Abstract
We simultaneously investigated the genetic landscape of ankylosing spondylitis, Crohn's disease, psoriasis, primary sclerosing cholangitis and ulcerative colitis to investigate pleiotropy and the relationship between these clinically related diseases. Using high-density genotype data from more than 86,000 individuals of European-ancestry we identified 244 independent multi-disease signals including 27 novel genome-wide significant susceptibility loci and 3 unreported shared risk loci. Complex pleiotropy was supported when contrasting multi-disease signals with expression data sets from human, rat and mouse, and epigenetic and expressed enhancer profiles. The comorbidities among the five immune diseases were best explained by biological pleiotropy rather than heterogeneity (a subgroup of cases that is genetically identical to another disease, possibly due to diagnostic misclassification, molecular subtypes, or excessive comorbidity). In particular, the strong comorbidity between primary sclerosing cholangitis and inflammatory bowel disease is likely the result of a unique disease, which is genetically distinct from classical inflammatory bowel disease phenotypes.
Introduction
Genome-wide association studies have revealed overlap in the genetic susceptibility to human diseases that affect a range of tissues. This overlap is most notable in immune-mediated diseases1,2 including the clinically related conditions ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis (PS), primary sclerosing cholangitis (PSC) and ulcerative colitis (UC). Co-morbidity of these conditions in the same individual and increased risk of any of these conditions in family members have been extensively documented3,4. Recently a large-scale discovery-driven analysis of temporal disease progression patterns using data from an electronic health registry covering the whole population of Denmark revealed substantial population-wide co-morbidity5. This raises the possibility of a hidden molecular taxonomy that differs from the traditional classification of disease by organ or system. Cross-disease genetic studies provide an opportunity to resolve overlapping associations into discrete pathways and explore details of apparently shared etiologies.
In this study, we combined Immunochip genotype data for 52,262 cases and 34,213 controls of European ancestry, the currently largest available genetic data sets in five clinically related seronegative immune-driven phenotypes (AS, CD, PS, PSC and UC) to explore the extent of sharing of genetic susceptibility loci. The aims of this cross-phenotype study were to: 1) identify subsets of the 5 phenotypes with shared genetic risk loci using a cross-phenotype meta-analysis approach, 2) to identify additional susceptibility loci, 3) to investigate co-morbidity and pleiotropy amongst these phenotypes and 4) to improve the understanding of shared pathways and biological mechanisms common to subsets of the phenotypes studied.
Results
Cross-phenotype association analysis
We analyzed Immunochip genotype data of 52,262 cases from AS (8,726), CD (19,085), PS (6,530), PSC (3,408) and UC (14,413) and 34,213 healthy controls (Supplementary Table 1) using variants with a minor allele frequency >0.1% to examine the shared and distinct genetic etiology between these diseases (see Methods). By utilizing Immunochip-only data, we were able to perform a uniform and central quality control of all batches, thus reducing potentially existing batch effects (see Methods). Next, we utilized a recently published subset-based meta-analysis approach (SBM)6 to exhaustively explore all subsets of disease combinations for the presence of association signals. The method identifies the best subset of non-null studies, while in parallel accounting for multiple testing and a fixed control group (see Methods). By performing primary SBM analyses, we identified 166 genome-wide significant (PSBM<5×10−8) loci outside the major histocompatibility complex (MHC, chromosome 6 region at 25–34 Mb) (Supplementary Fig. 1). Three of these 166 loci (rs2042011 at MIR1208; rs2812378 at CCL21/FAM205A; rs1893592 at UBASH3A) have not been reported previously for any of the five diseases under study and thus are novel shared risk loci. SNP associations at UBASH3A (chr21q22.3) and CCL21 (9p13.3) have been reported previously for other autoimmune disorders7,8. These three novel loci would have been missed using single disease analyses alone. To avoid any loss of power, where variants are only associated with a single phenotype, we looked up single disease vs. control subsearches on any SNPs that achieved PSBM<5×10−7 in the primary analysis. Using this SBM-directed approach, we identified 27 novel genome-wide significant disease associations (Pdisease<5×10−8) including 17 novel genome-wide significant loci for AS, 6 loci for CD, and 4 loci for PSC (Figure 1, Supplementary Table 2, Supplementary Fig. 2). 24 out of these 27 associations were also genome-wide significant in the primary SBM analyses (PSBM<5×10−8) thus leading to a total of 169 non-MHC risk loci. In order to identify additional independent association signals within the 169 non-MHC risk loci, we performed a stepwise conditional SBM analysis following a recently published stepwise conditional SBM fine-mapping approach9 (see Methods). In total, we identified 244 independent association signals with 187 signals being shared by at least two diseases for the five diseases under study (Supplementary table 3; Supplementary Fig. 3, 4 and 5). We estimated the heritability explained by these 244 variants for each disease (Figure 2) and for all pair-wise disease comparisons (Supplementary Fig. 6). The ten pair-wise comparisons of disease-associated alleles show diverse patterns of sharing with respect to size and direction of allelic effects and the number of unique associations (Supplementary Fig. 6).
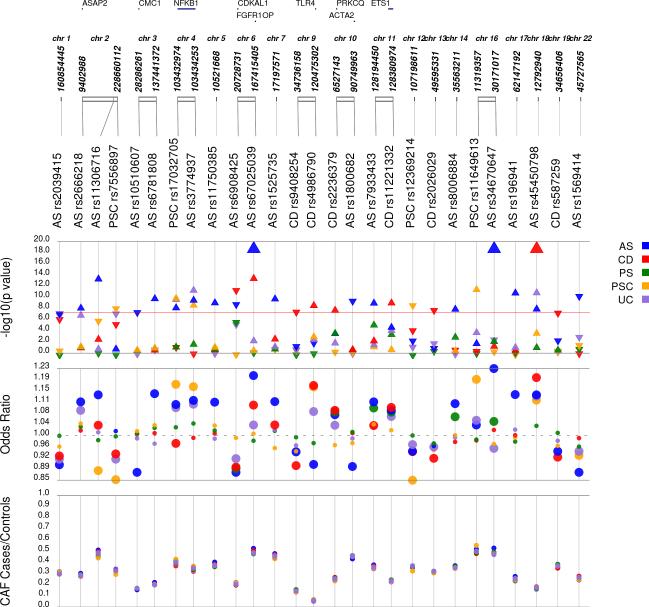
Figure 1. 27 novel genome-wide significant disease associations (Pdisease<5×10−8) for ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis (PS), primary sclerosing cholangitis (PSC) and ulcerative colitis (UC).
Single disease analyses were performed only on SNPs that achieved PSBM<5×10−7 in the primary (unconditioned) cross-disease subset-based association meta-analysis (SBM) approach (see Main Text). We identified 17 novel genome-wide significant susceptibility loci for AS, 6 loci for CD and 4 loci for PSC (Supplementary table 2). Corresponding P-values and ORs for each novel association are shown separately for each disease. With this, the number of known AS, IBD, and PSC risk loci increased to 48, 206, and 20, respectively. For 22 out of 27 gws associations, lead SNPs from the SBM approach (PSBM<5×10−8) and the single disease lookups (PSBM<5×10−7 and Pdisease<5×10−8) are identical, in five instances we have different lead SNPs between SBM and the single disease analyses (Supplementary table 2).
−log10 P-value: −log10 P-values (Pdisease) from Immunochip analysis (Supplementary Table 2) with regard to the physical location of markers; direction of triangle denotes direction of disease-individual effect; OR: odds ratio from the five single disease vs. control subsearches (OR(disease) in Supplementary Table 2). Large circles denote nominal significant disease-individual P-values (Pdisease<0.05); CAF cases/controls: case/control minor allele frequency; If available, the nearest gene within 10kb of the variant is depicted.
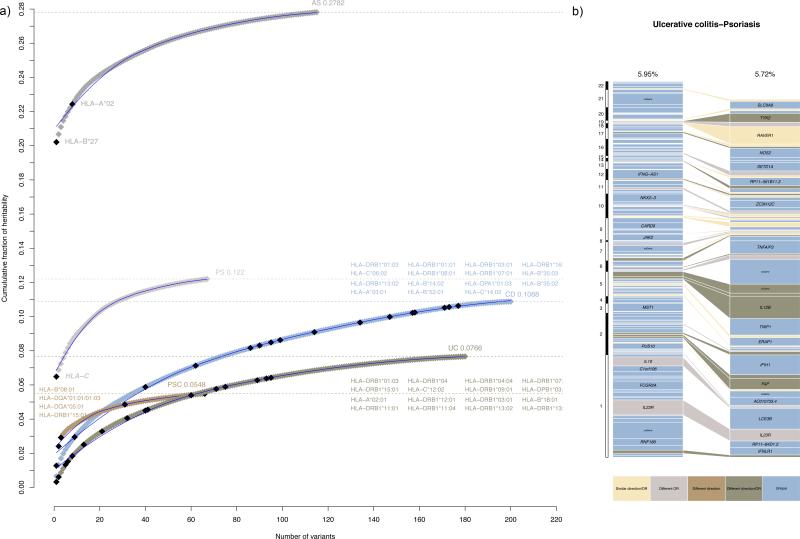
Figure 2. Heritability explained per risk variant from 244 independent multi-disease association signals identified through cross-disease subset-based association meta-analysis.
