ABSTRACT
Human DNA replication and repair is a highly coordinated process involving the specifically timed actions of numerous proteins and enzymes. Many of these proteins require interaction with proliferating cell nuclear antigen (PCNA) for activation within the process. The interdomain connector loop (IDCL) of PCNA provides a docking site for many of those proteins, suggesting that this region is critically important in the regulation of cellular function. Previous work in this laboratory has demonstrated that a peptide mimicking a specific region of the IDCL (caPeptide) has the ability to disrupt key protein-protein interactions between PCNA and its binding partners, thereby inhibiting DNA replication within the cells. In this study, we confirm the ability of the caPeptide to disrupt DNA replication function using both intact cell and in vitro DNA replication assays. Further, we were able to demonstrate that treatment with caPeptide results in a decrease of polymerase δ activity that correlates with the observed decrease in DNA replication. We have also successfully developed a surface plasmon resonance (SPR) assay to validate the disruption of the PCNA-pol δ interaction with caPeptide.
Keywords: Cancer therapeutics, competition kinetics, DNA polymerase δ, DNA replication, proliferating cell nuclear antigen (PCNA), surface plasmon resonance (SPR)
Abbreviations
- PCNA
proliferating cell nuclear antigen
- IDCL
interdomain connector loop
- caPeptide
cancer-associated peptide
- RF-C
replicating factor C
- FEN-1
flap endonuclease 1
- RPA
replication protein A
- PIP-box
PCNA-interacting protein box
- pI
isoelectric point
- caPCNA
cancer-associated PCNA
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- SV40
Simian virus 40
- T-ag
T antigen
- EC50,
50% effective dose
- SPR
surface plasmon resonance
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- pol δ
polymerase δ
- pol α
polymerase α
- scrPeptide
scrambled peptide
- KD,
equilibrium dissociation constants
- POLD3
partial recombinant pol δ of p68 subunit
- ka,
association constant
- kd,
dissociation constant
- DMEM
Dulbecco's Modified Eagle Media (DMEM)
- PBS
phosphate buffered saline
- DMSO
dimethyl sulfoxide
- TCA
trichloroacetic acid
- SDS
sodium dodecyl sulfate
- EDTA
ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- RNA
ribonucleic acid
- TBE
50 mM Tris/ 90 mM boric acid/ 2 mM EDTA
- PCR
polymerase chain reaction
- HBS-EP
HEPES buffered saline with EDTA and P20
Introduction
DNA replication is a well-regulated process requiring the coordinated effort of a number of proteins and enzymes, including: proliferating cell nuclear antigen (PCNA), replication factor C (RF-C), flap endonuclease 1 (FEN-1), replication protein A (RPA), DNA ligase I, topoisomerase I and II, RNAse H, and DNA polymerase α, δ, and ϵ.1-6 Many of these proteins, including polymerase δ, require PCNA interaction for activation and processivity within the cell cycle. Proteins bind to PCNA via a conserved protein-protein interaction domain known as the “PCNA-interacting protein” (or “PIP”)-box. Structural studies of PCNA-protein interactions reveal that the PIP-box is typically a single turn 310 helix displaying a side chain of residues that “plug” into the hydrophobic patch of the interdomain connector loop (IDCL) of PCNA.7 This common feature creates inherent competition among binding partners for this site, resulting in the observed regulation of cellular processes.
Mutagenic analyses reveal that the DNA replication machinery derived from malignant breast cell lines and actual tumor tissue replicated DNA in a significantly more error-prone manner, as compared to the replication machinery derived from nonmalignant counterparts.8 A structural comparison of these components from both normal and malignant cell lines revealed a unique form of PCNA present in malignant breast cells.9 Two-dimensional PAGE analysis illustrated that malignant breast cells harbored an additional isoform of PCNA with an acidic isoelectric point (pI), as opposed to the normal cell expressing PCNA with only a basic pI. The two distinct PCNA isoforms have been resolved in a variety of cancer cell lines, including neuroblastoma, 10 hepatic carcinoma, 11 high-grade prostatic intraepithelial neoplasia and prostate cancer.12 The acidic isoform associated with cancer (caPCNA) is not the result of genetic mutations or alternate splicing, but is the result of differential post-translational modification within the PCNA molecule.13 The specificity of caPCNA (amino acids 126–133) was identified and validated using antibody sequence mapping within the IDCL (amino acids 68–230).14 This uniquely exposed region (amino acids 126–133) was then used to develop a peptide (caPeptide). With the addition of a nona-D-arginine (R9) subunit (to facilitate nuclear transport), R9-caPeptide has demonstrated the ability to induce apoptosis in a triple-negative breast cancer (TNBC) cell line (MDA-MB-436).14 A variety of other cancer cell types tested (e.g. breast cancer, neuroblastoma, lymphoma, prostate, pancreatic) exhibit a range of sensitivity to R9-caPeptide treatment.14-16 R9-caPeptide has also demonstrated the ability to effectively inhibit tumor growth in mice bearing xenograft tumors derived from both TNBC (MDA-MB-436) and neuroblastoma (SK-N-BE(2)c) cell lines, further validating the therapeutic potential of the peptide.14,15
The observed cytotoxicity R9-caPeptide treatment induces in cancer cells is thought to be the direct result of caPeptide interference with caPCNA-protein interactions that are critical to the proper functioning of DNA replication and repair. Cancer cells are virtually unaffected when treated with the sequence either scrambled or in D-conformation, further validating this theory that the specific sequence is important.14 A complete alanine scan of the peptide (detailed in Smith, et. al.14) pinpointed the specific amino acids within the sequence that were critical to cytotoxicity, and these correlated to previously characterized PCNA interactions with DNA replication and repair proteins, including p21 and polymerase δ.17-19
The purpose of this study is to validate the disruption of DNA replication in cancer cells due to caPeptide treatment and to identify the interactions targeted by the caPeptide. The in vitro DNA replication model, using a plasmid DNA containing the SV40 replication origin (pSVO+), is a technique commonly used to study eukaryotic DNA replication.4,20 This cell-free system utilizes the host cell's DNA replication apparatus to support DNA synthesis, with large T antigen (T-ag) being the only viral-encoded protein required to initiate unwinding of the DNA helix.21,22 The technique is particularly useful in evaluating the effectiveness of chemotherapeutic agents thought to inhibit DNA replication.23-25 Previously, our laboratory successfully isolated a stable functioning multi-protein complex able to support DNA replication using this in vitro papovavirus replication model system.26-28 This isolated complex, referred to as the DNA synthesome, has successfully been isolated from numerous cancer cell lines, including: human cervical cancer (HeLa), 26,28 breast cancer (MCF 7 27 and MDA-MB 468 29,30), leukemia (HL-60), 31 and neuroblastoma (IMR-32).10,32 The DNA synthesome contains all the identified proteins necessary in the successful replication of DNA.
In this study, we validate the inhibition of DNA replication with caPeptide treatment using both intact cell and in vitro DNA replication assays in HeLa cells. The observed caPeptide activity was compared with a scrambled version of the peptide (scrPeptide) as the control. The 50% effective dose (EC50) of the caPeptide was evaluated in both assays (intact cell, in vitro). We use additional in vitro assays to identify the PCNA-protein interactions targeted by caPeptide. The ability of the caPeptide to disrupt the interactions identified is further confirmed and quantified using surface plasmon resonance (SPR).
