Abstract
Cellular immunity against viral infection and tumor cells depends on antigen presentation by the major histocompatibility complex class 1 molecules (MHC I). Intracellular antigenic peptides are transported into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP) and then loaded onto the nascent MHC I, which are exported to the cell surface and present peptides to the immune system1. Cytotoxic T lymphocytes recognize non-self peptides and program the infected or malignant cells for apoptosis. Defects in TAP account for immunodeficiency and tumor development. To escape immune surveillance, some viruses have evolved strategies to either down-regulate TAP expression or directly inhibit TAP activity. To date neither the architecture of TAP nor the mechanism of viral inhibition has been elucidated at the structural level. In this study we describe the cryo-electron microscopy (cryo-EM) structure of human TAP in complex with its inhibitor ICP47, a small protein produced by the herpes simplex virus I. We show that the twelve transmembrane helices and two cytosolic nucleotide-binding domains (NBDs) of the transporter adopt an inward-facing conformation with the two NBDs separated. The viral inhibitor ICP47 forms a long helical hairpin, which plugs the translocation pathway of TAP from the cytoplasmic side. Association of ICP47 precludes substrate binding and also prevents NBD closure necessary for ATP hydrolysis. This work illustrates a striking example of immune evasion by persistent viruses. By blocking viral antigens from entering the ER, herpes simplex virus is hidden from cytotoxic T lymphocytes, which may contribute to establishing a lifelong infection in the host.
Inside our body, every nucleated cell has surface “barcodes” that are surveyed by the immune system. These barcodes are peptides derived from intracellular proteins, presented on the surface by MHC I molecules to indicate whether the cell is healthy (reviewed in ref. 1). Peptides generated from normal cellular proteins are ignored by cytotoxic T cells, whereas viral-derived or malignant peptides will trigger an adaptive immune response, resulting in elimination of the infected or tumor cells. The peptide repertoire is generated in the cytoplasm, mainly by the proteasome, but also in part by cytosolic peptidases (Fig 1a). Peptide uploading onto MHC I molecules takes place inside the ER and is orchestrated by a macromolecular assembly collectively called the MHC class I peptide-loading complex (PLC). Cytosolic peptides are delivered across the ER membrane by the ATP-binding cassette (ABC) transporter TAP. The chaperones calnexin and calreticulin stabilize nascent MHC I molecules awaiting peptides. The tapasin/ERp57 heterodimer brings MHC I molecules and TAP within close proximity and catalyzes peptide loading. Peptide-loaded MHC I molecules are then released from the ER and transported to the cell surface for antigen presentation.
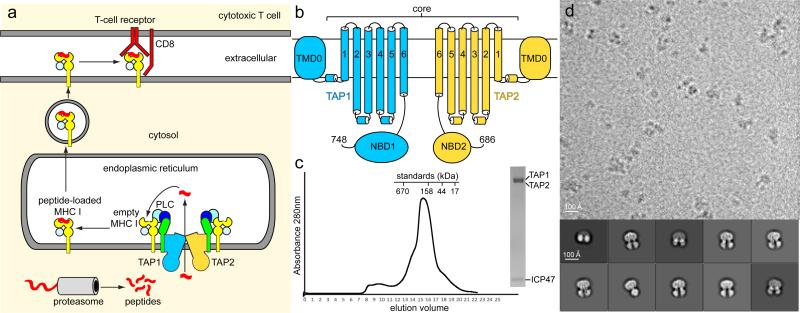
Figure 1. Purification and cryo-EM characterization of TAP.
a, The MHC class I antigen presentation pathway. PLC: peptide-loading complex. b, Topology diagram of TAP1 (blue) and TAP2 (gold). The residue numbers of the C termini are indicated. c, Gel-filtration profile of the TAP/ICP47 complex. Inset: SDS-PAGE gel of the peak fraction stained with Coomassie blue. d, A typical micrograph of the TAP/ICP47 complex after drift correction. Also shown are representative 2D class averages of the particles.
As the MHC I antigen presentation pathway plays a crucial role in eradicating intracellular pathogens, it is not surprising that some viruses have evolved the ability to interfere with this process (reviewed in ref. 2). The peptide transporter TAP in particular is a primary target for viral evasion (reviewed in ref. 3). TAP is a heterodimeric ABC transporter that contains two subunits, TAP1 and TAP2, which share 37% sequence identity and are predicted to have similar structures. Each subunit contains an N-terminal transmembrane region (TMD0) that interacts with tapasin, followed by six transmembrane (TM) helices that form the peptide translocation pathway and a canonical nucleotide-binding domain (NBD) that hydrolyzes ATP (Fig 1b)4. The core TAP, devoid of the TMD0s, is necessary and sufficient for peptide transport4. So far, five viral proteins have been identified as TAP inhibitors. Four are encoded by members of the herpes virus family and one by cowpox virus3. These viral inhibitors are valuable tools for selective immune suppression and for understanding the fundamental mechanism of antigen presentation.
