicata (St) and V to Dasysyprum villosum (V). The St subgenome originated from Pseudoroegneria diploid species has been found unequivocally present in Th. intermedium [32]. The Jr (= J = E) and Jvs (= Js = (V-J-R)s = (V-J-H)s) are closely related subgenomes and assumed to be the progenitor genomes in the lineages of Th. bessarabicum (Jb) and Th. elongatum (Je). The presence of repetitive sequences from V (Dasypyrum) and R (Secale) genomes in the Th. intermedium genome has been revealed by fluorescent in situ hybridization (FISH) [32]. Lu et al. [33] and Li et al. [34] studied chromosome distribution of the CRW (Cereba)-like elements in the genomes of Th. intermedium and related species. However, their studies did not contain comparison in the abundance of the elements between related species.
For the quantification of transposable elements (TE) in genome along with FISH, Southern hybridization, and dot blot analyses SYBR Green-based real-time quantitative PCR (qPCR) assay is applied [35, 36]. Using qPCR assay the dynamics and variation of the TE copy number has been shown in Aegilops and Triticum species [35–37]. However, the abundance of CRs using qPCR assay has not been studied in other Triticeae species so far.
Here, we characterize the reverse-transcriptase parts of the Ty3/gypsy centromeric retrotransposon (RT-CR) and demonstrate their abundance in Th. intermedium, Th. bessarabicum, P. spicata and D. villosum. The results provide valuable information to shed more light on the evolution and speciation in Triticeae.
Results
Sequencing and clusterization of RT-CR sequences
PCR with the TAIDF/R primers using genomic DNA of Th. intermedium, Th. bessarabicum, P. spicata and D. villosum resulted in the amplification of a pool of closely related ~ 490 bp fragments of the reverse transcriptase gene of centromeric retrotransposons (RT-CR). The PCR products were subsequently cloned and at least 20 clones were randomly selected for sequence analysis from each species. While some clones contained identical sequences only unique sequences were published in GenBank (accessions KR873398-KR873420). Homology-based searching (NCBI BLASTn and Conservative Domain Search) confirmed the similarity of the isolated sequences to the RT domains of the published centromeric RT-CR Ty3/gypsy retrotransposons of other species. The multiple alignment of the cloned RT-CR sequences revealed that they are highly homologous between each other (S1 Fig).
Phylogenetic relationships
Phylogenetic relationships were established using nucleotide and deduced amino acid sequences. The dendrogram from DNA sequences was generated by aligning the cloned RT-CR nucleotide sequences (numbers of RT-CR SNP variants: Th. bessarabicum– 4, Th. intermedium– 9, P. spicata– 5, and D. villosum– 6) with 964 homologous to them sequences from GenBank found using NCBI BLAST search (Nucleotide collection). All RT-CRs sequences of Th. intermedium, Th. bessarabicum, D. villosum and one sequence of P. spicata shared a cluster with T. aestivum, H. vulgare, and S. cereale (S2 Fig, group 1); another cluster was shared by three RT-CRs of P. spicata together with Z. mays and O. sativa (S2 Fig, group 2). Among RT-CR sequences found using NCBI Nucleotide collection search database a great majority (632) belong to Triticum aestivum chromosome 3B cultivar Chinese Spring and was represented both in group 1 (Th. intermedium, Th. bessarabicum, D. villosum and one sequence of P. spicata) and group 2 (P. spicata) (S2 Fig).
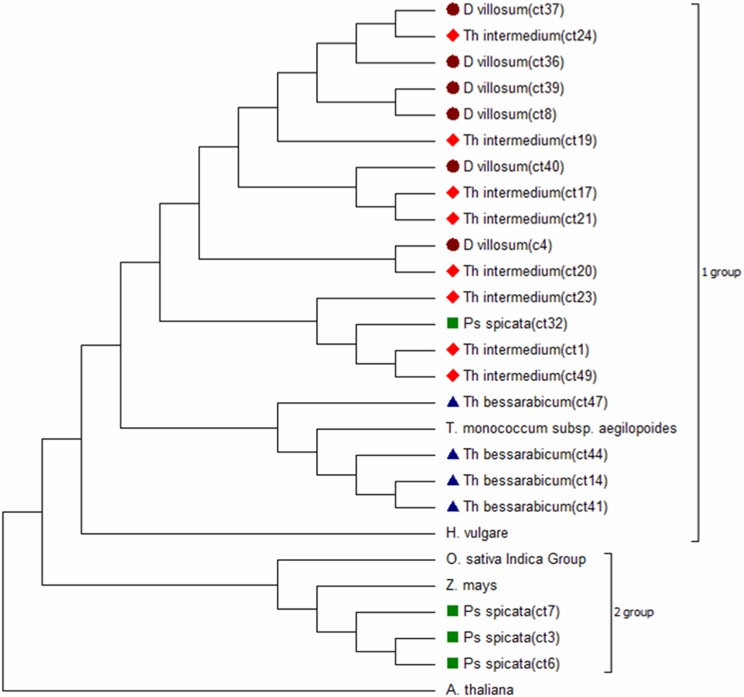
Thereafter, the phylogenetic tree from amino acid sequences was constructed by aligning RT-CR sequences obtained in this study with the RT domains of the known centromeric repeats of rice (DQ458289.1), maize (AAM94350.1), wheat (ABI96971.1) and barley (AAK94517.1) (Fig 1). The RT-CR element of A. thaliana (AAF79348.1) was used as an out-group. The RT-CRs of Th. intermedium, Th. bessarabicum, D. villosum and one of P. spicata formed one clade with RT sequences of wheat (CRW) and barley (Cereba) (Fig 1, group 1). Three sequences of the P. spicata RT-CR were more distant from others and formed their own clade with rice (CRR) and maize (CRM) (Fig 1, group 2).
