Abstract
Cancer cells often select for mutations that enhance signaling through pathways that promote anabolic metabolism1. Although the PI3K/Akt signaling pathway, which is frequently dysregulated in breast cancer2, is a well-established regulator of central glucose metabolism and aerobic glycolysis3,4, its regulation of other metabolic processes required for tumor growth is not well defined. Here we report that in mammary epithelial cells, oncogenic PI3K/Akt stimulates glutathione (GSH) biosynthesis by stabilizing and activating Nrf2 to up-regulate the GSH biosynthetic genes. Increased Nrf2 stability is dependent on the Akt-mediated accumulation of p21Cip1/WAF1 and GSK-3 inhibition. Consistently, in human breast tumors, up-regulation of Nrf2 targets is associated with PI3K pathway mutation status and oncogenic Akt activation. Elevated GSH biosynthesis is required for PI3K/Akt-driven resistance to oxidative stress, initiation of tumor spheroids, and anchorage-independent growth. Furthermore, inhibition of GSH biosynthesis with buthionine sulfoximine (BSO) synergizes with cisplatin (CDDP) to selectively induce tumor regression in PI3K pathway mutant breast cancer cells, both in vitro and in vivo. Our findings provide insight into GSH biosynthesis as a metabolic vulnerability associated with PI3K pathway mutant breast cancers.
To investigate how Akt regulates metabolism in breast cancer, we evaluated metabolic reprogramming induced by an oncogenic Akt mutation identified in breast cancer patients: the E17K mutation, found in both AKT1 and AKT25,6. We stably expressed wild-type AKT1/2 or AKT1/2(E17K) in non-tumorigenic mammary epithelial MCF10A cells in a doxycycline-inducible manner to assess acute metabolic changes that occur upon oncogenic Akt activation. As previously reported5,7,8, phosphorylation of Akt1(E17K) on S473 is only slightly elevated under serum-starved conditions and does not lead to increased phosphorylation of Akt substrates such as PRAS40 and GSK-3 (Figure 1a). By contrast, Akt2(E17K) is robustly constitutively active, as evidenced by high basal S473 phosphorylation, enhanced phosphorylation of downstream substrates, and increased in vitro kinase activity (Figure 1a and Supplementary Figure 1a). Although AKT1/2(E17K) does not enhance proliferation in the presence of growth factors (Supplementary Figure 1b), as reported7,8, only MCF10A AKT2(E17K) cells proliferate in a growth factor-independent manner (Figure 1b), demonstrating a functional consequence of constitutive Akt2(E17K) activity. Together, these results indicate that AKT2(E17K) is a more potent oncogenic mutation than AKT1(E17K).
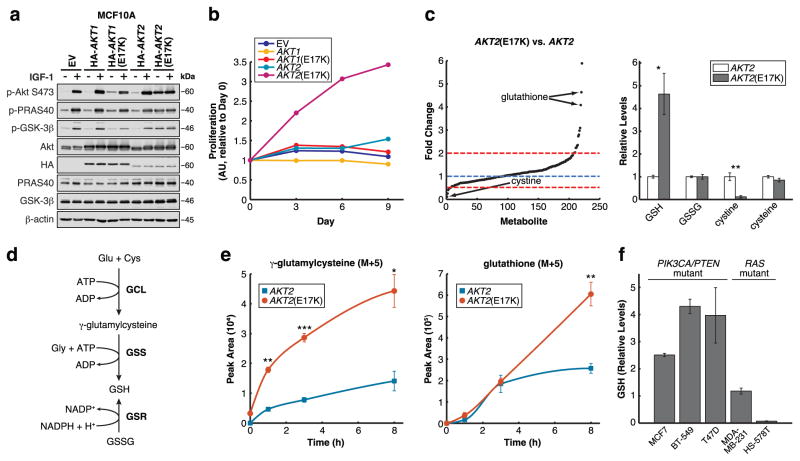
Figure 1. Oncogenic signaling through Akt2(E17K) stimulates GSH biosynthesis.
a, Cells serum-starved and stimulated with IGF-1 were immunoblotted for the indicated proteins (data is representative of three independent experiments). b, Proliferation of cells grown in the absence of serum and growth factors was determined using the sulforhodamine B (SRB) assay (data are from one experiment that was independently repeated five times with similar results (Supplementary Table 1)). c, Fold changes of metabolite abundances by LC-MS/MS in serum-starved MCF10A cells expressing AKT2(E17K) vs. AKT2 (left), and relative levels of GSH, GSSG, cystine, and cysteine (right) (n = 3 technical replicates from a single independent metabolomics experiment; the experiment was repeated twice with similar results (Supplementary Table 1)). d, Schematic of the glutathione biosynthetic pathway. e, Incorporation of U-13C5-glutamine into -GluCys and GSH over 1, 3, and 8 h in serum-starved cells (n = 3 technical replicates from a single metabolomics experiment (Supplementary Table 1)). f, GSH levels in breast cancer cell lines from the NCI-60 metabolomics data set (n = 3 biologically independent replicates). All error bars represent s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 by a two-sided Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Figure 6.
Oncogenic signaling through Akt2(E17K) may support enhanced growth factor-independent proliferation by reprogramming cellular metabolism. To test this idea, we used targeted liquid chromatography-based tandem mass spectrometry (LC-MS/MS) via selected reaction monitoring (SRM)9 to profile steady-state metabolite changes between serum-starved MCF10A AKT2 versus AKT2(E17K) cells and found significant differences between the two cell lines (Supplementary Figure 2a). Consistent with Akt-mediated regulation of glycolysis, Akt2(E17K) strongly up-regulates glycolytic intermediates, including the most up-regulated metabolite NADH (Supplementary Figure 2b–c). Interestingly, the second highest metabolite up-regulated by Akt2(E17K) is reduced glutathione (GSH), while levels of oxidized glutathione (GSSG) are unchanged, resulting in an increased GSH/GSSG ratio (Figure 1c).
Elevated GSH levels suggest that Akt2(E17K) controls cellular redox status by increasing the reducing potential of MCF10A cells, a conclusion supported by changes in levels of other metabolites. First, Akt2(E17K) selectively up-regulates metabolites in the oxidative pentose phosphate pathway (PPP), while metabolites exclusively in the non-oxidative PPP are unchanged or decreased (Supplementary Figure 2d). This is consistent with mTORC1 activation downstream of Akt that selectively up-regulates the oxidative PPP10. Importantly, the oxidative PPP generates reducing equivalents in the form of NADPH, which can be used to reduce GSSG to GSH (Figure 1d). To further support a change in redox status, the most down-regulated metabolite in MCF10A AKT2(E17K) cells is cystine, the oxidized form of cysteine taken up from growth media (Figure 1c). This is consistent with a more reducing intracellular environment wherein any cystine taken up is rapidly reduced to cysteine. Interestingly, steady-state cysteine levels do not change (Figure 1c), suggesting that flux through metabolic processes that utilize cysteine, such as GSH biosynthesis (Figure 1d), may be increased by Akt2(E17K) as well.