Here we focus our study on a TAP inhibitor encoded by herpes simplex virus (HSV). Both types of HSV, HSV-1 (oral herpes) and HSV-2 (genital herpes), somehow elude the human immune system and lead to a lifelong infection. The first clue as to how HSV bypasses the immune system came from observations that cells infected by HSV have reduced surface expression of MHC I molecules5 and are resistant to cytotoxic T cells6. Since this resistance develops within three hours of HSV infection, researchers narrowed their search for the responsible gene to those few expressed in the early stage of infection7,8. Out of these, an 88-residue protein, ICP47, was found to bind to TAP and prevent peptide translocation into the ER7,8. Consequently, empty MHC I molecules were retained in the ER and viral peptide presentation was suppressed. Subsequent studies have shown that ICP47 interacts with TAP from the cytosolic side of the membrane and somehow prevents peptide binding9,10. The functional domain of ICP47 has been mapped to the N-terminal 35 residues11,12, which form an extended helix-loop-helix structure in lipid bilayers13.
In this study, we pursued structural determination of a TAP/ICP47 complex using cryo-electron microscopy (cryo-EM). The small size of the complex (166 kDa total) and the predicted pseudo two-fold symmetry between TAP1 and TAP2 make it extremely challenging to accurately align particles for 3D reconstruction. To maximize the difference between the two TAP subunits, we used a shorter allele of TAP2 that lacks the last 17 amino acids in NBD2 (Fig 1b)14. Co-expression of TAP1 and TAP2 in Pichia pastoris (Extended Data Fig 1) produced a heterodimer that dissociates readily in detergents. However, by incubating the TAP-enriched membranes with ICP47 before detergent solubilization, the complex consisting of TAP1, TAP2, and ICP47 becomes more stable (Fig 1c and 1d). Cryo-EM analysis of this complex (Extended Data Fig 2 and Fig 1d) produced a density map (Fig 2) of an overall resolution of 6.5 Å, determined by the gold-standard refinement procedure (Extended Data Fig 3)15. In this reconstruction the TM helices and the connectivity between the helices are clearly resolved. The density corresponding to one NBD is significantly smaller than that for the other, allowing us to confidently differentiate TAP1 from TAP2 (Fig 2b). Most importantly, we observe strong density corresponding to the functional region of ICP47, which reveals how this viral protein inhibits peptide translocation (Fig 2b).
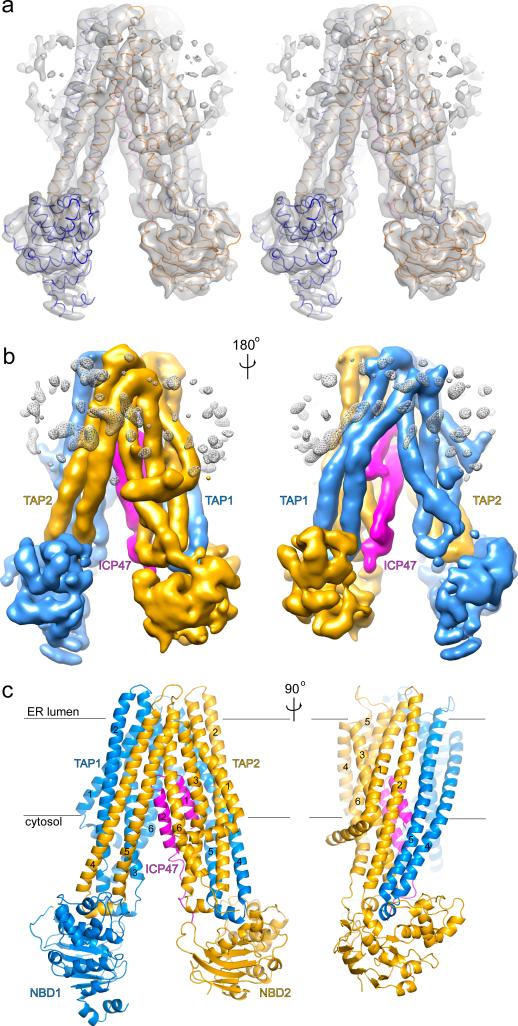
Figure 2. Three-dimensional reconstruction.
a, Stereo view of the overall density map, filtered to 6.5 Å. The α-carbon traces of the TAP core and the N-terminal 50 residues of ICP47 are also shown. b, Two views of the overall density map, colored by protein subunit. c, Ribbon diagram of the poly-alanine model presented in two orthogonal views. The TM helices in the core of the transporter are labeled according to Fig 1b. Color code: TAP1, blue; TAP2, gold; and ICP47, magenta.
