Abstract
Genomic approaches have predicted hundreds of thousands of tissue specific cis-regulatory sequences, but the determinants critical to their function and evolutionary history are mostly unknown1–4. Here, we systematically decode a set of brain enhancers active in the zona limitans intrathalamica (zli), a signaling center essential for vertebrate forebrain development via the secreted morphogen, Sonic hedgehog (Shh)5,6. We apply a de novo motif analysis tool to identify six position-independent sequence motifs together with their cognate transcription factors that are essential for zli enhancer activity and Shh expression in the mouse embryo. Using knowledge of this regulatory lexicon, we discover novel Shh zli enhancers in mice, and a functionally equivalent element in hemichordates, indicating an ancient origin of the Shh zli regulatory network that predates the chordate phylum. These findings support a strategy for delineating functionally conserved enhancers in the absence of overt sequence homologies, and over extensive evolutionary distances.
Organization of the vertebrate brain into discrete structural and functional regions begins early during embryonic development in response to signaling molecules secreted from localized brain organizing centers7–9. The zli is one such signaling hub in the posterior diencephalon of all vertebrates that specifies the thalamic and prethalamic territories through the release of Shh, but is divergent or absent in invertebrate chordate lineages9. Central to the understanding of zli formation is how Shh transcription is regulated in this crucial brain signaling center and the extent to which this regulatory mechanism is shared across species.
Comparative sequence analysis is often used to identify conserved noncoding regulatory elements10. However, it has become increasingly apparent that not all functionally conserved regulatory elements show clear evidence of DNA sequence homology11–15, which may confound interpretations of their evolutionary origin. Moreover, conventional methods of phylogenetic footprinting do not always capture individual binding sites at nucleotide resolution, especially when long stretches of regulatory sequence are under strong positive selection.
To decipher the regulatory logic of Shh expression in the zli, we adapted a strategy that does not rely on DNA sequence conservation alone, but instead, follows the premise that enhancers with similar spatiotemporal profiles often share common cis-regulatory features16–20. With this concept in mind, we surveyed the collection of experimentally validated regulatory elements in the Vista Enhancer Browser21 for patterns of reporter activity that overlapped with SBE1, an enhancer located in the second intron of the Shh gene that directs expression to the ventral midbrain, ventroposterior diencephalon and zli22 (Supplementary Fig. 1a–c). We identified 52 distinct SBE1-like enhancers scattered throughout the mouse and human genomes (Supplementary Fig. 2). Each of these enhancers is located in proximity to at least one gene transcribed in the region of the mid-diencephalic organizer according to the RNAseq profile of SBE1 positive cells at E10.5 (Supplementary Fig. 3 and Table 1).
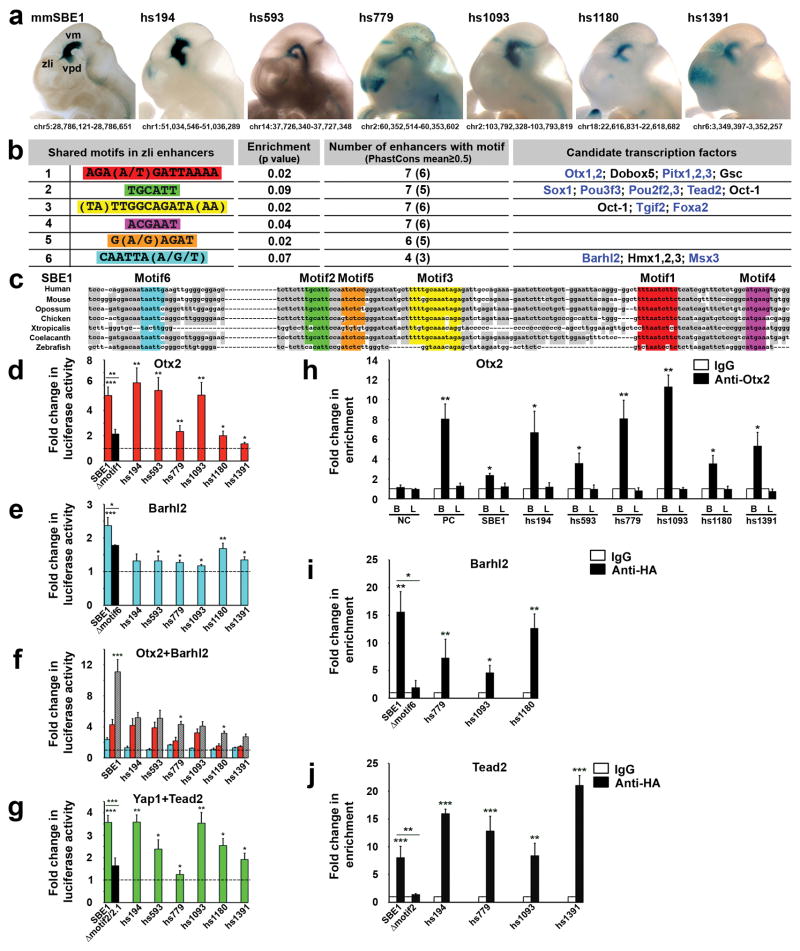
To determine if the SBE1-like enhancers possess a common cis-regulatory signature, we applied the Weeder algorithm23 and performed an unbiased search for shared DNA sequence motifs in seven of the most specific SBE1-like enhancers, including SBE1 (Fig. 1a). This approach identified five motifs that were enriched in the seven enhancers compared to random genomic sequence (Fig. 1b). Interestingly, the motifs showed significant sequence conservation across vertebrate phyla, suggestive of their functional importance. Within SBE1, the five motifs were clustered in a 116 bp homology block extending from human to zebrafish that was both necessary and sufficient for full enhancer activity (Fig. 1c and Supplementary Fig. 1).