Fig 1. The dendrogram of the deduced RT-CR amino acid sequences and their homologs from the NCBI Nucleotide collection search database.
The dendrogram was constructed using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 1000 replicates. The colored bullets indicate the RT-CRs obtained in the present study.
Thus, in both cases the dendrograms of amino and nucleic acid sequences show similar pattern of clusterization of our RT-CRs and RT-CR sequences from wheat, rye, barley and maize.
FISH signal on different genomes
To physically localize RT-CRs FISH was performed on the chromosomes of Th. bessarabicum, P. spicata, D. villosum, and Th. intermedium using the labeled PCR products obtained with TAID_F/R primers and the DNA of the corresponding species as a template. The PCR probes obtained from genomic DNA of Th. intermedium, P. spicata, D. villosum and Th. bessarabicum hereinafter are referred to as Thin, Pssp, Davi and Thbe, respectively. FISH resulted in clear signals located in the centromeric regions of all chromosomes of the studied species (Fig 2).
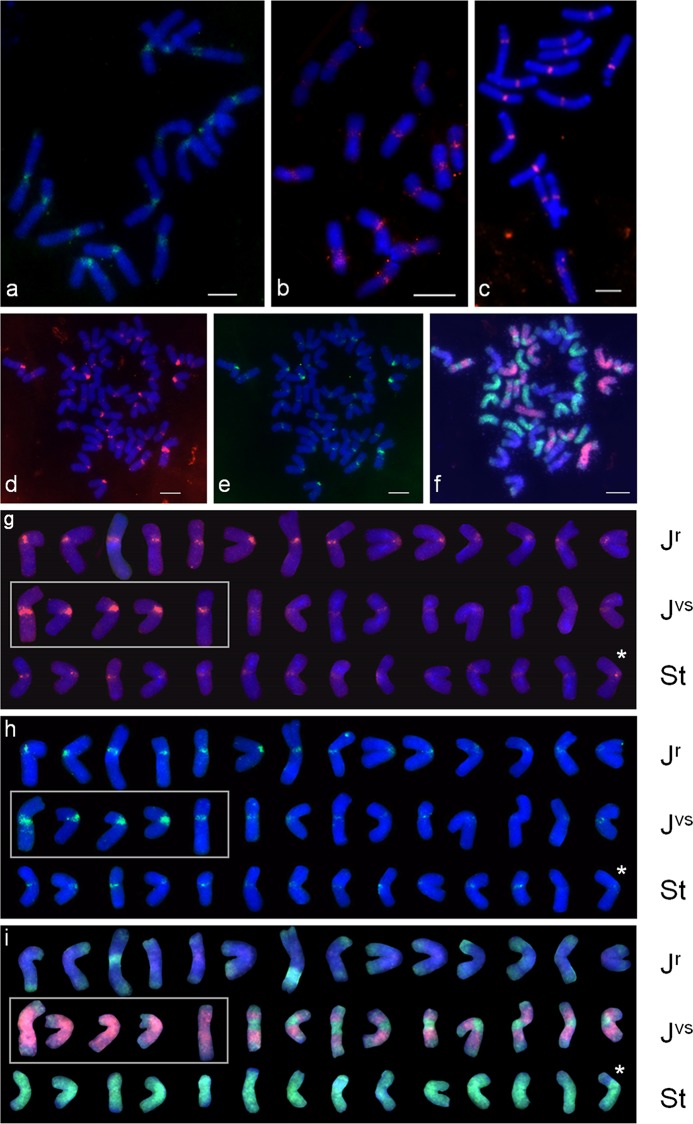
Fig 2. Hybridization of the RT-CR probes on chromosomes of D. villosum, P. spicata, Th. bessarabicum, and Th. intermedium.
