Abstract
Mosaic loss of the Y chromosome (mLOY) leading to gonosomal XY/XO commonly occurs during aging, particularly in smokers. We investigated whether mLOY was associated with non-hematologic cancer in three prospective cohorts (8,679 cancer cases and 5,110 cancer-free controls), and genetic susceptibility to mLOY. Overall, mLOY was observed in 7% of men and increased with age (per year OR=1.13, 95%CI=1.12–1.15; P<2×10−16), reaching 18.7% among men over age 80. mLOY was associated with current smoking (OR=2.35, 95%CI=1.82–3.03; P=5.55×10−11); however, the association weakened with years after cessation. mLOY was not consistently associated with overall or specific cancer risk (e.g. for bladder, lung, or prostate) nor with cancer survival after diagnosis (multivariate-adjusted hazard ratio=0.87, 95% CI=0.73–1.04, P=0.12). In a genome-wide association study, we observed the first example of a common susceptibility locus for genetic mosaicism, specifically mLOY, which maps to the T-cell leukemia/lymphoma 1A (TCL1A) gene on 14q32.13, marked by rs2887399 (OR=1.55, 95%CI=1.36–1.78; P=1.37×10−10).
Mosaic loss of the Y chromosome (mLOY) refers to the loss of the Y chromosome in a subset of cells, while the remainder of cells retains the normal chromosome. For more than four decades, it has been noted that a fraction of healthy men lose all or some portion of the Y chromosome over the course of their lifetime1. Moreover, several studies have reported mLOY in males of advanced age suggesting mLOY is associated with aging and increasing hypodiploidy1–4. Other studies suggested mLOY is associated with specific hematologic disorders including acute myelogenous leukemia (AML), myelodysplastic syndrome (MDS), and preleukemia5–9.
Single nucleotide polymorphism (SNP) genotyping arrays have become an important tool for discovering common variants that contribute to human diseases10. Two widely-used applications of this technology are genome-wide association studies (GWAS) and copy number variant (CNV) analyses for large-scale mosaic autosomal aberrations11–18. SNP microarray data has also been used to investigate mosaicism on the sex chromosomes19,20. Other studies have evaluated next-generation sequencing data to detect mosaic mutations at the base pair level21–23. While each of these studies of mosaic CNVs, mosaic uniparental disomies, mosaic single nucleotide variants (SNVs) and mLOY ascertain different aspects of the biological process of clonal expansion, taken together these studies suggest that mosaic events, both large and small, increase with age. This trend could reflect either a deterioration in the capacity to maintain a stable genome or, alternatively, a decline in stem-cell diversity22–25.
We investigated the association between mLOY and age at DNA collection, smoking status, DNA source (derived from blood or buccal material), inferred ancestry, genetic susceptibility to mLOY, non-hematologic cancer risk and cancer-specific survival in subjects from three prospective cohorts: the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial (PLCO), and Cancer Prevention Study-II (CPS-II) (Supplementary Table 1). SNP microarray data generated using Illumina Infinium arrays for genome-wide association studies (GWAS) that contained sufficient coverage of chromosome Y (Human Hap 610K, Hap1M, Omni1M, and Omni2.5M) were used to detect mLOY in DNA isolated from blood or buccal cells from 8,679 males with non-hematologic cancer and 5,110 cancer-free adult male controls. The hybridization data from all subjects were examined for deviations from expected log2 intensity ratio (see Online Methods) for evidence of loss or gain of the male specific region of chromosome Y (MSY) (chrY:6,671,498-22,919,969; hg18/build36) (Supplementary Figure 1A). We observed 970 men (7.03%) with detectable mLOY and the estimated fraction of cells was 22.7% to 73.4% (Supplementary Figure 1C and Online Methods), which differs from autosomes (7–95% mosaicism14,15). No significant difference in mLOY frequency was observed by DNA source, either blood or buccal origin (P=0.33), or by genotyping array (P=0.14, Supplementary Table 2).
The accuracy of our mLOY detection method was validated by quantitative PCR (qPCR) markers for 15 genes distributed across MSY (Supplementary Figure 1B)26. We selected 124 subjects from the cohorts with probable mLOY for validation by qPCR assays. The concordance rate between the SNP microarray and qPCR was 87.9% (Supplementary Table 3), with no differences observed by array type.
Among the 13,789 males scanned, we found evidence of chromosome Y gain for 133 males (0.96%). In the males with suspected Y gain, we had DNA available from 69 individuals for qPCR validation and the concordance rate was only 49% for mosaic gains (Supplementary Table 3), with validation corresponding to substantial LRR deviations from baseline. After removal of 34 men with validated Y gain, we excluded 26 participants with similarly large positive deviations from baseline after manual review. The resulting dataset included 8,632 cases and 5,097 controls for Y loss analysis. Of the 8,632 cancer cases, 5,545 had their blood or buccal cells collected at least one year prior to cancer diagnosis. The majority of these participants (n=5,369) had been diagnosed with bladder, lung or prostate cancer.