Figure 1. A common cis-regulatory signature in SBE1-like enhancers.
(a) Heads of transgenic embryos (E11.5) expressing mouse (mm) SBE1 and selected human (hs) SBE1-like reporter constructs from the Vista Enhancer Browser display similar patterns of X-gal staining in the ventral midbrain (vm), ventroposterior diencephalon (vpd) and zli. Chromosome positions are indicated (NCBI37/mm9, GRCh37/hg19). (b) Shared motifs identified by Weeder (motifs1–5) and JASPAR/UniPROBE (motif6) that are significantly enriched in SBE1-like enhancers. The transcription factors matching a particular motif that are also expressed in the SBE1 domain are indicated in blue. (c) DNA sequence alignment (MAFFT version 7) of a core region of SBE1 from representative vertebrate species showing the position of shared motifs. Deeply conserved SBE1 nucleotides are shaded in grey. (d–g) Luciferase reporter assays performed in COS-1 cells demonstrating responsiveness of SBE1-like enhancers to candidate transcription factors in comparison to empty vectors (dotted lines). Black bars indicate mutant SBE1 reporter constructs for which a particular motif was deleted. Lower concentrations of Otx2 and Barhl2 expression vectors were used in (f) compared to (d, e) to demonstrate the synergy between these transcription factors in the activation of some SBE1-like enhancers. Each bar represents the average of at least three experiments performed in triplicate. (h–j) ChIP-QPCR performed with chromatin isolated from E10.5 embryonic mouse brain (B) and limb bud (L) in (h), or transfected cells (i, j) and the specified antibodies. QPCR results represent an average of at least three biological replicates. Negative (NC) and Positive (PC) control primers in (h) amplify sequence upstream of SBE1 and within an Emx2 forebrain enhancer, respectively. Error bars in all graphs represent standard deviation of the mean (s.d.m). *P<0.05, **P<0.01, ***P<0.001, Student’s t-test.
We also searched the set of SBE1-like enhancers for overrepresented transcription factor binding sites present in the JASPAR and UniPROBE databases. Three of the five motifs identified by Weeder matched consensus binding sites for transcription factors, several of which are expressed in the SBE1 domain (Fig. 1b and Supplementary Fig. 4). This analysis also uncovered an additional overrepresented motif (motif6) that was missed by Weeder presumably due to its more stringent criteria for finding DNA sequence matches. The six motifs did not display any apparent order or spacing in the SBE1-like enhancers, suggesting that they follow a flexible arrangement model observed in other tissue specific enhancers19,24,25. Furthermore, the co-occurrence of motifs 1–6 is significantly higher in a larger set of SBE1-like enhancers (n=46) compared to random genomic sequence matched for GC content and length (P<0.05, Welch’s two-sample t-test), or a set of 172 heart enhancers from the Vista enhancer browser (P<2.2e-16).
Our benchmark for pairing motifs with their candidate transcription factors included expression in the SBE1 domain and prior indication for a role in Shh regulation and/or zli formation. Motifs 1 and 6 correspond to recognition sequences for homeodomain proteins of the Prd and NKL subclasses, respectively26. Otx1 and Otx2 are the best candidates to be recruited by motif1, given their roles in mid-diencephalic development and Shh expression27–29. Similarly, Barhl2 is a potential transcription factor for motif6 based on its requirement for zli formation in Xenopus embryos30. Foxa2 was previously shown to bind motif3, which is necessary for SBE1 activity in the ventral midbrain but not the zli22 and therefore, will not be discussed further here. Of several candidate motif2 binding factors we hypothesize that the TEA domain family member 2 (Tead2), a key mediator of Hippo signaling, is recruited to this site. Tead and its co-transcriptional activation partner Yap are dependent on Wnt and/or Shh signaling in various biological contexts31,32.
To determine whether a candidate transcription factor is capable of regulating SBE1-like enhancers through a given motif, we performed luciferase-reporter assays in COS-1 cells. Otx2 induced significant luciferase expression from all seven SBE1-like reporter constructs, but not when motif1 was deleted from SBE1 (Fig. 1d). The core Otx binding site in motif1 (AAGATTAAA) is preferentially flanked on either side by adenine nucleotides, which when mutated blocked Otx2 binding and activation of the SBE1-luciferase construct, suggesting a context dependent role in zli gene regulation (Supplementary Fig. 5).
Although co-transfection experiments with Barhl2 triggered only a modest response from SBE1-like enhancers (Fig. 1e), the combined action of Barhl2 and Otx2 resulted in a synergistic induction of reporter activity from most enhancers containing motifs1 and 6 (Fig. 1f). Therefore, crosstalk between Otx2 and Barhl2 may mobilize a subset of SBE1-like enhancers. We also observed that the Tead2/Yap1 coactivation complex stimulated transcriptional responses for most SBE1-like enhancers, including SBE1, which depended on motif2 and a second Tead binding site (motif2.1), located 141 bp downstream (Fig. 1g). No other transcription factor combinations tested showed synergistic interactions (Supplementary Fig. 6).
We next performed chromatin immunoprecipitation (ChIP) to examine the occupancy of candidate transcription factors on their respective binding sites in SBE1-like enhancers. Chromatin isolated from embryonic brain, but not limb bud extracts, was enriched for Otx2 at all seven SBE1-like enhancers (Fig. 1h). Barhl2 and Tead2 were also recruited to a subset of SBE1-like enhancers containing the corresponding motifs in cultured cells (Fig. 1i, j). These findings suggest that SBE1-like enhancers are directly regulated by a transcription factor collective comprising Otx2, Barhl2, and Tead2.
