Abstract
Acute metabolic changes of plasma membrane (PM) lipids, such as those mediating signaling reactions, are rapidly compensated by homeostatic responses whose molecular basis is poorly understood. Here we show that the Extended-Synaptotagmins (E-Syts), ER proteins which function as PI(4,5)P2 and Ca2+-regulated tethers to the PM, participate in these responses. E-Syts transfer glycerolipids between bilayers in vitro and such transfer requires Ca2+ and their SMP domain, a lipid-harboring module. Genome edited cells lacking E-Syts do not exhibit abnormalities in the major glycerolipids at rest, but display enhanced and sustained accumulation of PM diacylglycerol (DAG) upon PI(4,5)P2 hydrolysis by PLC activation, which can be rescued by expression of E-Syt1, but not by mutant E-Syt1 lacking the SMP domain. The formation of E-Syts-dependent ER-PM tethers in response to stimuli that cleave PI(4,5)P2 and elevate Ca2+ may help reverse accumulation of DAG in the PM by transferring it to the ER for metabolic recycling.
Introduction
The endoplasmic reticulum (ER) carries out a multiplicity of functions, including protein and lipid synthesis, lipid metabolism and Ca2+ storage for intracellular signaling. While membranes of the ER are functionally connected to all membranes of the secretory and endocytic pathways via vesicular transport, they only fuse with each other and with vesicles involved in retrograde transport to this organelle. However, close appositions between the ER and the membranes of all other membranous organelles, including the plasma membrane (PM), play major roles in cellular physiology. For example, ER membrane contact sites are involved in the control of Ca2+ homeostasis, in exchanges of lipids between bilayers, and in the function of ER-localized enzymes that act “in trans” on the apposed membrane1–7
One class of tethers that mediate ER-PM contacts are the three extended synaptotagmins (E-Syts) (tricalbins in yeast)8–11. E-Syts, which occur as homo and heterodimers, are embedded into the ER via an N-terminal membrane anchor that is followed by an SMP (synaptotagmin-like, mitochondrial-lipid binding protein) domain and multiple C2 domains (5 in E-Syt1 and 3 in E-Syt2 and E-Syt3). Their tethering function is mediated by C2 domain-dependent interactions with the PI(4,5)P2 in the PM and are additionally regulated by cytosolic Ca2+8, 12. The E-Syts resemble classical synaptotagmin13 in domain structure. However, while synaptotagmin is exported from the ER and tethers secretory vesicles to the PM in preparation for Ca2+-dependent fusion, the E-Syts are retained in the ER. Interestingly, all proteins previously considered synaptotagmin homologues in plants contain SMP domains and are ER proteins14–17.
SMP domains are members of a superfamily of modules, the TULIP domains, which are found in extra and intracellular proteins and whose shared property is to harbor lipids in a hydrophobic cavity18–22. One of the best-studied TULIP domain protein is CETP, which mediates transport of lipids between high and low density lipoprotein20. SMP domains comprise modules that are localized intracellularly at membrane contact sites9, 23. As revealed by crystallography, the SMP domain of E-Syt2 dimerizes to form a ~90 Å long cylinder traversed by a deep groove lined with hydrophobic residues that contains lipids22. Supporting these findings, mass spectrometry of an SMP domains detected presence of glycerolipids22, 24.
The precise role of the E-Syts in cell physiology remains unclear. A plausible scenario is that they function in the delivery of newly synthesized lipids from the ER to the PM and/or of lipid metabolites from the PM to the ER for metabolic recycling. As their tethering properties are regulated by Ca2+, their functions may be important in maintaining or resetting lipid homeostasis in response to acute physiological changes that produce PM lipid perturbations.
Goal of this study was to directly test the lipid transfer properties of the E-Syts and to begin determining their impact on lipid homeostasis at the PM in living cells.
Results
Endogenous E-Syt1 is rapidly translocated to ER-PM contacts upon cytosolic Ca2+ increase
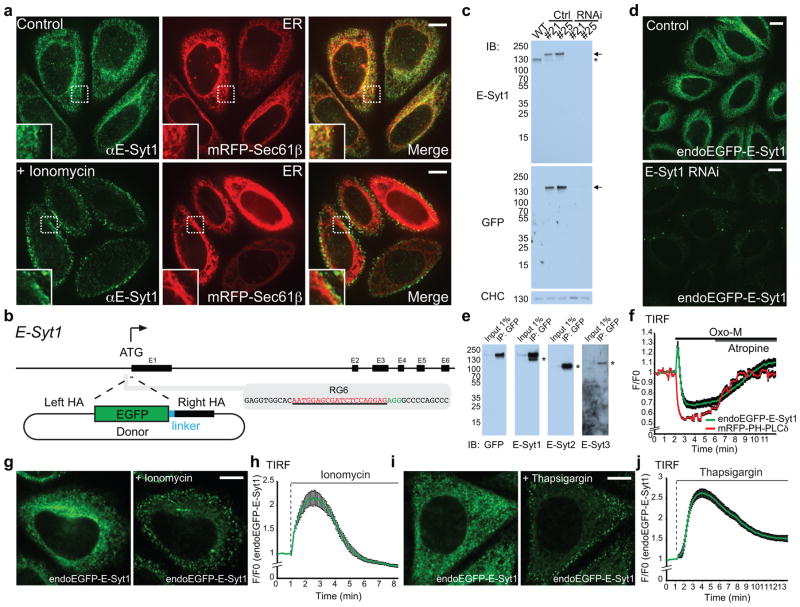
Previous E-Syts localization studies had capitalized upon exogenously expressed tagged proteins. We have now generated polyclonal antibodies against the N-terminus of human E-Syt1, which upon affinity-purification labeled specifically the endogenous protein in HeLa cells not only by western blotting (Supplementary Fig. 1a) but also by immunofluorescence (Fig. 1a). The signal generated by these antibodies colocalized with mRFP-Sec61β (a marker of the ER membrane) and was absent in genome-edited HeLa cells lacking E-Syt1 (see below; Supplementary Fig. 1b). The bulk of this immunoreactivity rapidly translocated to the cell cortex (peak of recruitment at about 2–3 min) after addition of the Ca2+-ionophore ionomycin25 (Fig. 1a), confirming results obtained with exogenous tagged E-Syt1 (Supplementary Fig. 1c,d and8).
Figure 1. Localization of endogenous E-Syt1.
(a) Confocal images of HeLa cells expressing mRFP-Sec61β. Localization of endogenous E-Syt1 was detected by an affinity-purified antibody. Insets show at higher magnification the area framed by a dotted line. Bottom images: HeLa cells stimulated with ionomycin (2μM) for 1 min. Note the translocation of the E-Syt1 fluorescent signals to the cortical region of the cell. (b) Strategy for the endogenous-tagging of E-Syt1 (see methods). (c) Lysates of control HeLa cells, two endogenously-tagged HeLa cell lines (#21 and #25), and the same two lines treated with RNAi against endogenous E-Syt1 were processed by SDS-PAGE and immunoblotting (IB) with anti-E-Syt1, anti-GFP and anti-clathrin heavy chain (CHC) antibodies. Arrows and an asterisk indicate endogenously-tagged EGFP-E-Syt1 and endogenous E-Syt1, respectively. (d) Confocal images of live #21 cells showing endogenously-tagged EGFP-E-Syt1 (endoEGFP-E-Syt1) fluorescent signals in control, but not in RNAi-treated cells. (e) Extracts of #21 cells were subjected to anti-GFP immunoprecipitation (IP) and then processed by SDS-PAGE and immunoblotting with indicated antibodies. Asterisks indicate coimmunoprecipitated bands corresponding to endogenous untagged proteins. Note the strong homo- and hetero-dimerization of endogenously-tagged E-Syt1 with endogenous E-Syt1, E-Syt2 and E-Syt3. (f) Time-course of normalized GFP signal, as assessed by TIRF microscopy, from cells expressing endogenously-tagged E-Syt1 as well as muscarinic M1 receptor (M1R) and the PI(4,5)P2 probe, mRFP-PH-PLCδ. Oxo-M (10μM) stimulation and atropine (50μM) application are indicated. The biphasic response of E-Syt1 reflects the increase in cytosolic Ca2+ immediately followed by massive PI(4,5)P2 depletion. (mean +/− SEM, n=16 cells pooled from 2 independent experiments.) (g–j) Confocal images of live clone #21 cells before and 1.5 min after stimulation with ionomycin (2μM) (g) and before and 4 min after stimulation with thapsigargin (2μM) (i). The time-course of ionomycin-induced and thapsigargin-induced recruitment of endoEGFP-E-Syt1 to the PM, as assessed by TIRF microscopy, is shown in (h) and (j) respectively [mean +/− SEM, n=8 cells (h) and 11 cells (j) assessed from 1 experiment, the experiment was repeated independently 3 times with similar results. Scale bars, 10μm. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To analyze the dynamics of E-Syt1 when expressed at endogenous levels, a HeLa cell line expressing tagged E-Syt1 from the endogenous locus was generated by inserting EGFP at its N-terminus using CRISPR/Cas9-mediated genome-editing (Fig. 1b). Expression of the fusion protein (endoEGFP-E-Syt1) was confirmed by western blotting, which revealed a band reactive for both anti-GFP and anti-E-Syt1 antibodies in extracts of these cells (Fig. 1c). This band, as well as EGFP fluorescence, disappeared upon RNA interference (RNAi) against endogenous E-Syt1 (Fig. 1c,d). Further analysis demonstrated that this protein had the expected properties and dynamics8 of E-Syt1. These included homodimerization as well as heterodimerization with other E-Syts (Fig. 1e) and Ca2+-dependent PM recruitment. This recruitment was very transient in response to the Ca2+ elevation produced by muscarinic M1 receptor (M1R) activation by oxotremorine M (Oxo-M) in cells overexpressing M1R26, as such elevation is the result of PLC dependent inositol 1,4,5-trisphosphate (IP3) generation27, 28 and thus correlates with very rapid and massive PI(4,5)P2 depletion (Fig. 1f). It was more persistent, as expected8, in response to Ca2+ elevations produced by ionomycin (Fig. 1g,h and Supplementary Movie 1), which induces a slower PI(4,5)P2 loss (see below; Fig. 5b) and to thapsigargin (Fig. 1i,j and Supplementary Movie 2), which does not have a major impact on PI(4,5)P2 levels.
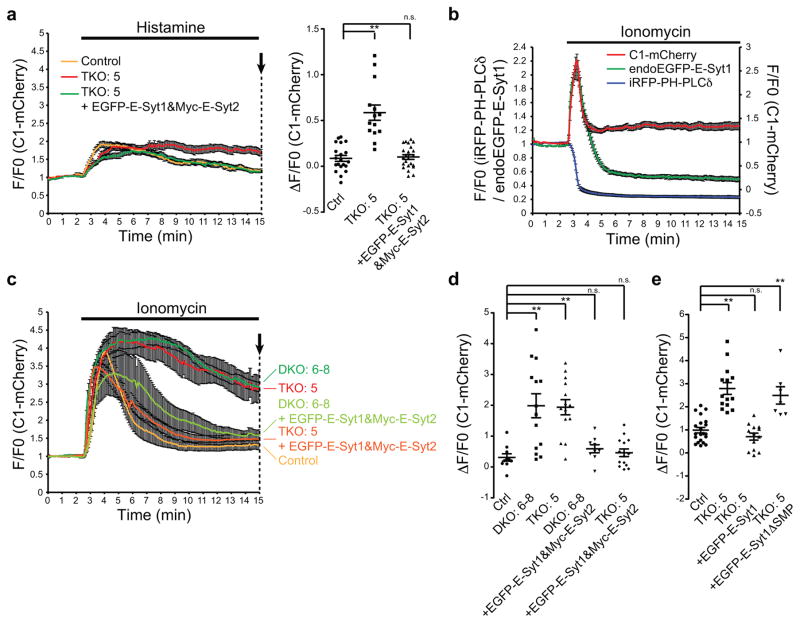
Figure 5. Prolonged accumulation of DAG in E-Syt KO cells upon phospholipase activation.