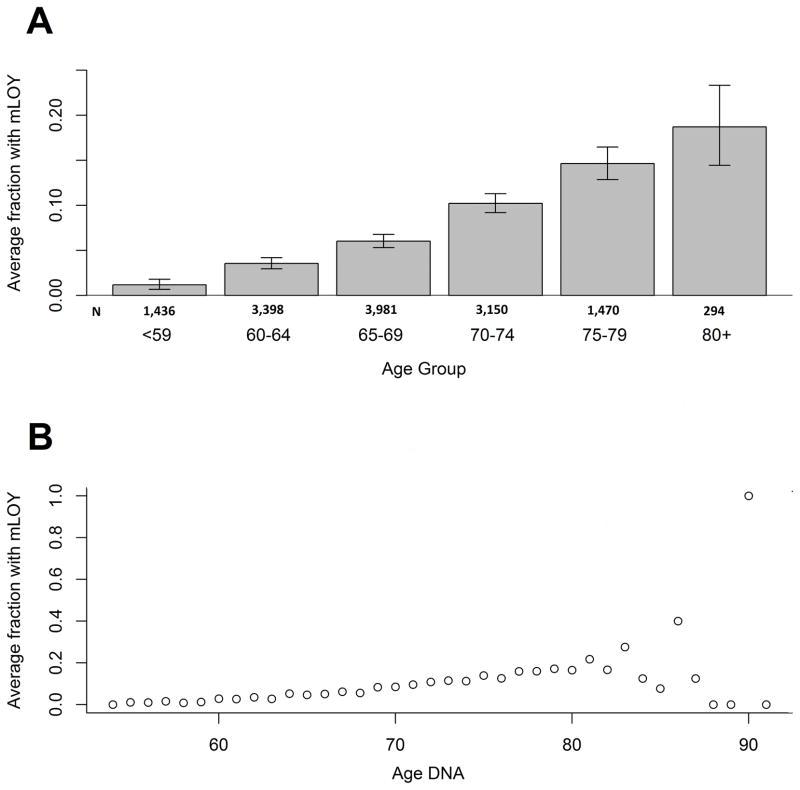
The most significant association for mLOY was with increasing age; in 13,729 men, the frequency of detectable mLOY increased from 1.18% under 60 to 18.71% over 80 years of age. The proportion of individuals with mLOY increased for every age stratum until 80 years of age (P<2.2×10−16); for older participants, the estimates became less stable due to small numbers (Figure 1A and 1B, Supplementary Table 4. Adjustment for smoking status (current smoking and for former smokers, years since cessation), ancestry, source of DNA and contributing studies, we observed evidence for an association between mLOY and age (OR per year of age=1.13, 95%CI=1.12–1.15; P<2.00×10−16) (Table 1, Supplementary Table 5). This association with age has a substantially greater magnitude than that observed previously for large-scale structural autosomal mosaic events (OR=1.05, 95%CI=1.04–1.07)15 and for mosaic SNVs (OR=1.08, 95%CI=1.07–1.09)22.
Figure 1. Fraction of men with mLOY across 5-year age groups for all subjects (n = 13,729).
Fraction of men with Y loss is calculated as men in the age group with Y loss divided by the total number of men in that age group. Error bars represent 95% Jeffery’s confidence intervals around the proportion estimate. (A) Mosaic chromosome Y loss is associated with older age at DNA collection, with frequencies of 1.18% in individuals less than 60 years and 18.71% in those 80 years or older (OR per year of age = 1.13, 95% CI = 1.12–1.15; P < 2×10−16). Chi-squared test for trend among 6 age groups shows there is significant evidence that the fraction of men with Y loss increases with age (P < 2.2×10−16). (B) Scatterplot for age versus fraction of men with Y loss. There is an overall increasing trend for the fraction of men with Y loss until age 80. After age 80, the trend became unstable reflecting the limited number of subjects in this age group in our study (Supplementary Table 4). All statistical tests are two-sided.
Table 1. Predictors of mLOY.
Age refers to the age at DNA collection and the odds ratio estimate is for a one-year increase in age. For the association between age and mLOY, the analysis was adjusted for smoking status (current smoking and for former smokers, the number of years since cessation), estimated admixture proportion, DNA source, and contributing study. For the association between mLOY and the number of years since cessation and number of cigarettes smoked per day, analyses were adjusted for age, estimated admixture proportion, DNA source, and contributing study.
| # of Y loss | # of Normal | Total | Proportion with mLOY | OR | 95% CI | p-value | |
|---|---|---|---|---|---|---|---|
| Age (ATBC+PLCO+CPSII) | 970 | 12759 | 13729 | 7.07% | 1.13 | 1.12–1.15 | <2.00×10−16 |
| Smoking use (PLCO+CPSII) | |||||||
| Never smoking | 209 | 3199 | 3408 | 6.13% | reference | ||
| Former smoking | 446 | 4964 | 5410 | 8.24% | 1.33 | (1.12–1.57) | 0.001 |
| Current smoking | 100 | 934 | 1034 | 9.67% | 2.35 | (1.82–3.03) | 5.55×10−11 |
| Years since quitting (PLCO+CPSII) | |||||||
| 1–4 years quit | 38 | 332 | 370 | 10.27% | 2.15 | (1.49–3.10) | 3.83×10−5 |
| 5–10 since quit | 64 | 567 | 631 | 10.14% | 1.92 | (1.43–2.58) | 1.26×10−5 |
| 11–20 since quit | 107 | 1167 | 1274 | 8.40% | 1.49 | (1.17–1.90) | 0.001 |
| > 20 since quit | 231 | 2809 | 3040 | 7.60% | 1.10 | (0.91–1.34) | 0.333 |
| Cigarettes per day in current smokers (ATBC+PLCO+CPSII) | |||||||
| 1 to 10 | 61 | 727 | 788 | 7.74% | reference | ||
| 11 to 20 | 154 | 2201 | 2355 | 6.54% | 1.04 | (0.76–1.43) | 0.800 |
| 21 to 30 | 66 | 1206 | 1272 | 5.19% | 0.92 | (0.63–1.33) | 0.648 |
| >30 | 26 | 429 | 455 | 5.71% | 0.95 | (0.58–1.55) | 0.835 |
The frequency of individuals exhibiting age-related mLOY was nearly 10-fold greater than that observed in the autosomes (0.7–2.0%)14–16. A detectable mosaic autosomal abnormality (>2 MB) was observed in 130 (0.95%) men16. This is substantially lower than what was observed for mLOY in the same individuals (7.07%), indicating that mLOY is the most frequent large-scale chromosomal somatic event. Of 970 mLOY males, 18 (1.86%) also had evidence of detectable autosomal mosaicism (Supplementary Table 6 and 7), suggesting men with mLOY are more likely to harbor large autosomal mosaic events than men without mLOY (OR=2.13, 95% CI=1.22–3.55; P=0.005).