Results
caPeptide exhibits cytotoxic behavior in HeLa cells
As described previously, our lab synthesized a peptide corresponding to a portion of the IDCL of PCNA that acts as the docking site for numerous proteins and enzymes required in DNA replication and repair. When the caPeptide was attached to a 9-arginine (R-conformation) chain (for cellular uptake), R9-caPeptide exhibited cytotoxic activity in numerous cancer cell lines, including: breast cancer (MDA-MB 436, MCF 7, and HCC 1937), pancreatic cancer (PaCa-2), lymphoma (U937), and neuroblastoma (SK-N-DZ, SK-N-BE(2)c, SK-N-AS, SK-N-SH, SK-N-FI) cell lines.14-16
The ultimate goal of this research is to identify and validate specific PCNA-protein interactions effectively blocked by caPeptide during DNA replication. The following studies were conducted in the well-characterized cervical cancer cell line (HeLa). To confirm its suitability, we treated HeLa cells with R9-caPeptide and used the MTT assay to assess effective cell cytotoxicity. When HeLa cells are treated with increasing concentrations of R9-caPeptide, observed cell proliferation is also reduced, and the concentration required to inhibit cell growth by 50% (EC50) is approximately 35 μM (Fig. 1A). When the scrambled (R9-scrPeptide, control) peptide was added at concentrations increasing from 1 to 100 μM, there was virtually no effect on cell growth (Fig. 1A). These patterns observed in HeLa cells are consistent with those observed in the treatment of other types of cancer cell lines, 14-16 and therefore is appropriate for use in this study.
Figure 1.
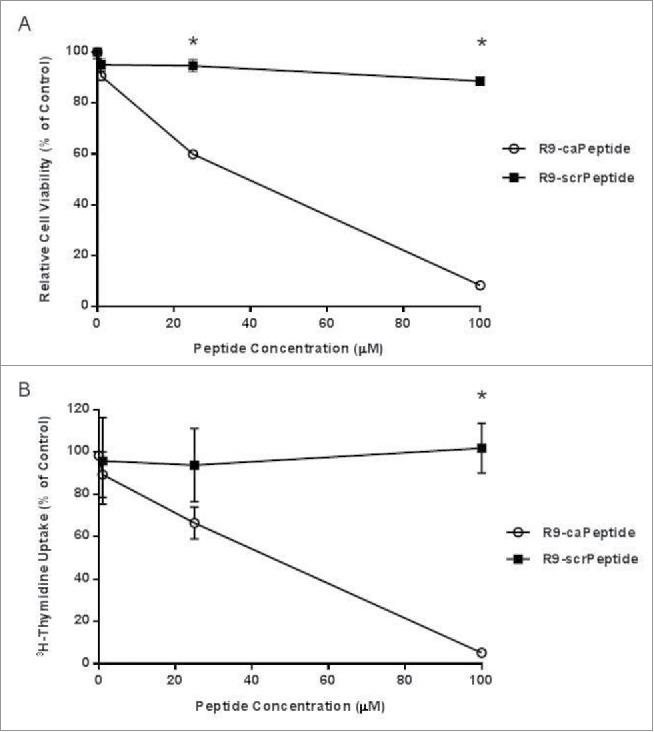
Cytotoxic activity of caPeptide. A, HeLa cells (4 × 103) were treated with increasing concentrations of R9- caPeptide or R9-scrPeptide for 24 hours, and cytotoxic activity was evaluated using the MTT assay. B, HeLa cells (5 × 105) were treated with increasing concentrations of R9-caPeptide and R9-scrPeptide. After 24 hours, the media was removed and 3H-Thymidine solution (1 nCi/mL) was added. Inhibition was analyzed as described previously in the Materials and Methods section of the paper. Each point on the graph represents the average ± standard deviation of 3 independent experiments. Cells analyzed in the absence of any peptide served as the control, and were compared to cells treated with caPeptide or scrPeptide. * represents p < 0.01.
caPeptide inhibits intact cell DNA synthesis
As mentioned previously, the premise for developing the R9- caPeptide was that this particular sequence within the IDCL was uniquely exposed in caPCNA, and therefore could be a novel target in cancer therapeutics. From preliminary data, we hypothesize the observed cytotoxicity from treatment with caPCNA was due to the effective disruption of PCNA-protein interactions key to DNA replication. To validate that DNA synthesis was, in fact targeted by caPeptide, we treated HeLa cells with R9-caPeptide and assessed the effect using 3H-Thymidine uptake. This type of assay is a common technique used to monitor DNA replication, as the measured amount of 3H-Thymidine correlates to the amount incorporated into newly synthesized DNA strands. For this study, R9-caPeptide treatment was compared to the scrambled control peptide (R9-scrPeptide) treatment. Here, we show, the amount of labeled thymidine incorporated into the newly formed DNA strand decreases with increasing R9-caPeptide treatment (Fig. 1B). The amount of R9-caPeptide required to inhibit 50% of 3H-Thymidine incorporation (EC50) was approximately 40 μM. This is consistent with the EC50 determined from the MTT cell proliferation assay (35 μM, Fig. 1A). When R9-scrPeptide was added in increasing concentrations (1 to 100 μM), virtually no effect on the uptake was observed (Fig. 1B).
caPeptide inhibits in vitro DNA replication
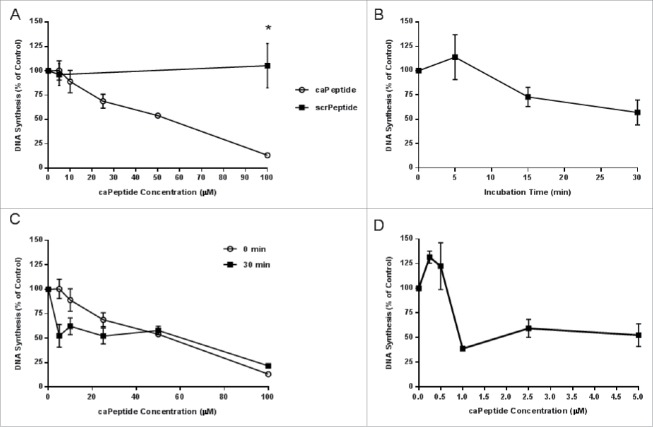
Once we were able to demonstrate the R9-caPeptide inhibition of DNA replication in whole cells (3H-Thymidine uptake), we then sought to prove the caPeptide sequence alone was responsible for the disruption of DNA replication. The activity of bare caPeptide (no R9- attachment) was then tested using the in vitro SV40 DNA replication assay, as described in the Materials & Methods section. Increasing caPeptide concentration resulted in a dose-dependent decrease of 32P incorporation into nascent DNA (DNA replication) (Fig. 2A) in a manner consistent with the observed effect in the 3H-Thymidine uptake assay (Fig. 1A and B). The EC50 of caPeptide required to inhibit 50% of in vitro DNA synthesis is approximately 55 μM (Fig. 2A). A neutral agarose gel analysis of the replication products from the 5 concentrations tested (Fig. S1) confirms caPeptide treatment blocks formation of both full-length DNA and replication intermediates. Replication intermediates are virtually eliminated with the addition of 100 μM of caPeptide, which is the highest concentration used in the study. When increasing concentrations of scrambled peptide (scrPeptide) was substituted for caPeptide in the reaction mixture, there was no observable effect on DNA synthesis (Fig. 2A).