The core region of TAP adopts an inverted “V”-shaped structure, with the two TMDs making close contact on the side corresponding to the ER lumen and the NBDs separated from each other inside the cytosol (Fig 2c). Domain swapping of TM helices 4 and 5 across the TAP1/TAP2 interface is a prominent structural feature (Fig 2c). Indeed, the overall structure of TAP is very similar to that of other ABC exporters, including the lipid flippase MsbA from Gram-negative bacteria16, the protein transporter PCAT1 from Gram-positive bacteria17, and the multidrug transporter P-glycoprotein in eukaryotes18,19. Although these transporters recognize very different substrates, they must share a common evolutionary origin and a common mechanism for coupling ATP hydrolysis to substrate translocation.
No density was observed for the N-terminal TMD0 domain of both TAP1 and TAP2 subunits (Fig 2). Studies have shown that the TMD0s are essential to the assembly of the large peptide-loading complex4,20 but dispensable in peptide translocation4. ICP47 inhibits both full-length TAP and the core construct4. Our results indicate that in the absence of tapasin the two TMD0s are flexibly tethered to the core region of TAP.
Biochemical data and homology modeling suggest that the peptide translocation pathway lies at the interface of the two TMDs21,22. Inside this pathway, we observe strong density consistent with the helix-loop-helix structure of ICP47 (Fig 2b and 3a). Guided by the NMR structure and secondary structure prediction13, we built residues 3-16 into the shorter helical density and residues 22-40 into the longer density (Fig 3). Additional density is packed along the cytosolic region of TAP2, into which we modeled residues 41-50. The C-terminal region, neither required for TAP inhibition nor conserved between HSV-1 and HSV-2 (Fig 3c), is not resolved in the EM map, suggesting high mobility.
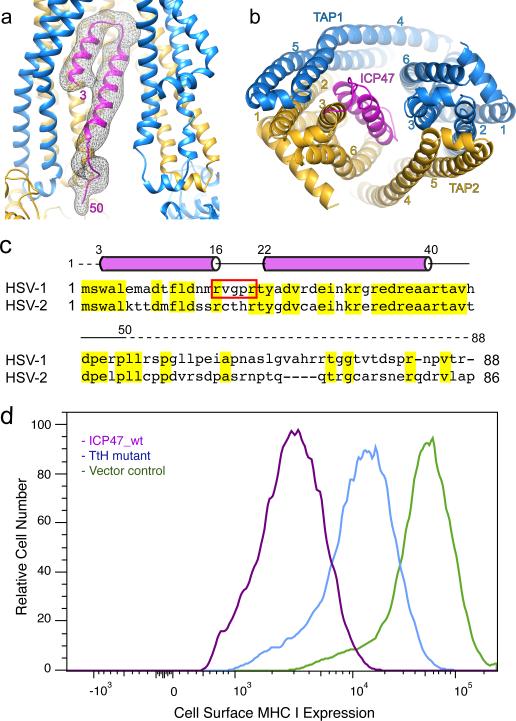
Figure 3. The viral inhibitor ICP47 plugs into the transmembrane pathway.
a, EM density corresponding to ICP47 (black mesh), viewed from the plane of the membrane. Terminal residues in the ICP47 model are indicated. b, Binding of ICP47 to TAP, viewed along the membrane normal from the cytoplasm. c, The helix-loop-helix structure of ICP47 is conserved between HSV-1 and HSV-2. Conserved residues are highlighted are in yellow. A red box highlights residues replaced by alanine in the “turn-to-helix” (TtH) mutant. d, Fluorescence-Activated Cell Sorting (FACS) analysis (three repeats) of MHC I surface expression in cells expressing wt and mutant ICP47.
On the basis of this model, the N-terminal half of ICP47 forms a hairpin-like structure pinned against the inner surface of TAP2 TM helices 2, 3, 6 and TAP1 TM helix 4 (Fig 3b). The two helices of ICP47 run anti-parallel to each other, connected by a sharp turn at the top of the TM cavity. The extensive packing between TAP and ICP47 is consistent with the nanomolar affinity of ICP47, orders of magnitude higher than those of the substrate peptides9,12.
The potency of ICP47 appears to come from its helical hairpin structure, which provides a greater interface with TAP than a typical substrate. To test whether the flexibility of the connecting loop is important, we constructed a “turn-to-helix” mutant by replacing residues 16-22 with alanine, which has the highest propensity to form an α-helix and thus would oppose (but not preclude) the formation of a turn. ICP47 activity was measured in human epithelial cells, in which cytosolic expression of ICP47 inhibits endogenous TAP and reduces the amount of MHC I molecules expressed on the cell surface. A GFP tag was fused to the C-terminus of ICP47 as a marker to select for cells expressing similar amounts of ICP47. Consistent with the EM structure and previous mutagenesis data11, the “turn-to-helix” mutant is much less potent than the wt construct. Specifically, mutant ICP47 reduced surface MHC I expression by only 5-fold as opposed to a 20-fold reduction by wt ICP47 (Fig 3d).
Although ICP47 competes for the same binding site, we do not believe it mimics the substrate binding process. Unlike substrates, ICP47 inhibits rather than stimulates ATP hydrolysis23. Furthermore, in contrast to ICP47, which separates the two NBDs, substrate binding induces partial closure of the NBDs24. More recently, electron paramagnetic resonance (EPR) studies showed that TAP binds its substrates in their extended conformation, comparable to how MHC I molecules present peptides25.