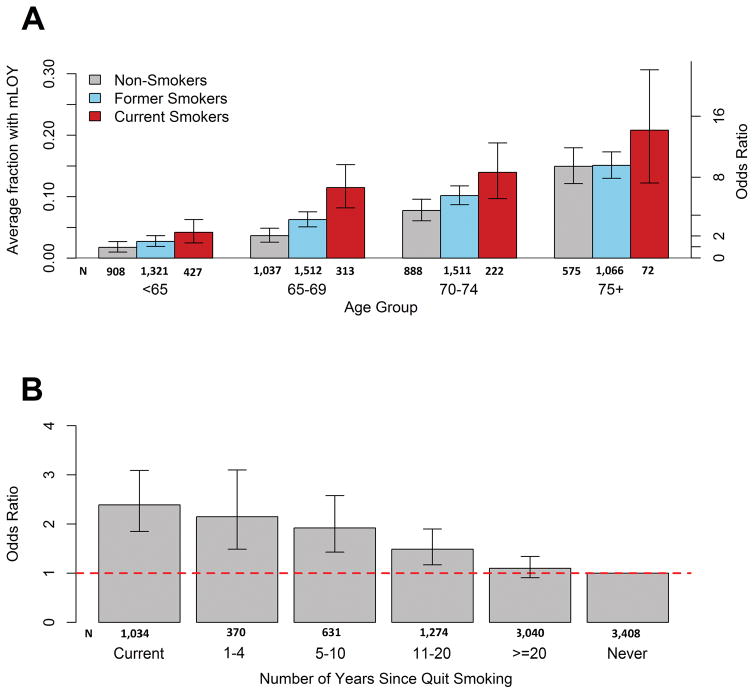
We investigated the association with smoking status in 9,859 subjects (1,034 current, 5,410 former, 3,408 never smokers, 7 unknown) from the PLCO and CPSII studies, excluding participants from ATBC, which recruited only current smokers. The frequency of mLOY was higher in ever smokers (65.4%) than never smokers (34.6%), with the highest frequency among current smokers. Relative to never-smokers who were less than 65 years old, current smokers over 75 years old had an increase of mLOY (OR=13.9, 95%CI=6.60–29.26; P=4.13 × 10−12) (Figure 2A, Supplementary Table 8). Adjusting for age, ancestry, DNA source and study, we observed an association of mLOY with current smoking (OR=2.35, 95%CI=1.82–3.03; P=5.55 × 10−11) and former smoking (OR=1.33, 95%CI=1.12–1.57; P=0.001) (Table 1, Supplementary Table 9). This result is consistent with a prior report that men who smoke are at greater risk of mLOY, which observed odds ratios for current versus non-smoking that ranged from 2.4 (95%CI=1.6–3.6) to 4.3 (95%CI=2.8–6.7)19 among three included studies. Mosaic SNVs have also been associated with smoking with comparable odds ratio estimates (OR=2.2)23. It is notable that prior studies have not reported an association between smoking and large-scale (>2 Mb) autosomal mosaicism14,15.
Figure 2. mLOY and Smoking Analysis.
(A) Proportion of males with mLOY across strata of 5-year age group and smoking status for subjects from PLCO and CPSII studies (n = 9,859). Non-smokers are in grey, former smokers are in blue, and current smokers are in red. Error bars represent 95% Jeffery’s confidence intervals around the proportion estimate. Current smoking men in the 75+ age group are at a 13.90 times increased odds of having mosaic Y loss as compared to non-smoking men less than 65 years old (95% CI =6.60–29.26, P = 4.13 × 10−12). (B) Association between current smoking and years since cessation and mosaic Y loss from adjusted logistic regression models (n=8,825). The dotted line reflects an odds ratio of 1.0 for the referent never smokers. All statistical tests are two-sided.
Due to differences in mLOY between current and former smokers, the risk of mLOY progressively declined with years after cessation: quitting smoking within 1 to 4 years (OR=2.15, 95%CI=1.49–3.10; P=3.83 × 10−5), within 5 to 10 years (OR=1.92, 95%CI=1.43–2.58; P=1.26 × 10−5) and within 11–20 years (OR=1.49, 95%CI=1.17–1.90; P=0.001), relative to never-smoking. By 20 years after cessation, there was no evidence for association (OR=1.10, 95% CI= 0.91–1.34, P=0.33) (Figure 2B, Table 1, Supplementary Table 10). Our findings suggest that smoking has a long lasting impact on mLOY, perhaps more than a decade after quitting, but the association wanes with long-term cessation. Among 4,904 current smokers, we observed no association for smoking intensity, measured as cigarettes per day (Table 1, Supplementary Table 11).