We next asked whether Akt2(E17K) increases flux through the GSH biosynthetic pathway. In this pathway (Figure 1d), glutamate-cysteine ligase (GCL), which consists of two subunits GCLC and GCLM, first ligates glutamate and cysteine to form -glutamylcysteine ( -GluCys). Glutathione synthetase (GSS) then catalyzes the condensation of -GluCys with glycine to form GSH. GSH is also formed from the reduction of GSSG by glutathione reductase (GSR). The three amino acids that constitute GSH are primarily derived from metabolites in growth media: glycine and cysteine are taken up exogenously or synthesized from extracellular glucose and methionine, respectively, and glutamate is derived from extracellular glutamine. Therefore, to compare pathway flux, we used LC-MS/MS11,12 to demonstrate that the rate of incorporation of exogenous U-13C5-glutamine into -GluCys and GSH is increased in MCF10A AKT2(E17K) cells (Figure 1e). By contrast, the uptake of U-13C5-glutamine and its incorporation into GSSG is comparable between the two cell lines (Supplementary Figure 2e–f). Therefore, oncogenic signaling through Akt2(E17K) stimulates flux through the GSH biosynthetic pathway. Finally, using the NCI-60 metabolomics data set (https://wiki.nci.nih.gov/display/NCIDTPdata/Molecular+Target+Data), which profiles five breast cancer cell lines, we find that relative to two Ras mutant cell lines, three cell lines that harbor PI3K pathway mutations have higher levels of GSH (Figure 1f), suggesting a correlation between elevated GSH levels and PI3K pathway mutation status in breast cancer.
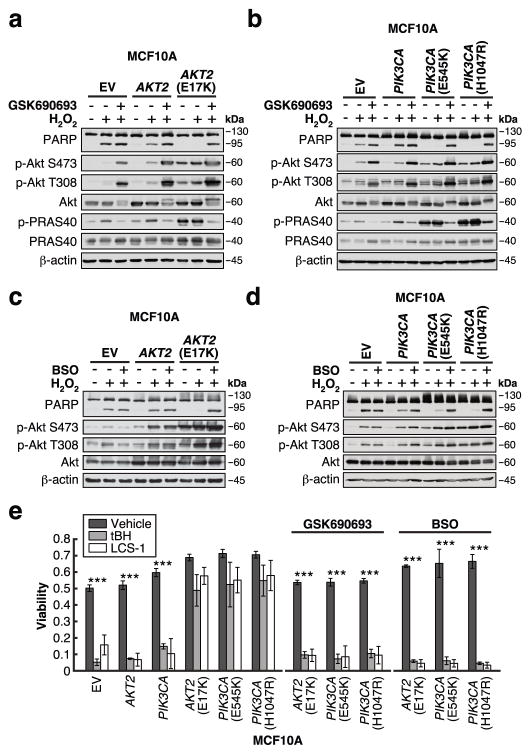
We next asked whether enhanced GSH biosynthesis mediated by oncogenic Akt signaling alters the intracellular redox status to increase resistance to oxidative stress. Compared to MCF10A AKT2 cells, apoptosis is suppressed in cells expressing AKT2(E17K) in response to H2O2, as demonstrated by reduced PARP cleavage (Figure 2a) and increased cell survival as measured by FACS (Supplementary Figure 3a). Resistance to oxidative stress is conferred by oncogenic Akt activity, since the Akt inhibitor GSK690693, which does not induce apoptosis on its own, sensitizes MCF10A AKT2(E17K) cells to H2O2 (Figure 2a, Supplementary Figure 3a,d). Similar results are observed in cells expressing oncogenic PIK3CA mutations (Figure 2b, Supplementary Figure 3b,e–f). Importantly, this oncogenic PI3K/Akt-driven resistance is dependent on enhanced GSH biosynthesis, since buthionine sulfoximine (BSO), a GCL inhibitor that reduces glutathione levels (Supplementary Figure 3c), selectively sensitizes the mutant cells to H2O2 (Figure 2c–d, Supplementary Figure 3d–f). Finally, the PI3K pathway mutant cells are also more resistant to two additional inducers of oxidative stress: tert-butyl hydroperoxide (tBH), and LCS-1, a superoxide dismutase 1 (SOD1) inhibitor that has been shown to inhibit the growth of breast cancer and lung adenocarcinoma cells by generating endogenous ROS13,14 (Figure 2e). Again, inhibition of either Akt or GSH biosynthesis sensitizes the mutant cells to these agents. Together, these data demonstrate that MCF10A cells harboring oncogenic PI3K pathway mutations are more resistant to oxidative stress, in part due to the stimulation of GSH biosynthesis by the PI3K/Akt pathway.
Figure 2. Enhanced GSH biosynthesis confers resistance to oxidative stress.
a–d, Cells were serum-starved for 20–24 h in the presence or absence of (a–b) 1 μM GSK690693 or (c–d) 50 μM BSO, followed by treatment with 500 μM H2O2 for 4 h. Cells were immunoblotted for the indicated proteins (data is representative of three independent experiments). e, Cells were serum-starved for 20–24 h in the presence or absence of 1 μM GSK690693 or 50 μM BSO, followed by treatment with 500 μM tert-butyl hydroperoxide (tBH) or 10 μM LCS-1 for 24 h. Cell viability was measured using a propidium iodide-based plate reader assay (EV, AKT2(E17K): n = 4; AKT2, PIK3CA: n = 3; PIK3CA(E545K), PIK3CA(H1047R): n = 5; n represents number of biologically independent replicates (Supplementary Table 1)). All error bars represent s.e.m. ***P < 0.001 by a two-sided Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Figure 6.