(a) Cumulative fraction of explained variance in disease liability (heritability) for each disease (see Methods). The loci are ordered from largest to smallest individual contribution. In total, 7.38, 10.30, 5.72, 2.53, and 5.95 percent of the heritability of ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis (PS), primary sclerosing cholangitis (PSC) and ulcerative colitis (UC), respectively, is explained by the 169 loci outside the extended MHC region (for a maximum of 244 independent signals from Supplementary Table 3a). When adding known risk alleles from the major histocompatibility complex (MHC)36-39 to the 169 loci, the cumulative variance increases to 27.82, 10.88, 12.20, 5.48 and 7.66 percent for AS, CD, PS, PSC and UC, respectively.
(b) Example of a pair-wise comparison of heritability explained per risk variant between PS and UC. See Supplementary Figure 6 for all ten pair-wise comparisons. Even if disease-associations account for approximately the same amount of variance explained in disease liability, e.g. as seen here for PS and UC, the pattern of sharing is complex in terms of size and direction of effect. Each box represents an independently associated SNP for the given disease. The size of each box is proportional to the amount of heritability for that variant. The colors of the boxes denote whether the difference in variance explained is due to different direction of effect (risk versus protective), significant heterogeneity of odds ratios (P<0.01) or both.
Functional annotation of associated variants
We functionally annotated the 244 risk alleles from the 169 distinct loci (see Methods). For 210 associations signals (86.1%) the lead variant was within 10 kb of a known gene and 34 signals were classified as intergenic regions (>10 kb distant to a gene) (Supplementary Table 4). The analysis identified 16 coding variants (14 missense, 1 frameshift and 1 splice donor) in genes that were previously implicated in immune-mediated diseases (Supplementary Table 5). Eight of these variants (located in PTPN22, GPR35, MST1, CD6, two in NOD2, TYK2 and CARD9) have been associated before with one of the traits included in this study, and six (GCKR, two in IFIH1, SH2B3, SMAD3, TYK2) with another phenotype, either listed in the GWAS catalog10 or in Immunobase. Two of the genes carrying a coding variant (TLR4, PRKCQ) have previously been suggested as candidate loci, but robust association signals were lacking yet (Supplementary Table 5).
We further checked for variants in high LD (r2>0.8) with the identified variants using 1000 Genomes haplotypes and found that in total 46 of the identified signals were highly correlated with 57 coding variants (48 missense, 2 stop_gain, 3 splice region variants, 1 frameshift, 3 regulatory variants, Supplementary Table 5). We found that 40 of the 57 coding variants, from 30 loci, had been described in previous GWAS or Immunochip studies involving one of the traits included in this study or another phenotype. Additionally, a further 9 variants have been mentioned as candidate variants in autoimmune disease publications. 8 coding variants (7 missense, 1 stop/gain, and all in high LD with our lead variants), located in EFNA1, FCGR2A, HSPA6, C7orf72, FAM118A, respectively, have not been described before in relation to any immune-mediated phenotype.
eQTL analysis in peripheral blood
Analyses of cis-eQTL microarray data from whole peripheral blood samples of an independent control cohort comprising 2,360 unrelated individuals11,12 (see Methods) identified cis-effects for 132 (PFDR<0.05; Supplementary Table 6) out of the 244 disease-associated SNPs from Supplementary Table 3a. Five of these represent the best eQTL SNP and another five represent best secondary eQTL SNPs independent from the best eQTL SNPs at a given locus.
Pathway, cell type, and annotation enrichment analyses
We tested for enrichment between SNPs in associated loci and various types of genomic annotations using GoShifter13. We used 620 different annotations from the NIH Roadmap Epigenomics14 and Fantom515 projects to look for enrichment of histone modifications and expressed enhancers, respectively (Supplementary Table 7 and 8). Results from the SBM association analysis were separated into groups to include all 244 identified variants, variants shared amongst 3 or more phenotypes, and those associated with a phenotype (Supplementary Table 9). For the Roadmap enrichment analysis, using a threshold of P<10−3, the inflammatory bowel disease (IBD) and PS phenotype subsets showed enrichment for H3K27ac modifications in CD3 primary cells and for H3K27ac in adipose tissue, respectively (Supplementary Table 10). The ‘all variant’ (n=244) analysis showed enrichment for H3K4me3 (for which the largest number of cell types were analyzed by the Roadmap consortium and which highlights transcribed promoters and TSS16) in HUES64 cell line as undifferentiated cells, CD34+ cells (bone marrow cells) and Natural Killer Cells (CD56). The Fantom5 data analysis shows enrichment for enhancers expressed in T cells (CD and ‘all_variants’ group) and also Natural Killer cells for CD. However, only the latter (T cells, ‘all_variants’ group) met the significance threshold of 0.05/620=8.06×10−5 needed for Bonferroni correction.
To test which candidate genes from the associated loci (Supplementary table 3a) are highly expressed in which tissues and to define disease relationships at the expression level, we conducted pathway and tissue/cell type enrichment analyses using DEPICT17, with 77,840 microarray expression profiles from human, rat and mouse and 209 tissue/cell type annotations18 (see Methods). Even when correcting for the biased Immunochip gene content, our DEPICT results confirmed that the genes from the 169 herein-reported non-MHC genome-wide significant susceptibility loci show greatest relevance for the regulation of immune response pathways (Supplementary Fig. 7) and the hematopoietic system (Supplementary Fig. 8).
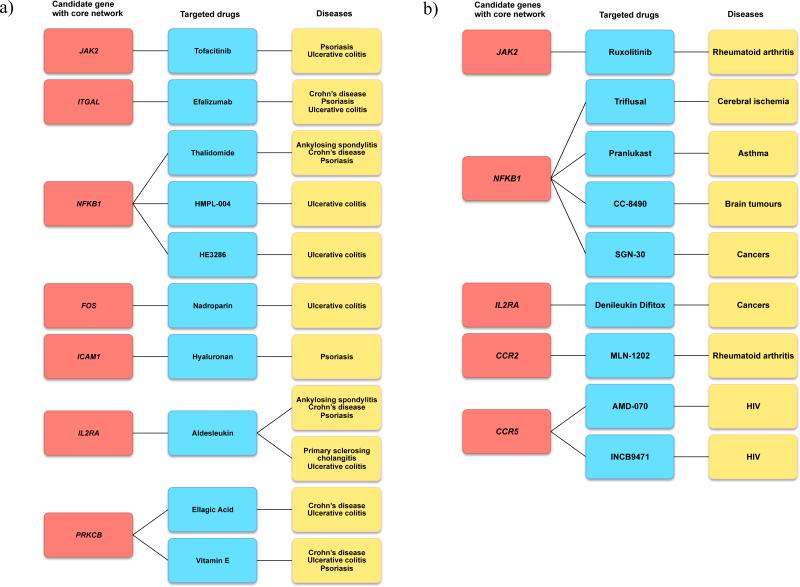
We further generated a protein-protein-interaction (PPI) network (111 gene nodes and 65 edges, see Supplementary Fig. 9) based on five prioritized gene sets of AS, CD, PS, PSC and UC SNP sets, respectively (Supplementary Table 9), from DEPICT analyses and a reference PPI data from ConsensusPathDB19 (see Methods). We observed that 36 gene nodes from this PPI network were connected in one single large component (Supplementary Fig. 9). Then we evaluated the potential role of these genes for their “druggability” by linking genes within this core network to drugs using Drugbank (see Methods). Since the nature and effect of the interaction between the drug and the encoded protein is mostly unknown, e.g some drugs we identified have effects opposite to the what we aim for, we performed a manual literature search to assess which of the identified drugs show evidence or could potentially be promising for any of the diseases under study by using PubMed (last search July 1st 2015) and ClinicalTrials.gov. All drugs were selected based on evidence from phase I/II/III randomized clinical trials (RCTs) or published animal studies. Nine drug target genes overlap with the 36 genes from the core network (Figure 3). Although further investigations are necessary, we propose that target genes/drugs selected by this approach could represent promising candidates for novel drug discovery for treatment of AS, CD, PS, PSC and UC. For example, novel CCR2-antagonists such as MLN-1202, and CCR5-antagonists INCB9471 and AMD-070 are potential new drugs for treatment of AS, CD, PS, PSC and UC.