It has been proposed that autosomal mosaicism may be associated with risk for certain solid tumors but not definitely established14–16. Moreover, others have suggested that mLOY is associated with cancer risk overall20. We investigated the frequency of mLOY in blood or buccal DNA and solid tumor risk in 5,545 cancer subjects from whom DNA was collected at least 1 year prior to cancer diagnosis and in 5,097 cancer-free individuals (Supplementary Table 12). Overall, mLOY was slightly more common in men who went on to develop cancer (6.67%) than in cancer-free controls (5.49%) (unadjusted OR=1.23, 95%CI=1.04–1.45; P=0.012; multivariate adjusted OR=1.19, 95%CI=1.00–1.42; P=0.047) (Table 2, Supplementary Figure 2, Supplementary Table 13). Furthermore, we investigated a possible association between cancer risk and mLOY for cancer types with sufficient sample size including bladder, lung, and prostate cancer in adjusted analyses (using continuous age; smoking status-current smoking and for former smokers, years since quitting; pack years; ancestry; source of DNA and study). In the cohort studies with DNA collected one year or more before diagnosis, we observed a possible association between mLOY and the risk of bladder cancer (OR=1.47, 95%CI=1.09–1.99; P=0.011) and risk of prostate cancer (OR=1.35, 95%CI=1.04–1.74; P=0.024), but no evidence for a relationship with lung cancer (OR=0.90, 95%CI=0.69–1.18, P=0.45) (Table 2). This latter point is striking given that lung cancer is more strongly associated with smoking than either bladder or prostate cancer. We examined cases diagnosed at or before biospecimen sampling. For bladder and prostate but not lung cancer, when examining DNA obtained contemporaneously with cancer diagnosis, we observed somewhat higher risk estimates that could reflect effects of treatment modalities (chemotherapy, surgery and/or radiation therapy). We additionally examined the possible association between mLOY and bladder cancer risk in three case-control studies (total of 2,062 cases and 2,064 controls), but found no association (OR=1.17, 95%CI=0.93–1.48; P=0.18; Supplementary Table 14). Together, these results provide limited support for the hypothesis that mLOY is a strong risk factor for common solid tumors and larger studies will be needed to investigate further based on current estimated effect sizes.
Table 2. Association between mLOY and incident cancer overall and stratified by date of DNA collection.
The analysis used mLOY as the predictor variable, cancer type as response variable, and was adjusted for continuous age, smoking status (current smoking and for former smokers, the number of years since cessation), pack years, estimated admixture proportion, DNA source, and contributing study. A total of 5,097 cancer-free controls were used as a reference. Other cancers in addition to bladder, lung, and prostate cancer were included when performing the combined cancer analysis.
| DNA 1 year before cancer diagnosis
|
DNA at and after cancer diagnosis
|
All
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | OR | 95% CI | P Value | N | OR | 95% CI | P Value | N | OR | 95% CI | P Value | |
| Bladder | 731 | 1.47 | (1.09–1.99) | 0.011 | 558 | 2.01 | (1.47–2.75) | 1.40×10−5 | 1289 | 1.69 | (1.33–2.13) | 1.26×10−5 |
| Lung | 1908 | 0.9 | (0.69–1.18) | 0.450 | 381 | 0.81 | (0.48–1.38) | 0.44 | 2289 | 0.90 | (0.70–1.17) | 0.44 |
| Prostate | 2730 | 1.35 | (1.04–1.74) | 0.024 | 2093 | 1.53 | (1.17–2.01) | 1.84×10−3 | 4823 | 1.43 | (1.19–1.73) | 1.40×10−4 |
|
| ||||||||||||
| Combined Cancer | 5545 | 1.19 | (1.00–1.42) | 0.047 | 3087 | 1.52 | (1.23–1.87) | 8.40×10−5 | 8632 | 1.31 | (1.13–1.53) | 4.79×10−4 |
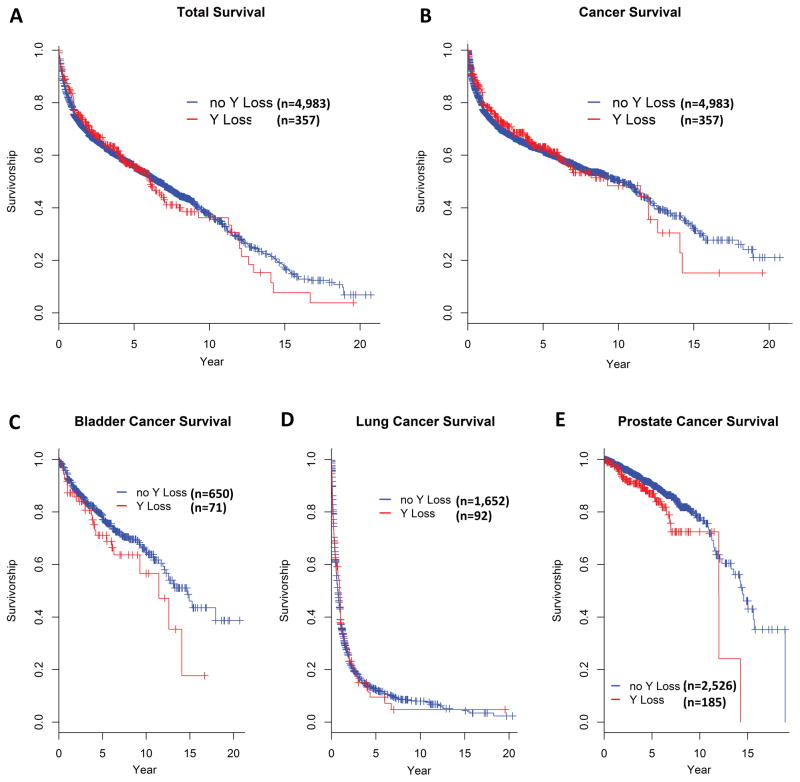
A recent report suggested that mLOY was associated with all-cause and cancer mortality among a cohort of 982 participants who were cancer-free at study baseline20. As cancer mortality reflects both developing cancer and dying from it, we examined whether mLOY may be associated with subsequent overall and cancer-specific mortality in our cancer cases, restricting our analysis to cases with DNA collected at least one year prior to diagnosis and available follow-up (N=5,340). We observed little evidence for an association with either endpoint, whether in Kaplan-Meier survival curves (Figure 3A–B) or in unadjusted or multivariate-adjusted Cox proportional-hazard models (Supplementary Table 15). After adjusting for age at diagnosis, smoking status (current smokers and for former smokers, the number of years since quitting), pack years, body mass index, and contributing study; the HR for mortality from all causes was 0.89 (95%CI=0.76–1.04; P=0.15) and the HR for mortality from cancer was 0.87 (95%CI=0.73–1.04; P=0.12). Similar findings were observed for bladder, lung, and prostate cancer separately (Figure 3C–E, Supplementary Table 15).