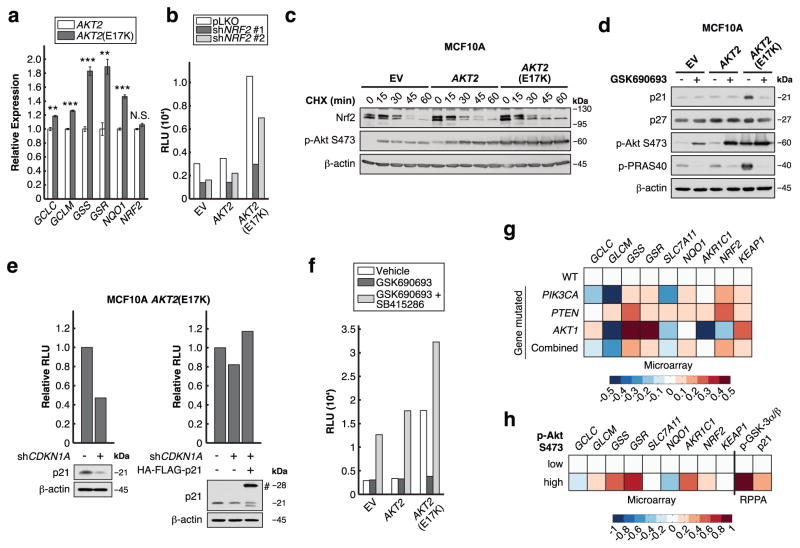
We determined that one mechanism by which Akt2(E17K) stimulates GSH biosynthesis is through the transcriptional up-regulation of the GSH biosynthetic genes GCLC, GCLM, and in particular GSS and GSR (Figure 3a). These genes are targets of the transcription factor Nrf2, which is the master regulator of the cellular antioxidant response15. Accordingly, NQO1, a canonical Nrf2 target, is also up-regulated by Akt2(E17K) (Figure 3a). In support of Nrf2 functioning downstream of Akt to stimulate GSH biosynthesis, knock-down of Nrf2 reduces total cellular glutathione levels and sensitizes AKT2(E17K) cells to oxidative stress induced by H2O2 (Supplementary Figure 4a–c). Using a luciferase reporter under the control of the antioxidant response element (ARE), which is recognized by Nrf2, we show that Akt2(E17K) strongly activates Nrf2-mediated transcription, in a manner dependent on both Nrf2 and oncogenic Akt activity (Figure 3b, f).
Figure 3. Akt2(E17K) activates Nrf2 to up-regulate the GSH biosynthetic genes.
a, mRNA levels were measured by qRT-PCR and are expressed as fold changes relative to MCF10A AKT2 cells (n = 3 biologically independent replicates (Supplementary Table 1)). b, Nrf2 was knocked down over 72 h, and ARE-luciferase activity was assayed in cells serum-starved for 20-24 h (data are from one experiment that was independently repeated two times with similar results (Supplementary Table 1)). c, Serum-starved cells treated with 20 μg/ml cycloheximide (CHX) were immunoblotted for the indicated proteins (data is representative of three independent experiments). d, Cells serum-starved in the presence or absence of 1 μM GSK690693 for 20–24 h were immunoblotted for the indicated proteins (data is representative of three independent experiments). e, p21 was knocked down in MCF10A AKT2(E17K) cells over 48 h, with or without the expression of shRNA-resistant HA-FLAG-p21 (#). ARE-luciferase activity was assayed in cells serum-starved for 20-24 h (data are from one experiment that was independently repeated five times with similar results (Supplementary Table 1)). f, ARE-luciferase activity was assayed in cells serum-starved for 20–24 h in the presence or absence of 1 μM GSK690693 and 25 μM SB415286 (data are from one experiment that was independently repeated two times with similar results (Supplementary Table 1)). g, h, Patient tumors from TCGA BRCA data set were stratified by (g) PI3K pathway mutation status and (h) Akt pS473 levels (See Supplementary Figure 4d). PIK3CA: PIK3CA mutation, PTEN: PTEN mutation or copy number loss, AKT1: AKT1 mutation, Combined: alterations in PIK3CA or PTEN or AKT1, WT: wild-type PIK3CA, PTEN, and AKT1. The heat map represents microarray/RPPA Z-scores for the indicated genes/proteins, which are expressed relative to values in the WT or Akt pS473 low group. All error bars represent s.e.m. **P < 0.01, ***P < 0.001 by a two-sided Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Figure 6.
Several mechanisms of Nrf2 activation in cancer have been reported16. In the context of oncogene-dependent signaling, mutations in oncogenes such as KRAS and BRAF activate Nrf2 by transcriptional up-regulation17. In MCF10A AKT2(E17K) cells, however, Nrf2 transcript levels do not change relative to wild-type cells (Figure 3a), suggesting Nrf2 activation through a post-transcriptional mechanism. Under basal conditions, the Keap1-Cul3 E3 ubiquitin ligase complex suppresses Nrf2 by ubiquitin-directed proteasomal degradation. Various oxidative cellular stresses disrupt Nrf2 ubiquitination by Keap1-Cul3, resulting in Nrf2 stabilization and activation15. Accordingly, by using cycloheximide (CHX), we find that Nrf2 is stabilized in MCF10A AKT2(E17K) cells (Figure 3c), indicating that oncogenic Akt signaling activates Nrf2 through stabilization of the Nrf2 protein.
We noted that two Akt substrates are known to regulate Nrf2 protein stability. First, p21Cip1/WAF1 has been reported to directly bind Nrf2 and disrupt the Keap1-Nrf2 interaction to induce Nrf2 activation18. Consistent with Akt-mediated enhancement of p21 stability19, we find that MCF10A AKT2(E17K) cells express higher levels of p21, in a manner dependent on Akt activity, while levels of p27Kip1, another cyclin-dependent kinase inhibitor, are unaffected (Figure 3d). Importantly, p21 is required for the induction of Nrf2 activity by Akt2(E17K) (Figure 3e). Another Akt substrate, GSK-3 , has been shown to phosphorylate and direct Nrf2 toward ubiquitination and proteasomal degradation after it is released from inhibition by Keap120. We therefore reasoned that Akt may activate Nrf2 through inhibition of GSK-3 . Indeed, GSK690693-mediated inhibition of Akt2(E17K)-induced Nrf2 activity is dependent on GSK-3 , since concomitant inhibition of Akt and GSK-3 with GSK690693 and SB415286, respectively, restores Nrf2 activity (Figure 3f). Therefore, GSK-3 is epistatic to and acts downstream of Akt with respect to Nrf2 activation. Based on these results, we propose that oncogenic PI3K/Akt signaling results in p21 accumulation, which disrupts the Keap1-Nrf2 interaction. Nrf2 stability is then further enhanced by Akt-mediated inhibition of GSK-3 , leading to Nrf2 activation and up-regulation of the GSH biosynthetic genes.