Figure 3. Identification of drug targeting genes from a core disease protein-protein interaction network.
We evaluated the potential role of genes in drug discovery by linking genes from a core protein-protein-interaction (PPI) network (Supplementary Figure 9) to drugs using Drugbank (see Methods). All drugs were selected based on evidence from phase I/II/III randomized clinical trials (RCTs) or published animal studies. Nine drug target genes overlap with the genes from the core network. (a) Connections between biological genes from core PPI (red), and drugs (blue) used for treatment of AS, CD, PS, PSC and UC (yellow). (b) Connections between biological genes from core PPI (red), and drugs (blue) used for treatment of other inflammatory disease and traits (yellow).
Bayesian multinomial regression for model selection
To compare different disease models for each of the 244 risk variants while accounting for the different sample sizes per diseases, we used Bayesian multinomial regression. The aim is to estimate the posterior probability (Probmodel) for each disease model conditional on the genotype and phenotype data that was observed (see Methods). A disease model is a set of diseases that a given locus is associated (i.e. has a non-zero log odds ratio) with, e.g. “associated with CD and UC, but not with AS, PS or PSC” is one disease model. There are a total of 32 possible disease models for the 5 phenotypes, which includes the null model (“not associated with any disease“). The Bayesian setting naturally handles the different uncertainties on the effect sizes for each disease due to their different sample sizes and powers.
We found 66 signals (59 non-MHC loci) with a best Probmodel≥60% including 14 Loci (with closest genes SH2B3, UBE2L3, TNP2, IL2RA, DNMT3B, CXCR2, CDKAL1, CARD9, MST1, ZMIZ1, ETS1) with Probmodel≥0.8 (Supplementary Table 3b) when assuming that each sharing model is given the same probability (uniform prior across all models, see Methods). However, because previous studies suggested that the structure of sharing of susceptibility is non-uniform2, we calculated posteriors for each model for each risk variant under six different priors and took a vote of the highest posterior models under each prior (see Methods). Then we counted how many priors voted for that model, and calculated the minimum, maximum and mean posterior (MeanProbmodel) for each risk variant (Supplementary Table 3c). Based on this consensus-finding process of merging results from six different priors, we identified 34 signals (31 non-MHC loci) with a best MeanProbmodel≥60% including 12 Loci (with closest genes SH2B3, IL2RA, IFIH1, NFKB1, TYK2) with Prob≥0.8 suggesting that we correctly identified the disease model (Table 1). Out of the 34 associations with MeanProbmodel≥0.6, 25 signals have 5 diseases involved, 6 signals have four diseases and 3 signals are unique to a single disease. Some of these disease sets show different directions of effect (risk versus protective), heterogeneity of odds ratios (P<0.01), or both, for the diseases being involved (Table 1 and Supplementary Fig. 6).
Table 1.
Bayesian logistic regression analysis identified 31 loci with 34 independent associations for which we determined a specific disease model constellation with high certainty (MeanProbmodel≥0.6). A disease model is a list of diseases that a given locus is associated with (i.e. has a non-zero log odds ratio). Mean posterior probabilities (MeanProb) were calculated on a consensus-finding process of merging results from six different priors (Supplementary Table 3c, see Methods). Loci with very high certainty (MeanProbmodel≥0.8) for the best disease model are shown in bold type. Out of the 34 associations with MeanProbmodel≥0.6, 25 signals have 5 diseases involved, 6 signals have four diseases and 3 signals are unique to a single disease.
| Locus | Signal | Chr | Locus_pos_L | Locus_pos_R | SNP | Nearby gene | OR(AS) | OR(CD) | OR(PS) | OR(PSC) | OR(UC) | Best model (VoteWinner) | VoteCount | MeanProb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119 | 1 | 12 | 111702182 | 113030487 | rs3184504 | SH2B3 | 0.915 | 1.062 | 1.055 | 1.189 | 1.047 | PS_AS_CD_UC_PSC | 6 | 0.978 |
| 130 | 1 | 16 | 11018622 | 11496579 | rs367569 | TNP2 | 0.947 | 0.906 | 0.893 | 0.920 | 0.929 | PS_AS_CD_UC_PSC | 6 | 0.963 |
| 166 | 1 | 22 | 21811991 | 22076405 | rs2266961 | UBE2L3 | 1.104 | 1.136 | 1.089 | 1.070 | 1.082 | PS_AS_CD_UC_PSC | 6 | 0.961 |
| 155 | 1 | 20 | 31201111 | 31588992 | rs6058869 | DNMT3B | 1.091 | 1.060 | 1.071 | 1.062 | 1.060 | PS_AS_CD_UC_PSC | 6 | 0.953 |
| 91 | 1 | 10 | 6028491 | 6197536 | rs61839660 | IL2RA | 1.079 | 1.198 | 1.152 | 0.719 | 1.022 | PS_AS_CD_UC_PSC | 6 | 0.943 |
| 8 | 1 | 1 | 67301096 | 67942593 | rs80174646 | IL23R | 0.602 | 0.449 | 0.717 | 0.872 | 0.620 | PS_AS_CD_UC_PSC | 6 | 0.915 |
| 133 | 1 | 16 | 28289243 | 29025978 | rs26528 | IL27 | 1.126 | 1.152 | 1.049 | 1.076 | 1.082 | PS_AS_CD_UC_PSC | 6 | 0.91 |
| 39 | 1 | 2 | 241553993 | 241664801 | rs3749171 | GPR35 | 1.179 | 1.081 | 1.044 | 1.202 | 1.164 | PS_AS_CD_UC_PSC | 6 | 0.878 |
| 151 | 1 | 19 | 10364404 | 10625796 | rs74956615 | RAVER1 | 0.809 | 0.778 | 0.627 | 0.831 | 0.887 | PS_AS_CD_UC_PSC | 6 | 0.841 |
| 95 | 2 | 10 | 64284517 | 64759410 | rs10761648 | ZNF365 | 1.087 | 1.115 | 1.042 | 1.106 | 1.161 | PS_AS_CD_UC_PSC | 6 | 0.837 |
| 32 | 3 | 2 | 162960873 | 163358537 | rs35667974 | IFIH1 | 1.140 | 1.175 | 0.710 | 1.323 | 1.377 | PS_AS_CD_UC_PSC | 6 | 0.83 |
| 22 | 1 | 2 | 24684352 | 25594432 | rs13407913 | ADCY3 | 1.060 | 1.127 | 1.063 | 1.078 | 1.072 | PS_AS_CD_UC_PSC | 6 | 0.811 |
| 153 | 1 | 19 | 49092430 | 49278082 | rs679574 | FUT2 | 1.065 | 1.114 | 1.078 | 1.100 | 1.027 | PS_AS_CD_UC_PSC | 6 | 0.798 |
| 126 | 1 | 14 | 75698304 | 75749875 | rs1569328 | FOS | 0.936 | 0.902 | 0.913 | 0.912 | 0.950 | PS_AS_CD_UC_PSC | 6 | 0.791 |
| 151 | 4 | 19 | 10364404 | 10625796 | rs35074907 | KEAP1 | 1.115 | 1.294 | 1.133 | 1.318 | 1.147 | PS_AS_CD_UC_PSC | 6 | 0.788 |
| 103 | 2 | 11 | 57887309 | 58457495 | rs10750899 | OR5B21 | 1.232 | 1.208 | 1.067 | 1.319 | 1.357 | PS_AS_CD_UC_PSC | 6 | 0.767 |
| 143 | 1 | 17 | 57487538 | 58119648 | rs1292035 | RPS6KB1 | 1.101 | 1.109 | 1.100 | 1.044 | 1.071 | PS_AS_CD_UC_PSC | 5 | 0.711 |
| 147 | 1 | 18 | 12516768 | 12926278 | rs12968719 | PTPN2 | 1.116 | 1.241 | 1.056 | 1.120 | 1.144 | PS_AS_CD_UC_PSC | 5 | 0.699 |
| 32 | 4 | 2 | 162960873 | 163358537 | rs72871627 | IFIH1 | 1.277 | 1.153 | 0.646 | 1.139 | 1.473 | PS_AS_CD_UC_PSC | 5 | 0.693 |
| 62 | 4 | 5 | 158496825 | 158948962 | rs6556411 | AC008697.1 | 0.913 | 0.913 | 1.092 | 0.951 | 0.907 | PS_AS_CD_UC_PSC | 5 | 0.681 |
| 52 | 1 | 5 | 38800374 | 39031577 | rs395157 | OSMR | 0.961 | 0.911 | 0.949 | 0.917 | 0.921 | PS_AS_CD_UC_PSC | 5 | 0.678 |
| 48 | 1 | 4 | 103388565 | 104010837 | rs3774937 | NFKB1 | 1.120 | 0.992 | 1.041 | 1.167 | 1.107 | PS_AS_UC_PSC | 6 | 0.673 |
| 151 | 3 | 19 | 10364404 | 10625796 | rs12720356 | TYK2 | 1.085 | 1.083 | 0.775 | 0.897 | 1.101 | PS_AS_CD_UC_PSC | 4 | 0.673 |
| 8 | 4 | 1 | 67301096 | 67942593 | rs183686347 | IL23R | 1.773 | 2.725 | 1.357 | 1.142 | 1.887 | PS_AS_CD_UC_PSC | 5 | 0.664 |