Figure 2.
Effect of caPeptide treatment on synthesome-mediated DNA replication reactions. A, caPeptide and scrambled peptide were added to the SV40 DNA replication reaction in increasing concentrations. B, 5 μM caPeptide was pre-incubated with HeLa cell extract for increasing time points at 37°C prior to the start of the SV40 DNA replication reaction. C, prior to the start of the reaction, HeLa cell extract was pre-incubated with the appropriate amount of caPeptide for 30 minutes at 37°C or run as previously described. D, Concentrations below 5 μM were pre-incubated with HeLa cell extract 30 minutes at 37°C prior to the start of the SV40 DNA replication reaction. Finalized reaction mixtures for all conditions were incubated for 1 hour at 37°C and analyzed as described previously in the Materials and Methods section. Control reactions were performed in the absence of any peptide. One unit of in vitro SV40 DNA synthesis activity is equivalent to 30 pmol of total radionucleotide incorporation per hour at 37°C. % Control is based on the control reaction performed in the absence of any peptide. Each point represents the average ± standard deviation of 3 to 5 experiments.* represents p < 0.01.
The EC50 determined from both intact cell and in vitro DNA replication assays were in close agreement with each other. It was, however, surprising that the amount of caPeptide required to inhibit 50% of the in vitro DNA replication was slightly higher than that determined from the intact cell experiment. We would assume the actual amount of caPeptide required to inhibit DNA replication in a cell to be significantly lower than the total amount a cell is treated with, as complex cellular processes limit the amount delivered to the nucleus. The caPeptide treatment employed in the SV40 assay has less effective contact time with replication proteins (1 hr), as compared to the intact cell assay, and this could be the flaw in a straightforward comparison of the 2 techniques. Therefore, we tested the effect of increasing the contact time of the caPeptide with the DNA replication machinery extracted from HeLa cells (nuclear extract) prior to the start of the in vitro DNA replication assay. Five μM of caPeptide was pre-incubated with HeLa nuclear extract in a 37°C water bath for increasing lengths of time (0 to 30 minutes), and then the mixture was combined with the remaining reaction components, to allow SV40 replication assay to continue as previously described. By increasing the interaction time of caPeptide with HeLa extract, we were able to enhance the inhibition of replication (Fig. 2B). When 5 μM of caPeptide was pre-incubated with HeLa nuclear extract up to 30 minutes, approximately 60% of DNA synthesis could be inhibited.
For a closer approximation of activity within the cell, we pre-incubated HeLa nuclear extract with increasing concentrations of caPeptide at 37°C for 30 minutes prior to the start of the reaction. DNA synthesis was significantly reduced with the addition up to 10 μM caPeptide, but the overall caPeptide inhibition of DNA synthesis did not significantly increase with the increased interaction time (30 minutes) (Fig. 2C). These results indicate the actual amount required to directly disrupt DNA replication is well below 5 μM, and we need to test lower concentrations. Therefore, we then pre-incubated HeLa nuclear extract at 37°C for 30 minutes with caPeptide concentrations below 5 μM. A thirty (30) minute incubation prior to the start of the SV40 DNA replication assay resulted in an EC50 of less than 1 μM (approximately .8 μM) (Fig. 2D). An analysis of the replication products on a neutral agarose gel (Fig. S3) confirms the observed inhibition.
caPeptide displays the ability to target PCNA-associated DNA polymerase activity
Thus far, we have demonstrated that caPeptide disrupts DNA replication in HeLa cells (Fig. 1), which was further validated using the in vitro DNA synthesis assays (Fig. 2). As mentioned previously, PCNA plays a vital role in DNA replication and repair. Numerous proteins, including DNA polymerase δ (pol δ), require PCNA binding to both synthesize and repair DNA during the replication cycle. We then tested the effect of caPeptide treatment on the normal interaction between the synthesome-associated DNA pol δ and PCNA. Pol δ enzymatic assays were carried out using increasing concentrations of caPeptide, as detailed in the Materials and Methods section. Increasing caPeptide treatment resulted in a decrease of pol δ activity (Fig. 3A and B), and this response corresponded to the observed decrease in SV40 origin-dependent in vitro DNA replication at both the high concentration (Fig. 3A) and low concentration (Fig. 3B) range.
Figure 3.
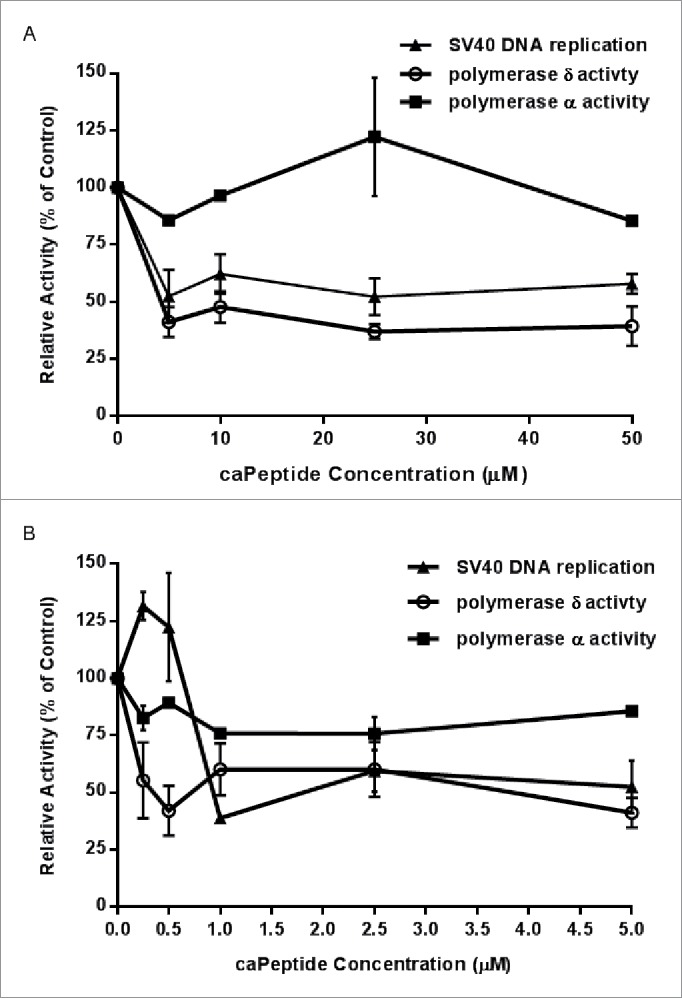
The effect of caPeptide on the relative activity of synthesome-associated DNA polymerase δ, DNA polymerase α, and SV40 DNA replication activity. Prior to each assay, HeLa nuclear extract was pre-incubated for 30 minutes at 37°C with either lower (A) or higher (B) concentrations of caPeptide. The assays (pol δ, pol α, and SV40 DNA replication) were performed as previously described in the Materials and Methods section. Control reactions performed in the absence of peptide were used to determine the relative activity (% of control) for each caPeptide concentration. Each point represents the average ± standard deviation of 3 to 5 experiments.