a-c FISH analysis using RT-CR probes on the root-tip cells at mitotic metaphase in a) D. villosum, b) P. spicata, c) Th. bessarabicum; the metaphase plates are probed with Davi (a, green), Pssp (b, pink) and Thbe (c, pink). Bar = 5 μm. d-f Sequential multicolor FISH and multicolor GISH on the root-tip cells at mitotic metaphase in Th. intermedium: d) the mcFISH results using the Thbe probe (red); e) the same cell in (d) with Pssp probe (green); f) GISH analysis for the same cell in (d) and (e) with the labeled genomic DNA of P. spicata (green) and D. villosum (pink) as probes and T. aestivum genomic DNA (ABD genome) as a block. Bar = 10 μm. g-i Karyotype of chromosomes of Th. intermedium from (d-f) with the results of sequential multicolor FISH and multicolor GISH. Chromosomes organized into genomes Jr, Jvs and St according to St- and V-genome DNA labeling and FISH signal intensity in centromeric region. The Jvs chromosomes grouped into Jv chromosomes (in frame) and Js chromosomes. The translocated chromosome marked with asterisk. g) Chromosomes with Thbe probe (red); h) The same chromosomes with Pssp probe (green); i) The same chromosomes labeled with genomic DNA of P. spicata (St genome, green) and D. villosum (V genome, pink) as probes and T. aestivum genomic DNA (ABD genome) as a block. Chromosomes counterstained with DAPI (blue).
In diploid species, although slightly varying between individual chromosomes within some metaphase plates, the signal intensity was uniformly bright on all chromosomes (Fig 2A, 2B and 2C). In Th. intermedium, the Thin probe hybridized to the centromeres of all chromosomes but the intensity of the FISH signals varied strongly among individual chromosomes within metaphase plates. Such differences were observed on all the analyzed metaphase plates. The strongest signals were observed on five chromosomes thereby visually distinguished from other chromosomes of the complement. The hardly visible signals were detected on eight chromosomes. The remained 29 chromosomes of Th. intermedium possessed FISH signals with the intermediate intensity.
The question appeared whether the differences in signal intensity in Th. intermedium are due to the (sub)genomic specificity of the probe used (for example, due to the presence of (sub)genome specific sequences) or it is a result of different RT-CR abundance on the chromosomes. To answer the question a cross multicolor FISH was performed on the metaphase plates of Th. intermedium with the probes Thbe, Pssp, and Davi. If all probes hybridized specifically to the same subgenomes from which they were obtained, this would mean that they were genome-specific. In case if the sample obtained by PCR with DNA from different species with the same fluorescence intensity would hybridize to the same chromosomes of Th. intermedium metaphase plates—this would mean that the difference was in the copy number of RT-CR in different chromosomes. Three multicolor FISH experiments were performed on the Th. intermedium chromosomes using simultaneously two probes, namely: i) Pssp and Thbe (Fig 2D and 2E); ii) Davi and Thbe (data not shown); iii) Davi and Pssp (data not shown). In all FISH experiments both probes demonstrated the same chromosomal pattern of hybridization (strong and faint signals on particular chromosomes, Fig 2D and 2E) and the intensity of FISH signal on individual chromosomes did not depend on the probes used.
To identify the chromosome sets belonged to each subgenome of Th. intermedium sequential FISH/multicolor GISH (mcGISH) was carried out on the same metaphase plates. McGISH analysis on mitotic chromosomes of Th. intermedium (Fig 2F) distinguished 14 chromosomes of Jr subgenome (blue with green subtelomeric signals), St subgenome (green, one of which with translocation) and Jvs subgenome (pink with green subtelomeric and/or pericentromeric signals). Five chromosomes of Jvs subgenome demonstrated green subtelomeric signal localization (designated Jv) while nine with both subtelomeric and pericentromeric signal localization (designated Js) (Fig 2F and 2I). Thus, five Jv- and nine Js-chromosomes were identified in Th. intermedium chromosome set. In previous studies, the number of Js chromosomes varied from eight to ten in Th. intermedium [23, 27, 38, 39]. We also found one St chromosome with translocation in the short arm from Jr chromosome (Fig 2I, asterisk). Previously, Kishii et al. [23] and Mahelka et al. [39] reported about translocation between chromosomes of different Th. intermedium subgenomes. These results suggest that the stabilization of the allopolyploid genome of Th. intermedium is still an ongoing process.
The comparison of FISH and GISH results showed that the strongest FISH signals from RT-CR PCR probe are located on all Jv chromosomes while the faintest signals were detected on four St chromosomes and four Js chromosomes. Other chromosomes carried the signal of intermediate intensity (Fig 2G and 2H).
On average, the total signal intensity of RT-CR probe on the Th. intermedium subgenomes can be ranged as follows: Jv>Jr> {Js≈St}.
qPCR
We performed qPCR with the primers TAID_F /R on the genomic DNA of Th. bessarabicum, P. spicata, and D. villosum to assess the relative quantity of RT-CR sequences. The analysis revealed that RT-CR content per genome in Th. bessarabicum is ~2 times and P. spicata is ~1,5 times higher than in genome of D. villosum.
Based on the results of qPCR analysis the genomes can be ranged according to the abundance of RT-CR as follows: Jb > St > V.