| 71 | 1 | 6 | 159322326 | 159545322 | rs2451258 | - | 1.086 | 1.114 | 1.111 | 0.918 | 0.990 | PS_AS_CD_PSC | 4 | 0.658 |
| 151 | 2 | 19 | 10364404 | 10625796 | rs35018800 | TYK2 | 0.598 | 0.641 | 0.576 | 0.799 | 0.723 | PS_AS_CD_UC_PSC | 5 | 0.653 |
| 163 | 1 | 21 | 40413101 | 40483777 | rs9977672 | - | 0.825 | 0.920 | 1.006 | 0.786 | 0.799 | AS_CD_UC_PSC | 4 | 0.652 |
| 165 | 1 | 21 | 45596207 | 45702354 | rs4456788 | AP001057.1 | 1.081 | 1.135 | 0.993 | 1.126 | 1.110 | AS_CD_UC_PSC | 4 | 0.652 |
| 11 | 1 | 1 | 152534954 | 152860452 | rs6693105 | LCE3B | 0.994 | 0.998 | 0.799 | 1.006 | 1.006 | PS | 6 | 0.648 |
| 35 | 1 | 2 | 218877398 | 219266204 | rs11676348 | CXCR2 | 1.055 | 1.081 | 0.980 | 1.136 | 1.065 | AS_CD_UC_PSC | 4 | 0.638 |
| 160 | 1 | 20 | 62180117 | 62488635 | rs6062496 | TNFRSF6B | 1.000 | 0.872 | 0.962 | 0.923 | 0.877 | PS_CD_UC_PSC | 6 | 0.628 |
| 110 | 1 | 11 | 114256749 | 114589971 | rs661054 | NXPE1 | 0.984 | 0.998 | 0.982 | 0.993 | 0.883 | UC | 6 | 0.614 |
| 4 | 3 | 1 | 20060965 | 20304744 | rs4655215 | RNF186 | 0.976 | 0.989 | 0.974 | 0.994 | 1.171 | UC | 6 | 0.603 |
| 32 | 1 | 2 | 162960873 | 163358537 | rs2111485 | FAP | 1.030 | 1.054 | 0.852 | 1.074 | 1.082 | PS_AS_CD_UC_PSC | 5 | 0.6 |
Locus: number of locus defined by annotation of association boundaries (see Methods); Signal: number of independent signal (from conditional analysis) within a certain locus; Chr: chromosome; Locus_pos_l/Locus_pos_r: left/right association boundaries for locus (see Methods section). Genomic positions were retrieved from NCBI's dbSNP build v142 (genome build hg19); SNP: rs ID; Nearby gene: gene candidate nearest to the index SNP as long as a gene was with 10kb of the SNP; OR: single disease odds ratio: Ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis (PS), primary sclerosing cholangitis (PSC) and ulcerative colitis (UC). Best model (VoteWinner): disease model with highest posterior probability under six different priors (Supplementary Table 3c); VoteCount: we counted how many priors voted for that model, and calculated the mean posterior from six different priors; MeanProb: Mean posterior probability (MeanProbmodel) for the proposed model and risk variant of six different priors.
Distinguishing pleiotropy from heterogeneity
Statistically significant temporal co-morbidity (disease A followed by disease B within a 5-year time frame of disease A, or vice versa) amongst the five diseases studied was confirmed for 8 out of 10 possible pairs of diseases (P<0.05/823606=1.21×10−9) after screening 823,606 directed pairs of diagnoses from an electronic health registry covering the whole population of Denmark5 (Supplementary Table 11, see Methods). Consistent with previous reports, we further observed high comorbidity rates among our patients (Supplementary Table 12), i.e. patients had more than one disease at the time of last diagnosis. This may occur due to pleiotropy (sharing of risk alleles between disease A and disease B) or heterogeneity (a subgroup of disease A cases has a higher loading of risk alleles for disease B). Heterogeneity can occur as the result of many different scenarios including diagnostic misclassifications, molecular subtypes, and excessive comorbidity. We evaluated whether pleiotropy or heterogeneity best explained the high comorbidity rates amongst the five diseases studied using BUHMBOX20 (see Methods). BUHMBOX detects heterogeneity by calculating the cross-locus correlation of disease B-associated loci among disease A cases; a non-zero correlation is indicative of heterogeneity20. We calculated the statistical power of BUHMBOX to detect various proportions of sample heterogeneity for all disease pairs (Online Methods). For 18 out of 20 pairs of diseases, we had >50% power to detect 20% sample heterogeneity (Supplementary Table 13, Supplementary Figure 10). Since BUHMBOX has high power for these pairs, non-significant BUHMBOX results strongly suggest that the genetic risk score (GRS) association is likely due to pleiotropy rather than heterogeneity.
First, to quantify genetic sharing for each of the 20 possible pairs of five diseases, we used a GRS approach (see Methods). We calculated GRSs for disease B (using known risk alleles, weighted by effect size) for all individuals in the disease A sample, and tested the association of the GRSs with disease A status using logistic regression. The GRSs test for enrichment of disease B alleles in disease A cases, and are expected to be significant both in the presence of pleiotropy and heterogeneity. As expected, we observed highly significant associations between disease B GRSs and disease A status for almost every possible pair (Supplementary Table 14), which demonstrated strong sharing of risk alleles between the different immune-mediated diseases.
We then tested if this observed genetic sharing was due to true pleiotropy or heterogeneity using BUHMBOX20. In the setting of pleiotropy, pleiotropic disease B risk alleles are shared across all disease A cases, whereas in heterogeneity, only a subset of disease A cases share disease B risk alleles. This leads to cross-locus correlations between disease B-associated loci being positive in the presence of heterogeneity, but not in the case of pleiotropy. BUHMBOX calculates the cross-locus correlation between disease B-associated loci in disease A cases, and determines if they are significantly non-zero. We calculated cross-locus correlations for all 20 disease-pairs (see Methods). We did not observe significant inter-locus correlations (Supplementary Table 14), despite high statistical power for many pairs (Supplementary Table 13 and Supplementary Fig. 10). Our findings suggest that the overall GRS association between the five immune diseases investigated is likely due to pleiotropy.
Immunochip-wide co-heritability analysis
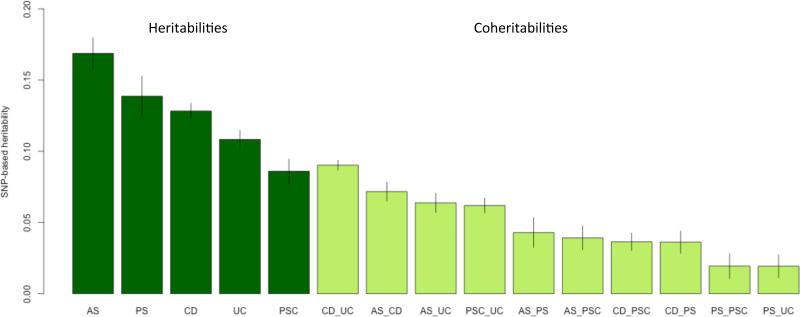
In order to estimate Immunochip-wide pleiotropy (the genetic variation and covariation between pairs of diseases in liability that is tagged by SNPs represented on the Immunochip), we applied univariate and bivariate linear mixed model heritability methods21,22 (see Methods). The relationships between disorders are expressed as SNP-based coheritabilities (Figure 4). When excluding SNPs from the MHC region, genetic correlation was highest between CD and UC (rG=0.78 ± 0.015 s.e., in concordance with previous estimates23), PSC and UC (rG=0.64 ± 0.027 s.e.), moderate (rG<0.5) between AS and CD (rG=0.49 ± 0.023 s.e.), AS and UC (rG=0.47 ± 0.026 s.e.), CD and PSC (rG=0.35 ± 0.030 s.e.), AS and PSC (rG=0.33 ± 0.035 s.e.), AS and PS (rG=0.28 ± 0.035 s.e.), CD and PS (rG=0.27 ± 0.029 s.e.), and low (rG<0.25) between PS and PSC (rG=0.18 ± 0.042 s.e.), and PS and UC (rG=0.16 ± 0.034 s.e.) (see Supplementary Fig. 11,12 and Supplementary Table 15). For correlation values including MHC variants see Supplementary Table 15. As a negative control, we conducted coheritability analyses between each immune-mediated disease under study and longevity, bipolar disorder, major depressive disorder and schizophrenia Immunochip studies (Supplementary Fig. 13). No coheritability was observed with the non-immune-mediated diseases studied here.