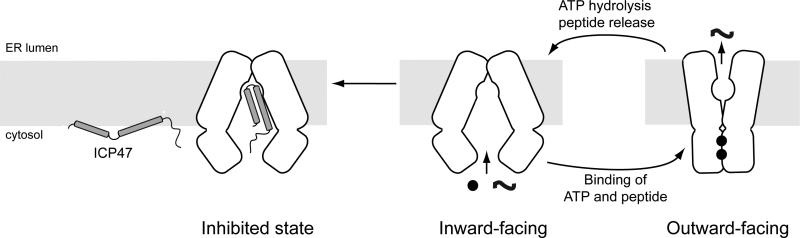
Comparison of our structure with the NMR structure13 suggests that ICP47 undergoes major conformational changes upon association with TAP (Fig 4). In isolation, the N-terminal two helices of ICP47 are flexibly linked and bind to the surface of the membrane at a slight tilt13. In the complex with TAP, ICP47 forms a straight hairpin and inserts perpendicularly into the membrane. Although exact determinations of the amino acid register cannot be made at the current resolution, the overall structure readily explains how ICP47 inhibits peptide transport into the ER. By plugging a long helical hairpin into the translocation pathway, ICP47 directly blocks substrates from binding. Furthermore, because ICP47 is too large to be transported by TAP, its high-affinity binding traps TAP in an inactive conformation. Like other ABC transporters, TAP functions by alternating between two major conformations, each exposing the translocation pathway to one side of the membrane (Fig 4). Binding of ICP47 stabilizes the inward-facing conformation, and thus prevents TAP from transitioning to an outward-facing state in which the NBDs form a closed dimer and the translocation pathway orientates towards the ER lumen (Fig 4).
Figure 4. ICP47 precludes peptide binding and traps TAP in an inward-facing conformation.
TAP functions via alternating access, cycling between two major conformations. In the absence of substrates, the transporter rests in an inward-facing state in which the two NBDs are separated and the translocation pathway is exposed to the cytosol. Upon association of substrates and ATP, the transporter undergoes a conformational change that reorients the TMDs and positions ATP at a closed NBD dimer interface for hydrolysis. ATP hydrolysis releases the substrate and resets the transporter to the resting state. ICP47 binds TAP stabilizes the inward-facing conformation.
In addition to viral inhibition discussed in this study, the structural basis of two cellular regulatory mechanisms has been elucidated26-28; both are relevant to nutrient uptake in bacteria. As a classic example of carbon catabolite repression, when a preferred carbon source is available, bacteria suppress the uptake of maltose through direct binding of a regulatory protein to the maltose transporter26. Methionine and molybdate transporters offer another example27,28. In both cases, at high intracellular concentration, the substrate binds and inhibits the corresponding transporter, a feedback mechanism that limits the amount of import into the cell27,28. Unlike viral inhibition, both cellular inhibitions are allosteric and reversible, regulated by the metabolic state of the cells. One common theme among all these inhibition mechanisms is that the inhibitor binds and stabilizes the transporter in the inward-facing state, a conformation unable to hydrolyze ATP. We speculate that this strategy may be advantageous in preserving cellular energy sources. It is also possible that for most ABC transporters, the inward-facing state is most common and thus naturally targeted by regulators.
Methods
Expression and purification of ICP47
The gene encoding human herpes simplex virus 1 (HSV-1) ICP47 was synthesized (BioBasic) and subcloned into the ligation-independent cloning vector pMCSG20 (AmpR), which contains an N-terminal glutathione S-transferase (GST) affinity tag preceding a tobacco etch virus (TEV) protease cleavage site. BL21(DE3)RIL E. coli cells containing the pMCSG20-ICP47 vector were grown to mid-exponential phase at 37°C in LB medium and expression was induced with 200 μM isopropyl β-D-thiogalactoside (IPTG) at 20°C for 24 h. The cells were harvested via centrifugation (4,000 xg for 12 min at 4°C) and broken by two passes through a high-pressure homogenizer (EmulsiFlex-C3; Avestin). Cell lysate supernatants were loaded onto glutathione sepharose 4B resin (GE Healthcare) equilibrated with PBS buffer, pH 7.4, containing 5 mM D,L-dithiothreitol (DTT). To remove the GST tag from ICP47, the column was equilibrated into a TEV protease cleavage buffer containing 50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 0.5 mM EDTA, and 5 mM DTT and incubated with TEV protease overnight. Untagged ICP47 was eluted and further purified by Superdex 75 gel-filtration chromatography (GE Healthcare) in TEV-cleavage buffer. Fractions containing the protein were pooled, concentrated to 3 mg/ml, and flash-frozen in liquid nitrogen.