Figure 3. Association between mLOY and overall and cancer survival among participants with DNA collected at least one year prior to cancer diagnosis.
Kaplan-Meier survival curves are for (A) all-cause mortality for all cancer cases (n=5,340), (B) overall cancer survival (died of cancer) for all cancer cases (n=5,340), (C) cancer survival of bladder cancer cases (n=721), (D) cancer survival of lung cancer cases (n=1744), and (E) cancer survival of prostate cancer cases (n=2711). Subjects without Y loss are in blue and with Y loss are in red. All statistical tests are two-sided.
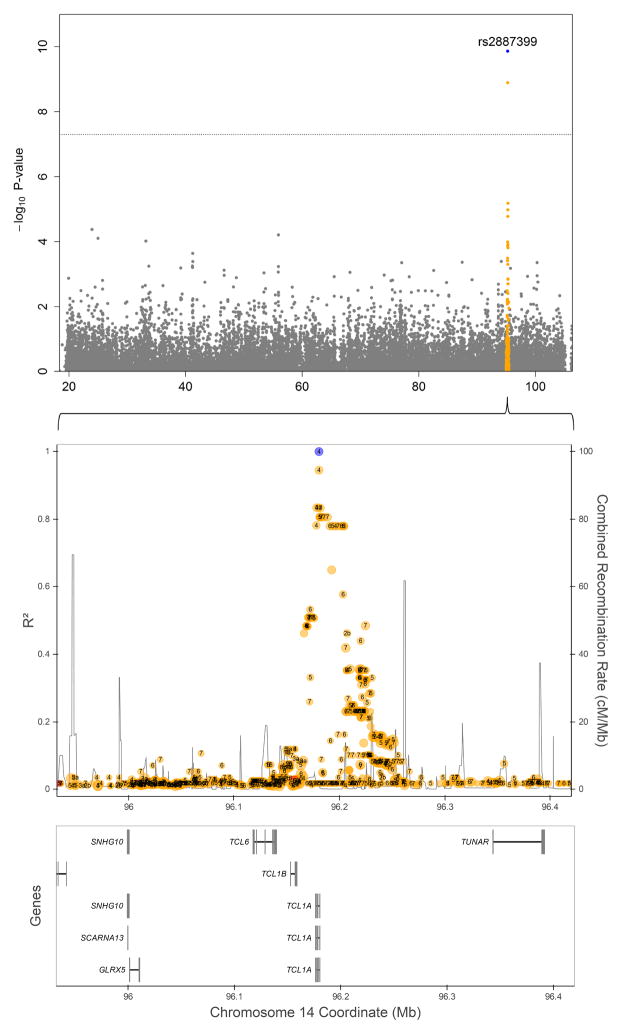
We conducted a genome-wide association study (GWAS) to identify regions associated with risk for mLOY in the three cohorts, analyzed separately and in a meta-analysis. We adjusted for smoking status (ever versus never smoker) and principal components significantly associated with mLOY in each cohort (reported previously to account for subtle differences in population substructure in participants of European background). The analysis included 895 men with detected mLOY and 11,474 men with no detected mLOY. The p-value distribution from the combined meta-analysis had a λGC of 1.015, as depicted in the quantile-quantile plot (Supplementary Figure 3). A significant association was observed with SNP rs2887399 at 14q32.13 (OR=1.55, 95%CI=1.36–1.78; p=1.37×10−10) (Figure 4, Supplementary Figure 4). The major risk allele (G) has a frequency of 0.77 in CEU. The effect estimates are consistent across the three studies and there was no evidence for heterogeneity (P=0.86, Supplementary Figure 5). The relationship remained robust when not adjusting for smoking status (OR=1.57, 95%CI=1.36–1.80; P=6.46×10−11). The rs2887399 variant maps to the 5′ end of the T-cell leukemia/lymphoma 1A gene (TCL1A), which functions as a coactivator of the cell survival kinase AKT and has been implicated in T-cell and B-cell hematological malignancies27, mainly because recurrent chromosomal rearrangements bring TCL1A in close proximity to the T-cell antigen receptors gene28. The rs2887399 variant is the first common variant associated with a clonal-expansion phenotype beyond the known association of a JAK2 haplotype with mosaicism for the common JAK2 V617F mutation29–32. This genetic finding linking germline variation to somatic mosaicism could lead to understanding how clonal hematopoiesis relates many chronic diseases; it is plausible that the susceptibility haplotype could contribute to actual loss of the Y chromosome or it could be permissive for clonal expansion. Further work is needed to fine map the region and investigate its biological underpinnings on development of mLOY.