Finally, since these results largely rely on the MCF10A system, we asked whether Nrf2 activation by oncogenic PI3K/Akt signaling functions in human breast tumors. We analyzed TCGA Invasive Breast Cancer (BRCA) microarray data21 by stratifying patient tumors according to mutation status: tumors harboring PIK3CA, PTEN, or AKT1 mutations were compared to ‘‘wild-type’’ tumors without these mutations. Analysis of several Nrf2 target genes indicates that many of these, and especially GSS and GSR, are up-regulated in PI3K pathway mutant tumors (Figure 3g), which closely reflects the qRT-PCR data from the MCF10A AKT2(E17K) cells (Figure 3a). Similarly, up-regulation of Nrf2 target genes is associated with tumors with high Akt S473 phosphorylation, as determined by TCGA reverse-phase protein array (RPPA) data (Figure 3h, Supplementary Figure 4d). Moreover, tumors with high Akt pS473 have elevated GSK-3 phosphorylation and p21 protein levels, consistent with our proposed mechanism of Nrf2 activation by oncogenic Akt signaling.
Next, we explored whether GSH biosynthesis is a metabolic vulnerability associated with PI3K/Akt-driven breast cancer progression. Recent studies demonstrated that in RAS mutant cancer cell lines, glutathione depletion by BSO or by inhibitors of the cystine-glutamate antiporter SLC7A11 leads to the inhibition of the glutathione peroxidase GPX4, subsequently inducing a form of cell death termed ferroptosis22,23. However, in MCF10A cells expressing AKT2(E17K), we do not observe an increase in SLC7A11 or GPX4 expression (Supplementary Figure 5a). Consistently, in 2D culture conditions, rather than causing a loss in viability, BSO enhances the growth factor-independent proliferation of MCF10A AKT2(E17K) cells (Supplementary Figure 5b). Similarly, PIK3CA mutant cells are unaffected by BSO (Supplementary Figure 5c). In full growth media, low BSO concentrations stimulate proliferation, and growth inhibition is not observed until ~1 mM BSO, at which off-target effects are expected (Supplementary Fig. 5d–e). This is consistent with observations that low BSO concentrations may stimulate proliferation in certain contexts24, perhaps due to the induction of milder levels of oxidative stress that stimulate proliferative signaling25,26.
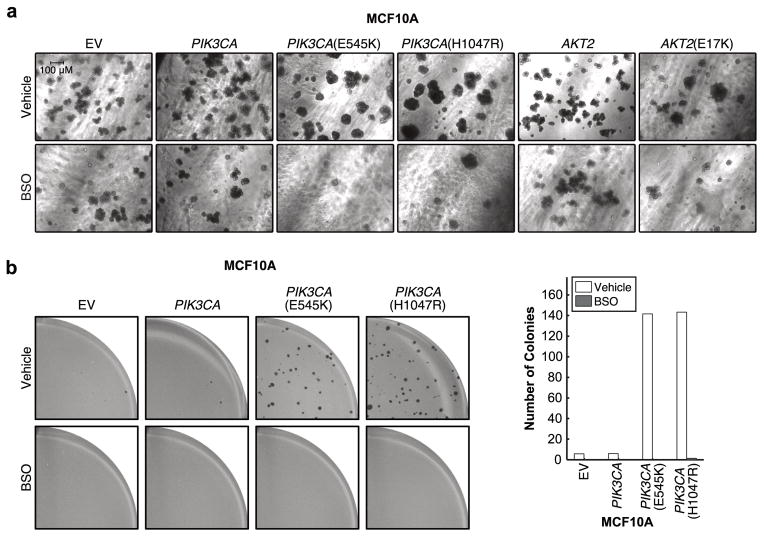
Since certain phenotypes differ in cells grown in 3D culture27, which more accurately recapitulates the morphology of tumors growing in vivo, we evaluated how BSO affects proliferation in 3D (Figure 4a). The number and morphology of spheroids formed by cells expressing empty vector control, wild-type PIK3CA, or wild-type AKT2 are only slightly affected by BSO. By contrast, the number of spheroids formed by cells expressing oncogenic PIK3CA or AKT2(E17K) is markedly reduced by BSO treatment, and spheroids that do grow are smaller and do not form multi-acinar structures observed in untreated cells. Therefore, in contrast to 2D culture, under 3D culture conditions that model both tumor initiation and maintenance we find that BSO selectively prevents tumor spheroid initiation by PIK3CA or AKT2 mutant MCF10A cells. This is consistent with the recent finding that inhibition of GSH biosynthesis in a mouse model by GCLM knockout markedly inhibits breast tumor initiation induced by the polyomavirus middle T oncogene, but has minimal effect on tumor maintenance28.
Figure 4. GSH biosynthesis is required for the PI3K/Akt-driven initiation of tumor spheroids.
a, Cells were grown as spheroids in 3D culture for 14 days in the presence or absence of 50 μM BSO. Representative images from two biologically independent experiments are shown. b, Cells were grown as colonies in soft agar in the presence or absence of 50 μM BSO for 4 weeks. Viable colonies were stained with iodonitrotetrazolium chloride. Representative images from two biologically independent experiments are shown (left). The number of colonies was counted with MatLab (right, data are from one experiment that was independently repeated two times with similar results (Supplementary Table 1)).
We also assessed whether enhanced GSH biosynthesis is required for mutant cells to resist anoikis and undergo anchorage-independent growth in soft agar. Although AKT2(E17K), like AKT1(E17K), does not drive colony formation7, PIK3CA mutant cells form colonies in a manner sensitive to BSO treatment (Figure 4b). Therefore, enhanced GSH biosynthesis is necessary, but not sufficient, for anchorage-independent growth. Since cells experience increased levels of oxidative stress when detached from their matrix29, stimulation of GSH production by oncogenic PI3K/Akt signaling is necessary for PIK3CA mutant cells to overcome this elevated oxidative stress and undergo anchorage-independent growth.