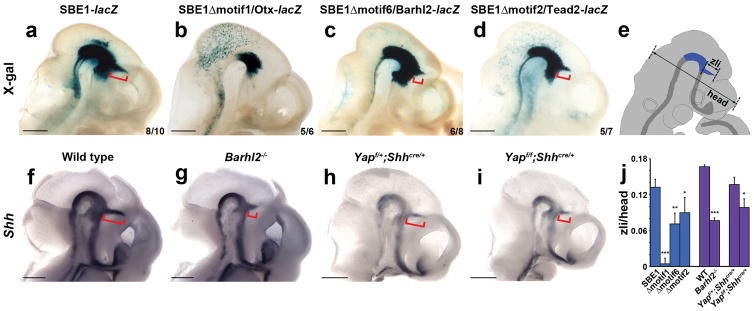
To assess the in vivo requirement of the SBE1 transcription factor collective, we performed transgenic mouse reporter assays with SBE1-lacZ constructs containing mutations in motif1 (Otx), motif2/2.1 (Tead), or motif6 (Barhl). X-gal staining was greatly compromised in the zli of embryos carrying the SBE1Δmotif1-lacZ transgene (96±7% reduction in staining along the zli length compared to SBE1-lacZ, p<0.0001; Fig. 2a–b and e). Deletions of motif6 and motif2/2.1 also resulted in a significant loss of staining in the zli compared to SBE1-lacZ control embryos (46±14%, p<0.001 and 32±19%, p<0.01, respectively; Fig. 2a, c–d and j). A similar reduction in zli staining was observed for constructs with deletions in the two orphan motifs 4 and 5 (Supplementary Fig. 7). These results are further supported by genetic studies, which showed a selective reduction in Shh zli expression in Barhl2−/−, conditional Yap, and as shown previously, Otx1/2 mutant embryos27–29 (Fig. 2f–j). Together, these data validate the in vivo contribution of the SBE1 transcription factor collective in the direct control of Shh transcription in the zli.
Figure 2. In vivo requirement of the SBE1 transcription factor collective.
(a–d) X-gal staining of transgenic embryos expressing wild type or mutant versions of SBE1-lacZ constructs at E10.5. The extent of zli staining is indicated by the length of the red bracket. Reduced zli staining was observed upon deletion of motifs 1, 2, or 6. The number of stained embryos out of the total carrying a given transgene is indicated. Scale bars: 1mm. (e) Schematic of E10.5 embryo demonstrating the approach for measuring the spatial distribution of X-gal staining or Shh expression in the zli with respect to head size. (f–i) Whole mount in situ hybridization for Shh in control, Barhl2−/−(n=4), and conditional Yap (Shhcre/+; Yaploxp/loxp, n=4) mutant embryos at E10.5, showing a partial, yet consistent, reduction in zli staining (red bracket) in both mutants. Scale bars: 1mm. (j) Quantification of the spatial distribution of X-gal staining (blue bars) or Shh expression (purple bars) in the zli normalized to head size. Error bars represent s.d.m. (*P<0.01, **P<0.001, ***P<0.0001, Student’s t-test).
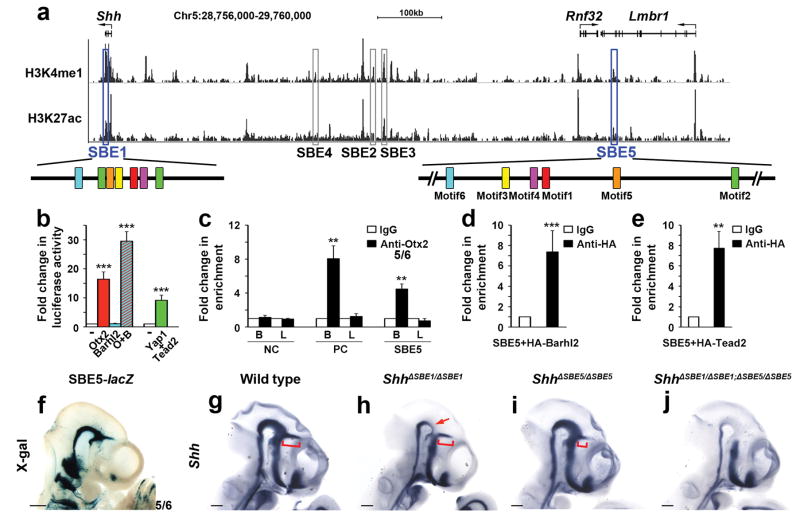
In earlier work, we reported that mouse embryos homozygous for a targeted deletion of SBE1 (ShhΔSBE1/ΔSBE1) failed to maintain Shh transcription in the basal plate of the rostral midbrain and caudal diencephalon after E10.0, yet retained expression in the zli33 (Fig. 3h). This implied the existence of another enhancer that functions independently of, or redundantly with, SBE1 to promote Shh expression in the zli. We sought to identify the missing Shh zli regulatory sequence using knowledge of the shuffled motif arrangement typified by SBE1-like enhancers.
Figure 3. Identification of SBE5 as a functional SBE1 homologue.