Previous studies have shown the pol δ-PCNA interaction to be critical for highly processive pol δ activity during DNA synthesis.33 From these preliminary activity assays, we have demonstrated that caPeptide inhibits pol δ –PCNA interaction, resulting in DNA replication inhibition. To determine if the observed inhibition was in fact due to the decrease in pol δ activity, we tested polymerase α (pol α) activity as a control, as pol α activity has been shown to act independently of PCNA interaction during replication.34 Consequently, caPeptide treatment did not significantly affect DNA pol α activity (Fig. 3A and B), verifying the observation that caPeptide does target specific PCNA interactions key to DNA replication. Additionally, when scrPeptide was used in place of caPeptide in both the DNA polymerase α and DNA polymerase δ assays, there was no observable effect on the activity of either DNA polymerases, further validating the specificity of caPeptide (Fig. S4).
caPeptide exhibits the ability to disrupt specific PCNA-pol δ interactions using surface plasmon resonance (SPR)
As shown in the previous figures, we have demonstrated through in vitro assays that caPeptide can disrupt PCNA-pol δ interactions during DNA replication (Fig. 3), resulting in a decrease of observed DNA replication (Figs. 1, 2, and 3). While results in the previous section were consistent with our hypothesis that caPeptide could effectively compete with PCNA for PCNA's naturally occurring binding partners, the data did not specifically demonstrate that the decrease in DNA synthesis resulted directly from the PCNA-binding partner disruption. Consequently, we sought to directly quantify the specific caPeptide disruption of PCNA-pol δ interaction using surface plasmon resonance (SPR). From previous pull-down 35 and co-crystallization 7 studies, the p68 subunit of DNA polymerase δ has been shown to interact with PCNA. Therefore, in this particular binding study, we selected the recombinant partial DNA pol δ protein (amino acids 357–466) of the p68 subunit with a GST-tag at the N-terminus (POLD3) containing the putative 22-amino acid PCNA binding sequence at the C-terminus (PIP-box).
The interaction of each analyte (PCNA, caPeptide, and scrPeptide) with POLD3 was first analyzed individually, with increasing concentrations of each flowed over the POLD3-captured CM5 chip. Both PCNA and caPeptide exhibited dose dependent binding to POLD3 (Fig. 4A and B), while scrPeptide displayed nonspecific binding to POLD 3 (Fig. 4D). The resulting PCNA and caPeptide binding curves could then be fit with parameters representing typical Langmuir binding (1:1 interaction). The equilibrium dissociation constants (KD) calculated for both PCNA (97.2 nM) and caPeptide (27.2 μM) corroborated the difference in binding magnitude observed in the PCNA and caPeptide binding curves (Fig. 4C).
Figure 4.
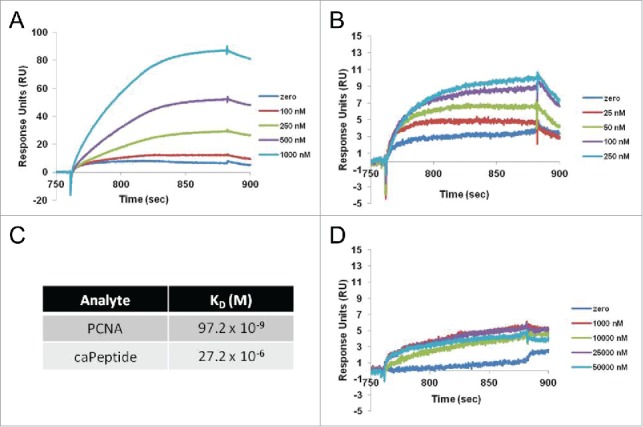
Interaction of recombinant PCNA (rPCNA), caPeptide, or scrambled peptide with GST-captured polymerase δ peptide (amino acids 357–466) (POLD3) using surface plasmon resonance (SPR). After POLD3 capture, each concentration of rPCNA (A), caPeptide (B), or scrambled peptide (D) was subsequently flowed over the capture surface at a rate of 5 μl/min for 2 minutes in HBS-EP buffer (10 mM HEPES pH7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% Surfactant P20), followed by dissociation and surface regeneration (10 mM glycine-HCl pH 2.1). All analyte solutions were serially diluted into HBS-EP and filtered prior to binding. C, the equilibrium dissociation constant (KD) of each analyte (PCNA, caPeptide) binding to captured POLD3 was calculated using BIAevaluation software from the resulting surface plasmon resonance (SPR) binding curves.
The caPeptide-mediated disruption of the PCNA-POLD3 interaction was then quantified using SPR competitive binding assays. For each set concentration of caPeptide added to the analyte solution (250, 500, and 1000 nM), dose-dependent PCNA binding curves (250, 500, and 1000 nM) were then evaluated and fit with binding parameters. For each PCNA concentration, an increase in the concentration of caPeptide added to the analyte solution resulted in a decrease of observed POLD3 binding. As PCNA concentration is increased, the resulting effect of caPeptide inhibition increases (Fig. 5A-D). To fit the binding curves to kinetic parameters, a constant caPeptide association and dissociation rate in a heterogeneous analyte solution was assumed. An increase of caPeptide added to the PCNA binding solutions results in a decrease of the association constant (ka) and increase of the dissociation constant (kd) fit for PCNA-POLD3 interaction (Fig. 5D).
Figure 5.
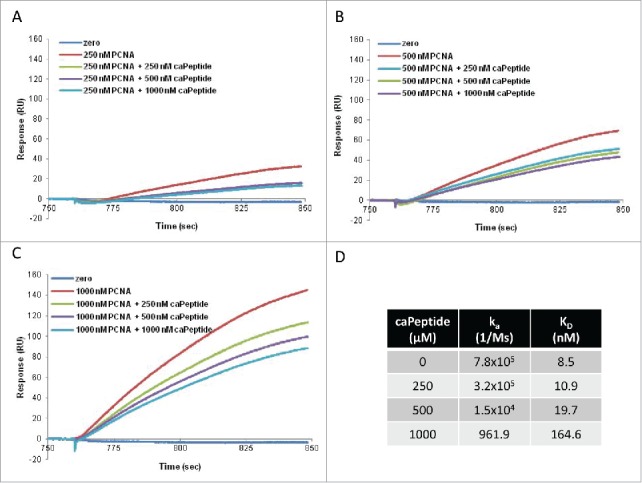
The competition of caPeptide with rPCNA for binding to GST-captured polymerase δ peptide (amino acid 357–466) (POLD3). After POLD3 capture on the chip, increasing concentrations of caPeptide were mixed with 250 nM (A), 500 nM (B), or 1000 nM rPCNA (C), and subsequently flowed through the chip. Each graph is a plot of the zero curve (blue), rPCNA alone at each concentration (red), and the resulting curve of the caPeptide-rPCNA competition (green, purple, and turquoise). D, the effect of POLD3-PCNA disruption by caPeptide was quantified using BIAevaluation software from the resulting surface plasmon resonance (SPR) binding curves.