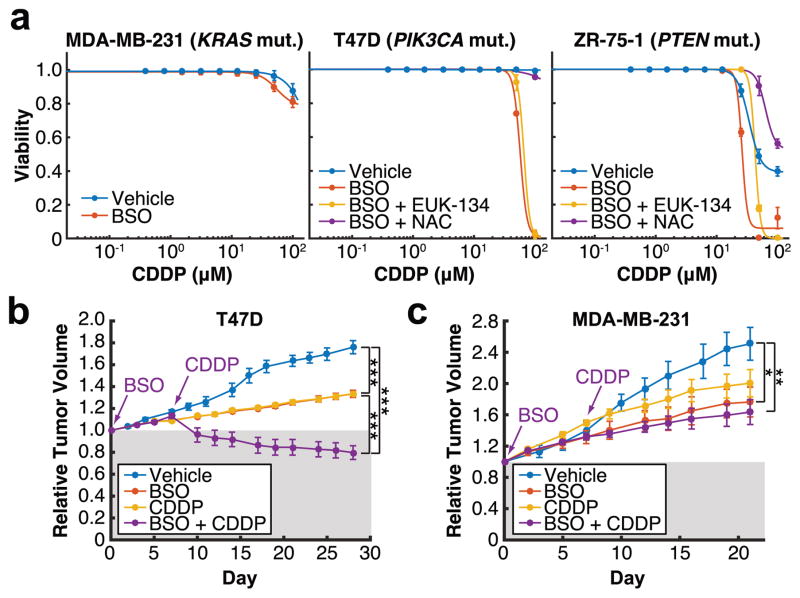
Finally, since BSO sensitizes PIK3CA and AKT2 mutant cells to oxidative stress (Figure 2), we hypothesized that targeting GSH biosynthesis in combination with agents that induce oxidative stress, such as chemotherapy or radiation, may be a viable therapeutic strategy for treating breast cancer cells harboring a PI3K pathway mutation. In particular, oxidative stress is a major mechanism involved in cisplatin (CDDP) toxicity30. Moreover, elevated GSH levels have been associated with CDDP resistance, and GSH has been suggested to form a conjugate with CDDP to facilitate its export and elimination from cells31. Therefore, we reasoned that BSO would sensitize PI3K pathway mutant breast cancer cells to CDDP. We identified three breast cancer cell lines that are resistant to CDDP as a single agent in vitro (Figure 5a). BSO potently synergizes with CDDP to induce cell death in T47D and ZR-75-1 cells, which harbor a PIK3CA and PTEN mutation, respectively. Importantly, this cell death is partially rescued by EUK-134, a hydrogen peroxide scavenger, and is fully rescued by the antioxidant N-acetyl cysteine (NAC), indicating that the synergistic effect is mediated by GSH depletion. In contrast, MDA-MB-231 cells, which lack a PI3K pathway mutation, are insensitive to this combination. These results were corroborated in vivo: While BSO and CDDP only inhibit the growth of T47D xenografts in nude mice as single agents, CDDP induces T47D tumor regression in mice that have been pre-treated with BSO for a week (Figure 5b). By contrast, tumor regression is not induced by the combination in MDA-MB-231 xenografts (Figure 5c), further suggesting a PI3K/Akt pathway-dependent effect of this drug combination. Given the renewed interest in using platinum-based agents for breast cancer treatment32, these results provide insight for inhibiting GSH biosynthesis to maximize the efficacy of platinum-based chemotherapies specifically for the treatment of breast tumors harboring a PI3K pathway mutation.
Figure 5. BSO synergizes with cisplatin to selectively induce cell death and tumor regression in PI3K pathway mutant breast cancer cells.
a, Cells were treated with vehicle or 50 μM BSO for 48 h prior to treatment with cisplatin (CDDP) for 48 h, in the presence or absence of 50 μM EUK-134 or 1 mM N-acetyl cysteine (NAC). Cell viability was measured using a propidium iodide-based plate reader assay (n = 3 biologically independent replicates (Supplementary Table 1)). b, c, T47D (b) or MDA-MB-231 (c) xenografts were grown in nude mice treated with vehicle (T47D: n = 5, MDA-MB-231: n = 5), BSO (T47D: n = 4, MDA-MB-231: n = 4), CDDP (T47D: n = 4, MDA-MB-231: n = 4), or a combination of one week of BSO pre-treatment followed by CDDP (T47D: n = 5, MDA-MB-231: n = 7) (n represents number of biologically independent tumors (Supplementary Table 1)). All error bars represent s.e.m. *P<0.05, **P<0.01, ***P < 0.001 by a two-sided Student’s t-test.
Taken together, our results show that the stimulation of GSH biosynthesis by Nrf2 activation constitutes a part of the metabolic reprogramming mediated by oncogenic PI3K/Akt signaling in breast cancer. By integrating analyses from a mammary epithelial cell line model, a panel of breast cancer cell lines, tumor xenografts, and TCGA breast cancer data, we provide pre-clinical evidence that may inform future therapeutic approaches for treating PI3K pathway mutant breast cancer cells by exploiting GSH biosynthesis as a metabolic vulnerability.
Methods
Cell Lines
MCF10A, MDA-MB-231, T47D, and ZR-75-1 cells were obtained from the American Type Culture Collection (ATCC) and authenticated using short tandem repeat (STR) profiling. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. MCF10A cells were maintained in DMEM/Ham’s F12 (CellGro) supplemented with 5% equine serum (CellGro), 10 mg/mL insulin (Life Technologies), 500 ng/mL hydrocortisone (Sigma-Aldrich), 20 ng/mL EGF (R&D Systems), and 100 ng/mL cholera toxin (Sigma-Aldrich). MDA-MB-231 cells were maintained in DMEM (CellGro) supplemented with 10% fetal bovine serum (FBS) (Gemini). T47D cells were maintained in RPMI 1640 (CellGro) supplemented with 10% FBS (Gemini) and 10 mg/mL insulin (Life Technologies). ZR-75-1 cells were maintained in RPMI 1640 (CellGro) supplemented with 10% FBS (Gemini). DMEM lacking glucose, glutamine, and pyruvate was obtained from CellGro. Cells were passaged for no more than 6 months and routinely assayed for mycoplasma contamination.
Inhibitors
Inhibitors were used as follows: GSK690693 (Selleck), 1 μM; L-S,R-buthionine sulfoximine (BSO) (Sigma-Aldrich), 50 μM; LCS-1 (Sigma-Aldrich), 10 μM; cycloheximide (Sigma-Aldrich), 20 μg/ml; SB415286 (Sigma-Aldrich), 25 μM; Cisplatin (Tocris), 0.39 - 100 μM; EUK-134 (Sigma-Aldrich), 50 μM; N-acetyl cysteine (NAC) (Sigma-Aldrich), 1 mM.
Antibodies
All antibodies except the anti-HA and anti-Nrf2 antibodies were purchased from Cell Signaling Technology: p-Akt S473 (#4060, 1:1000), pAkt T308 (#2965, 1:1000), Akt (#4691, 1:1000), p-PRAS40 T246 (#2997, 1:1000), PRAS40 (#2691, 1:1000), p-GSK-3β S9 (#9336, 1:1000), GSK-3β (#9315, 1:1000), PARP (#9542, 1:1000), p21 Waf1/Cip1 (#2947, 1:1000), p27 Kip1 (#3688, 1:1000), β-actin (#4970, 1:5000). The anti-HA monoclonal antibody was purified from the 12CA5 hybridoma (1:1000), and the anti-Nrf2 antibody was purchased from Abcam (ab62352, 1:1000).