(a) UCSC genome browser view (NCBI37/mm9) of a 1 Mb interval upstream of Shh showing ChIP-seq signal enrichment for chromatin marks (H3K27ac and H3K4me1) associated with enhancer activity in whole brain (E14.5)34. The location of SBE1 and SBE5 related peaks (outlined in blue), other Shh CNS enhancer related peaks (outlined in grey) and the shuffled arrangement of their motifs (colored boxes) are displayed. (b) Activation of an SBE5-luciferase reporter construct by members of the SBE1 transcription factor collective. (c–e) ChIP-QPCR shows SBE5 enrichment in Otx2 bound chromatin from embryonic brain (B) but not limb bud (L), as well as Barhl2 and Tead2 bound chromatin from tissue culture cells. Negative (NC) and Positive (PC) control primers in (c) amplify sequence upstream of SBE1 and within an Emx2 forebrain enhancer, respectively (**P<0.01, ***P<0.001, Student’s t-test). Error bars represent s.d.m. Each bar (b–e) represents the average of at least three experiments performed in triplicate. (f) X-gal staining of a transgenic embryo (E10.5) expressing SBE5-lacZ in a similar pattern to SBE1 (compare with Fig. 2a). (g–j) Whole mount in situ hybridization for Shh on control and SBE1/SBE5 single and double mutant embryos (E10.5). ShhΔSBE1/ΔSBE1embryos show reduced Shh expression in the ventral midbrain and caudal diencephalon (red arrow). ShhΔSBE5/ΔSBE5 embryos display a partial truncation in Shh zli expression (red bracket). ShhΔSBE1/ΔSBE1; ΔSBE5/ΔSBE5 embryos are devoid of Shh in the entire SBE1 domain. Scale bars: 0.5mm.
We surveyed a 1 Mb interval surrounding Shh for histone modifications (H3K4me1, H3K27ac) associated with active regulatory sequences using ENCODE data from E14.5 brain34 (Fig. 3a). Most of our previously identified Shh brain enhancers, including SBE1, showed significant H3K4me1 and H3K27ac enrichment. We searched the remaining peaks for evidence of the SBE1 motif signature and identified a single region located 784 kb upstream of Shh, within the penultimate intron of the Lmbr1 gene, that contains a cluster of permuted motifs compared to SBE1 in the absence of any other overt sequence homology (Fig. 3a and Supplementary Fig. 8a). We tested the 1.9 kb sequence under the peak in a transgenic reporter assay and observed embryos with a consistent pattern of X-gal staining in the ventral midbrain, ventroposterior diencephalon and zli that was reminiscent of SBE1 activity (Fig. 3f). We designated this regulatory element SBE5. SBE5 performed equivalently to SBE1 in all cell-based reporter and ChIP assays using components of the SBE1 transcription factor collective, demonstrating that SBE5 is also directly controlled by Otx2, Barhl2 and Tead2 (Fig. 3b–e).
Notably, Shh expression was only partially attenuated in the zli of mouse embryos homozygous for a 228 kb deletion encompassing SBE5 (Fig. 3g, i). Yet, in mutants lacking both SBE1 and SBE5, Shh transcription was completely eliminated from the ventral midbrain, ventroposterior diencephalon and zli (Fig. 3g, j). The rescue of Shh expression with a ShhP1 transgene suggests that this phenotype was caused by the loss of Shh enhancers rather than the deletion of other coding or non-coding sequence elements potentially involved in Shh regulation (Supplementary Fig. 9). In further support of this claim, we observed that a smaller (2 kb) deletion of SBE5 generated by CRISPR-Cas9 had the same effect on Shh zli expression as the larger (228 kb) SBE5 deletion allele (Supplementary Fig. 9). From these results, we conclude that SBE1 and SBE5 function in a partially redundant manner to regulate Shh zli expression, and that the activity of these two enhancers is achieved through similar cis and trans determinants (see model, Supplementary Fig. 10).
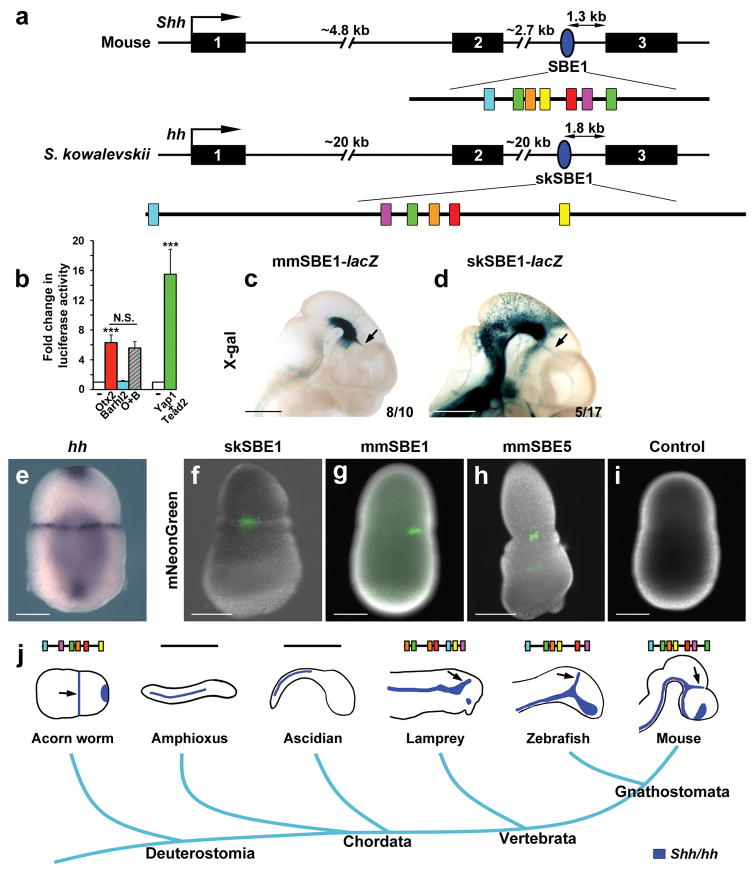
The origins of vertebrate brain signaling centers, including the zli, have been the subject of many studies with some proposing their first appearance in early vertebrates concurrent with increases in brain complexity, while others contended a more ancient deuterostome origin that predates the diversification of chordates35,36. Support for the latter hypothesis stems from studies performed in the hemichordate, Saccoglossus kowalevskii, which showed patterns of gene expression for many signaling ligands and transcription factors along the anteroposterior axis of the embryo that respected a similar distribution to those expressed in vertebrate brain signaling centers36. Of particular interest was the description of S. kowalevskii hedgehog (hh) expression in a narrow band of cells at the proboscis-collar boundary that appeared zli-like in character in relation to surrounding genes. To determine if this pattern of hh expression is governed by a similar cis-regulatory mechanism to its vertebrate counterpart, we searched the hh locus for evidence of the SBE1 motif signature and identified a 1.1 kb