Co-expression of TAP1/TAP2
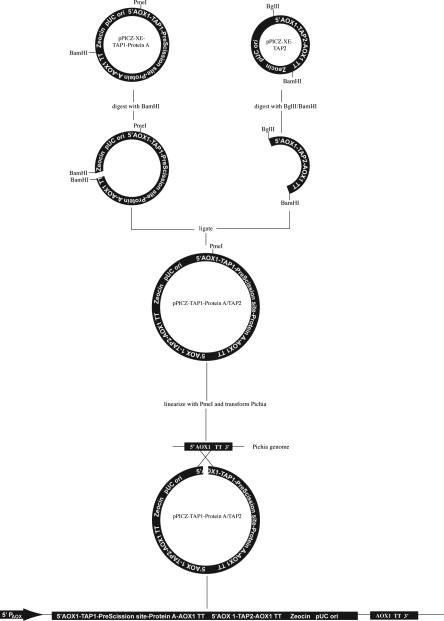
Synthetic human TAP1 and TAP2 genes were codon-optimized for expression in Pichia pastoris (BioBasic) and subcloned into pPICZ-C-XE-Protein A (ZeoR) and pPICZ-C-XE vectors (ZeoR), respectively. In order to ensure co-expression at a 1:1 molar ratio, we took advantage of the compatible cohesive ends of BamHI and Bgl II restriction sites to generate an expression cassette as outlined in Extended Data Figure 1. The resultant vector was linearized by PmeI digestion and transformed into a HIS+ strain of SMD1163 by electroporation (BioRad Gene Pulser II). Transformants were selected on yeast extract peptone dextrose sorbitol (YPDS) agar containing 800 μg/ml zeocin. Colonies were grown in yeast extract peptone dextrose (YPD) cultures at 28°C until they reached an OD 600 of 4 to seed flasks containing minimal glycerol medium (MGY), 13.4% yeast nitrogen base and 1% glycerol at a starting OD 600 of 0.5. The MGY cultures were grown at 28°C for 24 h until they reached an OD 600 of 20, at which point they were harvested by centrifugation (1,500 xg for 15 min at 4°C) and used to seed flasks containing minimal methanol medium (MMY), 13.4% yeast nitrogen base and 0.5% methanol at a starting OD 600 of 10. These MMY cultures were grown at 28°C for 24 h before harvesting (1,500 xg for 15 min at 4°C). Cell pellets were fractionated and flash-frozen in liquid nitrogen.
Purification of the TAP/ICP47 complex
Cells expressing TAP1-Protein A/TAP2 were lysed using a mixer mill (Retsch Mixer Mill 400) and incubated with purified ICP47 in a buffer containing 50 mM Tris-HCl, pH 8, 500 mM NaCl, 15% glycerol, DNase I, protease inhibitors, and 2 mM TCEP for 30 min. Cells were then solubilized with 1.5% n-dodecyl-β-D-maltoside (DDM; Anatrace) for 2 h. The solubilized fraction was isolated by centrifugation (70,000 xg for 40 min at 4°C). The TAP1-Protein A/TAP2/ICP47 complex was isolated using the Protein A affinity tag on TAP1 by lgG Sepharose 6 Fast Flow (GE Healthcare). After extensive washing, PreScission protease (GE Healthcare) was added to the column and incubated overnight to remove the Protein A tag. The complex was eluted with additional buffer and further purified using a Superose 6 column (GE Healthcare) equilibrated with 20 mM Hepes, pH 7.4, 150 mM NaCl, 2 mM TCEP, 1 mM DDM, and 1 mM octaethylene glycol monododecyl ether (C12E8; Anatrace). The peak fraction was used to prepare cryo-EM grids.
Initial cryo-electron microscopy imaging and generation of an initial model
Vitrified specimens of TAP/ICP47 complex were prepared on glow-discharged Quantifoil holey carbon grids by plunge-freezing into liquid ethane using a Vitrobot (FEI). Cryo-EM data were collected at liquid-nitrogen temperature using a K2 Summit direct electron detector camera (Gatan Inc.) on a Tecnai F20 electron microscope (FEI) operating at 200 keV. Dose-fractionated image stacks were recorded with UCSF Image 429 in super-resolution counting mode at a calibrated magnification of 40410x (nominal magnification of 29,000x) with a dose rate of 8 electrons/pixel/s (5.2 electrons/Å2/s). Frames were read out every 200 ms and 30 frames were collected, resulting in an exposure time of 6 s and a total dose of 31.2 electrons/Å2. Dose-fractionated image stacks were 2x binned and motion-corrected, as described29. The defocus was determined with CTER30. BOXER was used to interactively pick 28,813 particles from ~750 images31. The particle images were subjected to the iterative stable alignment and clustering (ISAC) procedure32 implemented in SPARX33. Four ISAC generations specifying 100 particles-per-group and a pixel error threshold of 0.7 yielded 324 averages. Of these, 270 averages were used to calculate an initial 3D density map with the validation of individual parameter reproducibility (VIPER) procedure in SPARX.