Plasmids
The AKT1(E17K) and AKT2(E17K) mutations were generated by site-directed mutagenesis (Qiagen) from pcDNA3-HA-AKT1 and pcDNA3-HA-AKT2 (Addgene). For doxycycline-inducible overexpression of AKT1/2 and AKT1/2(E17K), pTRIPZ-HA-AKT1, pTRIPZ-HA-AKT2, pTRIPZ-HA-AKT1(E17K), and pTRIPZ-HA-AKT2(E17K) were constructed. HA-AKT1/2 and HA-AKT1/2(E17K) cDNA were amplified by PCR, digested with the restriction enzymes AgeI and ClaI, and inserted into the pTRIPZ lentiviral vector (Thermo Scientific). Expression of each Akt variant was induced by treatment of cells with 150 ng/ml doxycycline for 48 h. pJP1520-HA-GFP, pJP1520-HA-PIK3CA WT, pJP1520-HA-PIK3CA(E545K), and pJP1520-HA-PIK3CA(H1047R) were generous gifts from Dr. Albert S. Baldwin33. pDONR221-p21 was obtained from the Dana-Farber/Harvard Cancer Center DNA Resource Core, and p21 was sub-cloned into the pHAGE-C-HA-FLAG lentiviral expression vector (W. Harper, Harvard Medical School). The shRNA-resistant pHAGE-C-HA-FLAG-p21 was generated by site-directed mutagenesis (Qiagen).
RNA Interference
Validated shRNA hairpins targeting NRF2 were obtained from Sigma Mission shRNA- shNRF2 #1: TRCN0000007555; shNRF2 #2: TRCN0000007558. The tet-on p21 shRNA sequence has been previously described and validated27. Cells expressing shRNA were cultured in medium containing puromycin (0.5---2 μg/mL). Gene knock-down was induced by incubating cells with 150 ng/mL doxycycline for 48 to 72 hours.
Immunoblotting
Cells were washed with PBS at 4°C and lysed in radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mmol/L NaCl, 50 mmol/L Tris---HCl (pH 7.5), proteinase inhibitor cocktail, 50 nmol/L calyculin, 1 mmol/L sodium pyrophosphate, and 20 mmol/L sodium fluoride) for 15 minutes at 4°C. Cell extracts were pre-cleared by centrifugation at 13,000 rpm for 10 minutes at 4°C, and protein concentration was measured with the Bio-Rad DC protein assay. Lysates were then resolved on acrylamide gels by SDS---PAGE and transferred electrophoretically to nitrocellulose membrane (Bio-Rad) at 100 V for 90 minutes. The blots were blocked in Tris-buffered saline (TBST) buffer (10 mmol/L Tris---HCl, pH 8, 150 mmol/L NaCl, and 0.2% Tween 20) containing 5% (w/v) nonfat dry milk for 1 hour, and then incubated with the specific primary antibody diluted in blocking buffer at 4°C overnight. Membranes were washed three times in TBST and incubated with HRP-conjugated secondary antibody for 1 hour at room temperature. Membranes were washed three times and developed using enhanced chemiluminescence substrate (EMD Millipore).
In Vitro Protein Kinase Assays
MCF10A cells expressing empty vector, HA-AKT1/2, or HA-AKT1/2(E17K) were serum starved for 16 hours and lysed as described above in EBC buffer (0.5% NP-40, 120 mmol/L NaCl, 2 mmol/L EDTA, 2 mmol/L EGTA, 50 mmol/L Tris---HCl (pH 7.4), proteinase inhibitor cocktail, 50 nmol/L calyculin, 1 mmol/L sodium pyrophosphate, and 20 mmol/L sodium fluoride). HA-AKT1/2 or HA-AKT1/2(E17K) were immunoprecipitated from cell extracts with an anti-HA antibody and incubated with 300 ng GSK-3 fusion protein peptide (Cell Signaling Technology) in the presence of 150 mmol/L cold ATP in a kinase buffer (Cell Signaling Technology) for 40 min at 30°C. The kinase reaction was terminated by the addition of SDS-PAGE sample buffer.
Proliferation Assays
For WST-1 assays, MCF10A cells were seeded into 96-well plates (Corning) at a density of 1500 cells per well in 100 μL medium. The medium was changed to either regular growth medium or serum-free medium after 16 h. Cell viability was measured at the indicated time points after the media change using the water soluble tetrazolium salt WST-1 assay (Clontech) according to the manufacturer’s protocol.
For sulforhodamine B (SRB) assays, MCF10A cells were seeded into 12-well plates (Corning) at a density of 5 x 104 cells per well in 1 mL medium. The medium was changed to either regular growth medium or serum-free medium every two days. Cell number was measured at the indicated time points using sulforhodamine B staining, as previously described34.
LC-MS/MS Metabolomic Profiling
Cells were maintained in the indicated growth media, and fresh media was added 3 h before the experiment. For metabolite extraction, media from biological triplicates was aspirated, and 80% (v/v) methanol at dry ice temperatures was added. Cells and the metabolite-containing supernatants were collected, and the insoluble material in lysates was centrifuged at 4,000xg for 5 min. The resulting supernatant was evaporated using a refrigerated SpeedVac. Samples were re-suspended using 20 μl HPLC grade water for mass spectrometry. Ten microliters were injected and analyzed using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM) of a total of 254 endogenous water soluble metabolites for steady-state analyses of samples. Some metabolites were targeted in both positive and negative ion mode for a total of 285 SRM transitions using positive/negative switching. ESI voltage was +4900 V in positive ion mode and ---4500 V in negative ion mode. The dwell time was 4 ms per SRM transition and the total cycle time was 1.89 s. Approximately 9---12 data points were acquired per detected metabolite. Samples were delivered to the MS via normal phase chromatography using a 4.6 mm i.d x 10 cm Amide Xbridge HILIC column (Waters Corp.) at 300 μl/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0–5 min; 42% B to 0% B from 5---16 min; 0% B was held from 16---24 min; 0% B to 85% B from 24---25 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate (pH = 9.0) in water:acetonitrile (95:5). Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v2.0 software (AB/SCIEX). For 13C-labeled experiments, SRMs were created for expected 13C incorporation in various forms for targeted LC-MS/MS. Data analysis was performed in MatLab. For steady-state profiling, metabolite peak area intensities represent the normalized (by the average of cell number and cellular protein content), integrated total ion current from a single SRM transition and are expressed as relative values across all samples. For flux analyses, metabolite abundances represent the normalized (by cell number), integrated total ion current from a single SRM transition.