Discussion
caPCNA has been previously identified as a novel acidic isoform of PCNA uniquely expressed in numerous types of cancer cell lines and tissue.36 This isoform has a uniquely exposed region within the IDCL, which is critical to the interaction of a number of proteins and enzymes within DNA replication and repair. We have successfully developed a peptide (caPeptide) to mimic the region identified as specific to caPCNA. R9-linked caPeptide has demonstrated the ability to target and kill cancer cells, while leaving normal cells virtually unaffected.14-16 Preliminary tests in animal models indicate its potential as a novel therapeutic.14,15 In this study, the mechanism of action for caPeptide was tested using intact cell (3H-Thymidine uptake) and in vitro (SV40 DNA replication assay) DNA synthesis experiments to measure the inhibitory ability of caPeptide within the replication process. As shown in both in vitro and intact cell experiments, caPeptide has the ability to block DNA replication in the selected cancer cell line (HeLa) in a sequence specific manner (Fig. 1).
We used the in vitro SV40 DNA replication assay, along with DNA polymerase α and δ activity assays in parallel with intact cell proliferation assays to compare the effect of caPeptide on in vitro DNA synthesis versus DNA synthesis of intact cells. We determined that amount of caPeptide required to inhibit 50% (EC50) of in vitro SV40 origin dependent DNA synthesis (55 μM) was comparable to 50% inhibition of intact DNA synthesis (40 μM) and intact cell proliferation (35 μM) (Fig. 2). The higher caPeptide concentration observed in the in vitro experiment is most likely due to the fact that the required association time for effective disruption of PCNA-protein interactions is greater than that which was allowed for during the in vitro SV40 DNA replication assay (1 hr vs. 24 hr). When the contact time of caPeptide with the DNA replication proteins was increased to 30 minutes prior to the start of the reaction, the EC50 decreases from 55 μM (no pre-incubation) to 0.8 μM (30 minute pre-incubation) (Fig. 2C). The EC50 determined from the 30 minute pre-incubation study is most likely a closer approximation to the actual amount of caPeptide required to effectively inhibit replication within an actual cell. Studies to quantify the effective amount reaching the nucleus and to improve nuclear targeting and delivery of caPeptide are currently underway.
The T antigen-dependent SV40 DNA replication assay is instrumental to understanding the underlying mechanisms involved in eukaryotic DNA replication.37 Numerous studies have verified that efficient replication requires the action of at least 3 DNA polymerases: pol α, pol δ, and pol ϵ.34 The DNA polymerase α/primase complex (pol α/primase) is involved in the initiation step of leading/lagging strand DNA synthesis. Pol α can synthesize small oligoribonucleotides (10–15 bp) and elongate short strand distances (˜35 nucleotides). Because pol α does not have any direct interaction with PCNA, 38 caPeptide is expected to have no direct effect on the pol α activity. The results of the DNA polymerase α activity assay (Fig. 3A and B) confirm this.
While pol α can act independently, PCNA acts as a tether for DNA polymerase δ to the primed DNA template, promoting highly processive DNA synthesis.39 Polymerase δ can elongate these segments on both the leading and lagging strands of the DNA molecule. PCNA recruits polymerase δ to elongate the Okazaki fragment in DNA replication, 40 as well as work in homologous repair.41 Numerous studies confirm the binding of DNA pol δ with the IDCL of the PCNA in order to function properly in DNA synthesis.42-45 By mimicking this region of PCNA, caPeptide has proven to be effective in blocking these key interactions.
In this study, we demonstrate the ability of caPeptide to inhibit pol δ-PCNA interaction, validating our hypothesis that caPeptide targets PCNA binding proteins interactions and recruitment. While DNA pol δ activity, and subsequently overall DNA replication activity, is significantly reduced, it is however, not completely eliminated upon the addition of caPeptide. This may be due to the level of residual DNA replication activity exhibited by pol δ in the absence of PCNA interaction. It is also possible that DNA pol α activity, which is not inhibited by caPeptide, has taken over for pol δ and extends short pieces of the primed DNA template. This, however, would suggest that short, Okazaki-like fragments would accumulate as the amount of caPeptide increases, but this is not observed (Fig. S1).
This inability to completely eliminate activity, however, is most likely due to the structure of pol δ itself. Polymerase δ consists of 4 separate subunits: p12, p50, p68, and p125. A proposed model for pol δ – PCNA interactions in vivo is that 3 of the 4 subunits of pol δ (p125, p68, and p12) interact with PCNA directly, with each subunit contacting a separate PCNA monomer of the same homotrimer.46 Experimental evidence has established the binding of PCNA with the p125, p12, and p68 subunits of pol δ and supports the model that multiple modes of pol δ – PCNA interactions exist.47
In the presence of replication stress or genotoxic agents, the pol δ heterotetramer can convert to a heterotrimer upon degredation of the p12 subunit.48 Because pol δ inherently has multiple modes of PCNA interaction, the observed loss of the p68 subunit of pol δ does not result in complete loss of function in processive DNA synthesis.49,50 Studies have shown that pol δ subassemblies lacking either p68 or p12 retained the ability to bind PCNA and maintained some degree of in vitro processive DNA synthesis.51,52 Polymerase δ complexes lacking either the p12 or p68 subunit allow for the p50 subunit to make a substitute interaction, helping to maintain the trivalent structure within the the complex.46 Therefore, when caPeptide disrupts the interaction of p68 with the IDCL of the PCNA molecule, this only partially renders the DNA pol δ molecule unable to synthesize DNA because pol δ can still retain some of the PCNA interaction with the 3 remaining subunits. In previous studies, this interaction multiplicity and resulting structural integrity has been verified by observations that loss of the p68 subunit of pol δ does not result in complete loss of function in processive DNA synthesis.49,50 Furthermore, pol δ subassemblies lacking either p68 or p12 retained the ability to bind PCNA and maintained some degree of in vitro processive DNA synthesis.51,52 Further studies will need to be conducted to fully characterize this complex interaction.
In this study, we were able to further validate caPeptide as a novel molecule that has the ability to disrupt the replication machinery of cancer cells. The targeting of the caPeptide to specific PCNA-protein interactions results in the inhibition of DNA replication. These experiments validate the use of the in vitro SV40 DNA replication assay and surface plasmon resonance in studying the mechanism of action of molecules that directly inhibit the DNA replication process.
Materials and methods
Cell lines
All cancer cell lines were cultured according to procedures established by the American Type Culture Collection (ATCC). HeLa cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Mediatech #10-017-CV) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All cell cultures were maintained at 37°C supplemented with 5% CO2.
Peptides
All peptides used in the study (caPeptide, R9-caPeptide, scrPeptide, R9-scrPeptide) were custom synthesized and isolated to >95% purity by Anaspec. All peptides were received in powder form and stored at −20°C. Prior to use, each peptide was dissolved in sterile PBS (Mediatech #20-031-CV) to a concentration of 10 mM, and aliquots were stored at −20°C.