(a) Time-course of normalized mCherry signal, as assessed by TIRF microscopy, from WT control (Ctrl) and E-Syt TKO cells expressing a DAG probe (C1-mCherry). Histamine (1mM) stimulation is indicated. Re-expression of EGFP-tagged E-Syt1 (EGFP-E-Syt1) together with Myc-E-Syt2 in E-Syt TKO cells rescued the accumulation of DAG, as monitored by C1-mCherry. (Right) Values of ΔF/F0 corresponding to the end of the experiment as indicated by an arrow [mean +/− SEM, n=20 cells (Ctrl), n=14 cells (TKO#5), n=23 cells (TKO#5 + EGFP-E-Syt1&Myc-E-Syt2); data are pooled from 2 independent experiments except for Ctrl that is pooled from 3 independent experiments; Bonferroni’s multiple comparisons test, ** denotes P=0.0005] (b) Time-course of normalized EGFP, iRFP (left axis) and mCherry (right axis) signal, as assessed by TIRF microscopy, from cells expressing E-Syt1 tagged with EGFP at the endogenous locus (endoEGFP-E-Syt1) as well as a PI(4,5)P2 probe (iRFP-PH-PLCδ) and a DAG probe (C1-mCherry). Ionomycin (2μM) stimulation is indicated. (mean +/− SEM, n=6 cells assessed from 1 experiment, the experiment was repeated independently 3 times, with similar results) (c,d) (c) Time-course of normalized mCherry signal, as assessed by TIRF microscopy, from WT control (Ctrl), E-Syt1/2 DKO cells and E-Syt TKO cells expressing C1-mCherry. Ionomycin (2μM) stimulation is indicated. Re-expression of EGFP-E-Syt1 together with Myc-E-Syt2 in E-Syt1/2 DKO and E-Syt TKO cells rescued the accumulation of DAG, as monitored by C1-mCherry. (d) Values of ΔF/F0 corresponding to the end of the experiment as indicated by arrow in c [mean +/− SEM, n=10 cells (Ctrl), n=14 cells (DKO#6-8), n=14 cells (TKO#5), n=9 cells (DKO#6-8 + EGFP-E-Syt1&Myc-E-Syt2), n=14 cells (TKO#5 + EGFP-E-Syt1&Myc-E-Syt2); data are pooled from 2 independent experiments for each condition; Bonferroni’s multiple comparisons test P=0.0001 (DKO#6-8); P=0.0002 (TKO#5)] (e) Values of ΔF/F0 corresponding to the end of the experiment as assayed in the same way as in c [mean +/− SEM, n=21 cells (Ctrl), n=14 cells (TKO#5), n=13 cells (TKO#5 + EGFP-E-Syt1), n=7 cells (TKO#5 + EGFP-E-Syt1ΔSMP); data are pooled from 2 independent experiments except for TKO#5 + EGFP-E-Syt1ΔSMP that is assessed from 1 experiment, the experiment was repeated independently 2 times, with similar results; Bonferroni’s multiple comparisons test, ** denotes P<0.0001]. n.s. = not significant (cut-off P>0.05).
Ca2+-regulated E-Syt1-dependent lipid transport in vitro
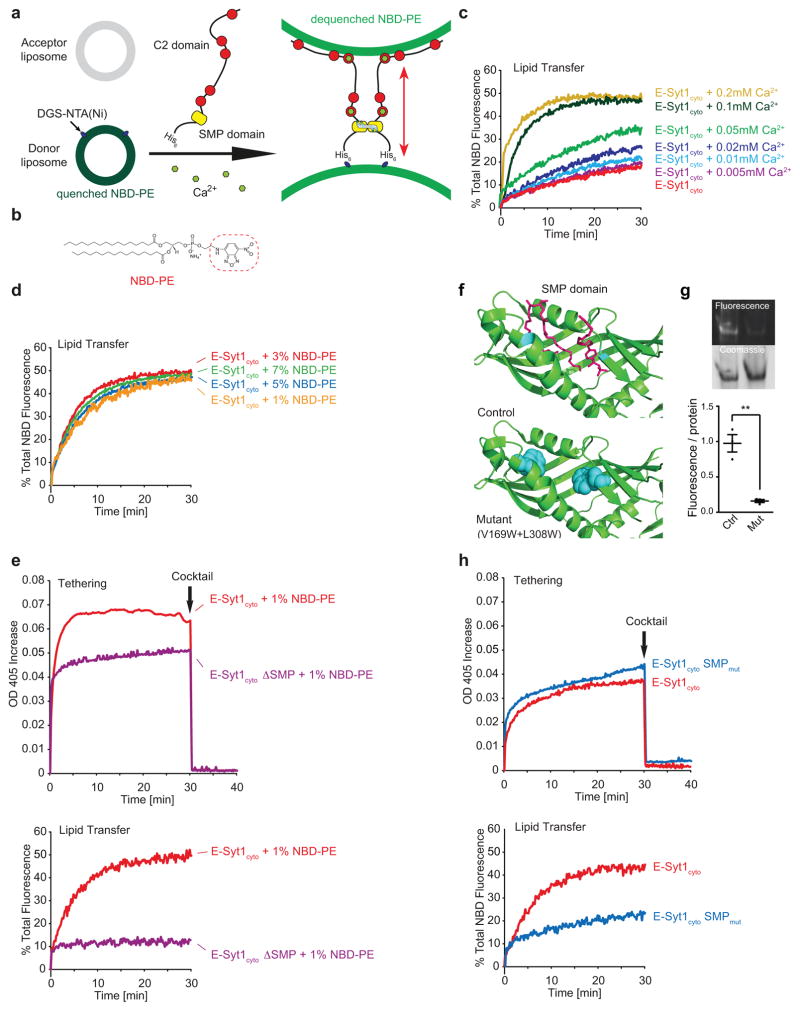
The presence of the SMP domain in the E-Syts suggested a role of these proteins in lipid transfer22. To provide direct evidence for such function, we tested the property of the cytosolic domain of E-Syt1 to transfer lipids between liposomes using a Fluorescence Resonance Energy Transfer (FRET)-based assay (Fig. 2a). In this assay, donor liposomes comprising phosphatidylcholine (PC), a fluorescent dye-labeled lipid [NBD-phosphatidylethanolamine (PE) (Fig. 2b)] and a nickel-conjugated lipid [DGS-NTA(Ni)], but no acidic phospholipids (and thus ER-like) were mixed with acceptor acidic phospholipid containing (PM-like) liposomes [PC, phosphatidylserine (PS) and PI(4,5)P2 ] and with the purified His-tagged cytosolic portion of E-Syt1 (E-Syt1cyto) (Fig. 2a). This construct becomes anchored to the donor liposomes by the His tag and binds PI(4,5)P2 containing acceptor liposomes in trans, and in a Ca2+-dependent way, via its C2 domains (Fig. 2a).
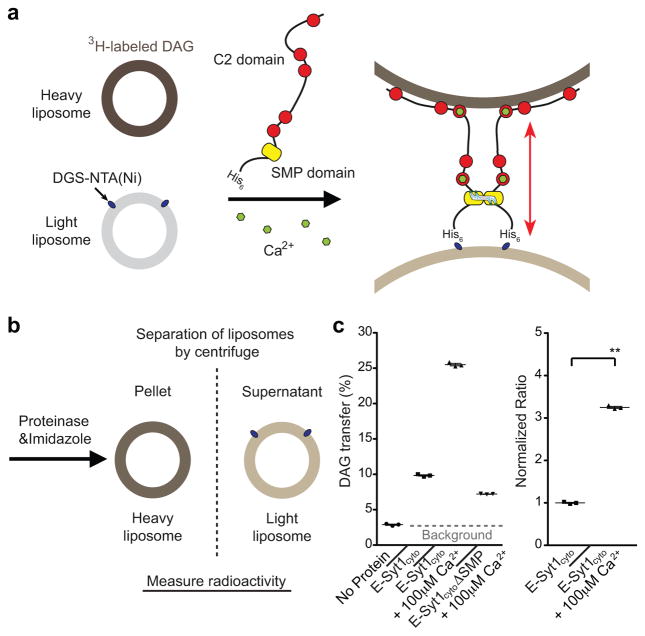
Figure 2. E-Syt1 is a Ca2+-dependent lipid transfer protein.
(a) Schematics showing the in vitro lipid transfer assay. Donor liposomes [PC, DGS-NTA(Ni), NBD-PE], and acceptor liposomes [PC, PS, PI(4,5)P2] were incubated with histidine (His)-tagged cytosolic portion of E-Syt1 protein (E-Syt1cyto). Dequenching of self-quenched NBD-PE fluorescence, i.e. transfer of the fluorescent lipids from donor to acceptor liposomes, was monitored using a fluorometer (see methods). (b) Structure of NBD-PE. (c) Time-course of normalized fluorescence signals from liposomes mixtures containing 1% NBD-PE in the donor liposomes at the indicated concentration of Ca2+ in the assay buffer. E-Syt1cyto was added at time 0. (d) Time-course of normalized fluorescence signals from E-Syt1cyto/liposome mixtures containing different moles percent of NBD-PE in the donor liposomes and incubated with 100μM Ca2+. (e) (top) Time-course of turbidity of the suspension (see methods). Turbidity reflects liposome clustering due to tethering of donor and acceptor liposomes. (bottom) Time-course of normalized fluorescence signals from liposome mixtures containing 1% NBD-PE in the donor liposomes and either E-Syt1cyto or E-Syt1cyto lacking the SMP domain (E-Syt1cyto ΔSMP). (f) Design of mutant SMP domain defective in lipid harboring. Hydrophobic amino acids lining the deep hydrophobic groove22 were mutated to tryptophan (W), thus creating steric hindrance to access of acyl chains to the SMP channel. Aromatic rings of tryptophan are shown as surface representation. (g) Lipid-binding of E-Syt1 SMP domain. (top) Purified WT SMP domain (Ctrl) and mutant SMP domain, carrying V169W and L308W mutations (Mut), were incubated with NBD-PE, run on native-PAGE and analyzed by fluorometry and coomassie blue staining; (bottom) Quantification of fluorescence signals of NBD-PE normalized to the total amount of protein (mean +/− SEM, n=3 independent experiments; two-tailed Student’s t-test with equal variance, P=0.0028). (h) (top) Time-course of turbidity of the suspension. (bottom) Time-course of normalized fluorescence signals from liposome mixtures containing 1% NBD-PE in the donor liposomes and either E-Syt1cyto or E-Syt1cyto with lipid-binding deficient SMP domain (E-Syt1cyto SMPmut). The transfer of NBD-PE is much reduced with E-Syt1cyto SMPmut. For all the liposome-based assays, data are from one experiment; three experiments that yielded similar results were performed
In the absence of E-Syt1cyto, NBD-PE was self-quenched in the donor liposomes, and solubilization of the liposomes with n-dodecyl-β-D-maltoside (DDM) resulted in an efficient dequenching (Supplementary Fig. 2a).