Figure 4. Estimation of Immunochip-wide pleiotropy (excluding the MHC region) between the five diseases under study.
Proportion of genetic variance in liability (SNP-based heritability) and proportion of genetic covariance in liability between diseases (SNP-based coheritability) with 95% error bars (see Supplementary Table 15a; estimates including MHC SNPs see Supplementary Table 15b). Disease-associated SNP markers from Supplementary Table 3a (at a maximum of 244 independent signals from 169 non-MHC risk loci, see also Supplementary Figure 6) explain 42.2% of AS-, 79.63% of CD-, 39.6% of PS-, 29% of PSC- and 55% of UC-Immunochip-wide SNP-heritability (excluding the MHC region) on the liability scale, respectively. As the Immunochip densely tags common variants but at the cost of losing genome-wide coverage, the estimated SNP-heritabilities are lower bounds for genome-wide SNP-heritabilities.
Discussion
By combined assessments of Immunochip genotyping datasets from 52,262 patients with five closely associated conditions (AS, CD, PS, PSC and UC; all seronegative inflammatory diseases as per clinical definition) and 34,213 healthy controls we were able to delineate the genetic overlap between the conditions. A key outcome of the overlap analysis is that despite the profound pleiotropy, clear demarcations of the genetic risk for the individual conditions exist. Implicit to this, hence conflicting an existing paradigm where a causal relationship between IBD and the involved extra-intestinal conditions exists24,25, our modeling rather supports (a) the presence of shared pathophysiological pathways as the basis for the clinical co-occurrence and (b) the hypothesis that patients with concomitant syndromes are genetically distinct from patients without concomitant syndromes.
Our cross-disease association framework also enabled the identification of novel coding variants and known eQTLs. One newly identified missense variant for CD, rs4986790, is located at exon three of toll-like receptor 4 (TLR4), which is an important mediator of innate immunity. This SNP has been shown to modulate TLR4 effector functions either by interfering with the binding capacity of TLR4 with its ligands or by controlling the extracellular deposition of functional TLR426,27. Another newly identified missense SNP for CD, rs2236379, which has not been previously associated with other disease traits, is located at exon nine of PRKCQ encoding protein kinase C-theta (PKC-θ). PKC-θ is essential in the signaling cascades that lead to NFkB, AP-1 and NFAT activation28 and is also critical for stabilizing Th17 cell phenotype by selective suppression of the STAT4/IFN-c/T-bet axis at the onset of differentiation29. Furthermore, PKC-θ inhibition enhances Treg function and protects Treg from inactivation by TNF-α, restores activity of defective Treg from rheumatoid arthritis patients, and enhances protection of mice from inflammatory colitis30. We also found that one of the AS/UC secondary signals rs61802846 is in perfect LD (r2=1.0) with a stop-gain SNP rs9427397, resulting in a premature stop codon in FCGR2A. This appears to be distinct (r2=0.12) from the known IBD-associated missense variant in FCGR2A rs18012741,31. Among the 10 strongest eQTL SNPs (PFDR<0.05; Supplementary Table 6, Supplementary Fig. 4) are the intronic SNP rs3766606 (at PARK7 shared by PS (risk) and CD,UC (protective)), the intronic variant rs2910686 (at ERAP2 shared by AS,CD,UC (risk only)), the intronic SNP rs1893592 (at UBASH3A shared by PSC, UC (protective)), the missense SNP rs12720356 (at TYK2 shared by AS,CD,UC (risk) and PS (protective)), and the intronic variant rs679574 (at FUT2 shared by AS,CD,PS and PSC (risk only)).
Most “shared” loci exhibit complex patterns of multi-disease associations suggesting multiple types of pleiotropy32. Through subsequent Bayesian multinomial regression modeling, we identified 31 loci with 34 independent associations (Table 1) for which we determined a specific disease model constellation with high certainty (MeanProbmodel≥60%). For example, at 12q24.12 (Locus 119; SH2B3) the single lead-SNP rs3184504 (Prob=0.98) is associated with decreased risk of AS (ORAS=0.92) but increased risk of the other diseases (ORCD=1.06; ORPS=1.06; ORPSC=1.19; ORUC=1.05), and has been associated before with >10 other phenotypes in the GWAS catalog10, thus suggesting that 12q24.12 is a common risk locus with heterogenous effect sizes for multiple complex diseases.
In addition to contrasting the genetic landscape of AS, CD, PS, PSC and UC, we investigated comorbidity and pleiotropy amongst these phenotypes. GRS and cross-locus correlation analyses20 suggest that the increased comorbidity rates among our patients are due to biological pleiotropy rather than heterogeneity. In other words, an individual with a pleiotropic risk variant is more likely to acquire both diseases. Among all non-zero comorbid rate pairs, the pair of PSC and IBD is particularly noticeable for its high frequency of comorbidity (Supplementary Table 12). PSC patients suffer from a highly increased frequency (62-83%) of IBD33 (called PSC with concomitant IBD, or PSC-IBD, although IBD is most often classified as UC). Interestingly, despite the high prevalence of IBD in PSC the loci encoding IL23R and IL10 (both of which are strongly associated with CD and UC) did not show any evidence of association with PSC. However, we found that many PSC risk variants are shared with UC and have similar effects both in terms of magnitude and direction (Supplementary Fig. 6). It is unlikely that pleiotropy with UC accounts for the comorbid IBD seen in PSC on its own, given the exceedingly higher prevalence of IBD in PSC patients compared to the population prevalence of UC. We therefore questioned whether PSC-IBD is a unique disease distinct from UC, or whether PSC-IBD is the result of UC that is prevalent among PSC patients due to a causal relationship between the two diseases (i.e. UC causes subsequent development of PSC, or vice versa). If the PSC-IBD phenotype is the result of a causal relationship between UC and PSC, there would be a subgroup of PSC cases with a higher loading of UC risk alleles (or vice versa). We tested UC loci in PSC cases with BUHMBOX, and found no evidence of a UC-driven subgroup (Supplementary Table 14) despite high power (99.9% power given the hypothesis that 62% of PSC are affected by UC). We also tested PSC loci in UC cases with BUHMBOX (Supplementary Table 14); while the result was negative (P=0.48), the test was underpowered to detect subtle heterogeneity proportions. Although we cannot completely rule out a causal relationship between PSC and UC at this time, we expect that these findings will become clearer as additional PSC-associated loci are identified in future studies, improving power to detect heterogeneity. At present, our findings are most consistent with the hypothesis that PSC-IBD is a unique disease that shares some genetic factors with UC, but is distinct from classical IBD phenotypes4,34. This hypothesis is further supported by the observation that PSC-IBD shows significant clinical differences from classical IBD, and requires specialized management; compared to IBD, PSC-IBD has a higher rate of pancolitis with ileitis and rectal sparing, as well as a higher incidence of colorectal cancer34.
Our results from testing of enrichment between multi-disease signals and large-scale expression data sets, epigenetic and expressed enhancer profiles further reflect this excessive pleiotropy and mainly highlight perturbations in immune response pathways and blood cell tissues. However, we could not pinpoint which genomic features and which cells a variant influences. We hypothesize that larger gene expression data sets for the disease-relevant tissues and cell types from affected individuals should be generated to allow for high-resolution and more eQTL studies since eQTLs are often cell-specific35. Further, the discovery of multiple further genetic associations increases the power of such analyses to define pathways and cell types involved in specific diseases.