Cell viability assay
HeLa cells (5 × 103) were seeded in 96-well plates and incubated overnight at 37°C with 5% CO2. Cells were then treated with increasing concentrations of R9-linked peptide (R9- caPeptide, R9 -scrPeptide). After 24 hours, the medium was removed; cells were washed with PBS and incubated with 500 μg/mL thiazolyl blue tetrazolium bromide (MTT) (Sigma #M5655) for 4 hours at 37°C with 5% CO2. The MTT solution was then removed, and cells were resuspended in dimethyl sulfoxide (DMSO) (J.T. Baker #9224-01). The optical density of the solubilized product was then read at 570 nm.
Measurement of intact HeLa cell DNA synthesis
HeLa cells (5 × 105) were seeded in 6-well plates and incubated overnight at 37°C with 5% CO2. Cells were then treated with increasing concentrations of R9-linked peptide. After 24 hours, the medium was removed, the cells were washed with PBS and then incubated in media containing a 1:1000 dilution of 3H-Thymidine (1 μCi/mL) (Perkin Elmer #NET027E001MC) for 4 hours at 37°C with 5% CO2. The thymidine-spiked media was removed; cells were washed 3 times with cold PBS, lysed with 0.1% SDS in PBS, and then transferred to individual eppendorf tubes. 100 μL of 100% (w/v) trichloroacetic acid (TCA) (Acros #42145-1000) was added to each tube and placed on ice for 30 minutes. Each tube of lysis solution was then transferred onto a separate glassmicrofibre filter (GF/C) (Whatman #1822025), and the filters were washed with 10% TCA and dried. The amount of 3H-Thymidine remaining on the filter (taken up into the cells) was then quantified using liquid scintillation counting.
Subcellular fractionation
HeLa cell pellets were prepared and subfractionated using previously published procedures.28 Briefly, pellets were fractionated into a low-salt (0.15 M KCl) extract of isolated nuclei (NE) and a postmicrosomal supernatant solution (S-3). These fractions were combined and layered onto a 2 M sucrose cushion and recentrifuged at 100,000g for 16 hours at 4°C. Following overnight centrifugation, the 2M sucrose interphase (P-4) fraction was then removed by aspiration. P-4 was then dialyzed in a 50 mM Tris-HCl (pH = 7.5), 0.15 M KCl, 1 mM EDTA, and 10% glycerol buffer, aliquoted, and stored at −80°C.
In vitro SV40 DNA replication assay
The in vitro replication of the pSVO+ plasmid containing the SV40 viral origin of DNA replication was carried out as previously published, 28 with modifications. A 25 μL volume reaction contained 30 mM HEPES (pH = 7.2), 7 mM MgCl2, 0.5 mM DTT, 5 μCi [α-32P]dCTP (Perkin Elmer #BLU513H), 1 μM dCTP, 100 μM each of dTTP, dCTP, and dGTP, 200 μM each of CTP, UTP, and GTP, 4 mM ATP, 40 mM of phosphocreatine, 50 μg of creatine phosphokinase, 50 ng of pSVO+, 0.1–1.0 μg T-Ag (optimal concentration determined by titration assays), and optimal amount of protein fraction (determined by titration assays). In an additional arm of the study, HeLa nuclear extract was first incubated with increasing concentrations of caPeptide or scrPeptide for 30 minutes (or for other specified time points) prior to starting the DNA replication reaction. The reactions were then incubated at 37°C for 1 hour and spotted on Whatman DE81 filters, and the filters were washed with 100 mM sodium pyrophosphate (pH 7.4) and 300 mM ammonium formate (pH 7.4), then dried. The amount of [α-32P]dCTP incorporated was then determined by liquid scintillation counting.
To analyze the replication products by agarose gel, the reaction was stopped with the addition of a 1% sodium dodecyl sulfate (SDS) and 1 mM ethylenediaminetetraacetic acid (EDTA), followed by digestion for 1 hour at 37°C with 100 ng/μl proteinase K. 100 μg yeast RNA coprecipitate was added, and the DNA replication products were isolated using the SpinPrep PCR Clean-up Kit (EMD Millipore #70852). The isolated DNA was then electrophoresed in 1% agarose gels containing TBE (50 mM Tris/ 90 mM boric acid / 2 mM EDTA). Gels were subsequently dried and imaged using autoradiography with a Typhoon phosphoimaging scanner (GE Healthcare).
DNA polymerase α assay
DNA polymerase α activity was analyzed using the procedure outlined previously by Malkas et al.28 Experiments were conducted in the presence of increasing peptide (caPep, scrPep) concentrations for 1 hour at 37oC. The reaction mixtures were spotted on Whatman DE81 filters, washed with 100 mM sodium pyrophosphate (pH 7.4) and 300 mM ammonium formate (pH 7.4), then dried. The amount of [3H]-TTP incorporated was quantified by liquid scintillation counting.
DNA polymerase δ assay
DNA polymerase δ activity was analyzed according to the procedure described by Xie et al., 49 with some modifications. The 25 μl reaction mixture contained 25 mM HEPES (pH 5.9), 10 mM MgCl2, 0.2 mg/ml BSA, 5% glycerol, 20 μg HeLa extract, 10 μM TTP, 2 units/ml of 10:1 poly[dA]/oligo[dT] template (GE Healthcare Biosciences AB #27-4110-01, Sigma #04387), and 0.5 μCi of [3H]-TTP (Perkin Elmer #NET221X005MC). The reaction mixture was incubated with increasing peptide concentrations (caPep, scrPep) for 15 minutes at 37°C. The reaction products were spotted on Whatman DE81 filter paper, washed with 100 mM sodium pyrophosphate (pH 7.4) and 300 mM ammonium formate (pH 7.4), and dried. The amount of [3H]-TTP incorporated was quantified by liquid scintillation counting.