Addition of E-Syt1cyto and of various Ca2+ concentrations (5 to 200μM) to the donor plus acceptor liposomes mixture induced rapid dequenching of NBD-PE in Ca2+ -dependent manner, consistent with the transfer of NBD-PE from donor to acceptor liposomes (Fig. 2c,d). 1% fluorescent lipids and 100μM Ca2+ were used in subsequent transfer assays. Absence of PI(4,5)P2 in the acceptor liposomes drastically slowed the dequenching of NBD-PE (Supplementary Fig. 2b). Furthermore, lipid transfer was bidirectional, as incorporating NBD-PE in either the ER-like or the PM-like liposomes, i.e. reverting donor and acceptor liposomes, resulting in dye dequenching with the same efficiency (Supplementary Fig. 2c). NBD-PE dequenching was not due to membrane fusion as a similar assay in which the fluorescent lipid tag in the donor liposomes was replaced by a water-soluble luminal self-quenching dye (Sulphorhodamine B) revealed no content mixing of the liposomes (Supplementary Fig. 2d). Potential lipid mixing due to hemifusion as a result of liposome tethering was ruled out: as revealed by turbidity assay, the potent liposome tethering produced by E-Syt1cyto could be completely reversed by the addition of a cocktail of EDTA, Imidazole and Proteinase K (“Cocktail”) (Fig. 2e, 2h top panels). Furthermore, a mutant E-Syt1cyto lacking the SMP domain (E-Syt1cyto ΔSMP) had negligible lipid transfer activity, while its liposome tethering properties were only slightly reduced relative to E-Syt1cyto (Fig. 2e). This minor reduction could have been explained by the lack of SMP-dependent dimerization (Fig. 2e). Finally, replacing two hydrophobic residues (V169 and L308) lining the lipid-binding groove of the SMP domain22 with the bulky hydrophobic amino acid tryptophan (W) strongly impaired lipid (NBD-PE) binding and transfer activity (Fig. 2f,g) without affecting liposome tethering (Fig. 2h).
Changing the ratio of donor and acceptor liposomes, from 1:9 to 1:1, significantly reduced the maximum dequenching efficiency achieved at the end of the assay (Supplementary Fig. 2e) indicating that E-Syt1 transfer glycerolipids along a concentration gradient. Furthermore, addition of non-labeled PE to acceptor liposomes did not affect NBD-PE transfer (Supplementary Fig. 2f), as PE is not preferred over other glycerolipids present in the liposome mixture (all glycerolipdis are transported bidirectionally).
Having validated the hypothesis that an E-Syt has lipid transfer activity, we explored the role of the E-Syts in lipid dynamics and homeostasis with a gene KO approach.
Generation of E-Syt1/2 double-knockout and E-Syt1/2/3 triple-knockout HeLa cells using TALEN and CRISPR
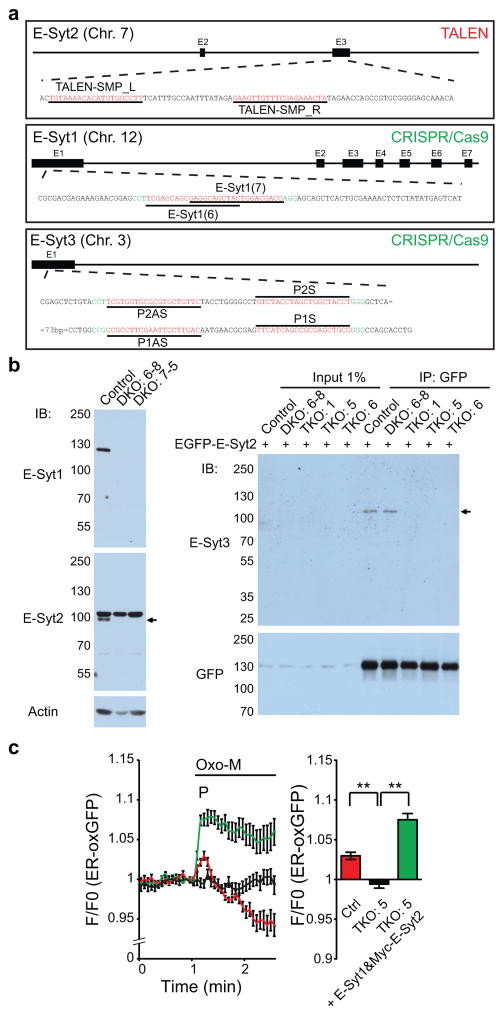
We first disrupted the E-Syt2 gene in HeLa cells using transcription activator-like effector nuclease (TALEN) and then sequentially the E-Syt1 and the E-Syt3 genes in these cells using CRISPR/Cas9 system (Fig. 3a, Supplementary Fig. 3a–c, and see methods for details).
Figure 3. Generation of E-Syt1/2 double knockout (DKO) and E-Syt1/2/3 triple knockout (TKO) HeLa cells using TALEN and CRISPR.
(a) Schematics of the TALEN- and Cas9/sgRNA-targeting sites in human E-Syt2, E-Syt1 and E-Syt3 loci. The targeting sequence is underlined and highlighted in red. The protospacer-adjacent motif (PAM) is labeled in green for E-Syt1 and E-Syt3 loci. (b) (left) Lysates of control HeLa cells and two independently isolated E-Syt1/2 DKO cell lines were processed by SDS-PAGE and immunoblotting (IB) with anti-E-Syt1, anti-E-Syt2 and anti-actin antibodies. Arrow indicates the specific band for E-Syt2. (right) Extracts of control HeLa cells, clone DKO#6-8 and three clones of E-Syt TKO cells transfected with EGFP-E-Syt2 were subjected to anti-GFP immunoprecipitation (IP) and then processed by SDS-PAGE and immunoblotting with anti-E-Syt3 and anti-GFP antibodies. Arrow indicates coimmunoprecipitated bands. Note the absence of IP-enriched E-Syt3 expression in E-Syt TKO cells. (c) (left) Time-course of normalized GFP signal, as assessed by TIRF microscopy, from WT control (Ctrl) and E-Syt TKO cells expressing a luminal ER marker (ER-oxGFP) and M1R. Oxo-M (10μM) stimulation is indicated. Re-expression of E-Syt1 together with Myc-tagged E-Syt2 (Myc-E-Syt2) in TKO cells rescued the loss of ER dynamics. (right) Quantification of fluorescence corresponding to the peak for each qunatification shown in the left graphs [for both left and right panels, mean +/− SEM, n=7 cells (Ctrl); n=18 cells (TKO#5); n=18 cells (TKO#5 + E-Syt1&Myc-E-Syt2; data are pooled from 3 independent experiments except for Ctrl that is from 1 experiment, the experiment was repeated independently 2 times with similar results); two-tailed Student’s t-test with equal variance, Ctrl: P=0.0028, TKO#5 + E-Syt1&Myc-E-Syt2: P<0.0001]. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Absence of the E-Syt1 and E-Syt2 bands in western blots of extracts of two E-Syt1/2 DKO clones (DKO#6-8 and DKO#7-5) is shown in Fig. 3b, while the loss of E-Syt1 immunofluorescence in the DKO#6-8 clone is shown in Supplementary Fig. 1b (See also Supplementary Fig. 3d–g). Given the low expression level of endogenous E-Syt3, validation of its absence in triple KO (TKO) cells required an enrichment procedure. To this end, we expressed EGFP-tagged E-Syt2 as a bait for E-Syt3 with which it heterodimerizes: anti-GFP immunoprecipitates obtained from these cells were enriched in the E-Syt3 immunoreactive band only when generated from cells where the E-Syt3 gene had not been edited, irrespective of the expression of the other endogenous E-Syts (Fig. 3b, See also Supplementary Fig. 3h), confirming successful KO of E-Syt3.
No obvious defects were observed in cell viability and overall morphology of KO cells, with the exception of a slower growth relative to parental HeLa cells. However, more detailed analysis revealed some changes, as described below. Initial experiments were carried out in E-Syt1/2 DKO cells. Subsequently, selected experiments were repeated in E-Syt TKO cells with similar results, most likely because of the only low expression level of E-Syt38.
Ca2+-enhanced apposition of the ER to the PM is impaired in E-Syt KO cells
As a premise to study the impact of the E-Syts on lipid dynamics, we assessed whether absence of the E-Syts had an impact on the ER in response to a rise in cytosolic Ca2+. As reported, in wild type (WT) cells, elevation of cytosolic Ca2+ results in increased fluorescence for the luminal ER marker (ER-oxGFP) in TIRF microscopy, most likely reflecting an expansion of areas of apposition between the ER and the PM8. To assess the impact of the lack of the E-Syts on such a response, changes of the ER-oxGFP fluorescence following muscarinic receptor stimulation by Oxo-M were monitored in control and TKO cells also transfected with the muscarinic M1 receptor (M1R). In E-Syt TKO cells, the Ca2+-dependent increase of ER-oxGFP fluorescence [which is very transient due to concomitant PI(4,5)P2 loss] was abolished and this effect could be “rescued”, in fact resulting in exaggeration of recruitment, by exogenous expression of E-Syt1 along with Myc-E-Syt2 (Fig. 3c) (note that the E-Syt1/E-Syt2 heterodimer is predominantly localized throughout the ER at resting Ca2+ levels, in contrast to the E-Syt2 homodimer that is already concentrated at the ER-PM contact sites at resting Ca2+ levels8). Near absence of the E-Syts does not inhibit SOCE8, 12, thus ruling out impaired Ca2+ elevations as the cause of the defective increase of ER-oxGFP fluorescence.
The steady state glycerolipid composition of the PM is not affected in E-Syt KO cells
The SMP domain of the E-Syts harbors glycerophospholipids22 and can transfer them in vitro between artificial membranes (see above). As the E-Syts form ER-PM tethers, they may exchange such lipids between these two membranes.
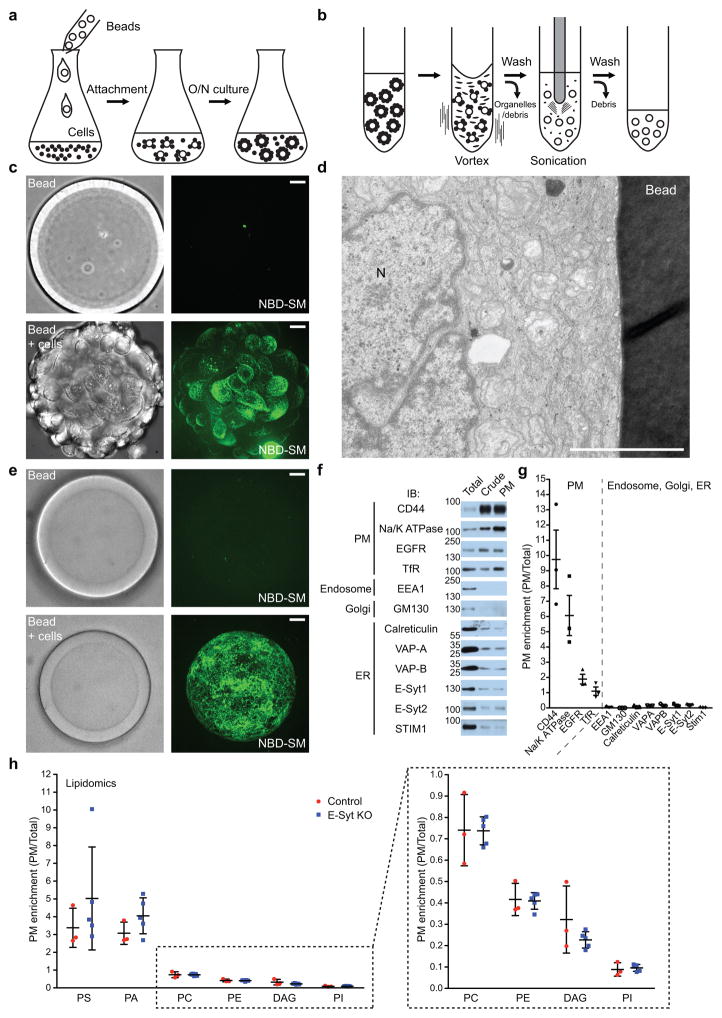
In order to assess the impact of E-Syts in the control of steady-state PM lipid levels, we applied with some modifications a method for the rapid separation and purification of PMs from cultured cells29. HeLa cells in suspension were mixed with poly-D-lysine coated dextran beads and the mixture was incubated overnight to allow cells adhesion to beads (Fig. 4a,c,d). Beads were then exposed to a hypotonic solution to induce osmotic lysis, vigorously vortexed and briefly sonicated to remove most organelles from the PM sheets that remained tightly attached to the bead surface and could be visualized by NBD-labeled sphingomyelin (SM) (Fig. 4b,e, and see methods). As shown by western blotting, the material that remained bound to the beads was very strongly enriched relative to the starting material in PM marker proteins (CD44 and Na+/K+-ATPase). Other proteins tested partitioned roughly as expected. PM receptors [epidermal growth factor receptor (EGFR) and transferrin receptor (TfR)] were also present but not equally enriched in the PM fraction, in agreement with the presence of large pools of these receptors on endosomal membranes. The endosomal marker protein (EEA1) was greatly depleted, and the Golgi marker proteins GM130 was absent, while small amounts of ER proteins recovered in the PM, such as calreticulin, VAP-A, VAP-B and STIM1, most likely reflect tightly attached cortical ER (Fig. 4f,g). Importantly, the recovery of endogenous E-Syt1 and E-Syt2 on bead-attached PM sheets was similar to that of other integral ER proteins (calreticulin, VAP and STIM1) (Fig. 4g), further supporting their localization in the ER8. The similar subcellular recovery of endogenous E-Syt1 and E-Syt2 in the bead bound material confirms that these two proteins are primarily present in cells as heterodimers localized throughout the ER, with only a small fraction of the heterodimer being already at ER-PM contact sites in control conditions8.