In summary, we performed the largest systematic cross-disease genetic study for chronic immune-mediated diseases to-date. Using novel cross-phenotype analytic methodologies we identified 17 novel genome-wide significant susceptibility loci for AS, 6 loci for CD and 4 loci for PSC, and 3 novel yet unreported risk loci for the diseases under study. With this, the number of known AS, IBD, and PSC risk loci increased to 48, 206, and 20, respectively. Due to lower coverage at unselected regions on Immunochip, imputed GWAS data would further increase statistical power to identify novel shared associations outside established risk loci in future studies. Future cross-disease studies of a wider range of phenotypes, in combination with more sophisticated fine-mapping studies on individual diseases and specific layers of multi-omics data sets are needed to provide another layer of information for a potential new disease classification based on molecular genetic profiles. While most cross-disease studies employ patient panels that were manually curated for single phenotypes and often rely on questionnaire data, future studies could employ even larger collections of hospitalized patients, for which exhaustive electronic medical patient records and array data exists. Moreover, longitudinal data from electronic health charts could pinpoint further comorbidities that should be included in a more systematic next-generation cross-disease approach.
Online Methods
Study subjects
All DNA samples included in the study (Supplementary Table 1) were genotyped using the Illumina Immunochip custom genotyping array40, a targeted high-density genotyping array with comprehensive coverage of 1000 Genomes Project SNPs41 within 186 autoimmune disease-associated loci. CD/UC case and control cohorts were collected from 15 countries across Europe, North America and Australia and have previously been described1. Initially, 19,761 Crohn's disease cases, 14,833 ulcerative colitis cases and 28,999 controls of European ancestries from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) were included in the study. Genotyping of the IIBDGC cohorts was performed in 31 different batches (34 batches before quality control) across 11 different genotyping centers. The initial AS case-control collection (2 main batches) consisted of 10,417 cases and 12,338 controls of European ancestry and were described previously.36 All AS case genotyping was performed at one centre (University of Queensland Diamantina Institute, Translational Research Institute, Brisbane, Australia). 6,577 Psoriasis case and 15,085 control samples (2 main batches) were collected from 13 countries across Europe and North America37. Recruitment of 3,789 PSC patients and 25,079 controls (2 main batches) was performed in 14 countries in Europe and North America.38 Since most control samples we shared between different disease consortia, we identified the set of non-overlapping (unique) control samples (Supplementary Table 1). 2019 schizophrenia cases, 1140 bipolar cases and 589 major depressive disorder cases were collected from different centers in Germany in the context of the MooDs consortium. All samples have been genotyped at the Life&Brain center in Bonn.
Written, informed consent was obtained from all study participants and the institutional ethical review committees of the participating centers approved all protocols.
Immunochip genotype calling and quality control
Initial genotype calling was performed with the Illumina GenomeStudio GenTrain 2.0 software and the custom generated cluster file of Trynka et al. (based on an initial clustering of 2,000 UK samples and subsequent manual readjustment of cluster positions)40. Based on normalized intensity information, we removed samples detected as intensity outliers (>4 s.d.). Based on initial genotype data, we further removed samples with <90% callrate using PLINK42. To identify ethnicity outliers (ie subjects of non-Europeans ancestry), we performed principal component analysis (PCA) with Eigenstrat43 and a set of 210 HapMap founder samples44 and projected Immunochip samples on the principal components axes on the basis of a set of 14,484 independent (minor allele frequency (MAF)>0.05) SNPs excluding X- and Y-chromosomes, SNPs in LD (leaving no pairs with r2>0.2), and 11 high-LD regions as described by Price et al.45. OptiCall genotype recalling was performed with a Hardy-Weinberg equilibrium P-value threshold of 10−15 for each batch, Hardy-Weinberg equilibrium blanking disabled and a genotype call threshold of 0.7. Hardy-Weinberg equilibrium was calculated with conditioning on predicted (European) ancestry, and related individuals were removed from this calculation.
After genotype calling a unified quality control procedure was conducted across 40 genotyping batches. We tested for significantly different allele frequencies of variants across the batches from a particular disease or the control group (with at most one batch being removed) with a false discovery rate (FDR) threshold of 0.01 (Supplementary Fig. 14). Variants that had >2% missing data, a minor allele frequency <0.1% in either of the different disease sets or in controls, had different missing genotype rates in affected and unaffected individuals (PFisher<10−5) or deviated from Hardy-Weinberg equilibrium (with a false discovery rate (FDR) threshold of 10−5 in controls (a) across the entire collection with at most one batch being removed (Supplementary Fig. 15a) or (b) falling below in two single batches (Supplementary Fig. 15b) were excluded. Samples that had >2% missing data and overall increased/decreased heterozygosity rates were removed (Supplementary Fig. 16). For robust duplicate/relatedness testing (IBS/IBD estimation) and population structure analysis, we used a pruned subset of 14,484 independent SNPs (see text above). Pair-wise percentage IBD values were computed using PLINK. By definition, Z0: P(IBD=0), Z1: P(IBD=1), Z2: P(IBD=2), Z0+Z1+Z2=1, and PI_HAT: P(IBD=2) + 0.5 * P(IBD=1) (proportion IBD). One individual (the one showing greater missingness) from each pair with PI_HAT>0.1875 was removed.
To resolve within-Europe relationships and to test for population stratification, the remaining QCed 52,262 cases and 34,213 unique controls were tested using the PCA method, as implemented in FlashPCA46. PCA revealed no non-European ancestry outliers (Supplementary Fig. 17a-c). We computed Tracy-Widom statistics to evaluate the statistical significance of each principal component identified by PCA and identified the top seven axes of variation being significant at PTW<0.05 (Supplementary Table 16). 130,052 QCed polymorphic variants with MAF>0.1% and 52,262 cases and 34,213 unique controls were available for analysis.
Cross-phenotype association analysis
We conducted primary association analysis based on subsets (ASSET) methodology6. Even after adjusting for the large number of comparisons, the SBM method maintains similar type-I error rates as for standard meta-analysis. This method offers a substantial power increase (sometimes approaching between 100-500%6) compared to standard univariate meta-analysis approaches, where the (heterogeneous) effect of a specific SNP is not exclusively restricted to a single disease. Under the assumption that association signals from shared risk loci based on positional overlap are tagging same causal variant for different diseases, the (unconditioned) subset-based meta-analysis (SBM) approach improves power compared to standard fixed-effects meta-analysis methodology. For the situation that distinct variants within shared susceptibility region may confer independent effects for individual diseases, the conditional SBM approach is well suited to reveal these independent (often multi-disease) associations signals (see stepwise subset-based conditional logistic regression).
The subset-based meta-analysis is a generalized fixed-effects meta-analysis and explores all possible subsets of diseases (or a restricted disease set if specified) for the presence of true association signals, while adjusting for the multiple testing required and a fixed control group shared by all diseases. To control for potential population stratification, we adjusted association test statistics by means of principal component analysis (PCA) using the top seven axes of variation (Supplementary Table 16). Adjusted two-tailed PSBM values (risk versus protective) were obtained using the discrete local maxima (DLM) method estimating tail probabilities of the Z score test statistic that is maximized over a grid of neighboring subsets6,47. The maximum (in absolute value) of the subset-specific Z statistics is a conservative variable selector in the sense that for large samples, it will select only non-null studies, but it is not guaranteed to select all of the non-null studies6. The genomic inflation factor (λ) calculated using 1,820 “null”-SNPs (outside the MHC region) associated with reading and writing ability, psychosis and schizophrenia was 1.082 (λ1000 for an equivalent study of 1,000 cases and 1,000 controls=1.002), indicating minimal evidence of residual population stratification in the overall data set of 52,262 cases and 34,213 controls.
Where a particular SNP is only associated with a single disease, the standard meta-analysis methodology has slightly higher power than the subset-based approach. To avoid loss of statistical power in such settings, we looked up every SNP with PSBM<5×10−7 within and outside the 166 non-MHC susceptibility loci, to see if gws (Pdisease<5×10−8) was achieved in any of the five single disease vs. control subsearches. Univariate association statistics (restricted to a single disease data sets versus the fixed control group) were obtained using the same DLM method. The increased statistical power of the single association test (Pdisease) in comparison to original individual disease Immunochip analyses1,36-38,48 is likely due to the fact that the larger sample-sized Immunochip data here (except for AS) was used as a screening tool instead of using it as a replication data set after screening smaller sized GWAS discovery data sets. The large number of novel AS loci can largely be attributed to an approximately 2.5× increased size control cohort compared to the original AS Immunochip study (13,578 controls in the original study vs. 34,213 controls in the current study). After “subtracting” the novel trans-ancestry CD/UC loci from the inflammatory bowel disease (IBD) trans-ancestry study49, 27 of 35 new gws non-MHC risk loci remain for the five diseases under study. Using an alternative method we identified more pleiotropic loci shared between UC and CD (Supplementary table 17, see Conjunctional False Discovery Rate analysis below).