region in the second intron that contained all six motifs in the absence of any other sequence homology (Fig. 4a and Supplementary Fig. 8b). Interestingly, the motif arrangement in S. kowalevskii (sk) SBE1 was once again shuffled compared to mouse (mm) SBE1, yet the sequence of a given motif differed by no more than a single nucleotide compared to its mouse equivalent. In cell-based reporter assays the skSBE1 element was activated by Otx2 and Tead2/Yap1, but not the Barhl2/Otx2 tandem, for unknown reasons (Fig. 4b). Remarkably, mouse embryos expressing the skSBE1-lacZ transgene displayed X-gal staining in the ventral midbrain, ventroposterior diencephalon and zli, similar to those expressing the mouse SBE1-lacZ transgene, albeit with less consistency (Fig. 4c–d). These results indicate that skSBE1 is a functional orthologue of mouse SBE1 and that the cis and trans determinants underlying Shh zli expression are of ancient origin, predating the last common chordate ancestor that existed over 500 million years ago.
Figure 4. Ancient origin of SBE1 predates the chordate phylum.
(a) Schematic of the Shh gene structure in mouse and Saccoglossus kowalevskii (acorn worm) showing the position of SBE1 (blue oval) in the second intron of both species. The shuffled arrangement of SBE1-like motifs (colored boxes) within the 1.1 kb skSBE1 sequence is shown. (b) Otx2 and Yap/Tead2, but not Barhl2, were sufficient to activate skSBE1 luciferase activity in co-transfection assays performed in COS-1 cells (***P<0.001, N.S., not significant, Student’s t-test). Error bars represent s.d.m. Each bar represents the average of at least three experiments performed in triplicate. (c–d) Transgenic mouse embryos expressing mmSBE1-lacZ and skSBE1-lacZ reporter constructs at E10.5 show similar patterns of X-gal staining in the ventral midbrain, ventroposterior diencephalon and zli (arrow). The ectopic staining outside of these domains is likely due to the site of transgene integration. Scale bars: 1mm. (e) Hh expression in a hemichordate embryo at 48 hours post fertilization. (f–i) Transgenic S. kowalevskii embryos expressing mNeonGreen in a narrow band of cells at the prospective proboscis-collar boundary from: skSBE1 (n=6 of 60 injected); mmSBE1 (n=5 of 55 injected); mmSBE5 (n=5 of 15 injected); and Control (gbx promoter only; n=0 of 60 injected) reporter constructs. Scale bars: 100μm. (j) The evolutionary trajectory of Shh expression in the zli, or zli-like structure (black arrows) correlates with the presence of the SBE1 motif cluster.
We next tested the activity of SBE1 and SBE5 in S. kowalevskii embryos. The skSBE1, mmSBE1 and mmSBE5 constructs each drove mosaic expression of mNeonGreen in a narrow line of cells at the prospective proboscis-collar boundary, partially recapitulating the endogenous domain of hh expression (Fig. 4e–h). It is this domain of hh that is proposed to play a homologous patterning role to Shh in the zli of vertebrates36. Embryos injected with a negative control construct lacking an enhancer showed no reporter activity (Fig. 4i). These data demonstrate that SBE1 from mice and hemichordates possess functionally conserved species-specific regulatory activity in non-homologous structures.
Our finding that SBE1-like enhancers have shuffled binding sites, prompted us to reevaluate SBE1 motif conservation in basal chordates37. We screened the second intron of hh for evidence of the shuffled motif arrangement in amphioxus (cephalochordate), ascidian (tunicate) and lamprey (basal vertebrate). Interestingly, organisms that possess the SBE1 motif cluster (lamprey and all jawed vertebrates) express Shh in a delineated domain of the CNS that defines the zli38, whereas organisms without this motif cluster (amphioxus and ascidians) lack hh expression in a homologous region, suggesting secondary loss of the zli37,39 (Fig. 4j). This observation is consistent with data showing that the second intron of the amphioxus hh gene lacks enhancer activity in the zli37.
These results help to clarify the controversy surrounding the origin of the zli, and support the hypothesis of deep homology40 of a zli regulatory cassette that used SBE1 in an ancient deuterostome to activate hh in a narrow band of ectodermal cells in the anterior half of the embryo. Early chordates would have inherited SBE1 from this deuterostome ancestor, which was subsequently lost in the invertebrate chordate lineages. Following the diversification of hh ligands in vertebrates, SBE1 was maintained in the second intron of the Shh gene and used to activate its transcription in a narrow band of cells in the caudal forebrain, thus establishing the zli as a brain signaling center. The gain of SBE5 in vertebrates, whether by duplication and subsequent rearrangement of SBE1, binding site turnover of preexisting sequence, or some other means, is thought to buffer Shh expression in the zli.