Electron microscopy sample preparation and imaging for the final 3D reconstruction
Cryo-EM grids were prepared by pipetting 3 μl freshly purified TAP/ICP47 (2 mg/ml) onto glow-discharged C-flat holey carbon CF-1.2/1.3-4C grids (Protochips) and letting the sample adsorb for 20 s. The grids were blotted for 4 s at 90% humidity using a Vitrobot Mark IV (FEI) and immediately plunge-frozen in liquid nitrogen-cooled liquid ethane. The grids were imaged using a FEI Titan Krios electron microscope operating at an acceleration voltage of 300 keV. Images were recorded using a K2 Summit direct electron detector (Gatan Inc.) set to super-resolution counting mode with a super-resolution pixel size of 0.675 Å using the program SerialEM34. In addition, a Gatan Imaging filter with a slit width of 20 eV was used to remove inelastically scattered electrons. Movie frames were recorded with an exposure time of 200 ms using a dose rate of 10 electrons/pixel/s or 5.5 electrons/Å2/s (1.35 Å at the image plane). Three datasets were recorded using different total doses and defocus ranges (Extended Data Table 1 and Extended Data Figure 2).
Image processing
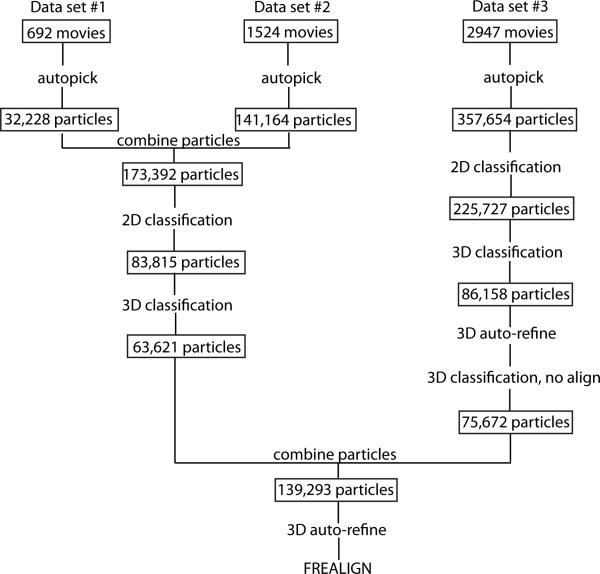
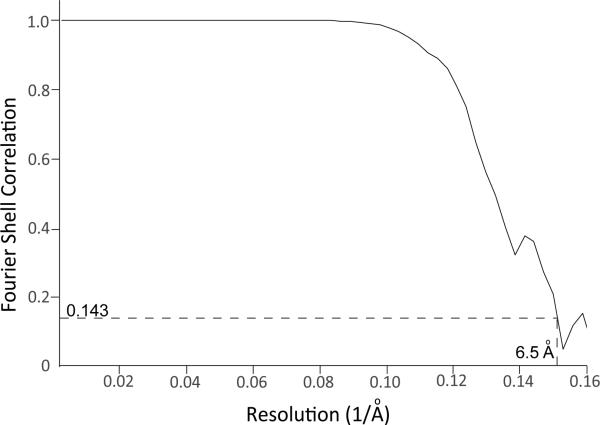
Movie frames were corrected for gain reference and binned by a factor of 2, giving a pixel size of 1.35 Å. Drift correction was performed using the program Unblur35,36. Next, the drift-corrected frames were summed into single micrographs, which were used to estimate the Contrast Transfer Function (CTF) using CTFFIND437. The program Summovie was used to recalculate the summed images, first with a low-pass filter for autopicking and then with the noise power restored after filtering for particle extraction38. Autopicking, particle extraction, 2D classification, 3D classification, and initial 3D refining were all performed in Relion39. In order to achieve a more robust classification of the extracted particles, 2D classification was performed with a particle mask diameter of 145 Å while ignoring the effects of the CTF until the first zero transition. 3D classification was performed on particles from selected 2D classes using the initial model calculated in SPARX as a reference map. Particles from selected 3D classes from both datasets were combined for 3D refinement. Using the orientation parameters determined by Relion, 3D refinement in FREALIGN was also performed40. The final map, reconstructed from 139,293 particles, had a resolution of 6.5 Å as determined by Fourier shell correlation (FSC) of independently refined half-datasets using the 0.143 cut-off criterion (Extended Data Figure 3).
Model building
We used a model of the TAP1 nucleotide-binding domain (NBD) from a previously reported structure of the isolated domain (PDBcode 1JJ7)41 to generate a homology model for the TAP2 NBD using the program Modeller42. We also generated a homology model of the TAP1 and TAP2 transmembrane domains (TMDs) using the half-transporter subunit of human ABCB10 (PDBcode 4AYT) as the source structure 43. We manually docked these poly-alanine models into our final cryo-EM map and rebuilt each model in Coot44.