Protein-protein binding and competition assays including kinetic analysis
All experiments were conducted on the Biacore T100 (GE Healthcare Biosciences AB) using HBS-EP (10 mM HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% Surfactant P20) (GE Healthcare Biosciences AB #BR-1006-69) as the running buffer. Anti-GST antibody (GE Healthcare Biosciences AB #BR-1002-23) was immobilized on a carboxymethylated dextran modified CM5 chip using carbodiimide covalent linkage procedures outlined by the manufacturer (GE Healthcare Biosciences AB). 100 nM of an N-terminal GST-tagged peptide corresponding to amino acids 357–466 of the p66 subunit of polymerase δ (POLD3) (Abnova #H00010714-Q01) was then captured on the chip at a 5 μl/min flow rate in HBS-EP running buffer. Both recombinant PCNA (rPCNA) (SurModics #A15400) and caPeptide were serially diluted in HBS-EP and flowed over the POLD3-captured CM5 sensor chip at a 5 μl/min flow rate with a contact time of 3 minutes, followed by dissociation and regeneration of the chip surface (10 mM glycine-HCl pH 2.0; GE Healthcare Biosciences AB #BR-1003-55). Binding curves were recorded for concentrations ranging from 100–1000 nM for rPCNA and 25–250 nM for caPeptide. Competition experiments between caPeptide and rPCNA for POLD3 binding were also performed using a similar approach. 250 nM caPeptide was incubated in solutions containing increasing concentrations of rPCNA (100–1000 nM). Kinetic parameters from Biacore binding data were determined using Biacore T100 Evaluation Software Version 2.0.3.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Department of Defense [Grant W81XWH-11-1-0786 to LHM]; and by the National Institutes of Health National Cancer Institute [Grants R01 CA121289 and P30CA033572]. Research reported in this publication included work performed in the X-Ray Crytallography and Macromolecular Characterization Core supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Alberts B. DNA replication and recombination. Nature 2003; 421:431-5; PMID:12540917; http://dx.doi.org/ 10.1038/nature01407 [DOI] [PubMed] [Google Scholar]
- 2.Ariga H, Sugano S. Initiation of simian virus 40 DNA replication in vitro. J Virol 1983; 48:481-91; PMID:6312104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decker RS, Yamaguchi M, Possenti R, Bradley MK, DePamphilis ML. In vitro initiation of DNA replication in simian virus 40 chromosomes. J Biol Chem 1987; 262:10863-72; PMID:3038899 [PubMed] [Google Scholar]
- 4.Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A 1984; 81:6973-7; PMID:6095264; http://dx.doi.org/ 10.1073/pnas.81.22.6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stillman BW, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol 1985; 5:2051-60; PMID:3018548; http://dx.doi.org/ 10.1128/MCB.5.8.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wobbe CR, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci U S A 1985; 82:5710-4; PMID:2994044; http://dx.doi.org/ 10.1073/pnas.82.17.5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure 2004; 12:2209-19; PMID:15576034; http://dx.doi.org/ 10.1016/j.str.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 8.Sekowski JW, Malkas LH, Schnaper L, Bechtel PE, Long BJ, Hickey RJ. Human breast cancer cells contain an error-prone DNA replication apparatus. Cancer Res 1998; 58:3259-63; PMID:9699652 [PubMed] [Google Scholar]
- 9.Bechtel PE, Hickey RJ, Schnaper L, Sekowski JW, Long BJ, Freund R, Liu N, Rodriguez-Valenzuela C, Malkas LH. A unique form of proliferating cell nuclear antigen is present in malignant breast cells. Cancer Res 1998; 58:3264-9; PMID:9699653 [PubMed] [Google Scholar]
- 10.Sandoval JA, Hickey RJ, Malkas LH. Isolation and characterization of a DNA synthesome from a neuroblastoma cell line. J Pediatr Surg 2005; 40:1070-7; PMID:16034747; http://dx.doi.org/ 10.1016/j.jpedsurg.2005.03.054 [DOI] [PubMed] [Google Scholar]
- 11.Venturi A, Piaz FD, Giovannini C, Gramantieri L, Chieco P, Bolondi L. Human hepatocellular carcinoma expresses specific PCNA isoforms: an in vivo and in vitro evaluation. Lab Invest 2008; 88:995-1007; PMID:18521065; http://dx.doi.org/ 10.1038/labinvest.2008.50 [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Hickey RJ, Malkas LH, Koch MO, Li L, Zhang S, Sandusky GE, Grignon DJ, Eble JN, Cheng L. Elevated Expression of Cancer-Associated Proliferating Cell Nuclear Antigen in High-Grade Prostatic Intraepithelial Neoplasia and Prostate Cancer. The Prostate 2011; 71:7; PMID:21031434; http://dx.doi.org/16888766 10.1002/pros.21291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoelz DJ, Arnold RJ, Dobrolecki LE, Abdel-Aziz W, Loehrer AP, Novotny MV, Schnaper L, Hickey RJ, Malkas LH. The discovery of labile methyl esters on proliferating cell nuclear antigen by MS/MS. Proteomics 2006; 6:4808-16; PMID:16888766; http://dx.doi.org/ 10.1002/pmic.200600142 [DOI] [PubMed] [Google Scholar]
- 14.Smith SJ, Gu L, Phipps EA, Dobrolecki LE, Mabrey KS, Gulley P, Dillehay KL, Dong Z, Fields GB, Chen YR, et al.. A Peptide mimicking a region in proliferating cell nuclear antigen specific to key protein interactions is cytotoxic to breast cancer. Mol Pharmacol 2015; 87:263-76; PMID:25480843; http://dx.doi.org/ 10.1124/mol.114.093211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu L, Smith S, Li C, Hickey RJ, Stark JM, Fields GB, Lang WH, Sandoval JA, Malkas LH. A PCNA-derived cell permeable peptide selectively inhibits neuroblastoma cell growth. PLoS One 2014; 9:e94773; PMID:24728180; http://dx.doi.org/ 10.1371/journal.pone.0094773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingeman RG, Hickey RJ, Malkas LH. Expression of a novel peptide derived from PCNA damages DNA and reverses cisplatin resistance. Cancer Chemother Pharmacol 2014; 74:981-93; PMID:25190177; http://dx.doi.org/ 10.1007/s00280-014-2574-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs E, Kelman Z, Gulbis JM, O'Donnell M, Kuriyan J, Burgers PM, Hurwitz J. The influence of the proliferating cell nuclear antigen-interacting domain of p21(CIP1) on DNA synthesis catalyzed by the human and Saccharomyces cerevisiae polymerase delta holoenzymes. J Biol Chem 1997; 272:2373-81; PMID:8999948; http://dx.doi.org/ 10.1074/jbc.272.15.10065 [DOI] [PubMed] [Google Scholar]
- 18.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell 2006; 24:279-91; PMID:17052461; http://dx.doi.org/ 10.1016/j.molcel.2006.08.015 [DOI] [PubMed] [Google Scholar]
- 19.Zhang P, Sun Y, Hsu H, Zhang L, Zhang Y, Lee MY. The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. J Biol Chem 1998; 273:713-9; PMID:9422722; http://dx.doi.org/ 10.1074/jbc.273.2.713 [DOI] [PubMed] [Google Scholar]
- 20.Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol 1985; 5:1238-46; PMID:2993858; http://dx.doi.org/ 10.1128/MCB.5.6.1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simanis V, Lane DP. An immunoaffinity purification procedure for SV40 large T antigen. Virology 1985; 144:88-100; PMID:2998049; http://dx.doi.org/ 10.1016/0042-6822(85)90308-3 [DOI] [PubMed] [Google Scholar]