Stepwise subset-based conditional logistic regression
Single and multiple disease-associated (independent) lead-SNPs were selected through stepwise regression to condition away lead-SNPs one at a time until no associations remain following a recently published stepwise conditional SBM fine-mapping approach9. It is an effective method for separating independent signals and assumes that LD between the independent causal variants is low. Significance was defined by Bonferroni correction of the number of LD-independent marker on the Immunochip (0.05/37,377 = 1.34×10−6).
Cluster plot inspection
Immunochip intensity cluster plots of all genome-wide significant SNP markers (PSBM<5×10−7 and Pdisease<5×10−8; PSBM<5×10−8) from Supplementary Tables 2 and 3 were manually inspected by three different persons using Evoker50 to ensure that they were well clustered.
Bayesian multinomial regression for model selection
To compare different disease models at each locus we used Bayesian multinomial regression. A disease model is a list of diseases that a given locus is associated (i.e. has a non-zero log odds ratio) with, e.g. “associated with CD and UC, but not with PS, AS or PSC” is one disease model, as is “associated with all diseases”. There are a total of 32 possible disease models for the 5 phenotypes, which includes the null model (“not associated with any disease“). Our aim is to infer the posterior probability for each of these disease models, conditional on the genotype and phenotype data we have seen. We do this under a Bayesian setting, as it naturally handles the different uncertainties on the effect sizes for each disease due to their different sample sizes and powers. The methods we describe below are implemented in the open source Trinculo software package.
The Bayesian multinomial logistic regression software calculates a marginal likelihood for each model, integrating out uncertainty in the effect size, as
Where β is a vector of log-odds ratios for each disease. The likelihood Pr(D | M) is given by the multinomial logistic likelihood. The prior distribution on the effect sizes is given by β|M ~ MVN(0,ΣM), where ΣM is the prior covariance matrix for model M. To enforce phenotypes that are not associated with the disease, we set ΣMij = 0 if either phenotype i or j is not associated with the locus. We use Newton's method to calculate the maximum a posteriori estimate (MAP) for the parameters, and calculate the marginal likelihood using a Laplace approximation around the MAP.
We calculate the posterior probability for each model as
The method thus requires two priors: the model covariance matrices ΣM and the per-model priors Pr(M). We analyze each variant using six different sets of priors, two different forms of the covariance matrix prior and three different forms of the per-model prior. For the covariance prior we use a) a simple independent covariance matrix where ΣMii = 0.2 if phenotype i is included in the model, and all other ΣMij = 0 and b) an empirical covariance prior where a single covariance matrix is learned by maximum likelihood assuming all loci are associated with all diseases (i.e. maximizing the product of the marginal likelihoods across all loci under the “associated with all phenotypes” model). For the per-model priors, we use a) a uniform prior across all models, b) a uniform prior across the number of phenotypes associated with the locus (so all models where there are the same number of associations sum to 1/6) with equal weight to each model with the same number of phenotypes and c) an empirical prior distribution on the number of phenotypes associated with the locus, inferred by maximum likelihood.
We calculated posteriors for each model for each risk variant under the six different priors. For each risk variant we then took a vote of the highest posterior models under each prior, such that we select whichever model was considered best by the largest number of priors. We also recorded how many priors voted for that model, and how much posterior each prior gives to the winning model.
If SNPs represent secondary independent association signals due to the results from stepwise conditional SBM analysis, then they were tested conditional on all other identified genome-wide significant independent signals within the same locus. Within the Bayesian logistic regression, we included the genotype at the lead SNP (and further preceding independent signals) as a covariate in the model.
Disease correlation measure and temporal comorbidity
To determine significant temporal co-occurrences (disease-pairs) for the five inflammatory diseases under study, we screened an independent data set covering ICD10 diagnose codes from 6,631,920 people of the entire Danish population in the period from 1996 to 20145. We used relative risk (RR) to measure the strength of the correlation between a pair of diagnoses (diagnosis A followed by diagnosis B). RR estimates and associated P-values were calculated using a sampling approach as described in the original study5. In brief, given a pair, diagnosis A followed by diagnosis B, RR of a temporal association was calculated as the ratio of the observed number of patients who had A then B within 5 years and the number of randomly matched control patients would get B within 5 years from a matched discharge. Each matched control has same age (birth decade) and gender as the case and has a discharge of same type (inpatient, outpatient or emergency room) within the same calendar-week as the case's A diagnosis (from which the 5 years to develop B is started). The significance threshold of P=0.05/823606=1.21×10−9 was applied using Bonferroni correction for testing 823,606 directed pairs in the original study5.
Distinguishing pleiotropy and heterogeneity
We used BUHMBOX v0.38 (Breaking Up Heterogeneous Mixture Based On Cross-locus correlations)20 to evaluate whether the sharing of risk alleles observed across pairs of diseases (disease A and disease B) was driven by true pleiotropy where there is pervasive sharing of risk alleles between two diseases, or by heterogeneity where a subgroup of disease A cases has a higher loading of risk alleles for disease B. The BUHMBOX approach has been described in detail elsewhere20. Briefly, a genetic risk score (GRS) approach is used to detect significant sharing of risk loci between disease A and disease B. If such genetic sharing is detected using GRSs, the BUHMBOX test statistic – which identifies heterogeneity by calculating the cross-locus correlation of disease B-associated loci among disease A cases – is applied to verify whether these associations are due to heterogeneity (e.g. sample misdiagnosis, excessive comorbidity) as opposed to biological pleiotropy. In the setting of pleiotropy, pleiotropic disease B risk alleles are shared across all disease A cases, whereas in heterogeneity, only a subset of disease A cases share disease B risk alleles. This leads to cross-locus correlations between disease B-associated loci being positive in the presence of heterogeneity, but not in the case of pleiotropy. To strictly control for false positives, BUHMBOX uses LD-pruning, the top seven principal components from PCA, and delta-correlations between cases and controls.
First, to quantify pleiotropy for each of the 20 possible pairs of five diseases, we calculated GRSs using known independent risk loci for disease B for each case and control in the disease A sample (based on disease B risk alleles, weighted by effect size) and tested the association of these GRSs with disease A status using logistic regression adjusted for the top seven principal components from PCA. The GRS P-values therefore test for enrichment of disease B risk alleles in disease A, and are expected to be significant both in the presence of pleiotropy and heterogeneity. We obtained the list of known associated loci from the previous literature1,36-38 for AS, CD, PS, PSC and UC.
Next, we evaluated the presence of heterogeneity by applying BUHMBOX20 to each of the 20 pairs of diseases. We estimated the statistical power of BUHMBOX to detect a certain proportion of sample heterogeneity by simulation (Supplementary Table 13, Supplementary Figure 10), using the effect sizes and allele frequencies of the disease B loci and randomly simulating the number of cases and controls in the disease A sample.
Functional annotation of associated variants
The variants identified in this study were annotated using the Ensembl variant effect predictor (VEP)51 (release-77) to determine genomic position annotations, including the closest gene, and functional consequences (using the most severe consequence due to SIFT52 and Polyphen53). The —assembly flag was set to GRCh37 and added the —pick flag to retrieve the most severe consequence for the variants. The UpDownDistance plugin was used to retrieve the nearest gene id within 10kb of the variant. TSS distance was retrieved using the TSSDistance plugin. We also included the —regulatory flag to annotate where a variant overlaps a regulatory feature. The DNA hypersensitivity sites (DHS) and promoter annotations were taken from 1KGP annotations54.
To determine whether any of the lead variants were in high LD (r2>0.8) with a functional variant, the 1000 genomes project v3 EUR haplotypes were used (1000 genomes Phase III 20130502 release). Pairwise LD was calculated between the lead SNPs and all other SNPs within this dataset using Plinkv1.0955. Only variants that occurred in 1000 genome dataset were included in this analysis. The GWAS-catalog10 was used to identify whether any lead variants or variants in high LD (r2>0.8) with the lead variants had been previously reported in other GWAS studies. Immunobase and Europe Pubmed Central were also used to determine whether variants had been previously associated with an auto-immune phenotype.