Figure 4. Rapid isolation and characterization of PM sheets.
(a) Schematics showing the attachment of HeLa cells to dextran beads. (left) Cells were dissociated and dextran beads were added drop by drop; (middle) A cell-bead mixture was incubated with gentle stirring to allow uniform attachment (right) and further incubated overnight. (b) Schematics showing the isolation of bead-attached PMs. Following a wash, the cell-bead mixture was exposed to a hypotonic buffer and vortexed to lyse cells and remove intracellular organelles. After extensive washes, a sonication pulse was applied to the beads to further remove intracellular organelles/debris from bead-attached PMs. (c) Confocal image of live HeLa cells attached to a bead. Top images: bead without cells. Bottom images: bead coated with HeLa cells as illustrated in a. Maximum projections of the serial Z-stack of confocal images are shown. The bead-attached PMs were stained with NBD-labeled Sphingomyelin (NBD-SM: green). Scale bars, 20μm. (d) Electron micrograph of a HeLa cell attached to a bead. N: nucleus. Scale bar, 5μm. (e) Confocal images of beads after sonication and staining with NBD-SM. Top images: bead not incubated with cells. Bottom images: bead with PM sheets. Scale bars, 20μm. (f) Bead attached material [total lysate (Total), crude PM sheets before sonication (Crude) and final PM sheets (PM)] was processed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. Note the enrichment of PM proteins and depletion of ER, endosomal and Golgi proteins in the PM fractions. (g) Quantification of protein abundance as analyzed in f. PM enrichment plots the ratio of “PM” over “Total” protein abundance (mean +/− SEM, n=3 extracts). (h) Comparisons of PM glycerolipid proliles of WT (Control) and E-Syt KO HeLa cells (pooled values of DKO and TKO cells) as analyzed by mass spectrometry. PM enrichment plots the ratio of PM lipids over total lipids (see methods) [mean +/− SD, n=3 extracts (Control), n=5 extracts (3 biologically independent samples from DKO#6-8 and 2 biologically independent samples from TKO#5 were pooled as DKO and TKO cells had similar properties in all functional assays consistent with the very low abundance of E-Syt3 in HeLa cells. PS, Phosphatidylserine; PA, Phosphatidic acid; PC Phosphatidylcholine; PE Phosphatidylethanolamine; DAG, Diacylglycerol; PI, Phosphatidylinositol. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Analysis by mass spectrometry of the major glycerolipid species in the PM fractions of WT and E-Syt KO cells did not reveal specific changes compared to total cellular lipids (individual extracts from DKO and TKO cells were independently analyzed and then pooled as they had similar properties in all functional assays consistent with the very low abundance of E-Syt3 in HeLa cells8). (Fig. 4h), thus speaking against a major role of the E-Syts in controlling the steady state distribution of these lipids in the PM.
Sustained accumulation of diacylglycerol at the PM upon activation of PLC in E-Syt KO cells
The E-Syts may play a role in homeostatic responses to conditions that acutely affect the lipid compositions of the PM. Transient increase in E-Syt1-dependent ER-PM tethers in response to activation of PLC-coupled receptors may be part of a mechanism to counteract PI(4,5)P2 loss in response to its hydrolysis. However, TIRF microscopy analysis of the PI(4,5)P2 probe mRFP-PHPLC to monitor PI(4,5)P2 depletion and resynthesis at the PM in response to Oxo-M-induced PLC activation of overexpressed M1R did not reveal obvious defects in the absence of the E-Syts at any time point (Supplementary Fig. 4a). These findings speak against a major role of these proteins in the regulation of PI(4,5)P2 dynamics.
Another potential function of E-Syts in response to PLC activation is the control of the dynamics of DAG generated by PI(4,5)P2 cleavage. Levels of PM DAG are very low at rest (Fig. 4h), transiently increase upon PLC activation and then gradually return to normal levels26. TIRF microscopy of the fluorescence signal of C1-mCherry (mCherry-C1 domains of PKCδ)30, a DAG probe, showed that the increase of this lipid in the PM upon stimulation of endogenous PLC-coupled histamine receptor was more sustained in the absence of the E-Syts. Importantly, the expression of EGFP-E-Syt1 together with Myc-E-Syt2 rescued the phenotype (Fig. 5a). While the increase of DAG occurs at the expense of PI(4,5)P2 cleavage, under these conditions (endogenous histamine receptor levels) only modest PI(4,5)P2 hydrolysis was observed (Supplementary Fig. 4b). Thus, a basal pool of E-Syts could still bind to the PM (Supplemental Fig. 4b) in contrast to what happens upon Oxo-M stimulation of M1R overexpressing cells, when PI(4,5)P2 was rapidly depleted (Supplementary Fig. 4c), leading to rapid and massive dissociation of the E-Syts from the PM.
We also tested the effect of ionomycin, whose stimulation of Ca2+ influx also results in massive, but somewhat delayed, PLC-dependent hydrolysis of PI(4,5)P231 so that E-Syt1 is recruited at the PM before complete PI(4,5)P2 depletion (see Fig. 5b for the recruitment of endogenously tagged E-Syt1, endoEGFP-E-Syt1). A very robust recruitment of C1-mCherry to the PM of these cells was observed in response to ionomycin (Fig. 5b,c), as detected by TIRF microscopy. Even with this drug, the accumulation of DAG (C1-mCherry fluorescence) in cells lacking E-Syts was more sustained in time relative to WT cells. Re-expression of either EGFP-E-Syt1 together with Myc-E-Syt2 (Fig. 5c,d) or EGFP-E-Syt1 alone (Fig. 5e) rescued the phenotype, while a mutant EGFP-E-Syt1 lacking the SMP domain did not (Fig. 5e), supporting a role of E-Syt1 in the regulation of DAG dynamics via an SMP-dependent mechanism. Prolonged accumulation of DAG upon activation of PLC in E-Syt KO was further confirmed by another DAG probes, YFP-DBD (YFP fused to C1b domain of PKCβII)32(Supplementary Fig. 5a).
Ca2+-dependent DAG transfer by E-Syt1 in vitro
The cell-free assays shown in Fig. 2 demonstrates that E-Syt1 can transfer a glycerolipid, and thus, most likely also DAG. However, the property of E-Syt1 to mediate Ca2+-stimultated DAG transfer was directly assessed. As DAG does not have a headgroup that can be labeled with NBD, a radioactivity-based in vitro assay was used. “Heavy” (sucrose loaded) liposomes composed of PC, PS, PI(4,5)P2 and supplemented with 3H-DAG were mixed in the presence or absence of 100μM Ca2+ with “light” (no sucrose) liposomes composed of PC and DGS-NTA(Ni) and with either the purified His-tagged cytosolic portion of E-Syt1 (E-Syt1cyto) or E-Syt1cyto lacking the SMP domain (E-Syt1cytoΔSMP) (Fig. 6a, and see methods). Subsequently, E-Syt1-tethered donor and acceptor liposomes were separated by the addition of a cocktail of imidazole and Proteinase K followed by centrifugation (Fig. 6b). Analysis of radioactivity in the two populations of liposomes revealed robust SMP domain-dependent and Ca2+-stimulated 3H-DAG accumulation in light liposomes, proving the property of E-Syt1 to transport DAG (Fig. 6c).
Figure 6. SMP domain-dependent and Ca2+-regulated diacylglycerol transfer by E-Syt1.
(a) Schematics of the 3H-DAG in vitro transfer assay. (b) A mixture of “heavy” donor liposomes containing 3H-DAG and of “light” acceptor liposomes was incubated for 10 min in the presence of 100μM Ca2+ and either E-Syt1cyto or E-Syt1cyto lacking the SMP domain (E-Syt1cytoΔSMP). Next the two populations of liposomes were separated by addition of proteinase K and imidazole followed by centrifugation. (c) Quantification of DAG transfer. (left) Values are plotted as percentage (%) of 3H radioactivity in the supernatant (i.e. light acceptor liposomes) over the sum of the total radioactivity in the supernatant and pellet (i.e. light and heavy donor liposomes). (right) Normalized Ca2+-dependence of E-Syt1-mediated DAG transfer was plotted after background subtraction (mean +/− SEM, n=3 independent experiments for all the conditions; two-tailed Student’s t-test with equal variance, ** denotes P<0.0001).
Ca2+-dependent recruitment of E-Syt1 in KO cells extracts DAG from the PM
A mechanism accounting for the clearance of excess DAG from the PM is its conversion to PA by DAG kinases (DGK)33. The lipid transport protein Nir2 is recruited to ER-PM contacts in DAG- and phosphatidic acid (PA)-dependent manner upon PLC-dependent PI(4,5)P2 hydrolysis and transfers PA to the ER for metabolic recycling34, 35 (Fig. 7a). E-Syt and Nir2-dependent mechanisms may cooperate in the recycling of these PI(4,5)P2 metabolites.