Discussion
The activation of CRs in Th. intermedium (JrJvsSt) might have happened after or as a result of allopolyploidization event. Although polyploidy-induced transposition burst of TE in plants is not a general rule it can be TE family- and species-specific and apparently fasten genome evolution in the long term [37, 40, 41]. In our study, qPCR revealed that D. villosum (V, related to Jvs subgenome) has significantly less copies of Ty3/gypsy CR than candidate donors for Jr (Th. Bessarabicum, Jb) and St (P. spicata, St) subgenomes. These results differ from those that could be expected based on the FISH results obtained for chromosomes of each subgenome of Th. intermedium, namely, that four Jv chromosomes are more abundant in CRs in centromeres than other chromosomes. Assuming that D. villosum, Th. bessarabicum, P. spicata are the donors for Jvs, Jr and St subgenomes of Th. intermedium (or very close to them), respectively, the observed differences in centromeric FISH signal intensity between chromosomes of different subgenomes in Th. intermedium cannot be explained by the differences in CR abundance in the genomes of donor species. Such difference may be associated with CR burst after polyploidization as it was shown in some Triticeae. Yakov et al. showed evolutionary- and revolutionary-scale increase in quantity of certain TEs in T. durum and T. aestivum in comparison to its candidate subgenome donors Ae. speltoides, Ae. taushcii, Ae. searsii and T. urartu [37]. In wheat, it was found that the time of burst of centromeric retrotransposons CRW and Quinta coincides with the time of the origin of tetraploid wheat [19, 42–44].
Although according to qPCR experiment in present study D. villosum contains the least amount of CRs, the polymorphism between different populations of diploids as well as that the absence of knowledge about the exact subgenome donors of Th. intermedium should be taken into consideration. Therefore, the alternative hypothesis also should be considered: the progenitor of Jv chromosomes had the CRs burst before polyploidization and its high abundance might have been a necessary condition for Jv to be incorporated into a newly formed polyploid genome and ensured its meiotic stability.
The stability of Th. intermedium as an allopolyploid may be associated with the expansion of CRs in the centromeric region of Jv chromosomes and recombinant nature of Js chromosomes. Centromeric and pericentromereic regions of the chromosomes are critical areas for the differentiation of subgenomes in polyploids during meiosis [45–48]. In Th. intermedium as an allopolyploid, the proper meiotic and mitotic behavior of chromosomes is crucial for its existence as a species. Our subsequent FISH/GISH experiments showed that the strong FISH signal belongs to all Jv chromosomes while the faintest signals were detected on four St chromosomes and four Js chromosomes. We can suggest that the expanded CR region in Jv chromosomes and recombined St-specific pericentromeric region in Js chromosomes prevents them from pairing with homologous chromosomes of St and Jr subgenomes in meiosis. Notably, the signal on Jr chromosomes on average was stronger than on St chromosomes that also may contribute to certain extent to their differentiation at meiosis. It was shown that genomic changes resulted from TE transposition may impede the homeologous chromosome pairing facilitating homologous pairing during meiosis [49, 50]. Similarly, Liu et al. showed that in T. aestivum that D subgenome chromosomes contained fewer copies of CRW than either the A or B subgenome chromosomes [18]. In our sequential GISH/FISH experiments on T. aestivum cv. Tulaykovskaya 100 (with 6Jr(6D) substitution [51]), the probes Thbe, Thin, Pssp, and Davi hybridized to the centromeric regions and the observed variation in FISH signal intensity among T. aestivum chromosomes was not as sharp as in Th. intermedium, the centromeric signal in 6Jr was fainter than in wheat chromosomes (S3 Fig). Therefore, the variation in CR abundance between Th. intermedium subgenomes exceeds that in wheat.
Based on the results of the analysis of nucleic acid and deduced amino acid sequences of RT-CR, we propose that there might have been a pool of different CR sequences in ancestral species. During speciation some of them predominantly amplified in different species while copy number of other RT variants did not undergo significant changes. This assumption is based on that although RT-CR variants of the studied species were included into two separate clades, the sequences of T. aestivum chromosome 3B were represented in both groups. This hypothesis is in agreement with “library” hypothesis that suggests the co-existence of different variants of centromere tandem repeats, that over time undergo through expansion and shrinkage and eventually the replacement of the most abundant variant with a different variant [52].
In conclusion, we demonstrated that the reverse transcriptase part of Ty3/gypsy centromeric retrotransposon (RT-CR) is highly conservative in segmental hexaploid Th. intermedium (JrJvsSt) and its possible diploid progenitors but the abundance of the Ty3/gypsy CR varied to a large extend. Our results may provide valuable information to shed more light on the evolution and speciation in Triticeae.