Flow Cytometry analysis of MHC class I surface expression
MHC I surface expression was analyzed using the phycoerythrin-coupled antibody W6/32 (Abcam), which recognizes a monomorphic epitope shared among MHC class I molecules. Genes encoding HSV-1 wild-type ICP47 and the “turn-to-helix” (TtH) mutant were cloned into a modified version of the pCDNA 3.1 vector (Life Technologies) that adds a C-terminal EGFP tag. HeLa cells (ATCC® CCL-2™) were seeded in 6-well plates at a density of 5 × 105 cells/well and transfected with wt ICP47, the TtH mutant, or the empty vector. Cells were detached from the plate using trypsin-EDTA (0.05%) at 72 h post-transfection and washed in ice-cold FACS buffer (Ca++/Mg++ free phosphate buffer, 10% FCS, 1% sodium azide) and centrifuged at 400 ×g for 5 min at 4°C. Nonspecific binding was blocked by incubating the cells with phosphate buffer containing 5% (w/v) bovine serum albumin (BSA) for 15 min on ice. Antibody was added at 5 μg/ml and incubated for 30 min at 4°C in the dark. Subsequently, the cells were washed 3 times in FACS buffer, resuspended at a density of 3 × 106 cells/ml, and counted using a BD LSR II Flow Cytometer (BD Biosciences). The cells were analyzed at wavelengths 405 nm for DAPI nuclear stain, 488 nm for GFP fluorescence, and 561 nm for phycoerythrin fluorescence. Only live, single cells with the same levels of GFP fluorescence were used in phycoerythrin gating to compare MHC class I expression. The flow cytometry data were analyzed using FlowJo 10.1 single cell analysis software (Tree Star). All experiments were repeated three times. The cell line was tested for mycoplasma contamination by PCR using the Universal Mycoplasma Detection Kit (ATCC® 30-1012K™)
Figure preparation
Figures were prepared using the programs PyMOL 45, Chimera 46, and FlowJo 10.1.
Extended Data
Extended Data Figure 1.
Cloning strategy for TAP1-Protein A/TAP2 co-expression
Extended Data Figure 2.
Cryo-EM data processing flowchart
Extended Data Figure 3. Fourier shell correlation (FSC) indicating the resolution of the density map.
FSC plots were generated between reconstructions from random halves of the data. The frequency at which the dashed line pass through FSC=0.143 indicate the reported resolution. Corresponding values are given in Extended Data Table 1.
| Data collection | |
| Microscope | Titan Krios I, 300 keV (FEI) |
| Detector | K2 Summit direct electron detector (Gatan) |
| Pixel size | 1.35 Å |
| Energy filter | 20 eV (Gatan) |
| Dataset # 1 | |
| Movies | 692 |
| Frames | 20 |
| Dose | 2.2 electrons/Å2/frame |
| Defocus range | −1.6 to −3.0 μm |
| Dataset # 2 | |
| Movies | 1524 |
| Frames | 40 |
| Dose | 2.2 electrons/Å2/frame |
| Defocus range | −2.0 to −5.0 μm |
| Dataset # 3 | |
| Movies | 2947 |
| Frames | 40 |
| Dose | 2.2 electrons/Å2/frame |
| Defocus range | −1.5 to −3.5 μm |
| Final Reconstruction | |
| Number of particles | 139,293 |
| Accuracy of rotations | 2.78 degrees |
| Accuracy of translations | 0.98 pixels |
| Overall resolution | 6.5 Å |
| B-factor correction | −500 Å2 |
Acknowledgements
We thank Z. Yu, C. Hong, and R. Huang at the Howard Hughes Medical Institute Janelia cryo-EM facility for assistance in data collection and processing. We also thank Sarah McCarry for editing the manuscript. R.K.H. is a Howard Hughes Medical Institute fellow of the Helen Hay Whitney Foundation and J.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Contributions
M.L.O. performed protein purification and cryo-EM experiments. R.K.H. provided guidance in data processing. A.M.S. provided assistance with protein expression and purification. E.D. performed the FACS experiments. Z.L. and T.W. collected preliminary cryo-EM data and generated the initial model. M.L.O. and J.C. prepared the manuscript with input from all co-authors.
The Authors declare that there is no conflict of interest.
The 3D cryo-EM density map has been deposited in the Electron Microscopy Data Bank under the accession number EMD-6533.