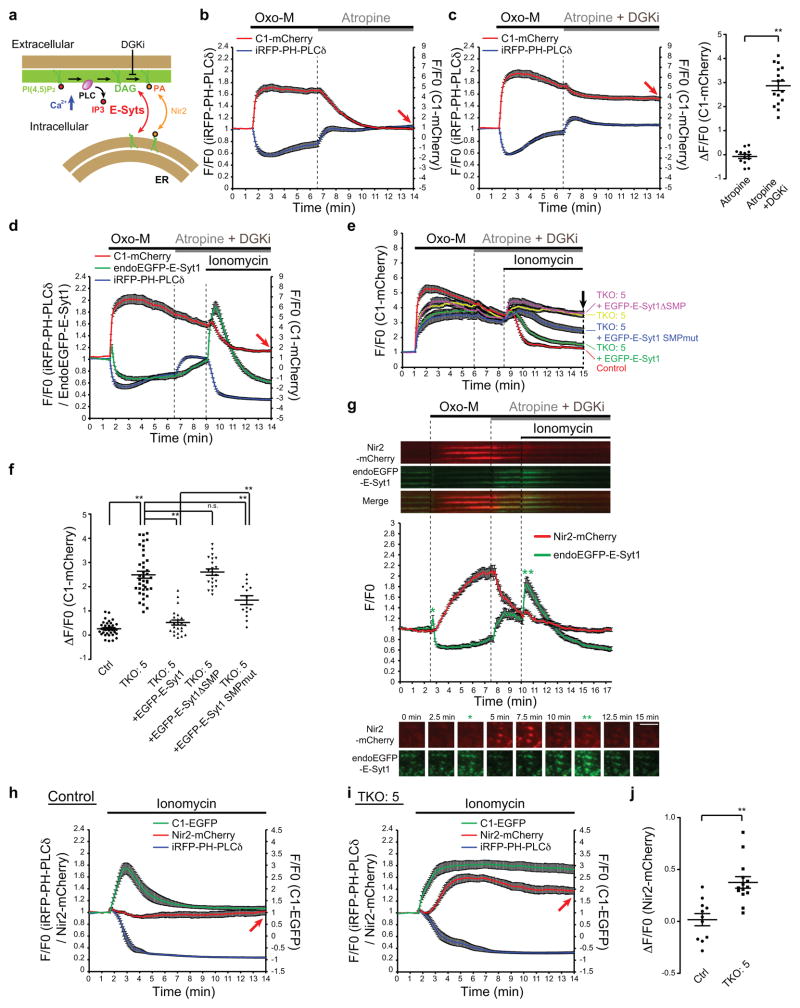
Figure 7. PM DAG extraction mediated by E-Syts.
(a) Schematics of possible role of E-Syts in the regulation of PM DAG dynamics during PLC activation. (b,c) Time-course of normalized fluorescence signal in response to (b) Oxo-M (10μM) and atropine (50μM) or (c) Oxo-M (10μM) and atropine plus DGKi (50μM each), as assessed by TIRF microscopy, from cells expressing M1R, iRFP-PH-PLCδ and C1-mCherry. (c,right) Values of ΔF/F0 corresponding to the end of the experiment as shown with red arrows in (b) and (c) [mean +/− SEM, n=13 cells (Atropine), n=16 cells (Atropine+DGKi);two-tailed Student’s t-test with unequal variance,** denotes P<0.0001]. Data are pooled from 2 independent experiments for each condition. (d) Time-course of normalized fluorescence signal, in response to the indicated compounds, as assessed by TIRF microscopy, from cells expressing endogenously-tagged E-Syt1 co-expressing M1R, iRFP-PH-PLCδ and C1-mCherry. Compare the C1-mCherry signals indicated by the red arrow with the values in c. (mean +/− SEM, n=20 cells pooled from 3 independent experiments) (e) Time-course of normalized mCherry signal, in response to the indicated compounds, as assessed by TIRF microscopy, from WT control and E-Syt TKO cells expressing C1-mCherry. (f) Values of F/F0 corresponding to the end of the experiment as shown in (e) by an arrow [mean +/− SEM, n=33 cells pooled from 5 independent experiments (Ctrl), n=39 cells pooled from 6 independent experiments (TKO#5), n=24 cells pooled from 4 independent experiments (TKO#5 + EGFP-E-Syt1), n=21 cells pooled from 3 independent experiments (TKO#5 + EGFP-E-Syt1ΔSMP), n=14 cells pooled from 2 independent experiments (TKO#5 + EGFP-E-Syt1ΔSMPmut);Bonferroni’s multiple comparisons test, ** denotes P<0.0001 except P=0.0002 for TKO#5 + EGFP-E-Syt1 v.s. TKO#5 + EGFP-E-Syt1ΔSMPmut]. (g) Time-course of normalized fluorescence signal, in response to the indicated compounds, as assessed by TIRF microscopy of cells expressing endogenously-tagged E-Syt1 co-expressing M1R, and Nir2-fused with mCherry(Nir2-mCherry). mean +/− SEM, n=20 Nir2 positive ER-PM contacts from 2 individual cells. Representative kymographs (top) and snap shots (bottom) of ER-PM contacts at different times are shown. Green asterisks indicate E-Syt1 recruitment. Scale bar, 5μm. n.s.=not significant. (h–j) Time-course of normalized fluorescence signal, as assessed by TIRF microscopy, from WT control (h) and E-Syt TKO cells (i) expressing Nir2-mCherry, iRFP-PH-PLCδ and C1-EGFP. Ionomycin (2μM) addition is indicated. (j) Values of ΔF/F0 corresponding to the end of the incubation as shown with red arrows [mean +/− SEM, n=11 cells pooled from 3 independent experiments (Ctrl), n=14 cells pooled from 4 independent experiments (TKO#5);two-tailed Student’s t-test with equal variance, P=0.0002].
In order to assess the interplay of E-Syts with Nir2 in the clearance of DAG, we inhibited its conversion to PA using the DGK inhibitor DGKi during the recovery phase from an Oxo-M dependent stimulus in M1R expressing cells. As assessed by TIRF microscopy, Oxo-M-triggered PLC activity resulted in a rapid rise of DAG, which was reversed by the M1R antagonist atropine (Fig. 7b). The rate of DAG removal during atropine treatment, i.e. when Ca2+ levels have returned to normal27, was not substantially affected by the absence of the E-Syts (Supplementary Fig. 6a). Addition of DGKi together with atropine, blocked the gradual DAG loss, as expected (Fig. 7c). However, addition of ionomycin 2.5 min following atropine/DGKi application to HeLa cells expressing E-Syt1 EGFP-tagged at the endogenous locus triggered robust recruitment of endoEGFP-E-Syt1 to the PM and rapid loss of the DAG reporter C1-mCherry (Fig. 7d). Both DAG removal and E-Syt1 recruitment required the presence of extracellular Ca2+ (Supplementary Fig. 6b). Importantly, in cells lacking E-Syts (TKO cells), ionomycin did not produce a loss of DAG signal in the PM and re-expression of EGFP-E-Syt1 alone, but not of E-Syt1 lacking the SMP domain (Fig. 7e,f) or its ER anchor8 (Supplementary Fig. 6c), was sufficient to rescue the phenotype. Mutations of the hydrophobic pockets of the SMP domain (see above) also significantly reduced rescuing activity (Fig. 7e,f).
These results support a role of E-Syt1 in exchanging lipids between the PM and the ER and the presence of a pathway that acts in parallel to Nir2 for the clearance of PI(4,5)P metabolites (Fig. 7a). However, further analyses suggested that the PM recruitments of 2 Nir2 and of E-Syt1 can be decoupled. Simultaneous imaging of Nir2-mCherry and endoEGFP-E-Syt1 showed that Nir2 is rapidly recruited to the PM upon triggered massive Oxo-M triggered PLC-dependent PI(4,5)P2 cleavage as reported34, 36, 37 (Fig. 7g). This rise occurred even when the E-Syts dissociated from the PM due to PI(4,5)P2 depletion after a very transient Ca2+-dependent recruitment before the drop of PI(4,5)P2. Conversely, during PI(4,5)P2 recovery upon atropine treatment, Nir2 dissociated from the PM while E-Syt1 (i.e. primarily reflecting the E-Syt1/E-Syt2 heterodimer in these experiments where tagged E-Syt1 was expressed at endogenous levels) returned to its basal levels at this membrane. Furthermore, the additional very robust recruitment of E-Syt1 triggered by ionomycin did not have an effect on Nir2 (Fig. 7g and Supplementary Movie 3). Nir2 and E-Syt1 recruitment occurred at the same ER-PM contact sites (see kymographs in Fig. 7g), suggesting the preexistence of contacts which are populated by different lipid transfer proteins depending upon the functional state of the cell.
Finally and importantly, robust recruitment of Nir2 to the PM was observed in cells lacking E-Syts upon ionomycin treatment, but not in control cells (Fig. 7h–j), indicating a strong functional connection between the functions of the two proteins (Fig. 7h–j). As recruitment of Nir2 to the PM is thought to be dependent, at least in part, upon DAG and PA presence in this membrane, these findings further support a role of the E-Syts in clearing PI(4,5)P2 lipid metabolites from the PM.
Altogether, these results reveal the occurrence of an E-Syt-dependent pathway for the removal of excess DAG from the PM upon PLC activation, which likely acts in cooperation with the pathway involving conversion to PA and Nir233, 34.
Discussion
Our present results provide direct evidence for the suggested property of the E-Syts to transfer lipids between bilayers and demonstrate the impact of the E-Syts on the regulation of lipid homeostasis in the PM. They further show the importance of the Ca2+-dependent regulation of E-Syt1 in its lipid transfer function.
The property of E-Syt1 to transport lipids was proven by in vitro lipid transfer between liposomes mimicking the ER (no acidic phospholipids) and the PM [presence of PI(4,5)P2 and PS] respectively. In these assays, the entire cytosolic domain of E-Syt1 was anchored to the ER-like liposomes liposomes via a histidine tag, and addition of Ca2+ led to its interaction “in trans” with PM-like liposomes to allow lipid transfer. Transfer was bidirectional, dependent on the lipid harboring properties of the SMP domain and on the presence of PI(4,5)P2 in the PM-like liposomes. Moreover, transfer was driven by the concentration gradient of the lipid and strongly potentiated by the presence of Ca2+ in the micromolar range, consistent with recent results describing the Ca2+-dependency of E-Syt1 recrutiment to the PM in semi-intact cells12. A major role of Ca2+ in lipid transfer is to promote the membrane tethering function of E-Syt1, which requires the Ca2+ binding properties of its C2C domain8, 11, 12, 37. However, it is also possible that Ca2+-dependent binding to lipid bilayers of its C2A domain, which also contains a Ca2+-binding site8, 13, 36, may assist lipid transfer by direct and local effect on the bilayer. As a Ca2+-sensitive C2A domain is present in E-Syt2 and E-Syt313, even these two proteins, whose ER to PM tethering function is not regulated by Ca2+, may in fact have a Ca2+-dependent role in lipid transfer. This hypothesis deserves further investigation. Collectively, in vitro studies demonstrate that the E-Syts facilitate passive transport of glycerolipids between bilayers along concentration gradients.
As we show here, the E-Syts are not essential for cellular life. Cells that lack expression of all the three E-Syts do not exhibit major obvious defects, although they grow more slowly than controls. This lack of major defects may be explained by overlapping functions with other SMP domain containing proteins in mammalian cells19. However, yeast cells that lack all three tricalbins, which appear to be the only SMP domain containing homologs of E-Syts in this organism, are also viable, suggesting that the function of this protein family is not essential10. Accordingly, mass spectrometry analysis of isolated WT and E-Syt KO cells PM generated by an efficient one-step procedure, did not reveal a difference in the composition of the PM in the major glycerolipid species. Our study suggests that one of E-Syts’ functions is to help produce adaptive changes in the PM following an acute perturbation. In cells lacking expression of all E-Syts, impaired disappearance from the cytosolic leaflet of the PM of newly generated DAG upon PLC-activation was observed. E-Syt1 alone could rescue the defect and rescue depended on the lipid-harboring property of its SMP domain and required its anchorage to the ER.
Another mechanism responsible for the rapid return to basal levels of DAG after its acute generation by PLC dependent PI(4,5)P2 cleavage is its conversion to PA in the PM, followed by PA transfer to the ER35 at least in part via the lipid transfer protein Nir2 (and possibly also by Nir3)34. As the E-Syts can harbor glycerophospholipids irrespective of the headgroup, a cooperation of E-Syts with Nir2 in PA transport is plausible. Like E-Syts, Nir2 is recruited to ER-PM contacts in response to PLC-activation34, 37. Our results show the recruitment of Nir2 and E-Syt1 to the PM, and thus their functions, can be decoupled. However, Nir2 and the E-Syt are functionally linked, as ionomycin stimulation induces a robust and prolonged accumulation of Nir2 at the PM in cells that lacks all three E-Syts but not in control cells.