Materials and Methods
Plant material
The following accessions were used in the experiments: Th. intermedium (PI 401200), Th. bessarabicum (PI 531711), D. villosum (W6 21717), P. spicata (PI 635993) obtained from Germplasm Research International Network; common wheat cv. Ivolga and cv. Tulaykovskaya 100 with 6Jr(6D) substitution [51].
DNA extraction, PCR amplification, cloning and sequencing
DNA was isolated from young leaves or seedlings according to Bernatzky and Tanksley (1986) [53]. The primers TAID_F 5’-TGGTACTTGGCGTCTGTGTG-3’ and TAID_R 5’-CTCGATTCCCTGTGGAGTA-3’ were designed based on conservative nucleic acid sequence of the reverse transcriptase gene of the CRM retrotransposon. Each PCR mixture (25 μl) contained 0.5 μl of DNA (50–300 ng) solution, 0.5 μl of each primer (10 pM/μL), 200 μM of each dNTP, 2.5 μM MgCl2, 2.5 μl of 10×Taq buffer, and 0.5 U of Taq polymerase. The PCR was performed at the following conditions: 5 min at 95°C; 35 cycles: 30 s at 95°C, 30 s at 58°C, 30 s at 72°C; 10 min at 72°C. The PCR products obtained in all analyzed species were run on agarose gel, and one distinct band approximately 500 bp was observed.
The primers WxF3 (5-TCTGGTCACGTCCCAGCTCGCCACCT-3) and WxVT1R (5-ACCCCGCGCTTGTAGCAGTGGAAGT-3) [54] were used for amplification and sequence exon I GBSSI gene and from species Th. bessarabicum (PI 531711), D. villosum (W6 21717), P. spicata (PI 635993) (S4 Fig). Each PCR mixture (25 μl) contained 1.0 μl of DNA (50–300 ng) solution, 1.0 μl of each primer (10 pM/μL), 200 μM of each dNTP, 2.5 μM MgCl2, 2.5 μl of 10×Taq buffer, and 0.5 U of Taq polymerase. The PCR was performed at the following conditions: 5 min at 95°C; 40 cycles: 30 s at 95°C, 60 s at 64°C, 120 s at 72°C; 10 min at 72°C.
The amplified products were cloned into pGEMT®-Easy Vector in E. coli strain DH10B. The individual clones were sequenced using ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations.
Sequence analysis and phylogenetic relationships
Alignment and translation of the sequences was performed using Mega6 software [55]. A search for homologous sequences was performed with a BLAST search (http://blast.ncbi.nlm.nih.gov). A search for conserved domains within a coding nucleotide sequences was performed using NCBI's CD-Search (http://ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Nucleic acid sequences of our RT-CRs were compared with 964 DNA sequences found using NCBI BLAST search (Nucleotide collection) by Mega6 software. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The bootstrap consensus tree inferred from 1000 replicates [55]. The sequences were analyzed without primer regions.
Deduced amino acid sequences of our RT-CRs were compared with the RT domains of known centromeric repeats of rice (DQ458289.1), maize (AAM94350.1), wheat (ABI96971.1) and barley (AAK94517.1) by Mega6 software. The dendrogram was constructed using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 1000 replicates [55]. The RT-CR element of A. thaliana (AAF79348.1) was used as an out-group.
In situ hybridization
Chromosome preparations were carried out as described in [56].
The PCR products obtained from Th. bessarabicum and Th. intermedium were labeled with biotin-16-dUTP, from P. spicata with digoxigenin-11-dUTP, from D. villosum with biotin-16-dUTP or digoxigenin-11-dUTP by PCR according to the manufacturer’s instructions (Roche, Germany); the probes were designated Thbe, Thin, Pssp, and Davi, respectively. One- and multicolor FISH experiments were performed as described in Karlov et al. (2003) [57]. The chromosomes were counterstained with 1 mg/ml DAPI and mounted in Vectashield (Vector laboratories, UK).
After post-hybridization washing of Th. intermedium chromosome mitotic preparations as described in Komuro et al. [58] multicolor genomic in situ hybridization (mcGISH) was conducted on the same slides as described in Kishii et al. [23] with modifications described in Salina et al. [51]. The probes were the P. spicata (St genome) and D. villosum (V genome) genomic DNA (50 ng/preparation) labeled with digoxigenin-11-dUTP and with biotin-16-dUTP, respectively, by nick translation according to the manufacturer’s instructions (Roche, Germany). The chromosomes were counterstained with 1 mg/ml DAPI and mounted in Vectashild (Vector laboratories, UK).
An AxioImager M1 fluorescent microscope (Zeiss) was used to observe chromosome preparations. The metaphase plates with fluorescent signals were photographed with a monochrome AxioCam MRm CCD camera and visualized using Axiovision software (Zeiss).