Figure 4. Regional plot for chromosome 14 mLOY meta-analysis association p-values.
The GWAS for mLOY included 895 men with detected mLOY and 11,474 men with no detected mLOY. The top panel is a Manhattan plot showing −log10 P-values on the Y axis and chromosomal position on the X axis. A 500 Kb region around the top SNP (rs2887399, blue) is highlighted (orange) and zoomed in using LDlink33 to investigate LD (middle panel) and nearby genes (bottom panel). Numbers encapsulated within points in the middle plot represent RegulomeDB values. All statistical tests are two-sided.
In summary, mLOY is the most common large-scale detectable mosaic chromosomal event in males and it has a striking association with aging and cigarette smoking, which is attenuated by years after cessation. We observed limited evidence for mLOY as a strong risk factor for three common cancers in men, and did not observe an association with survival after cancer diagnosis. Contrary to prior evidence suggesting that men with mLOY were at substantially higher likelihood of dying from cancer, our study provides little evidence for this hypothesis. Together, these data suggest that age and smoking have a substantial effect on the development of mLOY, but that there is insufficient evidence to conclude mLOY is a major risk factor for non-hematologic cancer in men. Lastly, our GWAS of mLOY identified a locus on 14q32.13, which may provide insight into the biological basis of mLOY in relation to smoking and aging in men.
ONLINE METHODS
Study overview
The analyzed data consists of 13,789 males drawn from cancer genome-wide association studies (GWAS) conducted within three prospective cohorts. The mean age at DNA collection was 67.26 years for all participants. The studies were approved by the institutional ethics committees of each participating hospital and the Institutional Review Board (IRB) of the United States National Cancer Institute (NCI). Written informed consent was obtained from all individuals.
DNA was extracted from peripheral circulating leukocytes (70.38%) and buccal samples (29.62%) for men drawn from the three cohorts (Supplementary Table 1). Genomic DNA was screened and analyzed at the NCI according to the standard sample handling process of the Cancer Genomics Research Laboratory (CGR), Division of Cancer Epidemiology and Genetics (DCEG). AmpFlSTR Identifiler assays confirmed each sample had gender concordance and removed samples with evidence for contamination. Genotyping was carried out on one of four Illumina Infinium SNP arrays (e.g., Hap610K, HapIM, Omni1M, and Omin2.5M), each of which has an adequate number of Y specific probes. Additional scanning on other microarray chips was not used because of the inadequate Y probe coverage. Of all participants, 94.99% were detected to have >=80% of European ancestry and 2.39% were detected to have >=80% of African ancestry. Case-control studies for bladder cancer were also included and drawn from two studies carried out in Spain and New England Bladder Cancer Study previously scanned with the Hap1M and Hap610K chip34, respectively. The MD Anderson bladder cancer study was initially scanned on a chip with inadequate Y chromosome probe coverage35, and so we detected mLOY in this study using the qPCR assay described below (Supplementary Table 14).
Intensity Analysis: Log2 Intensity Ratio (LRR) and B-Allele Frequency (BAF) generation
Sample intensity files (two files per sample, for red and green channels) were loaded into the Illumina GenomeStudio software. The intensity data were normalized using the Illumina five-step self-normalization procedures, which used information contained in the array itself to convert raw X and Y (allele A and allele B) signal intensities to normalized values. The LRR and BAF values for each assay were exported from GenomeStudio software using the “Genotype Final Report” (GFR) format.
Test region for detecting Y chromosome abnormality
We extensively examined loci across the Y chromosome for the four commercial chip types with adequate Y chromosome coverage and used the male specific region of chromosome Y between 6,671,498-22,919,969 (hg18/build36) as the test region for detecting Y chromosome mosaicism, since this region provides relatively stable signal intensity and was outside of majority of the regions containing genes with multiple copies26 (Supplementary Figure 1A).
Y Chromosome mosaicism detection method
The Y chromosome mosaicism was detected using log R ratio (LRR), which is the normalized measure of total signal intensity and provides data on relative copy number. Subjects were examined for deviations from expected log2 intensity ratio for evidence of loss of the male specific region of chromosome Y (MSY). A minimum mean threshold of LRR <= −0.15 was used for identifying a Y chromosome loss event. A minimum mean threshold of LRR >= 0.15 was used to define a Y chromosome gain event. Samples with mean LRR values falling below or above these thresholds were called as mosaic Y losses and gains, respectively. To minimize the false discovery of Y chromosome mosaicism, the ratio of the mean LRR to the standard deviation for the test region was calculated. A minimum threshold for the ratio was set to 0.25 to filter out samples with excessive noise in LRR values. For potential mLOY, each chromosome Y plot was then manually reviewed and suspect events were further excluded from subsequent analyses.
qPCR Validation
Quantitative PCR (qPCR) was used to evaluate the ratio of Y chromosome signal to an autosomal single copy gene signal. 15 qPCR gene assays spanning the p and q arms of the Y chromosome (Supplementary Figure 1B), were run in duplex with RNase P as the reference gene, known to be single copy26 (Supplementary Table 16).