If the E-Syts can transfer promiscuously glycerolipids (and possibly other lipids), one may wonder why they do not collapse the difference in glycerolipid composition between the ER and the PM. One explanation is that their equilibrating action is continuously conteracted by mechanisms that generate diversity. Another explanation, given the Ca2+-dependence of their transport, is that restoring normal lipid levels in response to acute perturbations, as shown here for PM localized DAG, may in fact be their major functions.
In summary, our study demonstrates a role of the ER localized E-Syts in the regulation of PM lipid homeostasis via in trans actions at ER-PM contact sites. Our findings emphasize a role of these proteins in mantaining lipid homeostasis in response to acute perturbations. However, additional functions of these proteins are plausible. Studies in yeast and plants have pointed to increased PM fragility in cells lacking tricalbins/extended-synaptotagmins17, 38–40. The potential occurrence of other changes in the PM bilayers of E-Syt TKO cells deserves further investigations. Further elucidating the physiological roles of these proteins as well as of other SMP domain containing proteins localized at membrane contact sites will be important to gain insight on how non-vesicular transport of lipids between bilayers contributes to maintain the specific lipid composition of each membrane.
Methods
Antibodies and chemicals
Primary and secondary antibodies used in this study are listed in Supplementary Table 1. Oxo-M, Atropine and Ionomycin (Sigma-Aldrich). Thapsigargin (Invitrogen/Life-technologies). DGK inhibitor (R 59–022, Tocris Bioscience). Proteinase K (Sigma-Aldrich). Following concentration of chemicals are used in all the experiments unless noted: Oxo-M, 10μM; Atropine, 50μM; Ionomycin, 2μM; Thapsigargin, 2μM; DGK inhibitor, 50μM. All non radiolabeled lipids were obtained from Avanti Polar Lipids; 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 850457; 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS), 840035; L-α-phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), 840046; (NBD)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 810144; 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] (DGS-NTA(Ni)), 790404; 1-2-dioleoyl-sn-glycerol (DAG), 800811. Radiolabeled lipids were purchased from American Radiolabeled Chemicals (St. Louis, MO); 1,2-Dioleoyl [9,10-3H] rac-glycerol, ART 2185.
Plasmids
Untagged E-Syt1, Myc-E-Syt2 (corresponding to the previously described Myc-E-Syt2S″8), EGFP-E-Syt1, iRFP-PH-PLCδ1 and mRFP-PH-PLCδ1 were previously described8, 41. The following reagents were kind gifts: mRFP-Sec61β from T. Rapoport (Harvard University)42, M1 muscarinic acetylcholine receptor (M1R) from B. Hille (University of Washington)28, ER-oxGFP from Erik L. Snapp (Albert Einstein College of Medicine), Nir2-mCherry from Rui Dong (our lab). YFP-DBD and C1-EGFP from Addgene (Plasmid #14874 and #21216)30. The sequences of primers used in this study are listed in Supplementary Table 2.
Cloning of EGFP-E-Syt1ΔSMP
EGFP-E-Syt1ΔSMP were generated by site-directed mutagenesis (QuickChange II-XL, Stratagene) using the primers, E-Syt1_SdeI_F and E-Syt1_Sdel_R, in EGFP-E-Syt1.
Cloning of EGFP-E-Syt1 V169W+L308W
2 hydrophobic amino acid residues (L308W and V169W) within the hydrophobic groove of the SMP domain were mutated to tryptophan using site-directed mutagenesis in EGFP-E-Syt1 using the primer sets, E-Syt1_L308W_MF, E-Syt1_ L308W _MR, E-Syt1_V169W_MF and E-Syt1_V169W_MR.
Cloning of GST-E-Syt1 SMP-C2A-C2B and GST-E-Syt1 SMP-C2A-C2B V169W+L308W
gBlock (IDT) containing E-Syt1 cDNA carrying V169W+L308W mutations were synthesized and amplified by PCR using the primer set, 5′ E1_SMP_EcoRI and 3′ E1_SMP_EcoRI.
PCR products were ligated in EcoRI site in the pDF6 vector containing E-Syt1 cDNA to generate GST-E-Syt1 SMP+C2A+C2B V169W+L308W.
Cloning of C1-mCherry
cDNA corresponding to C1 domains of C1-EGFP was excised and ligated into mCherry-N1 vector in the KpnI site. The vector was digested with BamHI and the 5′ overhung was filled by DNA Polymerase I, Large (Klenow) Fragment (NEB) in order to make C1 domains in-frame to mCherry.
Cell culture and transfection
HeLa cells were purchased from ATCC (Manassas, VA). They were cultured in Dulbecco’s modified essential Eagle medium (DMEM) (Life technologies) supplemented with 20% (v/v) fetal bovine serum (FBS) at 37°C and 5% CO2. Transfection of plasmids was carried out with Lipofectamine 2000 (Life Technologies) according to manufacturer’s instructions. RNAi against endogenous E-Syt1 was carried out as described in8. Wild-type as well as genome-edited HeLa cell lines were verified as free of mycoplasma contamination by a PCR-based method. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. All cell-based experiments were repeated at least 2 times.
Generation of anti-E-Syt1 antibodies
A peptide consisting of the N-terminal 19 amino acids of E-Syt1 (ERSPGEGPSPSPMDQPSAP) and an additional C-terminal cysteine was synthesized by GenScript, Inc. Keyhole limpet hemocyanin (KLH) was conjugated to the cysteine residue and rabbits were immunized with the peptide. Serum was collected and polyclonal antibodies were affinity purified on the peptide used for immunization with SulfoLink Immobilization Kit for Proteins (Thermoscientific). To further reduce non specific reactivity in immunofluorescence labeling of aldehyde fixed cells, the affinity purified anti-E-Syt1 antibodies were incubated with aldehyde fixed lysates of E-Syt1/2 DKO cells. Briefly, E-Syt1/2 DKO cells were fixed with 4% PFA in PBS at room temperature for 20 minutes, incubated with PBS containing 50mM glycine at room temperature for 10 minutes and scraped in PBS containing 1% BSA and 0.1% Triton X-100. Affinity purified antibodies were added to the resulting suspension and incubated at 4°C overnight. This material was then centrifuged at 21,000 g for 20 min at 4°C to pellet fixed cell debris, and the supernatant was used for immunofluorescence.
Fluorescence microscopy
For imaging experiments, cells were plated on 35mm glass bottom dishes at low density (MatTek Corp, Ashland, MA). Immunofluorescence and live cell imaging were carried out one day after transfection. Spinning disc confocal (SDC) microscopy was performed using the Improvision UltraVIEW VoX system (Perkin-Elmer) built around a Nikon Ti-E inverted microscope, equipped with PlanApo objectives (40x1.0-NA 1.0 and 60X1.49-NA) and controlled by Volocity (Improvision) software. Excitation light was provided by 488-nm/50-mW diode laser (Coherent) and 561-nm/50-mW diode laser (Cobolt), and fluorescence was detected by EM-CCD camera (C9100–50; Hamamatsu Photonics).
Total internal reflection fluorescence (TIRF) microscopy was performed on a setup built around a Nikon TiE microscope equipped with 60X1.49-NA. Excitation light was provided by 488-nm (for GFP), 561-nm (for mCherry/mRFP/mdsRed) and 640-nm (for iRFP) DPSS lasers coupled to the TIRF illuminator through an optic fiber. The output from the lasers was controlled by an acousto-optic tunable filter and fluorescence was detected with an EM-CCD camera (Andor iXon DU-897). Acquisition was controlled by Andor iQ software. Images were sampled at 0.20 Hz with exposure times in the 100–500 ms range. SDC microscopy was carried out at room temperature (20–25°C) and TIRF microscopy at 37°C.
Immunofluorescence
HeLa cells transfected with mRFP-Sec61β were washed twice with PBS and fixed with 4% PFA in PBS at room temperature for 20 minutes, washed with PBS and incubated with PBS containing 50mM glycine at room temperature for 10 minutes. Fixed cells were permeabilized with PBS containing 1% BSA and 0.1% Triton X-100 (blocking buffer) and incubated with primary antibodies for one hour at room temperature. Cells were then washed with blocking buffer twice and incubated with Alexa Fluor 488-conjugated anti-rabbit secondary antibodies for one hour at room temperature. After 4x washing with PBS, cells were examined under a SDC microscope. Images from a mid-focal plane are shown.
Live cell imaging
Cells were washed twice and incubated with either Ca2+ containing buffer (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, and 2 mM CaCl2 [pH 7.4]) or Ca2+-free buffer (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES, 10 mM glucose, and 3 mM EGTA [pH 7.4]) before imaging with either a SDC microscope or a TIRF microscope.
Image analysis
All fluorescent images were analyzed off-line using Image J (NIH). Changes in PM fluorescence over time (TIRF microscopy) were analyzed by manually selecting regions of interest covering the largest possible area of the cell footprint. Mean fluorescence intensity values of the selected regions were obtained and normalized to the average fluorescence intensity before stimulation after background subtraction. Quantification of fluorescence changes were performed using Excel (Microsoft) and all data are presented as mean ± SEM.
Electron microscopy
Cells attached Cytodex 3 microcarrier beads (Sigma-Aldrich) were fixed in 2% glutaraldehyde-0.1M sodium cacodylate buffer pH 7.4. Cells were post-fixed with 1% OsO4 in 1.5% K4Fe(CN)6-0.1M sodium cacodylate buffer, followed by en bloc staining with 2% uranyl acetate in 50mM sodium maleate buffer pH 5.2, dehydration and embedding in Embed 812. Electron microscopy reagents were purchased from Electron Microscopy Sciences (Hatfield, PA).
Biochemical analyses
Plasma membrane isolation and protein extraction
The procedure was modified from29. Briefly, 2g of Cytodex 3 microcarrier beads (Sigma-Aldrich) were reconstituted in 100ml PBS, autoclaved and coated by incubation with a poly-D-lysine solution overnight at 37°C. Cells were added to the reconstituted beads in sterile PETG flasks (Thermo scientific), allowed to attach to the beads for 4 hours with a gentle stirring every 30 min and then further incubated overnight with continuous stirring on rotating incubator at 37°C with 5% CO2. Beads were subsequently collected by spontaneous sedimentation and incubated with 220mM Sucrose, 40mM Sodium Acetate, pH5.0 for 5 min at room temperature. This step was necessary to maximize the area of plasma membrane adherent to the beads29. After the acid treatment, beads were collected, incubated with a hypotonic solution (10mM Tris-HCl, pH8.0) and vortexed for 10 seconds. A 10 seconds sonication pulse was then applied to the beads with Virsonic 550 (Virtis) in the same solution. Beads were finally washed three times again with the same solution and either once with PBS for subsequent protein extraction with SDS lysis buffer [10 mM Tris-HCl, 150 mM NaCl, 2% SDS, pH 8.0] or with 150 mM ammonium bicarbonate for lipidomic analysis.
Lipidomics
Lipid extraction, mass spectrometric analysis and data analysis were done by Lipotype GmbH (Germany). Briefly, lipids were extracted from either total cells still attached to the beads (total lipids), or from beads-bound PMs prepared as described above (PM lipids) using using Chloroform and Methanol43. Samples were spiked with lipid class-specific internal standards prior to extraction and lipid extracts were immediately subjected to mass spectrometric analysis. Mass spectra were acquired on a hybrid quadrupole/Orbitrap mass spectrometer (Q-Exactive, Thermo-Fisher) equipped with an automated nano-flow electrospray ion source (Triversa Nanomate, Advion) in both positive and negative ion mode. Lipid identification using Lipotype Xplorer was performed as previously described44.