Quantitative PCR
To estimate relative quantity of RT-CR sequence in the genomes of the studied species qPCR assay was performed according to Yaakov et al. (2013). Each template for qPCR analysis was run in triplicate reactions (technical replicates), each consisting of 2.5 μL of reaction mix containing SYBR Green I + ROX (Syntol LTD), 0.78, 1.56, 2.00, 3.12, 6.25, 12.50, 25.00 ng of DNA template, 1.0 μL of each forward and reverse primer (10 pM/μL). The relative quantity (RQ) of RT-CR sequences was calculated according to Yaakov et al. [37]; each reaction was compared to amplification of the GBSSI gene, as we suppose that it is found in one copy in each genome [39, 54, 59–61]. Primers for GBSSI gene sequence were developed on the basis of multiple alignment of the exon 1 sequences for species Th. bessarabicum, P. spicata, D. villosum received by us and validated using conventional PCR followed by gel electrophoresis (S1 Table, S4 and S5 Figs). The qPCR was performed at the following conditions: 10 min at 95°C, then 40 cycles of 15 s at 95°C and 20 s at 58°C and 30 s at 72°C. The normalized quantities were then compared to the quantity in D. villosum (W6 21717) as it showed the least quantity of RT-CR sequence.
Supporting Information
(TIF)
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The bootstrap consensus tree inferred from 1000 replicates.
(PDF)
a) Hybridization of the Thin probe on chromosomes of Tulaykovskaya 100. b) Identification of 6Jr chromosomes of Th. intermedium in Tulaykovskaya 100 by multicolor GISH with the labeled genomic DNA of P. spicata (green) and D. villosum (pink, signal absent) (blocked with genomic DNA of T. aestivum cv. Ivolga). Chromosomes counterstained with DAPI (blue). Chromosome 6Jr shown with asterisk. Bar = 10 μm.
(TIF)
The positions of primers DwaxyQ used in qPCR analysis are indicated in arrows and color highlighted.
(JPG)
As a negative control (NC), water was served as a template in the PCR reaction. A 100 bp size-ladder was used.
(JPG)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Jiang J, Nasuda S, Dong F, Scherrer CW, Woo SS, Wing RA, et al. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA. 1996;93: 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JT, Dong F, Jackson SA, Song J, Jiang J. Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 1998;150: 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Presting GG, Malysheva L, Fuchs J, Schubert I. A Ty3-gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 1998;16: 721–728. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu MH, et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14: 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36: 138–145. [DOI] [PubMed] [Google Scholar]
- 8.Dong F, Miller T, Jacson SA, Wang GL, Ronald PC, Jiang J. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci USA. 1998;95: 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagaki K, Song J, Stupar RM, Parokonny AS, Yuan Q, Ouyang S, et al. Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics. 2003;163: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagaki K, Neumann P, Zhang D, Ouyang S, Buell CR, Cheng Z, et al. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol. 2005;22: 845–855. [DOI] [PubMed] [Google Scholar]
- 11.Qi LL, Wu JJ, Friebe B, Qian C, Gu YQ, Fu DL, et al. Sequence organization and evolutionary dynamics of Brachypodium-specific centromere retrotransposons. Chromosome Res. 2013;21: 507–521. 10.1007/s10577-013-9378-4 [DOI] [PubMed] [Google Scholar]
- 12.Gorinsek B, Gubensek F, Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21: 781–798. [DOI] [PubMed] [Google Scholar]
- 13.Gorinsek B, Gubensek F, Kordis D. Phylogenomic analysis of chromoviruses. Cytogenet Genome Res. 2005;110: 543–552. [DOI] [PubMed] [Google Scholar]
- 14.Kordis D. A genomic perspective on the chromodomain-containing retrotransposons: chromoviruses. Gene. 2005;347: 161–173. [DOI] [PubMed] [Google Scholar]
- 15.Neumann P, Navrátilová A, Koblížková A, Kejnovský E, Hřibová E, Hobza R, et al. Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mobile DNA. 2011;2: 4 10.1186/1759-8753-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA. 2005;102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Miura A, Takashima K, Kakutani T. Ecotype-specific and chromosome-specific expansion of variant centromeric satellites in Arabidopsis thaliana. Mol Genet Genomics. 2007;277: 23–30. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Yue W, Li D, Wang RR, Kong X, Lu K, et al. Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma. 2008;117: 445–56. 10.1007/s00412-008-0161-9 [DOI] [PubMed] [Google Scholar]
- 19.Li B, Choulet F, Heng Y, Hao W, Paux E, Liu Z, et al. Wheat centromeric retrotransposons: the new ones take a major role in centromeric structure. Plant J. 2013;73: 952–65. 10.1111/tpj.12086 [DOI] [PubMed] [Google Scholar]