In summary, our study provides a framework for decoding coordinate enhancers that is generally applicable to other tissue specific regulatory sequences1–4. We demonstrate the feasibility of identifying enhancers with similar function that lack obvious sequence conservation, either in the same organism, or ones with disparate anatomies and separated by hundreds of millions of years of evolution. Applying our approach to other well-characterized cis-regulatory modules in diverse taxa may provide additional insights into genomic mechanisms underlying evolutionary change, or stasis, in gene regulation.
Methods
Cell based Reporter Assays
Mouse homologs of SBE1, SBE5 and SBE1-like enhancers from the VISTA Enhancer Browser (hs194, hs593, hs779, hs1093, hs1180, hs1391) were cloned into the pGL4.23 luciferase reporter vector (luc2/minP, Promega). The skSBE1 element was amplified from S. kowalevskii genomic DNA by PCR. SBE1 reporter constructs harboring deletions of motif1, 4 or 6 were generated by ligating two PCR products immediately flanking each motif. The QuikChange II XL Site-Directed Mutagenesis Kit was used to introduce the following SBE1 mutations: Δmotif2, Δ2.1, Δmotif5, SBE1MM1.1 and SBE1MM1.2. Mouse Barhl2, HA-Barhl2 or Flag-Otx2 cDNAs were cloned into the pcDNA3 (Life Technologies) mammalian expression vector. The pcDNA3-HA-Tead2 and pcDNA3-HA-Yap1 expression vectors were kindly provided by Dr. Duojia Pan (Johns Hopkins University, Baltimore, MD)41. The primers used to generate each of the reporter constructs are listed in Supplementary Table 2.
Luciferase reporter assays were performed in COS-1 cells by co-transfecting (FuGENE 6, Promega) 250ng of an enhancer driven reporter construct and 200–300ng of a transcription factor expression vector or empty vector, and 20 ng of pRL-TK (Promega) as an internal control. Cells were harvested 48 hours after transfection and assayed for firefly and renilla luciferase activities (Dual Luciferase Reporter Assay System, Promega). Enhancer activity was presented as fold induction relative to that of cells transfected with an empty pcDNA3 expression vector. At least three independent experiments were performed for each reporter construct in triplicate.
Chromatin Immunoprecipitation (ChIP)
The midbrain, caudal diencephalic region (including zli) and forelimb buds were dissected in DMEM (with 10% fetal bovine serum) from approximately 25–30 E10.5 embryos, pooled into separate brain and forelimb fractions, homogenized into small pieces, and crosslinked with 1% paraformaldehyde for 15 min at room temperature with shaking. ChIP was performed essentially as described42 using 6 μg of anti-Otx2 (Abcam), or anti-immunoglobulin G (IgG) (Cell Signaling Technology) antibodies. QPCR was conducted as described42 using primer sequences listed in Supplementary Table 2. Positive (PC) control primers in Fig. 1h and 3c amplify a DNA fragment from an Emx2 forebrain enhancer bound by Otx243.
A similar protocol was followed when performing ChIP-QPCR from COS-1 cells (107–108) cultured in 10cm plates and co-transfected with 3μg of SBE1-like enhancer constructs and 3μg of pcDNA3-Flag-Otx2, pcDNA3-HA-Barhl2 or pcDNA3-HA-Tead2 using anti-Flag (Sigma), anti-HA (kindly provided by Dr. Gerd Blobel, Children’s Hospital of Philadelphia, Philadelphia, PA) or anti-IgG (Sigma) antibodies.
Transgenic mouse reporter assay
SBE1-like enhancers were cloned into a vector containing the Shh promoter, lacZ gene and SV40 poly(A) cassette22. Transient transgenic embryos were generated by pronuclear injection into fertilized mouse eggs derived from the (BL6xSJL) F1 mouse strain (Jackson Laboratories) at the Transgenic and Chimeric Mouse Facility (Perelman School of Medicine, University of Pennsylvania).
Mouse Lines
Experiments were performed in accordance with the ethical guidelines of the National Institutes of Health and with the approval of the Institutional Animal Care and Use Committee of the University of Pennsylvania. The TRACER mouse deletion line Del(C1-Z), encompassing chr5:29,413,901–29,642,246 (mm9) including SBE5, was generated by CRE-mediated recombination between loxP sites carried by insertion alleles ShhSB144 and Z2D (Aktas and Spitz, unpublished) following a previously described strategy45. For simplicity, the Del(C1-Z) line is referred to herein, as ShhΔSBE5. To generate ShhΔSBE1/ΔSBE1;ΔSBE5/ΔSBE5 double mutant embryos, the ShhΔSBE5/+ line was first crossed with ShhΔSBE1/ΔSBE1 mutants33. ShhΔSBE1/+; ΔSBE5/+ males, carrying the SBE1 and SBE5 deletions in trans, were then bred to wild type CD1 females. The progeny from this cross were screened for recombination events that placed the SBE1 and SBE5 deletions in cis (1/600 offspring). The ShhΔSBE1/+; ΔSBE5/+ double heterozygous animals were then intercrossed to generate ShhΔSBE1/ΔSBE1; ΔSBE5/ΔSBE5 double homozygous embryos. Barhl2−/−46, conditional Yap (Yapf/f; ShhCre/+)47, and ShhP148 embryos were described previously.