- 22.Stillman B, Gerard RD, Guggenheimer RA, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J 1985; 4:2933-9; PMID:2998767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdel-Aziz W, Hickey RJ, Malkas LH. An in vitro model system that can differentiate the stages of DNA replication affected by anticancer agents. Biochem Pharmacol 2004; 68:11-21; PMID:15183113; http://dx.doi.org/ 10.1016/j.bcp.2004.03.021 [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Aziz W, Jiang HY, Hickey RJ, Malkas LH. Ara-C affects formation of cancer cell DNA synthesome replication intermediates. Cancer Chemother Pharmacol 2000; 45:312-9; PMID:10755320; http://dx.doi.org/ 10.1007/s002800050046 [DOI] [PubMed] [Google Scholar]
- 25.Wills PW, Hickey R, Malkas L. Ara-C differentially affects multiprotein forms of human cell DNA polymerase. Cancer Chemother Pharmacol 2000; 46:193-203; PMID:11021736; http://dx.doi.org/ 10.1007/s002800000119 [DOI] [PubMed] [Google Scholar]
- 26.Applegren N, Hickey RJ, Kleinschmidt AM, Zhou Q, Coll J, Wills P, Swaby R, Wei Y, Quan JY, Lee MY, et al.. Further characterization of the human cell multiprotein DNA replication complex. J Cell Biochem 1995; 59:91-107; PMID:8530540; http://dx.doi.org/ 10.1002/jcb.240590111 [DOI] [PubMed] [Google Scholar]
- 27.Jiang HY, Hickey RJ, Abdel-Aziz W, Tom TD, Wills PW, Liu J, Malkas LH. Human cell DNA replication is mediated by a discrete multiprotein complex. J Cell Biochem 2002; 85:762-74; PMID:11968016; http://dx.doi.org/ 10.1002/jcb.10182 [DOI] [PubMed] [Google Scholar]
- 28.Malkas LH, Hickey RJ, Li C, Pedersen N, Baril EF. A 21S enzyme complex from HeLa cells that functions in simian virus 40 DNA replication in vitro. Biochemistry 1990; 29:6362-74; PMID:2169868; http://dx.doi.org/ 10.1021/bi00479a004 [DOI] [PubMed] [Google Scholar]
- 29.Coll JM, Hickey RJ, Cronkey EA, Jiang HY, Schnaper L, Lee MY, Uitto L, Syvaoja JE, Malkas LH. Mapping specific protein-protein interactions within the core component of the breast cell DNA synthesome. Oncol Res 1997; 9:629-39; PMID:9563011 [PubMed] [Google Scholar]
- 30.Coll JM, Sekowski JW, Hickey RJ, Schnaper L, Yue W, Brodie AM, Uitto L, Syvaoja JE, Malkas LH. The human breast cell DNA synthesome: its purification from tumor tissue and cell culture. Oncol Res 1996; 8:435-47; PMID:9114436 [PubMed] [Google Scholar]
- 31.Lin S, Hickey RJ, Malkas LH. The isolation of a DNA synthesome from human leukemia cells. Leuk Res 1997; 21:501-12; PMID:9279361; http://dx.doi.org/ 10.1016/S0145-2126(96)00103-8 [DOI] [PubMed] [Google Scholar]
- 32.Sandoval JA, Grosfeld JL, Hickey RJ, Malkas LH. Structural analysis of the human neuroblastoma DNA replication complex: insights into faulty proliferation. J Pediatr Surg 2006; 41:266-70; PMID:16410145; http://dx.doi.org/ 10.1016/j.jpedsurg.2005.10.046 [DOI] [PubMed] [Google Scholar]
- 33.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 1987; 326:515-7; PMID:2882423; http://dx.doi.org/ 10.1038/326515a0 [DOI] [PubMed] [Google Scholar]
- 34.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem 2002; 71:133-63; PMID:12045093; http://dx.doi.org/ 10.1146/annurev.biochem.71.090501.150041 [DOI] [PubMed] [Google Scholar]
- 35.Ducoux M, Urbach S, Baldacci G, Hubscher U, Koundrioukoff S, Christensen J, Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J Biol Chem 2001; 276:49258-66; PMID:11595739; http://dx.doi.org/ 10.1074/jbc.M106990200 [DOI] [PubMed] [Google Scholar]
- 36.Malkas LH, Herbert BS, Abdel-Aziz W, Dobrolecki LE, Liu Y, Agarwal B, Hoelz D, Badve S, Schnaper L, Arnold RJ, et al.. A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. PNAS 2006; 103:19472-7; PMID:17159154; http://dx.doi.org/ 10.1073/pnas.0604614103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stillman B. Cell cycle control of DNA replication. Science 1996; 274:1659-64; PMID:8939847; http://dx.doi.org/ 10.1126/science.274.5293.1659 [DOI] [PubMed] [Google Scholar]
- 38.Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot 2010; PMID:21169293; http://dx.doi.org/ 10.1093/aob/mcq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem 1998; 67:721-51; PMID:9759502; http://dx.doi.org/ 10.1146/annurev.biochem.67.1.721 [DOI] [PubMed] [Google Scholar]
- 40.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 2009; 284:4041-5; PMID:18835809; http://dx.doi.org/ 10.1074/jbc.R800062200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol Cell 2009; 36:704-13; PMID:19941829; http://dx.doi.org/ 10.1016/j.molcel.2009.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol 1997; 17:6367-78; PMID:9343398; http://dx.doi.org/ 10.1128/MCB.17.11.6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem 2004; 279:1907-15; PMID:14594808; http://dx.doi.org/ 10.1074/jbc.M310362200 [DOI] [PubMed] [Google Scholar]
- 44.Podust VN, Podust LM, Goubin F, Ducommun B, Hubscher U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry 1995; 34:8869-75; PMID:7612628; http://dx.doi.org/ 10.1021/bi00027a039 [DOI] [PubMed] [Google Scholar]
- 45.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem 1999; 274:26647-53; PMID:10480866; http://dx.doi.org/ 10.1074/jbc.274.38.26647 [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Zhang Q, Chen H, Li X, Mai W, Chen K, Zhang S, Lee EY, Lee MY, Zhou Y. P50, the small subunit of DNA polymerase delta, is required for mediation of the interaction of polymerase delta subassemblies with PCNA. PLoS One 2011; 6:e27092; PMID:22073260; http://dx.doi.org/ 10.1371/journal.pone.0027092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahmeh AA, Zhou Y, Xie B, Li H, Lee EY, Lee MY. Phosphorylation of the p68 subunit of Pol delta acts as a molecular switch to regulate its interaction with PCNA. Biochemistry 2012; 51:416-24; PMID:22148433; http://dx.doi.org/ 10.1021/bi201638e [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of dna polymerase delta. J Biol Chem 2007; 282:15330-40; PMID:17317665; http://dx.doi.org/ 10.1074/jbc.M610356200 [DOI] [PubMed] [Google Scholar]
- 49.Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry 2002; 41:13133-42; PMID:12403614; http://dx.doi.org/ 10.1021/bi0262707 [DOI] [PubMed] [Google Scholar]
- 50.Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, Gao Y, Lee EY, Lee MY. Functional roles of p12, the fourth subunit of human DNA polymerase delta. J Biol Chem 2006; 281:14748-55; PMID:16510448; http://dx.doi.org/ 10.1074/jbc.M600322200 [DOI] [PubMed] [Google Scholar]
- 51.Xie B, Li H, Wang Q, Xie S, Rahmeh A, Dai W, Lee MY. Further characterization of human DNA polymerase delta interacting protein 38. J Biol Chem 2005; 280:22375-84; PMID:15811854; http://dx.doi.org/ 10.1074/jbc.M414597200 [DOI] [PubMed] [Google Scholar]
- 52.Zhang T, Brazhnik P, Tyson JJ. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle 2007; 6:85-94; PMID:17245126; http://dx.doi.org/ 10.4161/cc.6.1.3705 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.