- 20.Wang RR-C. Agropyron and Psathyrostachys In: Chittaranjan K, editor. Wild Crop Relatives: Genomic and Breeding Resources Cereals. Springer Heidelberg; Dordrecht London New York; 2011. pp. 77–108. [Google Scholar]
- 21.Mujeeb-Kazi A, Kazi AG, Dundas I, Rasheed A, Ogbonnaya F, Kishii M, et al. Genetic diversity for wheat improvement as a conduit to food security. Advances in Agronomy. 2013;122: 179–258. [Google Scholar]
- 22.Chen Q, Conner RL, Laroche A, Thomas JB. Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome. 1998;41: 580–586. [PubMed] [Google Scholar]
- 23.Kishii M, Wang RRC, Tsujimoto H. GISH analysis revealed new aspect of genomic constitution of Thinopyrum intermedium. Czech J Genet Plants Breed. 2005;41: 92–95. [Google Scholar]
- 24.Liu C, Yang ZJ, Jia JQ, Li GR, Zhou JP, Ren ZL. Genomic distribution of a long terminal repeat (LTR) Sabrina-like retrotransposon in Triticeae species. Cereal Res Commun. 2009;37: 363–372. [Google Scholar]
- 25.Chang ZJ, Zhang XJ, Yang ZJ, Zhan HX, Li X, Liu C, et al. Characterization of a partial wheat–Thinopyrum intermedium amphiploid and its reaction to fungal diseases of wheat. Hereditas. 2010;147: 304–312. 10.1111/j.1601-5223.2010.02156.x [DOI] [PubMed] [Google Scholar]
- 26.Hu LJ, Li GR, Zeng ZX, Chang ZJ, Liu C, Zhou JP, et al. Molecular cytogenetic identification of a new wheat-Thinopyrum substitution line with stripe rust resistance. Euphytica. 2011;177: 169–177. [Google Scholar]
- 27.Deng C, Bai L, Fu S, Yin W, Zhang Y, Chen Y, et al. Microdissection and Chromosome Painting of the Alien Chromosome in an Addition Line of Wheat–Thinopyrum intermedium. PLoS ONE. 2013;8: e72564 10.1371/journal.pone.0072564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahelka V, Kopecky D, Baum BR. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol Biol Evol. 2013;30: 2065–2086. 10.1093/molbev/mst106 [DOI] [PubMed] [Google Scholar]
- 29.Bao Y, Wu X, Zhang C, Li X, He F, Qi X, et al. Chromosomal constitutions and reactions to powdery mildew and stripe rust of four novel wheat-Thinopyrum intermedium partial amphiploids. J Genet Genomics. 2014;41: 663–666. 10.1016/j.jgg.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 30.Danilova TV, Friebe B, Gill BS. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet. 2014;127: 715–730. 10.1007/s00122-013-2253-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RR, Larson SR, Jensen KB, Bushman BS, DeHaan LR, Wang S, et al. Genome evolution of intermediate wheatgrass as revealed by EST-SSR markers developed from its three progenitor diploid species. Genome. 2015;58: 63–70. 10.1139/gen-2014-0186 [DOI] [PubMed] [Google Scholar]
- 32.Wang RRC, Lu B. Biosystematics and evolutionary relationships of perennial Triticeae species revealed by genomic analyses. Journal of Systematics and Evolution. 2014;52: 697–705. [Google Scholar]
- 33.Lu K, Xu Z, Liu Z, Zhang XY. FISH analysis of a CRW-homology sequence from Pseudoroegneria spicata in Thinopyrum ponticum and Th. intermedium [Article in Chinese]. Yi Chuan. 2009;31: 1141–1148. [DOI] [PubMed] [Google Scholar]
- 34.Li GR, Liu C, Wei P, Song XJ, Yang ZJ. Chromosomal distribution of a new centromeric Ty3-gypsy retrotransposon sequence in Dasypyrum and related Triticeae species. Journal of Genetics. 2012;91: 343–348. [DOI] [PubMed] [Google Scholar]
- 35.Belyayev A, Kalendar R, Brodsky L, Nevo E, Schulman AH, Raskina O. Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mobile DNA. 2010;1: 6 10.1186/1759-8753-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics. 2010;186: 801–812. 10.1534/genetics.110.120790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaakov B, Meyer K, Ben-David S, Kashkush K. Copy number variation of transposable elements in Triticum–Aegilops genus suggests evolutionary and revolutionary dynamics following allopolyploidization. Plant Cell Rep. 2013;32: 1615–24. 10.1007/s00299-013-1472-8 [DOI] [PubMed] [Google Scholar]
- 38.Tang S, Li Z, Jia X, Larkin PJ. Genomic in situ hybridization (GISH) analyses of Thinopyrum intermedium, its partial amphiploid Zhong 5, and disease resistant derivatives in wheat. Theor Appl Genet. 2000;100: 344–352. [Google Scholar]
- 39.Mahelka V, Kopecký D, Paštová L. On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol Biol. 2011;11: 127 10.1186/1471-2148-11-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salina EA, Sergeeva EM, Adonina IG, Shcherban AB, Belcram H, Huneau C, et al. The impact of Ty3-gypsy group LTR retrotransposons Fatima on B-genome specificity of polyploid wheats. BMC Plant Biol. 2011;11: 99 10.1186/1471-2229-11-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parisod C, Senerchia N. Responses of Transposable Elements to Polyploidy In: Grandbastien M-A, Casacuberta JM, editors. Plant Transposable Elements, Topics in Current Genetics 24 Springer-Verlag; Berlin Heidelberg; 2012. pp. 147–168. [Google Scholar]