Supplementary Material
Acknowledgments
We thank all patients with ankylosing spondylitis, Crohn's disease, primary sclerosing cholangitis, psoriasis and ulcerative colitis, their families, healthy control individuals and clinicians for their participation in this study. We thank Tanja Wesse, Tanja Henke, Sanaz Sedghpour Sabet, Rainer Vogler, Gunnar Jacobs, Ilona Urbach, Wolfgang Albrecht, Virpi Pelkonen, Kristian Holm, Hege Dahlen Sollid, Bente Woldseth, Jarl Andreas Anmarkrud and Liv Wenche Torbjørnsen for expert help. Dr. Felix Braun, Dr. Wolfgang Kreisel, Dr. Thomas Berg and Dr. Rainer Günther are acknowledged for contributing German PSC patients. Benedicte A. Lie and The Norwegian Bone Marrow Donor Registry at Oslo University Hospital, Rikshospitalet in Oslo and the Nord-Trøndelag Health Study (HUNT) are acknowledged for sharing the healthy Norwegian controls. This work was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (SysInflame grant 01ZX1306A). This project received infrastructure support from the DFG Excellence Cluster No. 306 “Inflammation at Interfaces” and the PopGen Biobank (Kiel, Germany). Andre Franke receives an endowment professorship by the Foundation for Experimental Medicine (Zuerich, Switzerland). EGCUT received targeted financing from Estonian Research Council grant IUT20-60, Center of Excellence in Genomics (EXCEGEN) and University of Tartu (SP1GVARENG). We acknowledge EGCUT technical personnel, especially Mr V. Soo and S. Smit. Data analyzes were carried out in part in the High Performance Computing Center of University of Tartu. We acknowledge support from the Department of Health via the NIHR comprehensive Biomedical Research Centre awards to Guy's and St Thomas’ NHS Foundation Trust in partnership with King's College London and to Addenbrooke's Hospital in partnership with the University of Cambridge. The study was supported by the German Federal Ministry of Education and Research (BMBF), within the context of the National Genome Research Network 2 (NGFN-2), the National Genome Research Network plus (NGFNplus), and the Integrated Genome Research Network (IG) MooDS (grant 01GS08144, grant 01GS08147). RKW is supported by a VIDI grant (016.136.308) from the Netherlands Organization for Scientific Research (NWO). JH was supported by the Swedish Research Council (521-2011-2764). This work is supported in part by funding from the National Institutes of Health (1R01AR063759 (SR), 5U01GM092691-05 (SR), 1UH2AR067677-01 (SR), U19 AI111224-01 (SR), 1R01DK084960-05 (KNL)) and the Doris Duke Charitable Foundation Grant #2013097. ABJ and SB acknowledge funding from the Novo Nordisk Foundation (grant NNF14CC0001) and the H2020 project MedBioinformatics (grant 634143). The study was supported by The Norwegian PSC Research Center . We thank Gosia Trynka for assistance in setting up GoShifter.
Footnotes
Author contributions
D.E., L.J., S.L.S., A.C., J.B., B.H., Y.R.P., J.G.P., S.R., Y.W., E.T., H.-J.W., L.F., T.H.P., R.K.W., V.C., O.A., A.B.J., S.B., M.D.A., performed statistical and computational analyses. M.H. performed computational analyses. T.F., A.M, M.D'A., J.H., W.L., F.D., A.J.F., A.H., S.S., U.M., B.D.J., K.N.L., R.C.T., S.W., M.W., E.E., J.T.E., J.N.W.N.B., M.A.B., were involved in study subject recruitment and assembling phenotypic data. D.E. wrote draft of manuscript. D.E., D.P.M., T.H.K., J.C.B., M.P., M.A.B., A.F. conceived, designed and managed the study. All authors reviewed, edited and approved the final manuscript.
URLs
PopGen biobank, http://www.popgen.de
GWAS catalog, www.genome.gov/gwastudies
Immunobase, www.immunobase.org
Buhmbox, https://www.broadinstitute.org/mpg/buhmbox/
GoShifter, https://www.broadinstitute.org/mpg/goshifter/
DEPICT, http://www.broadinstitute.org/mpg/depict/index.html
Trinculo, https://sourceforge.net/projects/trinculo/
ConsensusPathDB, http://cpdb.molgen.mpg.de/
Drugbank, www.drugbank.ca
PubMed, www.pubmed.gov
Europe Pubmed Central, http://europepmc.org
ClinicalTrials.gov, www.clinicaltrials.gov
Ensembl VEP, http://www.ensembl.org/info/docs/tools/vep/index.html
1000 Genomes, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502
Fantom5, http://fantom.gsc.riken.jp/5/
NIH Roadmap Epigenomics, http://www.roadmapepigenomics.org
Data access
Data access was granted by the management committees of the main disease consortia (The International IBD Genetics Consortium (IIBDGC), International Genetics of Ankylosing Spondylitis Consortium (IGAS), International PSC Study Group (IPSCSG), Genetic Analysis of Psoriasis Consortium (GAPC), Psoriasis Association Genetics Extension (PAGE). The genotype data is not freely accessible but access can obtained by submitting an application to the respective management committees, institutions or data owners.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 3.Najarian DJ, Gottlieb AB. Connections between psoriasis and Crohn's disease. J Am Acad Dermatol. 2003;48:805–21. doi: 10.1067/mjd.2003.540. quiz 822-4. [DOI] [PubMed] [Google Scholar]
- 4.Loftus EV, Jr., et al. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54:91–6. doi: 10.1136/gut.2004.046615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen AB, et al. Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat Commun. 2014;5:4022. doi: 10.1038/ncomms5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee S, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–35. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant SF, et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–5. doi: 10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum Mol Genet. 2014;23:6616–33. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehrmann RS, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelis M, et al. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trynka G, et al. Disentangling the Effects of Colocalizing Genomic Annotations to Functionally Prioritize Non-coding Variants within Complex-Trait Loci. Am J Hum Genet. 2015;97:139–52. doi: 10.1016/j.ajhg.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fantom Consortium et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrmann RS, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–25. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 19.Kamburov A, et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–7. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B, et al. Using genotype data to distinguish pleiotropy from heterogeneity: deciphering coheritability in autoimmune and neuropsychiatric diseases. bioRxiv. 2015 [Google Scholar]
- 21.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen GB, et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet. 2014;23:4710–20. doi: 10.1093/hmg/ddu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant AJ, Lalor PF, Salmi M, Jalkanen S, Adams DH. Homing of mucosal lymphocytes to the liver in the pathogenesis of hepatic complications of inflammatory bowel disease. Lancet. 2002;359:150–7. doi: 10.1016/S0140-6736(02)07374-9. [DOI] [PubMed] [Google Scholar]
- 25.Adams DH, Eksteen B. Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat Rev Immunol. 2006;6:244–51. doi: 10.1038/nri1784. [DOI] [PubMed] [Google Scholar]
- 26.Apetoh L, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–9. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 27.Prohinar P, Rallabhandi P, Weiss JP, Gioannini TL. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J Immunol. 2010;184:4362–7. doi: 10.4049/jimmunol.0903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isakov N, Altman A. PKC-theta-mediated signal delivery from the TCR/CD28 surface receptors. Front Immunol. 2012;3:273. doi: 10.3389/fimmu.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wachowicz K, Baier G. Protein kinase Ctheta: the pleiotropic T-cell signalling intermediate. Biochem Soc Trans. 2014;42:1512–8. doi: 10.1042/BST20140179. [DOI] [PubMed] [Google Scholar]
- 30.Zanin-Zhorov A, et al. Protein kinase C-theta mediates negative feedback on regulatory T cell function. Science. 2010;328:372–6. doi: 10.1126/science.1186068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14:483–95. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsen TH, Boberg KM. Update on primary sclerosing cholangitis. J Hepatol. 2013;59:571–82. doi: 10.1016/j.jhep.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 34.de Vries AB, Janse M, Blokzijl H, Weersma RK. Distinctive inflammatory bowel disease phenotype in primary sclerosing cholangitis. World J Gastroenterol. 2015;21:1956–71. doi: 10.3748/wjg.v21.i6.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Genetics of Ankylosing Spondylitis Consortium et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–8. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu JZ, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–5. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyette P, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–9. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 44.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9:e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor JEWKJGF. Maxima of discretely sampled random fields, with an application to ‘bubbles’. Biometrika. 2007;94:1–18. [Google Scholar]
- 48.Tsoi LC, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015;6:7001. doi: 10.1038/ncomms8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015 doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris JA, Randall JC, Maller JB, Barrett JC. Evoker: a visualization tool for genotype intensity data. Bioinformatics. 2010;26:1786–7. doi: 10.1093/bioinformatics/btq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.