5 ng of sample DNA, according to Quant-iT PicoGreen dsDNA quantitation (Life Technologies, Grand Island, NY), was transferred to LightCycler-compatible 384-well plates (Roche, Indianapolis, IN) and dried down. An internal standard curve (serial dilution, to 7 target ratios, of pooled male gDNA samples, with no detectable Y chromosome loss, with a pool of female gDNA samples) and assay control samples (3 target ratios, prepared similarly to the standard curve) were applied to the assay plates to guide analysis and indicate overall quality of assay performance. All experimental and control samples were assayed in triplicate on each plate.
PCR was performed using 5 uL reaction volumes consisting of: 2.5 uL of LightCycler 480 Probes Master Mix (Roche, Indianapolis, IN), 2.0 uL of MBG Water, 0.25 uL of 20X TaqMan® Copy Number Reference Assay, human, RNase P (Life Technologies, Grand Island, NY), and 0.25 uL of specific 20X TaqMan® Copy Number Assay (Life Technologies, Grand Island, NY). Thermal cycling was performed on a LightCycler 480 (Roche) where PCR conditions consisted of: 95°C hold for 5 min, denature at 95°C for 15 sec, anneal at 60°C for 30 sec, with fluorescence data collection, 45 cycles.
The LightCycler software (Release 1.5.0) was used for initial analysis of raw data. Utilizing absolute quantification analysis with the second derivative maximum method and high confidence detection algorithm, single target sequences were quantified and expressed as a ratio (Target/Reference) based on the internal standard curve of known ratios. The ratios of the 15 assays were averaged to yield an overall Y chromosome signal ratio.
Estimation of the proportion of cells with Y loss
A quadratic regression model was used to fit the average qPCR ratio and mean LRR data pairs with mean LRR as predictor variable (X) and the average qPCR ratio as the response variable (Y) to create a predictive polynomial equation: Y = ao + a1×X + a2×X2. Only the data points from subjects having consensus event calls between qPCR and chip data for Y loss and normal, with CV% <=10% from qPCR data were used to generate such relationship (n = 98 subjects). For qPCR, the standard curve utilized simulates known ratios of Y chromosome ranging from 10% to 100% (90% loss to no loss). This is achieved by diluting a pool of male samples with no mLOY with a pool of female samples to simulate these percentages of loss. Y gain data was not included when building the predictive model since the quantitative qPCR ratio for the estimated amount of gain is not precise since the data is extrapolated outside the experimental copy number range of 0.1–1 defined by the standard curve. For each mean LRR, the corresponding copy number can be predicted by inserting the mean LRR into the quadratic equation. The percent of cells with Y loss equals to 1- average Y signal ratio (Supplementary Figure 1C), for example a mean LRR of −0.15 corresponds to a frequency of Y loss of 22.7%. We also performed this analysis for the case-controls studies and found similar results (data not shown).
Logistic Regression Analysis
All of the logistic regression models were generated in R using the glm function with quasi-binomial family and logit as link function. To determine the relationship between individuals having Y chromosome loss and their age at DNA collection, smoking behavior, DNA source, ancestry, and study cohort, we fit several models that regressed the presence of Y loss for each individual on relevant covariates in a logistic model. To determine the relationship between individuals having Y chromosome loss and cancer diagnosis, we fit several models that regressed the presence of cancer diagnosis for each individual on relevant covariates in a logistic model. The following covariate terms were defined for each individual: (i) age of DNA (a continuous measure of age at DNA collection); (ii-a) smoking status (categorical variable with three levels: current, former, and never smokers (reference group)); (ii-b) number of years since stopped smoking (categorical variable with six levels: current smokers, 1–4, 5–10, 11–20, >20 years since quitting smoking, never smokers (reference group)); (iii) DNA source (categorical variable with 2 levels: individuals who contributed DNA derived from a buccal sample and for blood sample (reference group)); (iv) East Asian ancestry (a continuous measure of admixture estimate); (v) African ancestry (a continuous measure of admixture estimate); and (vi) study (categorical variable with 3 levels: ATBC, PLCO, CPSII (reference group)).
Survival analysis
Cox proportional-hazard models were generated in R using the Survival package. Using age as the time-scale, the start year is the year of cancer diagnosis, the end year is the year of death or censorship. For total survival analysis, the event indicator equals 1 for those who died during the study follow-up and 0 for those who did not. For cancer survival analysis, the event indicator equals 1 for those with a cancer cause of death during the period of the study and 0 for those who were alive at the end of the study or had a non-cancer cause of death. We observed similar associations when we started follow-up time at year of DNA collection. All case subjects used in the analysis had DNA collected at least 1 year before a cancer diagnosis. The following covariate terms were defined for each individual: (i) age of diagnosis (a continuous measure of age at diagnosis); (ii) BMI (a continuous measure of body mass index); (iii) pack years (categorical variable with 4 levels: >60, >60–40, >40–20, (>20 + never smokers (reference group)); (iv) mosaic Y loss (categorical variable with 2 levels: 1 for individuals identified as Y loss and 0 for individuals without Y loss. All other covariates were defined as in the logistic regression models.