The SBE5Δ2kb mouse line, referred as ShhSBE5Δ2kb, was generated with CRISPR-Cas9 genome editing tools. The Target Finder platform (Feng Zhang, MIT) was used to design two pairs of sgRNAs (Supplementary Table 2) flanking SBE5 with the lowest off-target specificity. Complementary guide sequences were annealed, phosphorylated, and cloned into the BbsI site of pX458 or pX459 vectors49. DNA from the two constructs was purified, mixed in a 1:1 ratio (2.5ng per construct) and injected into the male pronucleus of fertilized mouse eggs (BL6xSJL F1, Jackson Laboratories) at the Transgenic and Chimeric Mouse Facility (Perelman School of Medicine, University of Pennsylvania). F0 founder mice were screened by PCR for the expected 2kb deletion of SBE5 (5/44).
Whole-Mount β-Galactosidase Staining and In Situ Hybridization
For X-gal staining, whole embryos (E10.5) were fixed in 0.2% glutaraldehyde/1% formaldehyde at 4°C for 30 minutes, stained in a solution containing 1 mg/ml X-gal at 37°C overnight, washed in PBS, dehydrated in methanol, and cleared for imaging in a 1:1 ratio of benzyl alcohol:benzyl benzoate. The length of the stained portion of the zli, normalized to the width of the head, was quantified using ImageJ. For whole-mount RNA in situ hybridization, embryos were fixed in 4% paraformaldehyde at 4°C for overnight and hybridized with digoxygenin-UTP-labeled riboprobes according to a previously described protocol50.
RNA-seq
The 429M20eGFP BAC reporter line50 was used to guide the dissection of Shh expressing cells from the ventral midbrain, ventroposterior diencephalon and zli of E10.5 embryos under a fluorescent stereomicroscope. Total RNA was extracted from GFP+ brain tissue isolated from approximately 30 embryos using the miRNeasy Micro Kit (Qiagen, Valencia, CA). The RNA-seq library was prepared from 1μg of total RNA according to the manufacturer’s protocol for TruSeq RNA Sample Prep Kits (Illumina). Paired-end sequencing (100 bp) was performed on an Illumina HiSeq2000 platform at the Next Generation Sequencing Core (Perelman School of Medicine, University of Pennsylvania) to a depth of 62 million reads. Raw sequences were filtered to retain only high quality reads. Sequences were processed with RNA-Seq Unified Mapper (RUM)51 that aligns reads to the set of known transcripts in RefSeq, UCSC, ENSEMBL, and the mouse genome (mm9), and outputs feature-level quantitation (transcript, exon, and intron). To analyze global gene expression profiles, the number of uniquely aligning read counts to mRNA transcripts were extracted from the RUM output and processed using a custom script51. Transcripts with a fragment per kilobase of exon per million fragments mapped (FPKM) value >2 were considered as expressed.
Motif analysis
De novo motif discovery in mmSBE1 and the six human SBE1-like enhancers (hs194, hs593, hs779, hs1093, hs1180 and hs1391) was performed using Weeder (v1.4.2)23,52 on a MAC terminal. The Weeder parameter “HS/MM large S M T20” was employed to identify the top 20 over-represented motifs in each length category, ranging from six to 12 nucleotides, as the ‘interesting motifs’ (highest ranking). The number of mismatches in each length category is based on the default setting of the algorithm (motifs of 6 nucleotides allow 1 mismatch; 8 allow 2; 10 allow 3 and 12 allow 4). ‘Interesting motifs’ with overlapping sequences (≥4) were merged. Motif enrichment was calculated based on the probability of observing a given motif in 280 random genomic sequences (40 sets of 7 inputted to Weeder) matched for GC content and length, using Fisher’s exact test in R. Each of the motifs was used to query human and mouse transcription factor binding sites using the ‘search for Similar Motifs’ function in UniPROBE53 and ‘JASPAR CORE Vetebrata’54. The candidate transcription factors were further filtered based on their expression level in the SBE1 active region according to their RNA-seq profile. The seven SBE1-like enhancers were also screened for known human and mouse transcription factor binding sites using web based tools associated with the UniPROBE and JASPAR databases. In addition to transcription factor binding sites matching motifs 1–3, a Barhl1/2 binding site (motif6) was identified as significantly enriched in SBE1-like enhancers (30 out of 53) compared to random genomic sequence (p<0.1, Fisher’s exact test in R).
The enrichment of motifs 1–6 in SBE1-like enhancers was calculated using a random sampling approach to compare the co-occurrence of the six motifs in 46 SBE1-like enhancers with that in random genomic sequences (matched for GC content and length). Briefly, 20 random sequences were sampled from the 46 SBE1-like enhancers, and the number of sequences containing all six motifs was counted. The same calculation was also performed for 20 random sequences sampled from the human genome. After 1000 times of sampling, the two sets of counts were compared using the Welch’s two-sample t-test in R to determine the statistical significance of the enrichment.
Identification of SBE5 and skSBE1
The clustering of motifs 1–6 within a 2kb DNA sequence (the average length of enhancers in the VISTA Enhancer Browser) was used to predict the location of novel SBE1-like enhancers. To identify SBE5, we surveyed 1 Mb upstream and downstream of the Shh transcription start site for histone modifications (H3K4me1, H3K27ac) associated with regulatory sequences using Encode data from E14.5 mouse brain34. We next screened these putative regulatory sequences for the presence of motif1. This approach directed us to a region located 784 kb upstream of Shh, within the penultimate intron of the Lmbr1 gene. A sequence scan of the immediate area identified motifs 2–6 within a 1.9 kb region containing motif1. To identify the SBE1 ortholog in Saccoglossus kowalevskii, we searched the hh locus (Skow_1.1 scaffold44409) for the presence of the SBE1-like motif cluster and identified a ~1.1 kb region within intron 2 (303512–304562), close to the third exon, that contained all six motifs in a shuffled arrangement.