Isotope Labeling
DMEM lacking glucose, glutamine, and pyruvate (CellGro) was supplemented with 10 mM glucose and 2 mM U-13C5-glutamine (Cambridge Isotope Labs). Labeled media was added to the cells, and cellular metabolites were extracted as described above at 1, 3, and 8 h.
IGF-1, Hydrogen Peroxide, and Tert-Butyl Hydroperoxide Treatments
For IGF-1 stimulation, cells were serum-starved for 20–24 h and stimulated with 100 μg/ml IGF-1 (R&D Systems) for 10 min. For H2O2 and tert-butyl hydroperoxide treatments, cells were serum-starved for 20–24 h in DMEM lacking pyruvate and then stimulated with the indicated concentrations of H2O2 (Fisher) or tert-butyl hydroperoxide (Sigma-Aldrich) in Hank’s balanced salt solution (HBSS) (Gibco).
Cell Death Assay
Cells were trypsinized at 37°C, followed by staining with 7-AAD (BD Pharmingen). Briefly, cells were washed twice with PBS and stained with 5 μL 7-AAD in binding buffer for 15 minutes. FACS analysis was performed with FACSCalibur (Becton-Dickinson) and FlowJo (TreeStar Inc.) software.
Propidium Iodide Viability Assay
Cell viability was assayed with a propidium iodide-based plate reader assay, as previously described35. Briefly, cells in 96-well plates were treated with a final concentration of 30 μM propidium iodide for 60 min at 37° C. The initial fluorescence intensity was measured in a SpectraMax M5 (Molecular Devices) at 530 nm excitation/620 nm emission. Digitonin was then added to each well at a final concentration of 600 μM. After incubating for 30 min at 37° C, the final fluorescence intensity was measured. The fraction of dead cells was calculated by dividing the background-corrected initial fluorescence intensity by the final fluorescence intensity. Viability was calculated by (1 --- fraction of dead cells).
Total Glutathione Measurements
Cellular metabolites were collected by methanol extraction and concentrated by SpeedVac evaporation, as described above. Total glutathione levels within these samples were measured according to the manufacturer’s protocol by using the Glutathione Assay Kit from Cayman Chemical, which is based on the Tietze method36.
Quantitative Real-Time PCR
Total RNA was isolated with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription was performed using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative real-time PCR was performed using an ABI Prism 7700 sequence detector. GCLC primer: sense, 5’-TTGATCATCTCCTGGCCCAG-3’, antisense, 5’-TGTCTGAGTTTGGAGGAGGG-3’; GCLM primer: sense, 5’-CTCCTGCTGTGTGATGCCA-3’, antisense, 5’-CTCGTGCGCTTGAATGTCAG-3’; GSS primer: sense, 5’-ATGGCCAGGAAATTGCTGTG-3’, antisense, 5’-CACCTTCTTAGTCCCAGCCA-3’; GSR primer: sense, 5’-CAACGAGCTTTACCCCGATG-3’, antisense, 5’-TCGTTGCTCCCATCTTCACT-3’; NQO1 primer: sense, 5’-GCAGAAGGGAATTGCTCAGA-3’, antisense, 5’-CAAAAGCTGACCAAGAGTGG-3’; NRF2 primer: sense, 5’-CATGCCCTCACCTGCTACTT-3’, antisense, 5’-GTTCTGGTGATGCCACACTG-3’; SLC7A11 primer: sense, 5’-TGCTGGGCTGATTTATCTTCG-3’, antisense, 5’-GAAAGGGCAACCATGAAGAGG-3’; GPX4 primer: sense, 5’-GAAGCAGGAGCCAGGGAGTA-3’, antisense, 5’-GGTGAAGTTCCACTTGATGGC-3’. PCR reactions were carried out in triplicate. Quantification of mRNA expression was calculated by the CT method with 18S rRNA as the reference gene.
ARE-luciferase Assay
Luciferase assays were conducted using Promega’s luciferase assay system, as previously described37. Luciferase activity was normalized by cellular protein content. The lentiviral ARE-luciferase reporter plasmid (Qiagen) was a generous gift from Dr. Gina M. DeNicola.
TCGA Data Analysis
Mutation, copy number analysis, microarray, and RPPA data from the published TCGA Invasive Breast Cancer data set21 was downloaded from cBioPortal. Analysis of the data was conducted in MatLab.
3D Cultures
3D cultures were prepared as previously described38. Briefly, chamber slides were coated with growth factor---reduced, phenol-free Matrigel (Corning) and allowed to solidify for 30 minutes. Cells (2,000–4,000) in assay medium were seeded on coated chamber slides. Assay medium contained DMEM/Ham’s F12 supplemented with 5% equine serum (Gibco), 10 mg/mL insulin (Life Technologies), 500 ng/mL hydrocortisone (Sigma-Aldrich), 5 ng/mL EGF (R&D Systems), and 100 ng/mL cholera toxin (Sigma-Aldrich). The assay medium was replaced every 4 days. Doxycycline (150 ng/mL) was added every 2 or 3 days. The indicated concentrations of BSO (Sigma-Aldrich) were added one day after seeding and re-added every 2 or 3 days.
Soft Agar Assays
Cells (4 x 104) were added to 1 ml of growth media plus 0.4% noble agar (BD Biosciences), with or without BSO (Sigma-Aldrich). The resulting mixture was layered onto a 2 ml bed of growth media plus 0.8% noble agar in 6-well plates (Corning), with or without BSO. Cells were fed every 3–6 days with 0.5 ml of growth media (± BSO). After four weeks, the growth media was removed, and viable colonies were stained with iodonitrotetrazolium chloride (Sigma-Aldrich). Colony number was determined using MatLab.
In Vitro Combination Assays
MDA-MB-231, T47D, and ZR-75-1 cells were seeded at a density of 10000, 13000, and 10000 cells per well, respectively, in 96-well plates (Corning). Cells were treated with 50 μM BSO (Sigma-Aldrich) for 48 h, followed by treatment with cisplatin (Tocris) at a range of concentrations for 48 h, in the presence or absence of 50 μM EUK-134 or 1 mM N-acetyl cysteine (NAC). Cell viability was measured using a propidium iodide-based plate reader assay.