- 42.Huang S, Sirikhachornkit A, Su XJ, Faris J, Gill BS, Haselkorn R, et al. Genes encoding plastid acety1-CoA carboxylase and 3-phosphoglycerate kinase of Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002;99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dvorak J, Yang ZL, You FM, Luo MC. Deletion polymorphism in wheat chromosome regions with contrasting recombination rates. Genetics. 2004;168: 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gill BS, Appels R, Botha-Oberholster AM, Buell CR, Bennetzen JL, Chalhoub B, et al. A workshop report on wheat genome sequencing: international genome research on wheat consortium. Genetics. 2004;168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Dong Y, Wang RR- C. Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum—Thinopyrum ponticum by in situ hybridization, isozyme analysis and RAPD. Genome. 1996;39: 1062–1071. [DOI] [PubMed] [Google Scholar]
- 46.Jiang J, Birchler JA, Parrott WA, Dawe RK. A molecular view of plant centromeres. Trends Plant Sci. 2003;8: 570–575. [DOI] [PubMed] [Google Scholar]
- 47.Dawe RK. Centromere renewal and replacement in the plant kingdom. Proc Natl Acad Sci USA. 2005;102: 11573–11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci USA. 2005;102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levy AA, Feldman M. The impact of polyploidy on grass genome evolution. Plant Physiol. 2002;130: 1587–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eilam T, Anikster Y, Millet E, Manisterski J, Feldman M. Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome. 2008;51: 616–627. 10.1139/G08-043 [DOI] [PubMed] [Google Scholar]
- 51.Salina EA, Adonina IG, Badaeva ED, Kroupin PYu, Stasyuk AI, Leonova IN, et al. A Thinopyrum intermedium chromosome in bread wheat cultivars as a source of genes conferring resistance to fungal diseases. Euphytica. 2015;204: 91–101. [Google Scholar]
- 52.Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biology. 2013;14: R10 10.1186/gb-2013-14-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernatzky R, Tanksley SD. Toward a Saturated Linkage Map in Tomato Based on Isozyme and Random cDNA Sequences. Genetics. 1986;112: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guzman C, Caballero L, Alvarez JB. Molecular characterization of the Wx-Bl allelic variants identified in cultivated emmer wheat and comparison with those of durum wheat. Mol. Breeding. 2010;28: 403–411. [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroupin PYu Divashuk MG, Belov VI Glukhova LI, Aleksandrov OS Karlov GI. Comparative molecular cytogenetic characterization of partial wheat-wheatgrass hybrids. Russian Journal of Genetics. 2011;47: 432–437. [PubMed] [Google Scholar]
- 57.Karlov GI, Danilova TV, Horlemann C, Weber G. Molecular cytogenetic in hop (Humulus lupulus L.) and identification of sex chromosomes by DAPI-banding. Euphytica. 2003;132: 185–190. [Google Scholar]
- 58.Komuro S, Endo R, Shikata K, Kato A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome. 2013;56: 131–137. 10.1139/gen-2013-0003 [DOI] [PubMed] [Google Scholar]
- 59.Chen G, Zhu J, Zhou J, Subburaj S, Zhang M. Dynamic development of starch granules and the regulation of starch biosynthesis in Brachypodium distachyon: comparison with common wheat and Aegilops peregrina. BMC Plant Biology. 2014; 14:198 10.1186/s12870-014-0198-2/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Megan HD, Mason-Gamer RJ. The Evolution of North American Elymus (Triticeae, Poaceae) Allotetraploids: Evidence from Phosphoenolpyruvate Carboxylase Gene Sequences. Systematic Botany. 2004;29: 850–861. 10.1600/0363644042451017 [DOI] [Google Scholar]
- 61.Mathews S, Sharrock RA. The phytochrome gene family in grasses (Poaceae): A phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol. Biol. Evol. 1996;13: 1141–1150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The bootstrap consensus tree inferred from 1000 replicates.
(PDF)
a) Hybridization of the Thin probe on chromosomes of Tulaykovskaya 100. b) Identification of 6Jr chromosomes of Th. intermedium in Tulaykovskaya 100 by multicolor GISH with the labeled genomic DNA of P. spicata (green) and D. villosum (pink, signal absent) (blocked with genomic DNA of T. aestivum cv. Ivolga). Chromosomes counterstained with DAPI (blue). Chromosome 6Jr shown with asterisk. Bar = 10 μm.
(TIF)
The positions of primers DwaxyQ used in qPCR analysis are indicated in arrows and color highlighted.
(JPG)
As a negative control (NC), water was served as a template in the PCR reaction. A 100 bp size-ladder was used.
(JPG)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.