Transgenic Saccoglossus kowalevskii reporter assay
SkSBE1, mmSBE1 and mmSBE5 were cloned into an I-SceI flanked expression vector containing an sk gbx basal promoter upstream of the mNeonGreen reporter gene. The transgenes were digested with the I-SceI meganuclease and introduced into fertilized embryos by microinjection as previously described55. Embryos were cultured at 20°C and screened for expression beginning at 36 hours post fertilization.
Supplementary Material
Acknowledgments
We thank Dr. Jean Richa and his staff at the Transgenic and Chimeric Mouse Facility (Perelman School of Medicine, University of Pennsylvania) for assistance with transgenic mouse production. We also thank Dr. Steve Liebhaber, Dr. Klaus Kaestner, Dr. Casey Brown, Dr. Ken Zaret and members of the Epstein lab for helpful discussions and comments on the manuscript. This work was funded by grants from the National Institutes of Health, R01 NS039421 (DJE) and R21 EY023104 (LG), National Science Foundation, 1258169 (CJL), and a predoctoral fellowship from the Louis-Jeantet Foundation (OS).
Footnotes
URLs Target Finder platform (Feng Zhang, MIT): http://crispr.mit.edu/.
Accession codes RNA-seq data presented in this study were deposited at Gene Expression Omnibus (GEO) under accession GSE78005.
Author Contributions Y.Y. and D.J.E conceived the project, designed the experiments and wrote the manuscript. Y.Y. performed the co-transfection, transgenic mouse, gene expression and ChIP assays. P.J.M performed the transgenic hemichordate reporter assays. Y.J. performed the transgenic mouse reporter assays with CR constructs. Y.Z. performed the statistical analysis. Y.Y. and A.N.K. performed the motif analysis. A.P. and C.L. provided reagents and advice on the hemichordate experiments. Y.Y., L.G., O.S., W.G.C and F.S. generated mutant mouse lines and provided embryos.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- 6.Vieira C, Martinez S. Sonic hedgehog from the basal plate and the zona limitans intrathalamica exhibits differential activity on diencephalic molecular regionalization and nuclear structure. Neuroscience. 2006;143:129–140. doi: 10.1016/j.neuroscience.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 9.Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold CD, et al. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 2014;46:685–692. doi: 10.1038/ng.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 13.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 15.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busser BW, et al. A machine learning approach for identifying novel cell type-specific transcriptional regulators of myogenesis. PLoS Genet. 2012;8:e1002531. doi: 10.1371/journal.pgen.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Val S, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erives A, Levine M. Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci U S A. 2004;101:3851–3856. doi: 10.1073/pnas.0400611101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junion G, et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 20.Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci. 2012;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epstein DJ, McMahon AP, Joyner AL. Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development. 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- 23.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- 25.Smith RP, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland PWH, Booth HAF, Bruford EA. Classification and nomenclature of all human homeobox genes. Bmc Biology. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acampora D, Avantaggiato V, Tuorto F, Simeone A. Genetic control of brain morphogenesis through Otx gene dosage requirement. Development. 1997;124:3639–3650. doi: 10.1242/dev.124.18.3639. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai Y, et al. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev Biol. 2010;347:392–403. doi: 10.1016/j.ydbio.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Scholpp S, et al. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134:3167–3176. doi: 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juraver-Geslin HA, Gomez-Skarmeta JL, Durand BC. The conserved barH-like homeobox-2 gene barhl2 acts downstream of orthodentricle-2 and together with iroquois-3 in establishment of the caudal forebrain signaling center induced by Sonic Hedgehog. Dev Biol. 2014;396:107–120. doi: 10.1016/j.ydbio.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez LA, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbluh J, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong Y, et al. Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development. 2011;138:531–541. doi: 10.1242/dev.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irimia M, et al. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. Evodevo. 2010;1:7. doi: 10.1186/2041-9139-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pani AM, et al. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483:289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irimia M, et al. Comparative genomics of the Hedgehog loci in chordates and the origins of Shh regulatory novelties. Sci Rep. 2012;2:433. doi: 10.1038/srep00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugahara F, et al. Involvement of Hedgehog and FGF signalling in the lamprey telencephalon: evolution of regionalization and dorsoventral patterning of the vertebrate forebrain. Development. 2011;138:1217–1226. doi: 10.1242/dev.059360. [DOI] [PubMed] [Google Scholar]
- 39.Takatori N, Satou Y, Satoh N. Expression of hedgehog genes in Ciona intestinalis embryos. Mech Dev. 2002;116:235–238. doi: 10.1016/s0925-4773(02)00150-8. [DOI] [PubMed] [Google Scholar]
- 40.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 41.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, et al. Disruption of SoxB1-dependent Sonic hedgehog expression in the hypothalamus causes septo-optic dysplasia. Dev Cell. 2012;22:585–596. doi: 10.1016/j.devcel.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suda Y, et al. The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development. 2010;137:2939–2949. doi: 10.1242/dev.048843. [DOI] [PubMed] [Google Scholar]
- 44.Symmons O, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruf S, et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- 46.Ding Q, et al. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. J Neurosci. 2009;29:3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahoney JE, Mori M, Szymaniak AD, Varelas X, Cardoso WV. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 51.Grant GR, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pavesi G, Pesole G. Using Weeder for the discovery of conserved transcription factor binding sites. Curr Protoc Bioinformatics. 2006;Chapter 2(Unit 2):11. doi: 10.1002/0471250953.bi0211s15. [DOI] [PubMed] [Google Scholar]
- 53.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lowe CJ, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.