GWAS Analysis
A genome wide association scan (GWAS) was conducted adjusting for smoking status (ever versus never smoker) and principal components that were significantly associated with mLOY in each cohort. The combined meta-analysis of men from the three cohorts consisted of 928 men with detectable mLOY and 12,118 men without evidence of mLOY (Supplementary Figure 5). To ensure a robust association, a further analysis was carried out that did not adjust for smoking status.
Other analysis
Frequency plots were generated using R. Confidence intervals (CI) on frequencies were reported using 95% confidence bounds from the Jefferys interval method and were generated in R using the binom package. The 95% CI unadjusted analysis of count data and frequencies was performed using the Fisher’s Exact test for contingency tables, as implemented in the R software package. GLU software package was used to estimate admixture coefficients for each subject.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Data ACCESS
Provided in Supplementary Data
Conflict of Interest Statement
The authors declare no relevant conflicts of interest.
Author Contributions
M.D., M.Y., and S.J.C. conceived the analysis. W. Zhou, M.M., N.F., M.D., M.Y. and S.J.C. designed the study. W. Zhou, M.M., N.F., N.R., N.C., M.D., M.Y. and S.J.C. interpreted the primary results. W. Zhou, M.M., N.F., K. J., M.D., M.Y. developed the study methods. W. Zhou, M.M., N.F., C.D. and K.J. analyzed the data. W. Zhou, M.M. and A. H were responsible for production and analysis of the genotype data. W. Zhou, M.M., N.F. and J. S. performed statistical analysis. W. Zhou, M.M., N.F., M.D., M.Y. and S.J.C. drafted the manuscript. M.T., R.N.H. and S.J.C. provided vital programmatic and institutional support. N.M., L.T., M.M.G., S.M.G., V.L.S., D.A., S.W., J.V., S.B., A.B., D.S., J.F., M.G.C, F.X.R., J.E., G.M., B.R.S., M.K., A.J., M.S., X.W., J.G., Y.Y. and L.P.J. contributed data or samples. All authors contributed critical feedback, review and approval of the manuscript.
References
- 1.Pierre RV, Hoagland HC. Age-associated aneuploidy: loss of Y chromosome from human bone marrow cells with aging. Cancer. 1972;30:889–94. doi: 10.1002/1097-0142(197210)30:4<889::aid-cncr2820300405>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Loss of the Y chromosome from normal and neoplastic bone marrows. United Kingdom Cancer Cytogenetics Group (UKCCG) Genes Chromosomes Cancer. 1992;5:83–8. doi: 10.1002/gcc.2870050112. [DOI] [PubMed] [Google Scholar]
- 3.Guttenbach M, Koschorz B, Bernthaler U, Grimm T, Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am J Hum Genet. 1995;57:1143–50. [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs PA, Brunton M, Court Brown WM, Doll R, Goldstein H. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 1963;197:1080–1. doi: 10.1038/1971080a0. [DOI] [PubMed] [Google Scholar]
- 5.Abeliovich D, Yehuda O, Ben-Neriah S, Or R. Loss of Y chromosome. An age-related event or a cytogenetic marker of a malignant clone? Cancer Genet Cytogenet. 1994;76:70–1. doi: 10.1016/0165-4608(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 6.Herens C, et al. Loss of the Y chromosome in bone marrow cells: results on 1907 consecutive cases of leukaemia and preleukaemia. Clin Lab Haematol. 1999;21:17–20. doi: 10.1046/j.1365-2257.1999.00173.x. [DOI] [PubMed] [Google Scholar]
- 7.Wiktor A, et al. Clinical significance of Y chromosome loss in hematologic disease. Genes Chromosomes Cancer. 2000;27:11–6. [PubMed] [Google Scholar]
- 8.Wong AK, et al. Loss of the Y chromosome: an age-related or clonal phenomenon in acute myelogenous leukemia/myelodysplastic syndrome? Arch Pathol Lab Med. 2008;132:1329–32. doi: 10.5858/2008-132-1329-LOTYCA. [DOI] [PubMed] [Google Scholar]
- 9.Zhang LJ, Shin ES, Yu ZX, Li SB. Molecular genetic evidence of Y chromosome loss in male patients with hematological disorders. Chin Med J (Engl) 2007;120:2002–5. [PubMed] [Google Scholar]
- 10.Chanock S. Cancer biology: Genome-wide association studies. In: Stewart BWWC, editor. World Cancer Report 2014. International Agency for Research on Cancer; 2014. pp. 193–202. [Google Scholar]
- 11.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez JR, et al. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–8. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–50. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machiela MJ, et al. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet. 2015;96:487–97. doi: 10.1016/j.ajhg.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 18.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–48. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumanski JP, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–3. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg LA, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–8. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez LC, Torres M, Real FX. Somatic mosaicism: on the road to cancer. Nat Rev Cancer. 2016;16:43–55. doi: 10.1038/nrc.2015.1. [DOI] [PubMed] [Google Scholar]
- 25.Machiela MJ, Chanock SJ. Detectable clonal mosaicism in the human genome. Semin Hematol. 2013;50:348–59. doi: 10.1053/j.seminhematol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 27.Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000;6:395–407. doi: 10.1016/s1097-2765(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 28.Virgilio L, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci U S A. 1998;95:3885–9. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olcaydu D, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–4. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 30.Olcaydu D, et al. The ‘GGCC’ haplotype of JAK2 confers susceptibility to JAK2 exon 12 mutation-positive polycythemia vera. Leukemia. 2009;23:1924–6. doi: 10.1038/leu.2009.110. [DOI] [PubMed] [Google Scholar]
- 31.Jones AV, et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009;41:446–9. doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–9. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–5. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.