References
- 1.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. doi:10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van de Weijer ML, Luteijn RD, Wiertz EJ. Viral immune evasion: Lessons in MHC class I antigen presentation. Semin Immunol. 2015;27:125–137. doi: 10.1016/j.smim.2015.03.010. doi:10.1016/j.smim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Verweij MC, et al. Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog. 2015;11:e1004743. doi: 10.1371/journal.ppat.1004743. doi:10.1371/journal.ppat.1004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch J, Guntrum R, Heintke S, Kyritsis C, Tampe R. Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP). J Biol Chem. 2004;279:10142–10147. doi: 10.1074/jbc.M312816200. doi:10.1074/jbc.M312816200. [DOI] [PubMed] [Google Scholar]
- 5.Hill AB, Barnett BC, McMichael AJ, McGeoch DJ. HLA class I molecules are not transported to the cell surface in cells infected with herpes simplex virus types 1 and 2. J Immunol. 1994;152:2736–2741. [PubMed] [Google Scholar]
- 6.York IA, et al. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 7.Hill A, et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. doi:10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 8.Fruh K, et al. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. doi:10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 9.Tomazin R, et al. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J. 1996;15:3256–3266. [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn K, et al. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 1996;15:3247–3255. [PMC free article] [PubMed] [Google Scholar]
- 11.Galocha B, et al. The active site of ICP47, a herpes simplex virus-encoded inhibitor of the major histocompatibility complex (MHC)-encoded peptide transporter associated with antigen processing (TAP), maps to the NH2-terminal 35 residues. J Exp Med. 1997;185:1565–1572. doi: 10.1084/jem.185.9.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann L, Kraas W, Uebel S, Jung G, Tampe R. The active domain of the herpes simplex virus protein ICP47: a potent inhibitor of the transporter associated with antigen processing. J Mol Biol. 1997;272:484–492. doi: 10.1006/jmbi.1997.1282. [DOI] [PubMed] [Google Scholar]
- 13.Aisenbrey C, et al. Structure and dynamics of membrane-associated ICP47, a viral inhibitor of the MHC I antigen-processing machinery. J Biol Chem. 2006;281:30365–30372. doi: 10.1074/jbc.M603000200. doi:10.1074/jbc.M603000200. [DOI] [PubMed] [Google Scholar]
- 14.Powis SH, et al. Polymorphism in a second ABC transporter gene located within the class II region of the human major histocompatibility complex. Proc Natl Acad Sci U S A. 1992;89:1463–1467. doi: 10.1073/pnas.89.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. doi:10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. doi:10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin DY, Huang S, Chen J. Crystal structures of a polypeptide processing and secretion transporter. Nature. 2015;523:425–430. doi: 10.1038/nature14623. doi:10.1038/nature14623. [DOI] [PubMed] [Google Scholar]
- 18.Aller SG, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. doi:10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490:566–569. doi: 10.1038/nature11448. doi:10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonhardt RM, Keusekotten K, Bekpen C, Knittler MR. Critical role for the tapasin-docking site of TAP2 in the functional integrity of the MHC class I-peptide-loading complex. J Immunol. 2005;175:5104–5114. doi: 10.4049/jimmunol.175.8.5104. [DOI] [PubMed] [Google Scholar]
- 21.Nijenhuis M, Hammerling GJ. Multiple regions of the transporter associated with antigen processing (TAP) contribute to its peptide binding site. J Immunol. 1996;157:5467–5477. [PubMed] [Google Scholar]
- 22.Corradi V, Singh G, Tieleman DP. The human transporter associated with antigen processing: molecular models to describe peptide binding competent states. J Biol Chem. 2012;287:28099–28111. doi: 10.1074/jbc.M112.381251. doi:10.1074/jbc.M112.381251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbulev S, Abele R, Tampé R. Allosteric crosstalk between peptide-binding, transport, and ATP hydrolysis of the ABC transporter TAP. Proc Natl Acad Sci U S A. 2001;98:3732–3737. doi: 10.1073/pnas.061467898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng J, Sivaramakrishnan S, Raghavan M. Analyses of conformational states of the transporter associated with antigen processing (TAP) protein in a native cellular membrane environment. J Biol Chem. 2013;288:37039–37047. doi: 10.1074/jbc.M113.504696. doi:10.1074/jbc.M113.504696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herget M, et al. Conformation of peptides bound to the transporter associated with antigen processing (TAP). Proc Natl Acad Sci U S A. 2011;108:1349–1354. doi: 10.1073/pnas.1012355108. doi:10.1073/pnas.1012355108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Oldham ML, Davidson AL, Chen J. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature. 2013;499:364–368. doi: 10.1038/nature12232. doi:10.1038/nature12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science. 2008;321:250–253. doi: 10.1126/science.1157987. doi:10.1126/science.1157987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 2008;321:246–250. doi: 10.1126/science.1156213. doi:10.1126/science.1156213. [DOI] [PubMed] [Google Scholar]
- 29.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penczek PA, et al. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy. 2014;140:9–19. doi: 10.1016/j.ultramic.2014.01.009. doi:10.1016/j.ultramic.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. doi:10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Yang ZF, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative Stable Alignment and Clustering of 2D Transmission Electron Microscope Images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. doi:10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. doi:10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. doi:10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Brilot AF, et al. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. doi:10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell MG, et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. doi:10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.08.008. doi:10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. doi:10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. doi:10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. doi:10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Gaudet R, Wiley DC. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. doi:10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswar N, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5 6, doi:10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shintre CA, et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci U S A. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. doi:10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. doi:10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. 2002 on the World Wide Web < http://www.pymol.org> (2002)
- 46.Pettersen EF, et al. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]