Lipid-binding to purified E-Syt1
GST-tagged E-Syt1 fragments containing the SMP domain and C2A and C2B domains (predicted moleular weight: 53.6kDa) were generated and purified using a bacterial expression system and tested for in vitro lipid binding assay as described22.
Western blotting and immunoprecipitation
HeLa cells were lysed in SDS lysis buffer [10 mM Tris-HCl, 150 mM NaCl, 2% SDS, pH 8.0]. Cell lysates were incubated at 60°C for 20min followed by incubation at 70°C for 10 min. Benzonase Nuclease (Novagen) was added, and the samples were further incubated at room temperature for 20 min. Cell lysates were loaded and separated in 8% or 4–20% SDS-PAGE gels and immunoblotting was carried out as described8. Immunoprecipitation of endogenously-tagged EGFP-E-Syt1 (Fig. 1e), overexpressed EGFP-E-Syt2 (Fig. 3b) and EGFP-E-Syt1 (Supplementary Fig. 3f) was carried out using GFP-Trap beads as previously described8.
Liposome-based experiments
Cloning, Expression and Purification of an E-Syt1 fragment
The region coding for residues 93–1104 of human E-Syt1 was cloned into the pCMV6-AN-His vector with an N-terminal His6-tag. The protein was expressed in Expi293 cells for 3 days. Cells were harvested and lysed by three freeze-thaw cycles (liquid N2 and 37°C water bath) in buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.5 mM TCEP) supplemented with protease inhibitors (Complete EDTA-free; Roche). The protein was purified from the cell lysate by a Ni-NTA column (Clontech), and then further purified by gel filtration (Superdex 200, GE Healthcare) in buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.5 mM TCEP). Fractions containing E-Syt1 were pooled and concentrated to ~1 mg/ml.
Liposome Preparation
Lipids in chloroform were dried under a stream of N2 gas followed by further drying in the vacuum for 2 hours. Moles percent of lipids used for the acceptor and donor liposomes in FRET-based lipid transfer experiments were as indicated in Supplementary Table 3.
The dried lipid films were hydrated with buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.5 mM TCEP). Liposomes were then formed by ten freeze-thaw cycles (liquid N2 and 37°C water bath) followed by extrusion through polycarbonate filters with a pore size of 50 nm (Avanti Polar Lipids).
Dequenching (loss of self-quenching) Efficiency Assays
Dequencing assays of NBD were performed in 50 μL volumes containing 0.5 mM total lipids [50 μM donor liposomes containing either 1%, 3%, 5% or 7% fluorescent lipids plus 450 μM acceptor liposomes]. NBD was excited at 460 nm. Self-quenching was determined by recording emission intensity at 538 nm with or without 0.5% n-dodecyl-β-D-maltoside (DDM). Experiments were carried out at room temperature on a SpectraMax M5 Microplate Reader (Molecular Devices).
FRET-based Lipid Transfer Assays
Lipid transfer reactions were performed in 50 μL volumes. The final lipid concentration in the reaction was 0.5 mM, with donor and acceptor liposomes added at a 1:9 ratio (or 1:1 for Supplementary Figure 2e). Reactions were initiated by the addition of protein to a final concentration of 0.5 μM in a 96-well plate (Corning). The fluorescence intensity of NBD was monitored with an excitation of 460 nm and emission of 538 nm every 10 sec over 30 min at room temperature by using a SpectraMax M5 Microplate Reader (Molecular Devices). All data were corrected by setting to zero the value at the time of protein addition, and subtracting the baseline values obtained in the absence of the protein. The data were expressed as a percentage of the maximum fluorescence, determined after adding 10 μL of 2.5% DDM to the reactions after 30 min.
Liposome Tethering Assays
Liposome tethering assays were performed as for lipid transfer assays, except that the absorbance at 405 nm was measured every 10 sec over 40 min at room temperature by using a SpectraMax M5 Microplate Reader (Molecular Devices). 10 μL cocktail (1.8M imidazole, 15mM EGTA, 1.15mg/mL proteinase K) was added to stop the reaction after 30 min. The absorbance prior to protein addition was set to zero.
Content Mixing Assays
For content mixing assays, donor liposomes (85:15 mole percent POPC: DGS-NTANi) and acceptor liposomes [85:10:5 mole percent POPC: DOPS: PI(4,5)P2] were formed as for lipid transfer assays, except that 50 mM sulforhodamine B (Sigma-Aldrich) was included during the hydration of donor liposomes. The donor liposomes were pelleted in a TLA-100 centrifuge (Beckman Coulter) for 10 min at 55,000 rpm (20°C) to remove unencapsulated sulforhodamine B and the pellets were resuspended in buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.5 mM TCEP). The reactions were performed as for lipid transfer assays, except that the fluorescence intensity of sulforhodamine B was monitored with an excitation of 565 nm and emission of 585 nm.
Radio-labeled Lipid Transfer Assays
“Heavy” donor and “light” acceptor iposomes were as follows. “Heavy” liposomes: 300 μmol lipids (84:10:5:0.9:0.1 mole percent POPC: DOPS: PI(4,5)P2: DAG: tritium-labeled DAG for donor liposomes) in chloroform were dried in the vacuum for 1.5 hours. The lipid films were hydrated with buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP, 0.75M sucrose) and incubated at 37°C for 30 min. Liposomes were then formed by five freeze-thaw cycles (liquid N2 and 37°C incubation), pelleted and resuspended in buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP). “Light liposomes”: they were composed of 85% POPC and 15% DGS-NTA(Ni) and were prepared as for FRET-based lipid transfer assays.
Lipid transfer reactions were performed in 100 μL volumes. The final lipid concentration in the reaction was 0.5 mM, with “heavy” donor and “light” acceptor liposomes added at a 1:2 ratio. Reactions were initiated by the addition of protein to a final concentration of 0.5 μM and terminated by the addition of 10 μL a solution containing 1.8M imidazole and 1.15mg/mL proteinase K. “Heavy” and “light liposomes were then separated by centrifugation at 16,000g for 15 min and “heavy” liposomes recovered in the pellets were resuspended in 100-μL buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP). Radioactivity present in the supernatant and in the resuspended pellets was then determined at room temperature with a Liquid Scintillation Counter 1409DSA (Wallac).
CRISPR/Cas9 mediated endogenous tagging of E-Syt1
This procedure was performed by PNAbio (Thousand Oaks, CA). Briefly, the donor vector comprising cDNA encoding EGFP with a linker sequence (SGLRSRAQASNSAVD) flanked by ~500 bp homologous recombination arms covering the 5′ end of exon1 of human E-Syt1 gene was synthesized and co-transfected in HeLa cells with the guide RNA (RG6: AATGGAGCGATCTCCAGGAGAGG) and Cas9. EGFP-positive cells were FACS enriched and individual cells were isolated. PCR-based genotyping of ~150 clones identified several candidate clones with genomic integration of the donor sequence at the endogenous E-Syt1 locus. These clones were further analyzed by PCR-based genotyping, sequencing and Western Blotting. The endogenously-tagged cells were further characterized by RNAi and microscopy as shown in the results.
Generation of E-Syt knockout HeLa cell lines
A transcription activator-like effector nuclease (TALEN) pair that targets the region encoding the SMP domain of human E-Syt2 and two CRISPR guide RNAs, E-Syt1(6) and E-Syt1(7), that target the region encoding the hydrophobic hairpin stretch of human E-Syt1 were designed (Fig. 3a). The efficiency of the TALEN pair and CRISPR/Cas9 were estimated as10.9%, 2.5% and 4.5% respectively using Surveyor nuclease assay (Supplementary Fig. 3a). To enrich genome-edited cells that carry insertions or deletions (indels) at the targeted loci, the surrogate vector system45 and fluorescence activated cell sorting (FACS) were used to enrich for cells expressing the EGFP encoded by the surrogate vector. In brief, the inserted target sequences for TALEN or Cas9 are flanked by mCherry and EGFP. Without non-homologous end joining (NHEJ)-mediated repair of DNA double stranded break, stop codon (shown in red) allows only mCherry expression. After NHEJ, frame-shift in the target sequence leads to expression of mCherry-EGFP fusion protein, resulting in increase in EGFP signals (Supplementary Fig. 3b,c). First, KO HeLa cells for E-Syt2 were generated. The E-Syt1 and E-Syt3 genes were subsequently targeted in these cells by CRISPR/Cas9.
For the generation of HaLe cells lacking E-Syt2, WT cells were transfected with the plasmids encoding the E-Syt2-targeting TALEN pair and the surrogate vector, followed by enrichment of genome-edited cells via FACS and isolation of individual clones by dilution cloning. PCR analysis of the genomic DNA isolated from 39 individual clones verified candidate indels in the region encoding the SMP domain of E-Syt2 (Supplementary Fig. 3d). Two clones (line 10 and line 38) were further characterized with sequencing and Western blotting. These analyses revealed predicted deletions within the TALEN pair-binding sites, frame-shift and early termination in the open-reading frame of E-Syt2 gene, and the loss of E-Syt2 protein expression (Supplementary Fig. 3e,f). To generate E-Syt1/2 DKO cell lines, a subclone of the E-Syt2 knockout cell line 38 (38-2) was transfected with the plasmids encoding hCas9, E-Syt1 specific guide RNA expression vectors [E-Syt1 (6) or E-Syt1 (7)] as well as the surrogate vector, followed by the FACS-mediated enrichment of fluorescent cells (see above) (Supplementary Fig. 3b,c). Individually isolated clones, line 6-8 and line 7-5 (hereafter E-Syt1/2 DKO#6-8 and E-Syt1/2 DKO#7-5; in all figures and texts, E-Syt1/2 DKO denotes E-Syt1/2 DKO#6-8 unless stated), showed insertion of one nucleotide within the guide RNA-targeted loci, resulting in the lack of E-Syt1 protein expression (Fig. 3b and Supplementary Fig. 3g). Immunostaining of E-Syt1/2 DKO cells with the newly generated anti-E-Syt1 antibody (see above) confirmed the absence of E-Syt1 signals in these cell lines (Supplementary Fig. 1b).
To generate E-Syt triple-knockout (TKO) cell lines, the E-Syt1/2 DKO#6-8 cell line was transfected with a plasmid encoding hCas9 and four PCR-amplified guide RNAs targeting 5′ region of E-Syt3 (Fig. 3a). Surveyor nuclease assay showed robust cutting efficiency (~28%), allowing isolation of knockout cells without further enrichment (Fig. 3b). Three clones that showed large indels in exon1 of E-Syt3 by sequencing were isolated and the absence of E-Syt3 protein was confirmed by IP-enrichment followed by Western Blotting (Fig. 3b).
Construction of TALEN, CRISPR and surrogate vectors
TALEN for E-Syt2 knock-out:
A TALEN pair targeting human E-Syt2 was assembled using Golden-Gate ligation method as described46.
Target sequence, RVDs and vector used were:
TALEN-SMP_L: TGTAAAACACATGTGGCCTT, (NG) NN NG NI NI NI NI - HD NI HD NI NG NN - NG NN NN HD HD NG (NG), pTALEN_v2(NG)
TALEN-SMP_R: TAGTTTCTCGAAACAACTTC, (NG) NI NN NG NG NG HD - NG HD NN NI NI NI - HD NI NI HD NG NG (HD), pTALEN_v2(HD)
Surrogate vector was designed following the strategy previously published45. To generate a surrogate vector for the TALEN-SMP pair, DNA oligo, TALEN, was synthesized and used as a template for PCR reaction.