Xenograft Studies
Female nude mice (5–6 weeks old) were purchased from Taconic and maintained and treated under specific pathogen-free conditions. All procedures were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center (BIDMC; Boston, MA) and conform to the federal guidelines for the care and maintenance of laboratory animals. No statistical method was used to predetermine sample size. These experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. The mice were injected subcutaneously with 5 x 106 T47D cells (n = 20 mice) or 2.5 x 106 MDA-MB-231 cells (n = 20 mice) in media with 50% growth factor-reduced, phenol red-free Matrigel (Corning). For mice bearing T47D xenografts, 17 -estradiol was administered daily using a validated peroral method39. Tumor formation was examined every 2 to 3 days for the duration of the experiment. When tumors reached a size of 5–6 mm in diameter, they were divided into a control group and treatment groups of BSO alone, cisplatin (CDDP) alone, and BSO in combination with CDDP (at least n = 4 per group). L-S,R-BSO (Fisher) was administered using a validated continuous oral delivery method28,40 in drinking water at a concentration of 20 mM. CDDP (Tocris) was administered by I.P. injection at 5 mg/kg/week. For the combination treatment group, mice were pre-treated with BSO for 7 days before treatment with CDDP. Tumor volume = ( /6)(W2)(L); W = width, L = length.
Statistics and Reproducibility
Sample sizes and reproducibility for each figure are denoted in the figure legends, and raw data from independent replicate experiments can be found in the Statistics Source Data. Unless otherwise noted, all western blots and microscopy images are representative of at least three biologically independent experiments. Statistical significance between conditions was assessed by two-tailed Student’s t-tests. All error bars represent s.e.m., and significance between conditions is denoted as *P < 0.05; **P < 0.01; and ***P < 0.001. Statistical data for each quantitative experiment, including mean, s.e.m., and exact p-values can be found in the Statistics Source Data.
Supplementary Material
Acknowledgments
We thank Joan Brugge, Brendan Manning, John Blenis, Andrew Beck, Isaac Harris, and members of the Toker and Cantley labs for suggestions; Albert Baldwin, Y. Rebecca Chin, and Gina DeNicola for critical reagents; and Min Yuan and Susanne Breitkopf for technical assistance with mass spectrometry. Research support was derived in part from National Institutes of Health (R01CA177910 (A.T.), P01CA120964 (J.M.A.), P30CA006516 (J.M.A.), R01GM041890 (L.C.C.)). E.C.L. is a pre-doctoral fellow of the NSF graduate research fellowship program (NSF DGE1144152). C.A.L. is funded in part by the Pancreatic Cancer Action Network as a Pathway to Leadership Fellow and through a Dale F. Frey Breakthrough award from the Damon Runyon Cancer Research Foundation.
Footnotes
Author Contributions
E.C.L., C.A.L., L.C.C., and A.T. designed the study and interpreted the results. E.C.L. and A.T. wrote the manuscript. E.C.L. performed the experiments. J.M.A. and C.A.L. assisted with the LC-MS/MS metabolomic studies and data interpretation. A.J. and H.H. assisted with the in vivo xenograft studies.
Competing Financial Interests
L.C.C. owns equity in, receives compensation from, and serves on the Board of Directors and Scientific Advisory Board of Agios Pharmaceuticals. Agios Pharmaceuticals is identifying metabolic pathways of cancer cells and developing drugs to inhibit such enzymes to disrupt tumor cell growth and survival.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 3.Elstrom RL, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 4.Robey RB, Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpten JD, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 6.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauring J, et al. Knock in of the AKT1 E17K mutation in human breast epithelial cells does not recapitulate oncogenic PIK3CA mutations. Oncogene. 2010;29:2337–2345. doi: 10.1038/onc.2009.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salhia B, et al. Differential effects of AKT1(p.E17K) expression on human mammary luminal epithelial and myoepithelial cells. Hum Mutat. 2012;33:1216–1227. doi: 10.1002/humu.22100. [DOI] [PubMed] [Google Scholar]
- 9.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papa L, Hahn M, Marsh EL, Evans BS, Germain D. SOD2 to SOD1 switch in breast cancer. J Biol Chem. 2014;289:5412–5416. doi: 10.1074/jbc.C113.526475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somwar R, et al. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci U S A. 2011;108:16375–16380. doi: 10.1073/pnas.1113554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Dowbenko D, Lasky LA. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J Biol Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 20.Rada P, et al. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang YJ, Emery D, Enger MD. Buthionine sulfoximine induced growth inhibition in human lung carcinoma cells does not correlate with glutathione depletion. Cell Biol Toxicol. 1991;7:249–261. doi: 10.1007/BF00250979. [DOI] [PubMed] [Google Scholar]
- 25.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 26.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 27.Chin YR, Yuan X, Balk SP, Toker A. PTEN–Deficient Tumors Depend on AKT2 for Maintenance and Survival. Cancer Discov. 2014;4:942–955. doi: 10.1158/2159-8290.CD-13-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris IS, et al. Glutathione and Thioredoxin Antioxidant Pathways Synergize to Drive Cancer Initiation and Progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen HHW, Kuo MT. Role of glutathione in the regulation of Cisplatin resistance in cancer chemotherapy. Met Based Drugs. 2010;2010 doi: 10.1155/2010/430939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tung NM, Winer EP. Tumor-infiltrating lymphocytes and response to platinum in triple-negative breast cancer. J Clin Oncol. 2015;33:969–971. doi: 10.1200/JCO.2014.59.6031. [DOI] [PubMed] [Google Scholar]
- 33.Hutti JE, et al. Oncogenic PI3K mutations lead to NF-kappaB-dependent cytokine expression following growth factor deprivation. Cancer Res. 2012;72:3260–3269. doi: 10.1158/0008-5472.CAN-11-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, et al. Quantitative determination of apoptotic death in cultured human pancreatic cancer cells by propidium iodide and digitonin. Cancer Lett. 1999;142:129–137. doi: 10.1016/s0304-3835(99)00107-x. [DOI] [PubMed] [Google Scholar]
- 36.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 37.Yiu GK, Toker A. NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. J Biol Chem. 2006;281:12210–12217. doi: 10.1074/jbc.M600184200. [DOI] [PubMed] [Google Scholar]
- 38.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 39.Ingberg E, Theodorsson A, Theodorsson E, Strom JO. Methods for long-term 17beta-estradiol administration to mice. Gen Comp Endocrinol. 2012;175:188–193. doi: 10.1016/j.ygcen.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe T, et al. A novel model of continuous depletion of glutathione in mice treated with L-buthionine (S,R)-sulfoximine. J Toxicol Sci. 2003;28:455–469. doi: 10.2131/jts.28.455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.