Primers used were 5′ BglII_SMP-1 Surro and 3′ BamHI_SMP-1 Surro.
PCR produces were ligated between BglII and BamHI in mCherry-E-Syt2-EGFP8 to generate mCherry-SMP-1 Surro-EGFP.
PCR genotyping was carried out using the primer set 5′ SMP-1 N and 3′ SMP-1 Ex, which generates 311bp fragment in control.
TALEN-SMP_L, TALEN-SMP_R and mCherry-SMP-1 Surro-EGFP were co-transfected in HeLa cells, and EGFP-positive cells were sorted with iCyt SY3200 (Sony) 72 hours after transfection. Sorted-cells were individually cloned by dilution cloning and assessed by genotyping PCR and Western blotting.
CRISPR/Cas9 for E-Syt1 knock-out and E-Syt3 knock-out
E-Syt1 knock-out:
gBlock (IDT) comprising U6 promoter, human E-Syt1 target sequence and guide RNA was synthesized as described47. gBlock was incubated with Taq polymerase and ligated into pCR2.1 TOPO vector using TOPO-TA cloning kit (Life Technologies); target sequence used were E-Syt1 (6) and E-Syt1 (7). To generate a surrogate vector for the CRISPR/Ca9 targeting E-Syt1, DNA oligos, E-Syt1(6,7)_F and E-Syt1(6,7)_R, were synthesized, phosphorylated using T4 PNK and annealed,
Produces were ligated between BglII and BamHI in mCherry-E-Syt2-EGFP8 to generate mCherry-E-Syt1(6,7)-EGFP.
PCR genotyping was carried out using the following primer set, 5′ ESYT1_TM_F2 and 3′ ESYT1_TM_R2, which generates 489bp fragment in control.
Either TOPO CRISPR E-Syt1(6) or TOPO CRISPR E-Syt1(7) together with hCas947 and mCherry- E-Syt1(6,7)-EGFP were co-transfected in E-Syt2 KO (line 38 described above) HeLa cells, and EGFP-positive cells were sorted with iCyt SY3200 (Sony) 72 hours after transfection. Sorted cells were individually cloned by dilution cloning and assessed by genotyping PCR and Western blotting to obtain E-Syt1/2 DKO cells.
E-Syt3 knock-out:
gBlock (IDT) comprising U6 promoter, human E-Syt3 target sequence and guide RNA was synthesized as described47. gBlock was amplified using gRNA_F and gRNA_R primers47; target sequence used were P1S, P2AS, P2S and P2AS
PCR genotyping was carried out using the primer set, 5′ ESYT3_TM_F2 and 3′ ESYT3_TM_R2, which generates 573bp fragment in control:
4 PCR products containing the individual guide RNA expression system together with hCas947 were co-transfected in E-Syt1/2 DKO (line 6-8 described in the results) HeLa cells. Cells were then individually cloned by dilution cloning 72 hours after transfection and assessed by genotyping PCR to obtain E-Syt1/2/3 TKO cells.
The sequences of oligos and primers used are listed in Supplementary Table 2
Sequencing of mutant alleles
Sequencing of mutated alleles was carried out by cloning PCR products into the pCR4 Blunt-TOPO vector using the Zero Blunt TOPO PCR cloning Kit for sequencing (Life technologies). Biallelic insertions/deletions were confirmed by sequencing at least 10 individual colonies. The same primers were used as genotyping primers.
Statistics & Reproducibility
No statistical method was used to predetermine sample size. The experiments were not randomized. Comparisons of data were carried out by either the two-tailed Student’s t-test or the t-test with Bonferroni corrections for multiple comparisons (multiplicity adjusted P values are shown in the case of multiple comparisons) as appropriate with Prism 6 (GraphPad software).
Immunofluorescence experiments were repeated 3 times with similar results, and representative images are shown in Fig. 1a, and Supplementary Fig. 1b.
Live cell imaging experiments were repeated 3 times with similar results, and representative images are shown in Fig. 1d, 1g, 1i, 4c, 4e, 7g and Supplementary Fig. 1c, 3b.
Image analysis data in Fig. 1h, 1j and 5b are from a single experiment, but representative of 3 independent experiments with similar results. All other assays were repeated at least 2 times.
Multiple electron micrographs were acquired from different beads and a representative image is shown in Fig. 4d.
For biochemical analyses, when n was less than 5 experiments, individual data points are shown: Fig. 2g, 4g, 4h, 6c; Supplementary Fig. 2a.
Western blotting and immunoprecipitation experiments shown in Fig. 1c and 1e were done once, and the results were confirmed by fluorescence imaging experiments shown in Fig. 1d and 1f, respectively. Experiments shown in Fig. 3b were repeated 2 times with similar results, and representative images are shown; these results were confirmed by additional experiments shown in Supplementary Fig. 3e, 3f and 3g. Experiments of Supplementary Fig. 1a were repeated 2 times with similar results, and representative images are shown. Experiments shown in Fig. 4f were repeated 3 times with similar results, and representative images are shown. All the uncropped images are shown in Supplementary Fig. 7.
Supplementary Material
Acknowledgments
We thank Hoy Shen, Shawn Ferguson and Susumu Tomita, Rui Dong, William Hancock-Cerutti, Joshua Lees and Yiying Cai for discussion and/or sharing reagents. We thank Frank Wilson, Heather Czapla and Louise Lucast for superb technical assistance. This work was supported in part by NIH grants R37NS036251, DK45735 and DA018343 to PDC. Y.S. was supported by fellowships from the Uehara Memorial Foundation and the Japanese Society for Promotion of Science and X.B. from a Human Frontier Science Program long-term fellowship.
Footnotes
Author contributions
All authors participated in the design of experiments, data analysis and interpretation. Y.S. designed and performed all the genetic manipulations, all the imaging and biochemical studies and the isolation of plasma membrane sheets. X.B, F.P. K.M.R. participated in the design of lipid transfer assays that were performed by X.B. C.M.S. and K.M.R. designed SMP domain mutations and performed lipid binding assays. M.A.S and C.K. performed the lipidomic analysis. Y.S and P.D.C wrote the manuscript that was then reviewed by all authors.
References
- 1.Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nature reviews. Molecular cell biology. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 2.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends in biochemical sciences. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends in cell biology. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toulmay A, Prinz WA. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Current opinion in cell biology. 2011;23:458–463. doi: 10.1016/j.ceb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefan CJ, et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Mesmin B, et al. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 7.Prinz WA. Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics. The Journal of cell biology. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giordano F, et al. PI(4,5)P2-Dependent and Ca(2+)-Regulated ER-PM Interactions Mediated by the Extended Synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. Journal of cell science. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Developmental cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Busnadiego R, Saheki Y, De Camilli P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1503191112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idevall-Hagren O, Lu A, Xie B, De Camilli P. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. The EMBO journal. 2015 doi: 10.15252/embj.201591565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min SW, Chang WP, Sudhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craxton M. Genomic analysis of synaptotagmin genes. Genomics. 2001;77:43–49. doi: 10.1006/geno.2001.6619. [DOI] [PubMed] [Google Scholar]
- 15.Craxton M. Evolutionary genomics of plant genes encoding N-terminal-TM-C2 domain proteins and the similar FAM62 genes and synaptotagmin genes of metazoans. BMC genomics. 2007;8:259. doi: 10.1186/1471-2164-8-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy A, Zheng JY, Lazarowitz SG. Synaptotagmin SYTA Forms ER-Plasma Membrane Junctions that Are Recruited to Plasmodesmata for Plant Virus Movement. Current biology : CB. 2015 doi: 10.1016/j.cub.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Sancho J, et al. The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant physiology. 2015;168:132–143. doi: 10.1104/pp.15.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopec KO, Alva V, Lupas AN. Bioinformatics of the TULIP domain superfamily. Biochemical Society transactions. 2011;39:1033–1038. doi: 10.1042/BST0391033. [DOI] [PubMed] [Google Scholar]
- 20.Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nature structural & molecular biology. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 21.Oram JF, Wolfbauer G, Vaughan AM, Tang C, Albers JJ. Phospholipid transfer protein interacts with and stabilizes ATP-binding cassette transporter A1 and enhances cholesterol efflux from cells. The Journal of biological chemistry. 2003;278:52379–52385. doi: 10.1074/jbc.M310695200. [DOI] [PubMed] [Google Scholar]
- 22.Schauder CM, et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.AhYoung AP, et al. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3179–3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan AJ, Jacob R. Ionomycin enhances Ca2+ influx by stimulating store-regulated cation entry and not by a direct action at the plasma membrane. The Biochemical journal. 1994;300(Pt 3):665–672. doi: 10.1042/bj3000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willars GB, Nahorski SR, Challiss RA. Differential regulation of muscarinic acetylcholine receptor-sensitive polyphosphoinositide pools and consequences for signaling in human neuroblastoma cells. The Journal of biological chemistry. 1998;273:5037–5046. doi: 10.1074/jbc.273.9.5037. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz LF, et al. Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. The Journal of general physiology. 2005;126:243–262. doi: 10.1085/jgp.200509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suh BC, Horowitz LF, Hirdes W, Mackie K, Hille B. Regulation of KCNQ2/KCNQ3 current by G protein cycling: the kinetics of receptor-mediated signaling by Gq. The Journal of general physiology. 2004;123:663–683. doi: 10.1085/jgp.200409029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen CM, Kalish DI, Jacobson BS, Branton D. Membrane isolation on polylysine-coated beads. Plasma membrane from HeLa cells. The Journal of cell biology. 1977;75:119–134. doi: 10.1083/jcb.75.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Codazzi F, Teruel MN, Meyer T. Control of astrocyte Ca(2+) oscillations and waves by oscillating translocation and activation of protein kinase C. Current biology : CB. 2001;11:1089–1097. doi: 10.1016/s0960-9822(01)00326-8. [DOI] [PubMed] [Google Scholar]
- 31.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. The Journal of cell biology. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. The Journal of biological chemistry. 2006;281:30947–30956. doi: 10.1074/jbc.M603741200. [DOI] [PubMed] [Google Scholar]
- 33.Merida I, Avila-Flores A, Merino E. Diacylglycerol kinases: at the hub of cell signalling. The Biochemical journal. 2008;409:1–18. doi: 10.1042/BJ20071040. [DOI] [PubMed] [Google Scholar]
- 34.Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Developmental cell. 2015 doi: 10.1016/j.devcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whatmore J, Wiedemann C, Somerharju P, Swigart P, Cockcroft S. Resynthesis of phosphatidylinositol in permeabilized neutrophils following phospholipase Cbeta activation: transport of the intermediate, phosphatidic acid, from the plasma membrane to the endoplasmic reticulum for phosphatidylinositol resynthesis is not dependent on soluble lipid carriers or vesicular transport. The Biochemical journal. 1999;341(Pt 2):435–444. [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CL, et al. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013;5:813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 37.Chang CL, Liou J. Phosphatidylinositol 4,5-Bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. The Journal of biological chemistry. 2015;290:14289–14301. doi: 10.1074/jbc.M114.621375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schapire AL, et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. The Plant cell. 2008;20:3374–3388. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamazaki T, Kawamura Y, Minami A, Uemura M. Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. The Plant cell. 2008;20:3389–3404. doi: 10.1105/tpc.108.062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilar PS, Engel A, Walter P. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Molecular biology of the cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2316–2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. The Journal of biological chemistry. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampaio JL, et al. Membrane lipidome of an epithelial cell line. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzog R, et al. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome biology. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, et al. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nature methods. 2011;8:941–943. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- 46.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.